Abstract
No outbreaks caused by Escherichia coli–producing heat-labile enterotoxin LT2 have been reported to date. Here, we revealed that the E. coli O8:H8 strains isolated from patients in 2 independent diarrhea outbreaks were negative for any known virulence determinants in routine microbiological tests, were very closely related, and carried a prophage-encoded gene for a novel LT2 variant (LT2d) and the genes for colonization factor antigen III. We also showed that LT2d has a cytotonic activity similar to LT1. These data indicate the importance of E. coli strains producing LT2d as a human pathogen.
Keywords: colonization factor antigen, diarrhea outbreak, heat-labile enterotoxin, phage, scherichia coli
Two Escherichia coli O8-associated outbreaks of diarrhea were identified in Japan. Genome analysis revealed that both outbreaks were caused by closely related E. coli O8:H8 strains carrying a prophage that encodes a novel variant of heat-labile enterotoxin LT2, named LT2d.
Escherichia coli is a commensal intestinal inhabitant, but several strains that have acquired specific virulence factors can cause diverse diseases in healthy humans [1]. Enterotoxigenic E. coli (ETEC) causes watery diarrhea in both children and adults worldwide. ETEC is characterized by the production of heat-labile (LT) and/or heat-stable (ST) enterotoxins, along with diverse colonization factors (CFs), but includes a wide range of genetically diverse strains that express a variety of O antigens [1, 2]. LT is an AB5 toxin homologous to the cholera toxin (CT) produced by Vibrio cholerae and is genetically and antigenically divided into subtypes LT1 and LT2, both of which include several variants [3]. Although LT1 and LT2 have been shown to possess similar biological activities [4], LT2-producing strains rarely cause human disease [1], and no outbreaks caused by LT2-producing strains have been reported thus far. Known LT1 genes (elt1) are exclusively encoded on large plasmids, while LT2 genes (elt2) have been reported to be encoded by phage [5, 6]. Among the various CFs described so far (>25 variants), CF antigen I (CFA/I) and coli surface antigens 1–6 (CS1-CS6) are most prevalent in ETEC [7]. Here, we report 2 E. coli O8:H8–associated outbreaks of diarrhea and the results of the genome analysis of the isolates. Our results show that the 2 outbreaks were caused by very closely related E. coli O8:H8 strains that carry a prophage-encoded gene for a novel LT2 variant (named LT2d) and the genes for CFA/III. Unique features of the LT2d phage and the cytotonic effect of LT2d on CHO cells are also described.
The 2 outbreaks occurred in 2 different dormitories in Oita prefecture, Japan, in April 2014 (outbreak 1: OB1) and September 2016 (OB2). Among the 300 and 120 residents who lived in each dormitory and shared food and water, 13 and 39 developed gastrointestinal symptoms in OB1 and OB2, respectively (Table 1). Most patients in both outbreaks had diarrhea (mainly watery diarrhea) and abdominal pains. Fever, vomiting, and headache were also recorded in some patients in both outbreaks. In routine microbiological tests at Oita Prefectural Institute of Health and the Environment, 8 intestinal bacterial pathogens (Campylobacter spp., Salmonella spp., Shigella spp., Vibrio spp., Yersinia spp., Staphylococcus aureus, Bacillus cereus, Clostridium perfringens) were not isolated from any stool specimens tested in both outbreaks by using selective media for each pathogen. Known virulence determinants for diarrheagenic E. coli (afaD, aggR, astA, eae, elt, estA, invE, stx1, and stx2) and norovirus were also not detected in all stool specimens tested by routine polymerase chain reaction (PCR) screening. In OB2, Aichi virus, astrovirus, and sapovirus were additionally examined by PCR, but were negative in all specimens. We did not test for diarrheagenic parasites (Cryptosporidium, Giardia, and Cyclospora). No suspected infection sources or transmission routes were epidemiologically inferred for either outbreak. However, E. coli O8 was isolated from most tested stool specimens from patients (Table 1).
Table 1.
Summary of the Epidemiological Information of the 2 Escherichia coli O8 Outbreaks
| Outbreak | OB1 | OB2 |
|---|---|---|
| Year/month | 2014/08 | 2016/09 |
| Place | Oita prefecture, Japan | |
| No. of patients | 13 | 39 |
| No. of patients suffering from: | ||
| Abdominal pain | 9 | 35 |
| Diarrheaa | 12 (7) | 38 (28) |
| Fever | 7 | 2 |
| Vomiting | 3 | 1 |
| Headache | 1 | 3 |
| No. of patient fecal samples | ||
| Tested | 7 | 14 |
| E. coli O8-positive | 6 | 13 |
| Genome sequenced strain | Oita14070 | 16F5M1D1 |
aNumbers in parentheses indicate the numbers of patients with watery diarrhea.
E. coli O8 strains isolated from each outbreak displayed nearly identical XbaI-digested DNA banding patterns in pulsed-field gel electrophoresis (PFGE) analysis, with 2 faint additional bands observed in the strains from OB1 (data not shown). However, as described above, no known virulence determinants for diarrheagenic E. coli were detected in any of the 22 E. coli O8 isolates from the 2 outbreaks. To characterize the E. coli O8 strains, we sequenced 2 strains, Oita14070 and 16F5M1D1, isolated from patients with diarrhea from OB1 and OB2, respectively, by 300 × 2 paired-end sequencing using an Illumina MiSeq followed by assembly using a Platanus assembler [8]. The serotypes of the strains were both found to be O8:H8 by in silico analysis [9]. Only 49 SNPs and an 8-bp indel were detected between the 2 genomes. These results, together with the data from the PFGE analysis, indicate that the strains that caused the 2 outbreaks were very closely related, that is, very recently separated from a common clonal ancestor. The complete genome sequence determination of strain 16F5M1D1 using an Oxford Nanopore MinION sequencer revealed that the genome consisted of a 4 800 098-bp chromosome and 1 large and 2 small plasmids (Supplementary Table 1). The chromosome contained 5 prophages and 2 tandemly integrated integrative elements (Supplementary Figure 1). All sequence data generated in this study are available in the DDBJ/EMBL/GenBank BioProject database (PRJDB8539).
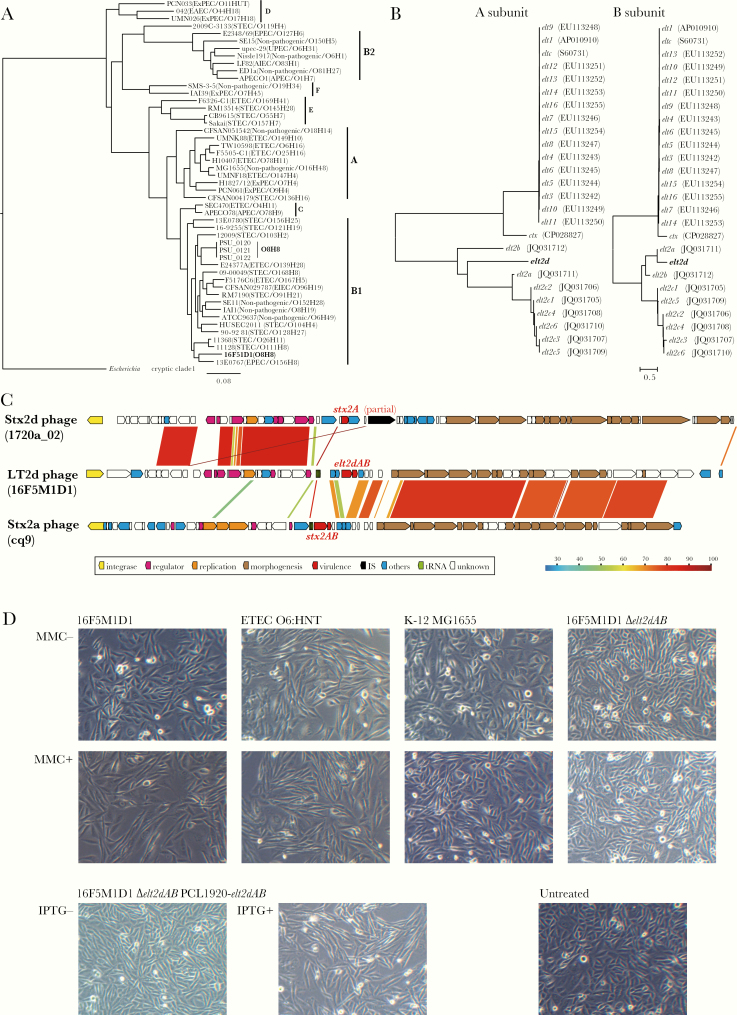
Phylogenetic analysis of strain 16F5M1D based on the core genes identified by Roary [10] revealed that 16F5M1D1 belongs to phylogroup B1 and was most closely related to the EPEC O156:H8 strain 13E0767 in the E. coli reference strain set used (Figure 1A; Supplementary Table 2). In the public database (accessed on 16/07/2019), the genome information of 3 E. coli O8:H8 strains isolated from spinach in 2011 in the United States (PSU_0120 to PSU_0122) was available. These strains had the O8:H8 serotype, as confirmed by in silico analysis [11], but were phylogenetically distinct from 16F5M1D1 (Figure 1A).
Figure 1.
The phylogenetic analyses, genetic structure of the LT2d-encoding phage, and CHO cell elongation assay. A, A core gene–based maximum likelihood (ML) tree of Escherichia coli O8:H8 strain 16F5M1D1 and an E. coli reference strain set. A cryptic Escherichia clade I strain TW15838 was included as an outgroup. The tree was constructed based on 225 254 SNP sites located on 2569 core genes. Phylogroups, pathotypes, and serotypes are indicated. B, Neighbor-joining (NJ) trees based on the nucleotide sequences of the A and B subunit genes of elt2d and other known elt1 and elt2 variants. The cholera toxin genes (ctx) were also included in this analysis. Accession numbers of each gene are indicated in parentheses. C, The genetic structure of the LT2d-encoding phage is shown. In panel, the genome sequence of the LT2d phage was compared with that of 2 Stx2 phages, to which the LT2d phage genome showed the highest similarity in the early and late regions, respectively. Sequence identities are indicated by different colors. D, The results of the CHO cell elongation assay. CHO cells (2×105 cells/well/500 µL) in a 24-well plate were treated with 100-fold diluted bacterial cell lysates for 48 hours at 37°C and visualized under a light microscope (×100). Cell lysates were prepared by sonicating bacterial cultures incubated with the presence or absence of 500 ng/mL of mitomycin C or 0.1 mM of Isopropyl β-D-1-thiogalactopyranoside (IPTG) for 5 hours at 37°C. Enterotoxigenic E. coli O6:HNT (LT1-positive) and E. coli K-12 MG1655 were used as positive and negative controls, respectively. Untreated: CHO cells untreated with bacterial lysate.
A search of the 16F5M1D1 genome sequence using the virulence factor database [12] identified the elt2 gene and a gene cluster for CFA/III biosynthesis. The elt2 gene was not targeted in our routine PCR screening because its contribution to diarrheal diseases in humans remains unknown. The elt1-detection PCR was unable to detect the elt2 gene due to the low sequence homology between elt1 and elt2 [6]. Genes for ETEC colonization factors were also not included in our routine PCR screening. The above-mentioned O8:H8 strains in the public database were all negative for both LT2 and CFA/III.
In the phylogenetic analysis of the A and B subunit genes of elt2, the 16F5M1D1 gene belonged to the elt2 cluster but formed a branch distinct from known elt2 variants (elt2a, b, and c) (Figure 1B). We therefore propose a designation of LT2d for the LT2 variant found in 16F5M1D1. By PCR analysis using newly designed elt2d-specific primers (elt2d-F: 5’-CTTTTTTCTCTGTATCTTCCAG-3’; and elt2d-R: 5’-CAGAAGCACACGGCGAATC-3’), elt2d was detected in all O8 strains isolated in the 2 outbreaks. LT2d is encoded by a lambda-like phage integrated into the prfC gene on the 16F5M1D1 chromosome (Figure 1C). The LT2d phage genome shows interesting similarity to Shiga toxin (Stx)–transducing phages; the early region of the LT2d phage showed the highest similarity to that of the Stx2d phage of the Stx-producing E. coli (STEC) strain 1720a_02, whereas the late region was most similar to that of the Stx2a phage of STEC cq9. This genetic organization suggests that the elt2d gene is under the control of the late gene promoter; thus, its expression is induced by phage-inducing agents, such as mitomycin C (MMC), as observed for stx2 genes [13]. As expected, in the CHO cell elongation assay [14], clear elongation of CHO cells was induced by the lysate prepared from the 16F5M1D1 culture treated with MMC but not by the lysate of the untreated 16F5M1D1 culture. In contrast, clear elongation was induced by the lysates of an LT1-producing ETEC strain, O6:HNT, irrespective of MMC treatment (Figure 1D). This result indicates that LT2d has a cytotonic activity similar to LT1 and LT2d production is dependent on phage induction. For further confirmation, we constructed an elt2d deletion mutant of strain 16F5M1D1 by the Wanner method [15] and its derivative, complemented with the Isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible elt2d gene, using a low copy number plasmid, pCL1920 [16], and performed the CHO cell elongation assay. CHO cell elongation was not induced by the lysate prepared from the elt2d deletion mutant even with MMC treatment. In contrast, that of the IPTG-induced complemented strain induced CHO cell elongation, confirming that the cytotonic effect on CHO cells observed was directly related to LT2d.
The CFA/III gene cluster was found in the 103-Kb plasmid in 16F5M1D1 (Supplementary Figure 2). It has been reported that CFA/III was identified in 8% of the ETEC strains isolated from patients with travelers’ diarrhea in Japan [17] and detected in several strains in a long-term global distribution study of ETEC [2]. This plasmid contained the repFIB replicon and a set of genes for conjugal transfer but no additional known virulence genes.
In conclusion, we identified 2 outbreaks of diarrhea caused by very closely related clones (strains) of E. coli O8:H8 that carry a gene for a novel variant of LT2, named LT2d, and the genes for CFA/III. The elt2d gene and the CFA/III gene cluster are encoded by a prophage and a large plasmid, respectively. Our findings indicate that more attention should be paid to infections by E. coli strains producing LT2d with colonization factors.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank M. Horiguchi, Y. Sato, and K. Ozaki for providing technical assistance.
Financial support. This work was funded by JSPS KAKENHI under grant numbers 16H06279 and 17H04077 to Y.O. and 16H05190 to T.H. and by AMED under grant numbers JP17efk0108127h0002 to Y.O. and 18fk0108065h0801 to T.H.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Croxen MA, Law RJ, Scholz R, et al. . Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 2013; 26:822–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. von Mentzer A, Connor TR, Wieler LH, et al. . Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution. Nat Genet 2014; 46:1321–6. [DOI] [PubMed] [Google Scholar]
- 3. Joffré E, von Mentzer A, Abd El Ghany M, et al. . Allele variants of enterotoxigenic Escherichia coli heat-labile toxin are globally transmitted and associated with colonization factors. J Bacteriol 2015; 197:392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Connell TD, Holmes RK. Characterization of hybrid toxins produced in Escherichia coli by assembly of A and B polypeptides from type I and type II heat-labile enterotoxins. Infect Immun 1992; 60:1653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jobling MG. The chromosomal nature of LT-II enterotoxins solved: a lambdoid prophage encodes both LT-II and one of two novel pertussis-toxin-like toxin family members in type II enterotoxigenic Escherichia coli. Pathog Dis 2016; 74:ftw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jobling MG, Holmes RK. Type II heat-labile enterotoxins from 50 diverse Escherichia coli isolates belong almost exclusively to the LT-IIc family and may be prophage encoded. PLoS One 2012; 7:e29898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gaastra W, Svennerholm AM. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol 1996; 4:444–52. [DOI] [PubMed] [Google Scholar]
- 8. Kajitani R, Toshimoto K, Noguchi H, et al. . Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res 2014; 24:1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ingle DJ, Valcanis M, Kuzevski A, et al. . In silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb Genom 2016; 2:e000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Page AJ, Cummins CA, Hunt M, et al. . Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015; 31:3691–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joensen KG, Tetzschner AM, Iguchi A, et al. . Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol 2015; 53:2410–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen L, Yang J, Yu J, et al. . VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 2005; 33:D325–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Waldor MK, Friedman DI. Phage regulatory circuits and virulence gene expression. Curr Opin Microbiol 2005; 8:459–65. [DOI] [PubMed] [Google Scholar]
- 14. Guerrant RL, Brunton LL, Schnaitman TC, et al. . Cyclic adenosine monophosphate and alteration of Chinese hamster ovary cell morphology: a rapid, sensitive in vitro assay for the enterotoxins of Vibrio cholerae and Escherichia coli. Infect Immun 1974; 10:320–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 2000; 97:6640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lerner CG, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res 1990; 18:4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Honda T, Wetprasit N, Arita M, Miwatani T. Production and characterization of monoclonal antibodies to a pilus colonization factor (colonization factor antigen III) of human enterotoxigenic Escherichia coli. Infect Immun 1989; 57:3452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.