Abstract
The tetraspanin CD82 is a potent suppressor of tumor metastasis and regulates several processes including signal transduction, cell adhesion, motility, and aggregation. However, the mechanisms by which CD82 participates in innate immunity are unknown. We report that CD82 is a key regulator of TLR9 trafficking and signaling. TLR9 recognizes unmethylated cytosine-phosphate-guanine (CpG) motifs present in viral, bacterial, and fungal DNA. We demonstrate that TLR9 and CD82 associate in macrophages, which occurs in the endoplasmic reticulum (ER) and post-ER. Moreover, CD82 is essential for TLR9-dependent myddosome formation in response to CpG stimulation. Finally, CD82 modulates TLR9-dependent NF-κB nuclear translocation, which is critical for inflammatory cytokine production. To our knowledge, this is the first time a tetraspanin has been implicated as a key regulator of TLR signaling. Collectively, our study demonstrates that CD82 is a specific regulator of TLR9 signaling, which may be critical in cancer immunotherapy approaches and coordinating the innate immune response to pathogens.—Khan, N. S., Lukason, D. P., Feliu, M., Ward, R. A., Lord, A. K., Reedy, J. L., Ramirez-Ortiz, Z. G., Tam, J. M., Kasperkovitz, P. V., Negoro, P. E., Vyas, T. D., Xu, S., Brinkmann, M. M., Acharaya, M., Artavanis-Tsakonas, K., Frickel, E.-M., Becker, C. E., Dagher, Z., Kim, Y.-M., Latz, E., Ploegh, H. L., Mansour, M. K., Miranti, C. K., Levitz, S. M., Vyas, J. M. CD82 controls CpG-dependent TLR9 signaling.
Keywords: tetraspanins, TLRs, myddosome, macrophages
Tetraspanins are a highly conserved family of proteins expressed on the cell surface and intracellular membranes of most cell types. As their name suggests, members of this protein family have 4 transmembrane domains, 2 extracellular loops, and short intracellular amino and carboxy termini (1). Tetraspanins influence diverse biologic functions including cell migration, adhesion, protein trafficking, and signal transduction. However, their primary role is to provide lateral organization to the plasma membrane through interactions with multiple proteins (1–4). A critical feature of tetraspanins is their ability to interact with diverse transmembrane and intracellular proteins, forming functional multimeric complexes known as tetraspanin-enriched microdomains (TEMs) (2, 5). Spatial organization of these complexes is essential for optimal signaling (6–9). Many tetraspanin-protein interactions are weak; thus, the composition and localization of TEMs are highly dynamic, explaining their broad influence on biologic processes including cancer progression and immunity. Down-regulation of CD82 is associated with poor prognosis of several malignancies, including breast, colon, lung, ovarian, and pancreatic cancers (10–12). In the context of immunity, CD82 expressed by macrophages and dendritic cells associates with class II major histocompatibility complex (MHC) and other components of the antigen-processing and presentation pathway (13–17) and has been reported to maintain dormancy of long-term hematopoietic stem cells (7). CD82 selectively traffics to fungal and bacterial-containing phagosomes in macrophages (15), suggesting that CD82 plays an important role in innate immune signaling. However, the exact role of tetraspanins during host defense to pathogens is not completely understood.
Similar to tetraspanins, TLRs are expressed on the plasma and endosomal membranes of immune cells (18, 19), including macrophages. Upon recognition of microbial ligands, TLRs dimerize (20), leading to recruitment of myeloid differentiation primary response 88 (MyD88), a core adaptor protein. This interaction licenses recruitment of interleukin-1 receptor-associated kinase (IRAK) family kinases (i.e., IRAK-2 and IRAK-4), forming the myddosome complex (21–23). Myddosome assembly is critical for activating the intracellular signaling cascade that promotes NF-κB nuclear translocation and proinflammatory cytokine production (e.g., TNF-α) (23). The intracellular localization of endosomal TLRs is critical for initiating a potent innate immune response against pathogens, as well as preventing recognition of self-DNA and autoimmunity (18, 19). The initial trafficking of endosomal TLRs from the endoplasmic reticulum (ER) to endolysosomal compartments is regulated by Unc-93 homolog B1 (UNC93B1) (24–27). However, the specific regulators of trafficking and signaling of individual endosomal TLRs have not been elucidated.
TLR9 is an endosomal innate immune receptor that binds unmethylated cytosine-phosphate-guanine (CpG) motifs in viral, bacterial, and fungal DNA. TLR9 is translated in the ER in its full-length form and passes through the Golgi to the endolysosomal compartments where the N terminus is proteolytically cleaved (28, 29). TLR9 then binds CpG motifs, which initiates myddosome assembly and downstream signaling leading to NF-κB activation (20, 25, 28, 30–32). We previously demonstrated that TLR9 recruitment to fungal-containing phagosomes requires the C-type lectin receptor, dectin-1 (33). Additionally, CD82 regulates dectin-1 clustering and subsequent downstream signaling in response to Candida albicans (34). TLR9 signaling bifurcates between NF-κB and interferon regulatory factor 7activation depending on the type of regulators involved (26, 27, 35). The notion that TLR9 and CD82 are rapidly and specifically recruited to fungal phagosomes (15, 33, 36) suggests a possible link between CD82 and TLR9 signaling.
To date, defining the precise function of individual tetraspanins has proven challenging because of a high degree of functional redundancy and lack of intrinsic receptor or catalytic activity (12). In this study, we demonstrate a nonredundant role for CD82 in the regulation of TLR9 signaling in response to CpG DNA. The dynamic association between TLR9 and CD82 occurs in macrophages. We further demonstrate that TLR9 trafficking to acidified CpG-containing compartments requires CD82. We also show that CD82 controls TLR9–dependent myddosome assembly but is dispensable for myddosome formation triggered by other TLRs. Finally, we show that CD82 regulates TLR9-dependent activation of NF-κB and TNF-α production. Together, these findings demonstrate a critical role of CD82 as a key regulator of TLR9 trafficking and signaling in innate immunity. Discovery of the specific regulators of TLR9 signaling will provide critical insight in the development of TLR9 agonist immune therapies in the context of cancer, infectious diseases, and autoimmune diseases.
MATERIALS AND METHODS
Reagents
The class B CpG phosphorothioate oligodeoxynucleotide 1826 (5′-TCCATGACGTTCCTGACGTT-3′) with or without Alexa Fluor 647 conjugated to the 3′ end was purchased from Integrated DNA Technologies (Coralville, IA, USA). LPS, imiquimod, and synthetic triacylated lipopeptide Pam3CsK4 were purchased from InvivoGen (San Diego, CA, USA).
Mice and cell lines
C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA), and CD82 knockout (KO) mice were obtained from Cindy Miranti (The University of Arizona Health Sciences, Tucson, AZ, USA). TLR9KO mice were a gift from Shizuo Akira (Osaka University, Osaka, Japan) and bred thereafter at Massachusetts General Hospital (MGH). Mice used in this study were housed and cared for in the MGH Hospital Thier Specific Pathogen Free barrier facility (MGH, Boston, MA, USA) according to Institutional Animal Care and Use Committee guidelines. The following immortalized cell lines were used for experiments: RAW264.7 (RAW), immortalized wild-type (WT) bone marrow–derived macrophages (BMDMs), CD82KO macrophages, TLR9KO macrophages, and Unc-93 homolog B1 (UNC93B1)-KO macrophages. The mouse macrophage-like cell line, RAW, was purchased from American Type Culture Collection (Manassas, VA, USA). CD82KO immortalized macrophages were generated by isolating bone marrow from CD82KO mice. The methods for immortalizing the cells were performed as previously described by Halle et al. (37).
Immortalized UNC93B1-KO macrophages were created in the laboratory of Eicke Latz. The TLR9KO cell line was a gift from Douglas Golenbock (University of Massachusetts Medical School). RAW and UNC93B1-KO macrophages were cultured in DMEM complete medium containing 10% heat-inactivated fetal bovine serum, 1% penicillin/streptomycin, and 1% l-glutamine (Thermo Fisher Scientific, Waltham, MA, USA). WT, TLR9KO, and CD82KO immortalized macrophages were cultured in Roswell Park Memorial Institute (RPMI) 1640–GlutaMax complete medium (Thermo Fisher Scientific) containing 10% heat-inactivated fetal bovine serum, 1% penicillin/streptomycin, 1% 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer, and 2 µM 2-ME.
Viral transduction and plasmids
The retroviral pMSCV vector containing murine TLR9 fused at the C terminus to green fluorescent protein (GFP) (pMSCV-TLR9-GFP). Retroviral transduction was performed as previously described (36). Hemagglutinin (HA) epitope was appended on CD82 fused at the N terminus (HA-CD82). The HA-CD82 cDNA was placed in the pMSCV vector, and retrovirus was produced. RAW macrophages were infected with the HA-CD82 retrovirus. Previously, CD82 cDNA was amplified by PCR from BALB/c mouse spleen and fused at the C terminus to murine (m) red fluorescent protein (RFP)1 (CD82-mRFP1) (14). This construct was inserted into the lentiviral pHAGE vector (14). Lentivirus generation in HEK293T cells and transduction were performed as previously described (14).
The TLR7-3xFLAG construct was a gift from Dr. Gregory Barton’s laboratory (University of California, Berkeley, CA, USA). TLR7-3xFLAG was generated as previously described by Newman et al. (38). The retroviral vector pMSCV-Thy1.1 (IRES-Thy1.1) containing the murine TLR7 cDNA fused to a 3xFlag tag at its C terminus was used to generate retrovirus in GP2-293 cells (Takara Bio, Kyoto, Japan) for subsequent transduction of immortalized BMDM. In brief, GP2-293 cells were plated in 6-well plates with a density of 0.5 × 106 cells per well. The following day, they were transfected with 1.7 µg TLR7 pMSCV and 0.83 µg vesicular stomatitis virus G-protein (VSV-G) using TransIT-293 transfection reagent (Mirus, Madison, WI, USA). The next day, the cells were moved to 32°C, and immortalized BMDMs were seeded at 3.0 × 105 cells per well of a 6-well plate. The supernatant containing retroviral particles was harvested and filtered through 40-µm filters. PolyBrene was added at a 1:1000 dilution to the virus, and the target cells were then transduced at 32°C overnight.
TNF-α production in mice sera
The protocol for serum TNF-α production in mice was adopted from previously described protocols in refs. 31 and 39. Age-matched WT, CD82KO, and TLR9KO mice (n = 3/group) on C57BL/6 background were intraperitoneally injected with 20 mg d-galactosamine (D-GalN; CNH Technologies, Woburn, MA, USA) +/− 20 nmol CpG (n = 3) for 1 and 3 h, or WT and CD82KO mice were intraperitoneally injected with 300 µg of LPS (MilliporeSigma, Burlington, MA, USA) for 1.5 h. Mice were euthanized, and sera were collected at indicated times. Serum TNF-α concentrations were measured by ELISA kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s specifications. Results are the means of sera samples from 3 mice, and control mice were used as baseline values.
ELISA
A total of 1 × 105 macrophages were plated in triplicate on tissue culture–treated 48-well plates and stimulated with ligands for the indicated times. Supernatants were collected for cytokine analysis (TNF-α) with ELISA per the manufacturer’s instructions (R&D Systems).
Immunoprecipitation assay
Macrophages at 1 × 106 were plated on 15-cm tissue culture–treated plates overnight. Cells were stimulated with TLR ligands as indicated for 1 h. Macrophages at 1 × 106 were plated on 15-cm tissue culture–treated plates overnight. Cells were stimulated with TLR ligands as indicated for 1 h. Except for myddosome experiments, the cells were then lysed in 700 µl lysis buffer containing 1% Brij-58, 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 10% glycerol, and protease/phosphatase inhibitors (MilliporeSigma). Lysis buffer containing 1% Nonidet P-40 (NP-40) was used for the myddosome immunoprecipitation Western blots. A total of 600 µl of cleared lysate was incubated with the 1 µg anti–myeloid differentiation primary response 88 (MyD88; R&D Systems) or anti-GFP (MilliporeSigma) with 50 µl of protein G Dynabeads (Thermo Fisher Scientific) overnight. The beads were washed 3 times in the lysis buffer prior to overnight incubation with cell lysate. For HA-CD82 pull-down, 600 µl of cell lysate was incubated with 100 µl of the anti-HA affinity matrix beads (3F10; Roche, Basel, Switzerland) overnight. The following day, beads were washed 3 times with wash buffer containing 1% NP-40 or 1% Brij-58, 200 mM Tris-HCL (pH 7.4), 600 mM NaCl, and 20 mM EDTA. Proteins were extracted by adding 50 µl Laemmli buffer with reducing agent and boiled at 95°C for 5 min. The proteins were then resolved by SDS-PAGE in 4–12% gels (Thermo Fisher Scientific), and methanol-activated PVDF membrane (Perkin Elmer, Waltham, MA, USA) was applied to the gel in transfer buffer (0.025 M Tris, 0.192 M glycine, 20% methanol). All buffer components were purchased from National Diagnostics (Atlanta, GA, USA) or MilliporeSigma. The gel and PVDF membrane were sandwiched between transfer sponge and blotting paper and were subjected to electrophoretic transfer at 100 V for 1 h. For detection of proteins, PVDF-immobilized gel transfers were blocked with 5% milk in PBS-0.01% Tween 20 (PBS-T; MilliporeSigma) overnight at 4°C or 1 h at room temperature. Blots were incubated with anti–GFP-horseradish peroxidase (HRP) (Santa Cruz Biotechnology, Dallas, TX, USA), anti-HA-HRP (3F10; MilliporeSigma), anti–interleukin-1 receptor-associated kinase (IRAK)2 (ProSci, San Diego, CA, USA), or anti–vesicle associated membrane protein 3 (VAMP3; Novus Biologicals, Centennial, CO, USA) in 1% milk in PBS-T for 1 h at room temperature. The membranes were washed and visualized using the Western Lighting Plus ECL chemiluminescent substrate (Perkin Elmer) on Kodak BioMax XAR Film (MilliporeSigma). Films were then scanned and cropped, and contrast was adjusted evenly to the entire image using Adobe Photoshop CC 2015 (Adobe Systems, San Jose, CA, USA).
TLR9 immunoprecipitation
Macrophages at 2 × 107 were plated on 15-cm tissue culture–treated plates overnight. The cells were then lysed in 1 ml lysis buffer containing 1% NP-40, 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 10% glycerol, and protease/phosphatase inhibitors (MilliporeSigma). The lysate was incubated with 100 µl of protein G Dynabeads and 25 µg of the TLR9 monoclonal (NaR9) antibody (40) overnight. The beads were washed 3 times in the lysis buffer prior to overnight incubation with cell lysate. The following day, beads were washed 3 times with wash buffer containing 1% NP-40 or 1% Brij-58, 200 mM Tris-HCL (pH 7.4), 600 mM NaCl, and 20 mM EDTA. Proteins were processed as described in the Materials and Methods section. For detection of proteins, PVDF-immobilized gel transfers were blocked with Block Ace (Bio-Rad, Hercules, CA, USA) in PBS-T for 1 h at room temperature. Blots were incubated with TLR9 pAb (41) in CanGet Signal Reagent 1 (Toyobo, Osaka, Japan) for 1 h at room temperature. The membranes were washed and then incubated with anti-rabbit HRP antibody (Agilent Technologies, Santa Clara, CA, USA) in CanGet Signal Reagent 2 for 1 h at room temperature. The membranes were washed and visualized as previously described.
Nuclear extraction
NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Fisher Scientific) was used to separate and prepare nuclear extracts from immortalized macrophages per the manufacturer’s instructions.
Confocal microscopy
Macrophages were plated overnight onto 8-chambered coverslips (Thermo Fisher Scientific). Cells were stimulated with Alexa Fluor 647–labeled CpG at 37°C for specified times. For live-cell imaging, coverslips were mounted on a Nikon Ti-E inverted microscope equipped with CSU-X1 confocal spinning-disk head (Yokogawa, Tokyo, Japan). An excitation light source, 4-W laser (Coherent, Santa Clara, CA, USA), was used to produce excitation wavelengths of 488, 568, and 647 nm using an acoustic optical tunable tuner. A high-magnification, high–numerical aperture objective (1003, 1.49 numerical aperture, oil immersion; Nikon, Tokyo, Japan) was used to acquire high-quality fluorescence images. To acquire differential interference contrast images, a polarizer (MEN 51941; Nikon) and Wollaston prisms (MBH76190; Nikon) were used. Emission light from the sample was collected after passage through the appropriate emission filters (Semrock, Rochester, NY, USA). Images were acquired using an electron-multiplying charge-coupled device camera (C9100-13; Hamamatsu, Hamamatsu, Japan). Image acquisition was performed using MetaMorph software (Molecular Devices, Sunnyvale, CA, USA). Raw image data files were processed using Adobe Photoshop CC 2015 and assembled in Adobe Illustrator CC 2015. Pearson correlation coefficient (PCC) analysis was performed on ∼100 cells to quantify colocalization between proteins.
Cell staining and flow cytometry
Cells were harvested from bone marrow or spleen in PBS + 0.5% bovine serum albumin + 2 mM EDTA and depleted of red cells [ACK lysis buffer (Thermo Fisher Scientific) or red blood cell lysis buffer (MilliporeSigma)]. The single-cell suspension was blocked with Fc Block (1:100 dilution) (eBioscience, San Diego, CA, USA) for 20 min at room temperature. Surface staining was performed in the dark for 30 min at 4°C in fluorescence-activated cell sorting (FACS) buffer. Cells were then washed twice with FACS buffer followed by fixation in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA, USA). A list of surface markers for these experiments (1:200 dilution; eBioscience) include B220-APC, CD4-Qdot 655, CD8a-PE, CD45-Cy7, CD30-APC, IgM-PE, IgD-FITC, CD21-FITC, CD23-PE, CD138-PE, CD19-APC, CD90-Pacific Orange, and CD11b-Pacific Orange.
All experimental samples were acquired using an LSR II Flow Cytometer (BD Biosciences, San Jose, CA, USA) located in the MGH Center for Regenerative Medicine Flow Cytometry Core Facility and analyzed using FlowJo software (Treestar, Ashland, OR, USA). Compensation was performed using single-color controls prepared from BD Comp Beads (BD Biosciences) for cell surface staining and live/dead cell staining (Thermo Fisher Scientific) for viability. Compensation matrices were calculated and applied using BD FACS Diva software.
Real-time quantitative PCR
Total RNA was extracted using the RNeasy Kit and DNase treated according to the manufacturer’s protocol (Qiagen, Germantown, MD, USA), and each sample was reverse transcribed using multiscribe reverse transcriptase (Applied Biosystems, Foster City, CA, USA). The 25-µl quantitative PCR reaction contained 2 µl of cDNA, 12.5 µl of 2× Sybr Green Master Mix (Applied Biosystems), and 500 nM of sense and antisense primers. Oligonucleotide primer sequences designed on the PrimerBank website obtained from Integrated DNA Technologies were as follows: mTLR4 forward 5′-ACCTGGCTGGTTTACACGTC-3′, reverse 5′-CTGCCAGAGACATTGCAGAA-3′; mTLR7 forward 5′-AATCCACAGGCTCACCCATA-3′, reverse 5′-AGGTACCAAGGGATGTCCT-3′; and mTLR9 forward 5′-ACTGAGCACCCCTGCTTCTA-3′, reverse 5′-AGATTAGTCAGCGGCAGGAA-3′.
Emitted fluorescence for each reaction was measured 3 times during the annealing-extension phase, and amplification plots were measured using Roche LightCycle 96 (Roche). Data were analyzed with MX4000 software, v.3.0 (Stratagene, San Diego, CA, USA). The quantity of gene expression was generated by comparison of the fluorescence generated by each sample with standard curves of known quantities and normalized by dividing by the number of copies of the housekeeping gene β2-microglobulin. Normalized values from each cell line were then divided by the normalized WT value to calculate the expression ratio.
Statistics
For ELISA, PCC, and densitometry analyses, statistical calculations were performed using Prism 7 software (GraphPad Software, La Jolla, CA, USA). Data were analyzed by a 2-tailed, unpaired Student’s t test or 2-way ANOVA Tukey’s multiple comparisons test and were considered significantly different when P ≤ 0.05.
RESULTS
TLR9 signaling is dependent on CD82
CD82 interacts with class II MHC molecules and chaperone proteins that regulate antigen processing and presentation (14, 16, 42). We previously demonstrated that CD82 is specifically recruited to bacterial- and fungal-containing phagosomes in macrophages (15). Additionally, CD82 organizes dectin-1 clustering on the phagosomes and the signaling machinery in response to fungi (34). Based on these findings, we hypothesized that CD82 plays a critical role in innate immunity. To define the role of CD82 in the innate immune system, we identified proteins associated with CD82 in resting macrophages by mass spectrometry, which revealed multiple TLR proteins including TLR1, TLR2, TLR7, and TLR9.
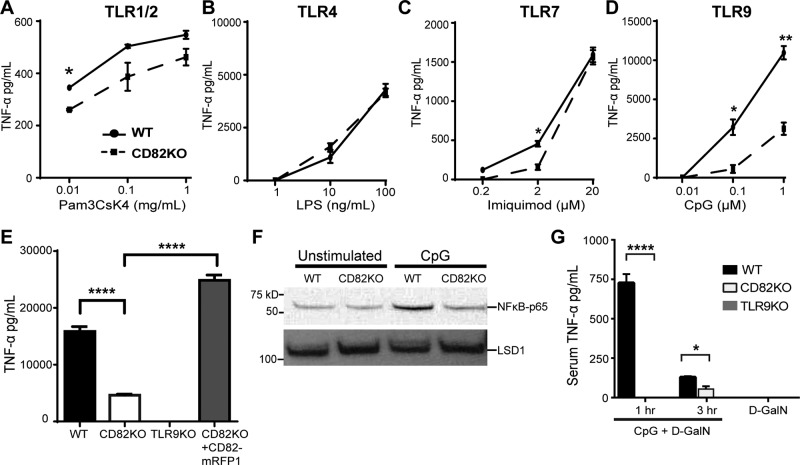
Upon ligand recognition, TLR signaling pathways activate NF-κB and trigger cytokine production (23). To determine whether CD82 regulates TLR signaling, we assessed TNF-α production in WT and CD82-knockout (CD82KO) macrophages in response to specific TLR agonists, including Pam3CsK4 (TLR1/2 agonist), LPS (TLR4 agonist), imiquimod (TLR7 agonist), and CpG (TLR9 agonist). All experiments with CpG used the class B CpG. In CD82KO macrophages stimulated with TLR1/2- and TLR7-dependent agonists, we observed minor differences in TNF-α secretion compared with WT (Fig. 1A, C). We observed a comparable dose-dependent increase in TNF-α secretion in response to TLR4 activation in both WT and CD82KO macrophages (Fig. 1B). In contrast, CD82KO macrophages stimulated with CpG produced significantly less TNF-α than WT macrophages (Fig. 1D), indicating a critical role for CD82 in TLR9 signaling. To confirm this observation, we tested whether restoration of CD82 expression in CD82KO macrophages would enable these cells to produce TNF-α in response to CpG. We stimulated CD82KO macrophages expressing CD82-mRFP1 with CpG and observed robust TNF-α production similar to WT macrophages (Fig. 1E). TLR9KO macrophages did not produce TNF-α, as expected (Fig. 1E). Our results demonstrate that CD82 controls TLR9-dependent TNF-α production in macrophages.
Figure 1.
A–D) CD82 regulates TLR9 signaling in macrophages and in vivo in response to CpG. TNF-α production in WT or CD82KO immortalized macrophages was measured by ELISA in response to increasing doses of Pam3CsK4 (A), LPS (B), imiquimod (C), or CpG (D) for 6 h. E) WT, CD82KO, TLR9KO, or CD82KO + CD82-mRFP1 macrophages stimulated with 1 µM CpG for 16 h and assessed for TNF-α production. F) WT and CD82KO macrophages stimulated with 1 µM CpG for 2 h or unstimulated were assessed for NF-κB translocation in nuclear lysates by immunoblot. LSD1, lysine-specific histone demethylase 1A. G) Sera of WT, CD82KO, and TLR9KO mice (n = 3 mice/group) intraperitoneally injected with 20 nmol CpG and/or 20 mg D-GalN and assessed for TNF-α production. ELISA results are presented as means of technical (A–E) or biologic (G) triplicates ± sd. *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001 (unpaired Student’s t test).
NF-κB activation plays a pivotal role in inflammatory and immune responses (43). Upon TLR activation, NF-κB translocates into the nucleus for targeted gene transcription, including TNF-α. To establish whether CD82 is involved in TLR9-mediated NF-κB activation, we compared nuclear translocation of NF-κB in WT and CD82KO macrophages stimulated with CpG or solvent control (PBS). Proteins from nuclear lysates were resolved by SDS-PAGE and probed for the p65 subunit of NF-κB. We detected baseline levels of NF-κB-p65 in the nuclear lysates of unstimulated WT and CD82KO macrophages. CpG stimulation of WT macrophages induced NF-κB–p65 translocation to the nucleus, whereas NF-κB translocation in CpG-stimulated CD82KO macrophages was similar to unstimulated macrophages (Fig. 1F, top blot). We confirmed equal loading of protein by probing for lysine-specific histone demethylase 1A, a nuclear protein (Fig. 1F, bottom blot). This result provides additional evidence that CD82 is required for NF-κB–p65 activation in response to CpG.
Despite the involvement of CD82 in a wide variety of biologic processes (12, 44, 45), the role of CD82 in an in vivo stimulation model has not been tested. To determine whether CD82 controls TLR9 signaling in vivo, we tested a mouse model of CpG-induced lethal shock in D-GalN–sensitized mice (46, 47). CD82KO mice are phenotypically normal (9). A detailed analysis of the immune system revealed equivalent T-cell, B-cell, neutrophil, and monocyte counts in CD82KO mice compared with WT controls (Supplemental Fig. S1). Additionally, there were comparable Ig levels and subsets. Architecture and size of lymph nodes and spleen were also similar. We injected intraperitoneally WT and CD82KO mice with D-GalN and CpG and assessed serum TNF-α levels 1 and 3 h after injection. As previously demonstrated by Hemmi et al. (31), WT mice injected with CpG + D-GalN demonstrated elevated levels of TNF-α in sera at 1 h, which was reduced at 3 h (Fig. 1G). In sharp contrast, we did not detect TNF-α in sera from CD82KO mice injected with CpG + D-GalN at 1 h, indicating a role for CD82 in TLR9-dependent signaling in vivo. Although a modest amount of TNF-α was present in sera from CD82KO mice after 3 h (Fig. 1G), these levels were significantly less than WT mice. As expected, TLR9KO mice did not produce TNF-α in response to CpG + D-GalN (Fig. 1G). Mice injected with D-GalN alone did not produce TNF-α. To determine whether CD82 regulation of serum TNF-α was TLR9 specific, we tested response to the activation of TLR4. We injected intraperitoneally WT and CD82KO mice with LPS and D-GalN only and assessed serum TNF-α levels 1.5 h postinjection. WT mice injected with LPS produced elevated serum levels of TNF-α (Supplemental Fig. S2), as previously shown by Blanqué et al. (39). Similarly, CD82KO mice injected with LPS produced comparable amounts of TNF-α as the WT mice (Supplemental Fig. S2), demonstrating that CD82 is not required for TLR4 signaling in vivo. Our data indicate that CD82 controls TLR9-induced TNF-α production in macrophages and in vivo (Fig. 1D, G). Our data demonstrate a critical role for CD82 in modulating TLR9 signaling.
CD82 and TLR9 associate in macrophages
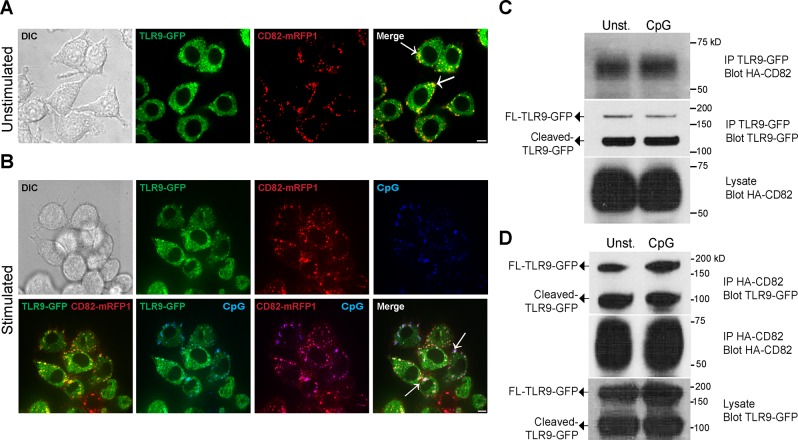
Tetraspanins interact with diverse transmembrane and intracellular proteins, including pattern recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (45, 48). Our finding that CD82 regulates TLR9 signaling led us to hypothesize that CD82 associates with TLR9 in macrophages. To test this hypothesis, we stimulated RAW macrophages stably transduced with TLR9-GFP and CD82-mRFP1 with CpG. The addition of GFP onto TLR9 or mRFP1 onto CD82 did not alter their function or subcellular distribution (15, 30). Using confocal microscopy, we observed that CD82-mRFP1 and TLR9-GFP colocalized in both unstimulated and CpG-stimulated cells (Fig. 2A, B). To quantify these findings, we analyzed TLR9-GFP and CD82-mRFP1 colocalization using PCC analysis. We did not observe differences in TLR9-GFP and CD82-mRFP1 colocalization between CpG-stimulated and unstimulated cells (Supplemental Fig. S3). Thus, TLR9 and CD82 associate in macrophages independent of CpG stimulation.
Figure 2.
CD82 and TLR9 associate in macrophages. A, B) Confocal imaging of RAW macrophages expressing TLR9-GFP (green) and CD82-mRFP1 (red). Macrophages incubated with PBS (A) or 1 µM Alexa Fluor 647–conjugated CpG (B) (blue) for 1 h. White arrows indicate colocalization. Scale bars, 5 µm. C, D) Immunoprecipitation (IP) of TLR9-GFP or HA-CD82 in WT macrophages stimulated with either PBS (Unst.) or 1 µM CpG. C) Western blot of lysates immunoprecipitated with anti-GFP and probed for HA-CD82 (top blot) or TLR9-GFP (second blot). Lysates probed for HA-CD82 (third blot). Western blot of lysates immunoprecipitated with anti-HA and probed for TLR9-GFP (top blot) or HA-CD82 (second blot) (D). Lysates probed for TLR9-GFP (third blot). DIC, differential interference contrast.
To confirm our microscopy results, we used a biochemical approach to test the interaction between TLR9 and CD82. WT macrophages stably expressing TLR9-GFP and CD82 with an amino-terminal HA tag (HA-CD82) were stimulated with CpG. Macrophages were lysed using a mild detergent (Brij-58) to preserve both weak and strong tetraspanin-protein interactions (1). To determine whether TLR9 enters into TEMs, TLR9-GFP immunoprecipitates were resolved by SDS-PAGE and probed for HA-CD82. TLR9-GFP and HA-CD82 interacted in both CpG-stimulated and unstimulated macrophages (Fig. 2C, top blot). We probed the TLR9-GFP immunoprecipitates with anti-GFP to confirm the presence of full-length and cleaved TLR9-GFP immunoprecipitated from the cell lysates, and no difference was observed between unstimulated and CpG-treated cells (Fig. 2C, second blot). The overall levels of HA-CD82 in the whole-cell lysates were comparable in the unstimulated and CpG-stimulated macrophages (Fig. 2C, third blot). To verify that the interaction between CD82 and TLR9 is specific, we performed the reverse experiment; we immunoprecipitated HA-CD82 and then immunoblotted for TLR9-GFP. We found that both full-length and cleaved TLR9-GFP interacted with HA-CD82 in CpG-stimulated and unstimulated macrophages (Fig. 2D, top blot). We confirmed that HA-CD82 immunoprecipitation was equal by probing the HA-CD82 immunoprecipitates with anti-HA (Fig. 2D, second blot). The expression of TLR9-GFP (full length and cleaved) was also comparable in unstimulated and CpG-stimulated macrophages (Fig. 2D, third blot). Consistent with live-cell imaging, these results indicate that CD82 and TLR9 interact in macrophages, independent of CpG stimulation. The use of a more stringent detergent (NP-40) abrogated the CD82-TLR9 interaction, suggesting that CD82 may interact indirectly with TLR9 in TEMs. To determine whether CD82 interacts with other intracellular TLRs, we tested the interaction between CD82 and TLR7. We immunoprecipitated HA-CD82 from WT macrophages stably expressing HA-CD82 and TLR7-3xFLAG (TLR7-FLAG) or TLR9-GFP. Immunoprecipitates were separated by SDS-PAGE and then immunoblotted for FLAG. We found both the full-length and cleaved fraction of TLR7-FLAG in HA-CD82 immunoprecipitates (Supplemental Fig. S4A), indicating that TLR7-FLAG and HA-CD82 interact in macrophages. These data demonstrate that CD82 not only associates with TLR9 but also interacts with TLR7. As a negative control for our immunoprecipitation experiments, we examined Rab7, a membrane protein in the endosome and a marker of phagosomal maturation. Consistent with previous findings by Koh et al. (49), we did not find an interaction between HA-CD82 and Rab7 (Supplemental Fig. S4B).
CD82 and TLR9 interaction occurs in the ER and post-ER
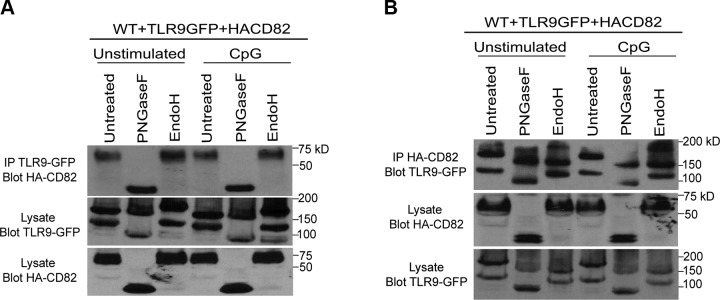
We next sought to determine where CD82 and TLR9 interaction occurs (i.e., ER or post-ER). To test this, we stimulated WT macrophages expressing TLR9-GFP and HA-CD82 with CpG. We immunoprecipitated TLR9-GFP and treated these immunoprecipitates with peptide-N-glycosidase F (PNGaseF) or endoglycosidase H (EndoH), enzymes that cleave glycans (acquired through the secretory pathway). N-linked glycans are acquired in the ER, and these glycans are further modified in the Golgi, rendering them resistant to EndoH. The immunoprecipitates are then subjected to SDS-PAGE, followed by immunoblot for HA-CD82. We found that CD82 recruited to TLR9-GFP immunoprecipitates is sensitive to PNGaseF, indicated by the decrease in the MW of CD82, but acquired resistance to EndoH in both unstimulated and CpG-stimulated macrophages (Fig. 3A, top blot), indicating a post-ER interaction. The amount of TLR9-GFP was comparable across all conditions (unpublished results). When TLR9-GFP was probed in the lysates, we found both full-length and cleaved TLR9-GFP (Fig. 3A, second blot). We observed that full-length TLR9-GFP is sensitive to both PNGaseF and EndoH, as demonstrated by the decrease in MW of full-length TLR9-GFP as compared with untreated, indicating that full-length TLR9-GFP resides in the ER. However, cleaved TLR9-GFP is only sensitive to PNGaseF and EndoH resistant, indicating that cleaved TLR9-GFP is found post-ER (Fig. 3A, second blot). The expression of HA-CD82 in lysates was equal across all conditions (Fig. 3A, third blot). Overall, these data demonstrate that full-length and cleaved TLR9 interact with CD82 post-ER.
Figure 3.
Full-length TLR9 and CD82 interact in the ER, whereas cleaved TLR9 and CD82 interact post-ER. A, B) WT macrophages expressing TLR9-GFP and HA-CD82 stimulated with 1 µM CpG for 1 h or unstimulated. A) Western blot of lysates immunoprecipitated with anti-GFP and probed for HA-CD82 (top blot). Cell lysates probed for TLR9-GFP (second blot) and HA-CD82 (third blot). B) Western blot of lysates immunoprecipitated with anti-HA and probed for TLR9-GFP (top blot). Cell lysates probed for HA-CD82 (second blot) and TLR9-GFP (third blot). FL, full length; IP, immunoprecipitation.
We performed the reverse immunoprecipitation (i.e., immunoprecipitation HA-CD82; blot TLR9-GFP) to further elucidate the locations of CD82 interaction. We performed PNGaseF and EndoH digestion on HA-CD82 immunoprecipitates obtained from WT macrophages. As expected, we found both full-length and cleaved TLR9-GFP in untreated HA-CD82 immunoprecipitates (Fig. 3B, top blot). The full-length TLR9-GFP is sensitive to both PNGaseF and EndoH in unstimulated and CpG-stimulated macrophages, indicated by the decrease in full-length TLR9-GFP MW as compared with untreated HA-CD82 immunoprecipitates (Fig. 3B, top blot). Moreover, the MW of TLR9-GFP found in HA-CD82 immunoprecipitates is decreased with PNGaseF but acquired resistance to EndoH as compared with untreated (Fig. 3B, top blot). These findings indicate that the interaction between CD82 and full-length TLR9-GFP occurs in the ER, whereas CD82 and cleaved TLR9 interact post-ER. The amount of HA-CD82 immunoprecipitated was comparable across all conditions (unpublished results). We also found comparable amounts of TLR9-GFP and HA-CD82 in the lysate (Fig. 3B, second and third blot). It should be noted that a faint band of HA-CD82 was observed in the EndoH-treated lysate (Fig. 3B second blot), suggesting that a small amount of EndoH-sensitive CD82 is present. Collectively, full-length TLR9 and CD82 interact in the ER, whereas the interaction between cleaved TLR9 and CD82 occurred post-ER.
CD82 controls TLR9 trafficking to acidified CpG-containing compartments
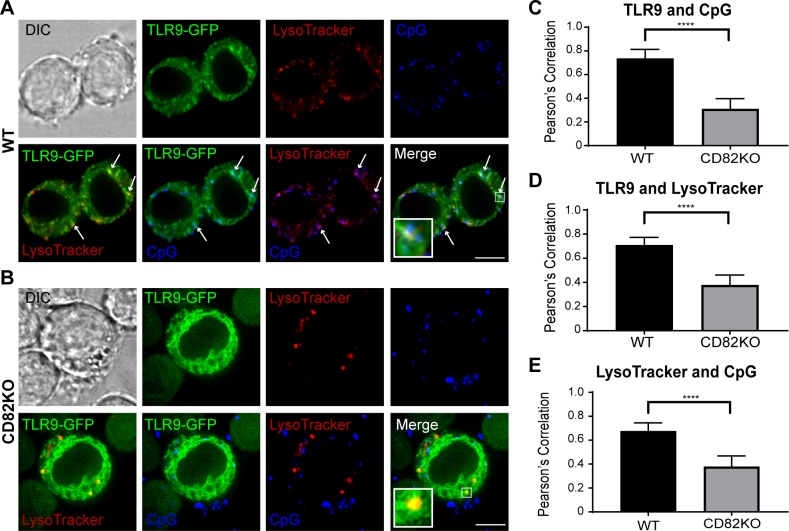
Acidification plays a key role in receptor activation and antigen processing. TLR9 activation requires proteolytic cleavage of the ectodomain by cathepsin L, cathepsin S, and insulin-regulated aminopeptidase in endolysosomal compartments (24, 29, 32, 35, 50, 51). CD82 is present on pathogen-containing phagosomes (15) and endolysosomal tubules that deliver peptide-loaded class II MHC to the cell surface (14), suggesting a possible role for CD82 in the regulation of protein trafficking in the endolysosomal pathway. Based on these findings, we hypothesized that CD82 functions as a regulator of TLR9 trafficking to CpG-containing acidified compartments. We pretreated WT and CD82KO macrophages expressing TLR9-GFP with LysoTracker to visualize intracellular acidified compartments, followed by stimulation of the cells with fluorescently labeled CpG. Using confocal microscopy, we visualized the subcellular distribution of TLR9-GFP, LysoTracker, and Alexa Fluor 647–labeled CpG in WT and CD82KO macrophages. In WT macrophages, we observed that TLR9-GFP colocalized with LysoTracker-positive and CpG-positive compartments. Additionally, CpG-containing compartments overlapped with LysoTracker-positive compartments (Fig. 4A). In contrast, the colocalization of TLR9-GFP with LysoTracker-positive and CpG-containing compartments in CD82KO macrophages appeared reduced (Fig. 4B). Likewise, colocalization between CpG-containing compartments with LysoTracker-positive compartments was diminished compared with WT. PCC analysis revealed that TLR9-GFP–positive compartments colocalized with CpG compartments in WT macrophages to a greater degree than CD82KO macrophages (Fig. 4C), indicating that CD82 facilitates trafficking of TLR9 into CpG-positive compartments. Furthermore, TLR9 colocalized with LysoTracker-positive compartments to a greater degree in WT macrophages compared with CD82KO macrophages in response to CpG (Fig. 4D). Finally, less accumulation of CpG in LysoTracker-positive compartments was observed in CD82KO macrophages when compared with WT macrophages (Fig. 4E). We did not observe differences in the overall acidification of the compartments in WT and CD82KO macrophages. Our confocal imaging analysis indicates that CD82 regulates TLR9 trafficking to acidified CpG-containing compartments.
Figure 4.
CD82 controls TLR9 trafficking to acidified CpG-containing compartments. Confocal imaging of WT (A) and CD82KO (B) macrophages expressing TLR9-GFP (green) stimulated with 1 µM Alexa Fluor 647–conjugated CpG (blue) and stained with LysoTracker (red). White arrows indicate colocalization. Scale bars, 5 µm. C–E) Grap represents mean PCC ± sd (n ∼ 100 images) for colocalization between TLR9-GFP and CpG (C), TLR9-GFP and LysoTracker (D), and CpG and LysoTracker (E). DIC, differential interference contrast. ****P ≤ 0.0001 (unpaired Student’s t test; n = 100 macrophages).
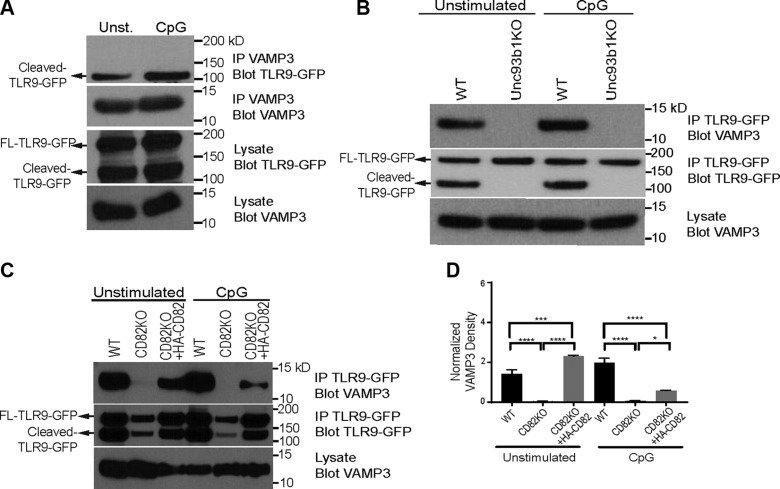
CD82 controls the interaction between TLR9 and VAMP3
Recruitment of specific proteins to the acidified compartments determines the fate of downstream signaling (26). VAMP3 is a member of the N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) family, which is involved in intracellular vesicle fusion (52) and downstream signaling. VAMP3 traffics to TLR9-positive compartments in CpG-stimulated macrophages (26). We sought to determine whether TLR9 and VAMP3 interact in macrophages. We stimulated WT macrophages stably transduced with TLR9-GFP with CpG. The lysates were immunoprecipitated for VAMP3, resolved by SDS-PAGE, and probed for TLR9-GFP. We observed an interaction between cleaved TLR9 and VAMP3 in WT macrophages, independent of CpG stimulation (Fig. 5A, top blot), suggesting this interaction occurs in the acidified compartments where the proteolytic cleavage of TLR9 occurs. The expression level of VAMP3 in the immunoprecipitates was comparable in all of the conditions (Fig. 5A, second blot). The overall amounts of TLR9-GFP and VAMP3 expressed in these macrophages were also similar (Fig. 5A, third and fourth blots). These data demonstrate that VAMP3 specifically associates with the cleaved isoform of TLR9.
Figure 5.
TLR9 and VAMP3 interaction is mediated by CD82. A) WT macrophages expressing TLR9-GFP stimulated with 1 µM CpG for 1 h or unstimulated. Western blot of lysates immunoprecipitated with anti-VAMP3 and blotted for TLR9-GFP (top blot) or VAMP3 (second blot). Lysates probed for TLR9-GFP (third blot) and VAMP3 (fourth blot). Unst., unstimulated. B) WT and UNC93B1-KO macrophages expressing TLR9-GFP stimulated with 1 µM CpG for 1 h or unstimulated. Western blot of lysates immunoprecipitated with anti-GFP and probed for VAMP3 (top blot) or TLR9-GFP (second blot). Lysates probed for VAMP3 (third blot). C) WT and CD82KO macrophages expressing TLR9-GFP or CD82KO macrophages expressing TLR9-GFP and HA-CD82 stimulated with 1 µM CpG for 1 h or unstimulated. Western blot of lysates immunoprecipitated with anti-GFP and probed for VAMP3 (top blot) or TLR9-GFP (second blot). Lysates probed for VAMP3 (third blot). D) Densitometry analysis of VAMP3 found in TLR9-GFP immunoprecipitates, using the blot from panel C. FL, full length; IP, immunoprecipitation. *P ≤ 0.05, ***P ≤ 0.01, ****P ≤ 0.0001 (2-way ANOVA Tukey’s multiple comparisons test).
Proteolytic cleavage of TLR9 by cathepsins in the endolysosomal compartment is required for its activation and downstream signaling in antigen presenting cells (APCs) (28–30). UNC93B1 is an ER resident protein that delivers endosomal TLRs, including TLR9, to endolysosomal compartments for signaling (24, 50). We hypothesized that UNC93B1 is required for TLR9 and VAMP3 interaction. TLR9-GFP immunoprecipitates from unstimulated or CpG-stimulated WT and UNC93B-KO macrophages were probed for VAMP3. We observed an interaction between VAMP3 and TLR9-GFP in both unstimulated and CpG-stimulated WT macrophages, but this interaction was absent in UNC93B1-KO macrophages, despite robust VAMP3 expression in both cell types (Fig. 5B, top blot and third blot). In the WT macrophages, we observed both full-length and cleaved TLR9-GFP in the TLR9-GFP immunoprecipitates, whereas only the full-length TLR9-GFP was found in the UNC93B1-KO macrophages (Fig. 5B, second blot). These data indicate that UNC93B1 controls the trafficking of TLR9 to VAMP3-positive compartments.
We next sought to determine whether the interaction between TLR9 and VAMP3 requires CD82. We stimulated WT and CD82KO macrophages stably transduced with TLR9-GFP with CpG. The lysates were immunoprecipitated for TLR9-GFP, resolved by SDS-PAGE, and probed for endogenous VAMP3. We observed an interaction between TLR9 and VAMP3 in WT macrophages, independent of CpG stimulation (Fig. 5C, top blot). In contrast, unstimulated CD82KO macrophages showed a reduced TLR9-VAMP3 interaction (Fig. 5C, top blot). Moreover, the interaction between TLR9 and VAMP3 was completely absent in CD82KO macrophages stimulated with CpG (Fig. 5C, top blot), suggesting that CD82 is important for the interaction between TLR9 and VAMP3. To confirm this observation, we stimulated CD82KO macrophages expressing HA-CD82 with CpG and observed that TLR9 and VAMP3 interaction was restored (Fig. 5C, top blot). Although our TLR9-GFP immunoprecipitates were positive for both full-length and cleaved TLR9-GFP, the expression and proteolytic cleavage of TLR9-GFP in CD82KO- and HA-CD82–expressing macrophages were not comparable to WT macrophages (Fig. 5C, second blot). Similarly, the expression of VAMP3 in the lysates of WT and CD82KO macrophages was not comparable (Fig. 5C, third blot). To address this issue, densitometry was used to quantitate the total amount of VAMP3 interacting with TLR9-GFP by normalizing the pixel density against VAMP3 expression levels in lysates (Fig. 5D). CD82 expression restores TLR9 and VAMP3 interaction in unstimulated and CpG-stimulated macrophages as compared with CD82KO macrophages (Fig. 5D). However, in response to CpG, the amount of VAMP3 and TLR9 interaction is significantly reduced in comparison to WT (Fig. 5D). Collectively, these data indicate that CD82 is required for delivery of TLR9 to VAMP3-positive acidified compartments.
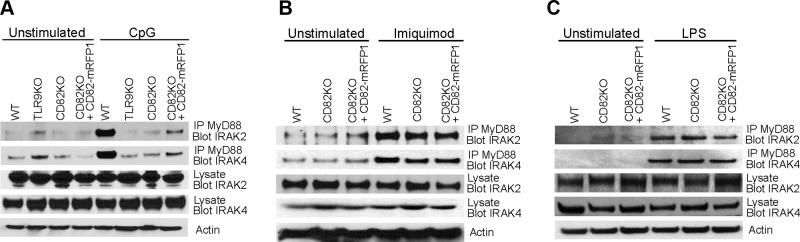
CD82 controls TLR9-dependent myddosome assembly
MyD88 is a TLR adaptor protein critical for the activation of NF-κB and cytokine production. MyD88 forms a complex with IRAK4, which sequentially recruits IRAK2 and forms a myddosome complex (21, 23). Based on our data demonstrating that CD82 regulates TLR9-dependent NF-κB activation (Fig. 1G), we sought to determine whether CD82 affects TLR-dependent myddosome assembly. To assess the role of CD82 in TLR9-induced myddosome assembly, we stimulated WT, TLR9KO, CD82KO, and CD82KO + CD82-mRFP1 macrophages with CpG and immunoprecipitated endogenous MyD88. The proteins from the immunoprecipitates were resolved by SDS-PAGE and probed for IRAK2 and IRAK4. In unstimulated cells, the presence of IRAK2 and IRAK4 in MyD88 immunoprecipitates indicated baseline levels of myddosome formation (Fig. 6A, top and second blots), despite robust expression of IRAK2 and IRAK4 in the lysate controls (Fig. 6A, third and fourth blots). In CpG-stimulated WT macrophages, we observed an increased interaction between IRAK2 or IRAK4 and MyD88, indicative of myddosome assembly (Fig. 6A, top blot). In contrast, CD82KO macrophages stimulated with CpG showed minimal interaction between MyD88 with IRAK2 and IRAK4, similar to unstimulated CD82KO macrophages (Fig. 6A, top and second blots), suggesting that CD82 controls TLR9-dependent myddosome formation. Expression of CD82-mRFP1 in CD82KO macrophages restored the interaction between MyD88 and IRAK2 or IRAK4 in response to TLR9 activation with CpG (Fig. 6A, top blot). We confirmed comparable protein loading in all conditions with actin (Fig. 6A, fifth blot). Additionally, we detected TLR9 gene and protein expression in CD82KO and CD82-mRFP1 expressing macrophages, which were comparable to WT macrophages (Supplemental Fig. S5A, B). We found that overexpression of TLR9-GFP in WT or CD82KO macrophages does not alter TNF-α production, and the requirement of CD82 persists (Supplemental Fig. S5C). The amount of TNF-α produced in WT macrophages is similar to that of WT + TLR9-GFP macrophages. Similarly, expression of TLR9-GFP in CD82KO macrophages did not affect TNF-α production as compared with CD82KO macrophages. These data indicate that CD82 regulates TLR9-dependent myddosome formation.
Figure 6.
CD82 promotes TLR9-induced myddosome assembly and is dispensable for TLR7- and TLR4-induced myddosome formation. WT, TLR9KO, CD82KO, and CD82KO + CD82-mRFP1 macrophages stimulated with 1 µM CpG (A), 20 µM imiquimod (B), or 100 ng/ml LPS (C) for 2 h. MyD88 was immunoprecipitated, resolved by SDS-PAGE, and immunoblotted for IRAK-2 and IRAK4. Cell lysates were probed for actin as a loading control. IP, immunoprecipitation.
We tested whether CD82 regulates myddosome formation in response to activation of other TLRs. We stimulated WT, CD82KO, and CD82KO + CD82-mRFP1 macrophages with TLR7 and TLR4 agonists, imiquimod, and LPS, respectively. Unstimulated macrophages demonstrated baseline levels of interaction between MyD88 and IRAK2 or IRAK4 (Fig. 6B, C, top and second blots). We detected equal amounts of IRAK2 and IRAK4 in the whole-cell lysates in all conditions (Fig. 6B, C, third and fourth blots). The interaction between MyD88 and IRAK2 or IRAK4 in response to imiquimod and LPS was comparable, regardless of the presence or absence of CD82 (Fig. 6B, C, top and second blots). These data suggest that CD82 is dispensable for TLR7- or TLR4-dependent myddosome assembly. We confirmed comparable gene expression of endogenous TLR7 and TLR4 in WT, TLR9KO, CD82KO, and CD82KO + CD82-mRFP1 macrophages (Supplemental Fig. S4A). We also tested whether CD82 regulates myddosome formation in response to TLR1/2 activation by Pam3CsK4 and observed that CD82 is not required for TLR1/2-dependent myddosome formation (Supplemental Fig. S6). Therefore, CD82 specifically controls TLR9-dependent myddosome formation and is dispensable for TLR7- and TLR4-dependent myddosome assembly.
DISCUSSION
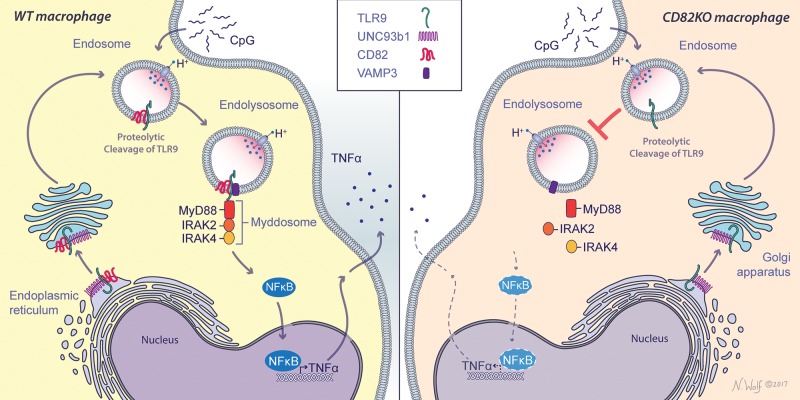
Compartmentalization of cell surface and endosomal membrane proteins is crucial in coordinating cellular signaling. Tetraspanins play a key role in the clustering, organization, and spatial distribution of membrane proteins in order to optimize signal transduction for a wide range of cellular processes. In response to pathogenic ligands, TLRs multimerize or cluster to initiate innate immune signaling (53). However, the role of tetraspanins in the regulation of TLR trafficking and signaling is poorly understood. In this study, we demonstrate a nonredundant role for the tetraspanin CD82 in the regulation of TLR9 trafficking and signaling in response to CpG (Fig. 7). Our results highlight the key role of CD82 in innate immunity and reveal a novel regulatory mechanism for TLR9 signaling.
Figure 7.
Schematic representation of CD82 controls CpG-dependent TLR9 signaling. CD82 interacts with TLR9 in WT macrophages in response to CpG. CD82 controls the assembly of TLR9-dependent myddosome assembly and subsequent NF-κB translocation to the nucleus and TNF-α production (left). In the absence of CD82, TLR9-mediated myddosome formation is absent, resulting in reduced TNF-α secretion (right). Illustration by Nicole Wolf, MS, ©2017. Printed with permission.
Most bacterial, viral, and fungal pathogens engage multiple PRRs, inducing a collaborative response that is unique from an immune response generated by an individual receptor. TEMs facilitate clustering of PRRs to integrate downstream signaling complexes. Dectin-1, a fungal β-glucan receptor, clusters to form a phagocytic synapse in myeloid cells that initiates the innate immune response (54–56). We recently reported that CD82 mediates dectin-1 clustering and enables robust downstream signaling (34). The tetraspanin CD37 interacts with dectin-1 to regulate surface expression in immune cells (48). Similarly, the tetraspanin CD9 regulates TLR4 signaling and distribution in TEMs (45). These data provide insight on the role of tetraspanins in influencing the trafficking and organization of cell surface PRRs; however, the regulation of endosomal PRRs is incompletely understood. We demonstrate for the first time a functional interaction between the endosomal PRR, TLR9, and the tetraspanin CD82. The interaction between CD82 and TLR9 is preserved with a mild detergent (Brij-58); however, it is disrupted with a stringent detergent (NP-40), suggesting that TLR9 enters into a TEM that requires CD82 in order to signal properly. Although we found an interaction with TLR7 and CD82, it is striking to see that CD82 only specifically regulates TLR9-mediated signaling. We also confirmed that the interaction between CD82 and TLR9 is not an artifact by demonstrating that CD82 and Rab7, another endosomal protein, do not interact with Brij-58, whereas TLR9 and CD82 interaction is maintained.
TLR9 activation and signaling results from the precise coordination of TLR9 subcellular distribution and integration of signal transduction machinery. Proteolytic cleavage of TLR9 by insulin-regulated aminopeptidase and lysosomal cysteine proteases (cathepsin L and cathepsin S) in acidified endosomes initiates the recruitment of signal transduction machinery (24, 29, 30, 32, 35, 51). In the absence of CD82, we observed that TLR9 trafficking to acidified CpG-containing compartments was significantly reduced. Our studies further revealed that CD82 interacts with both full-length and cleaved fractions of TLR9, indicating that this interaction occurs prior to the egress to acidified endosomes. These findings are consistent with our previous observation that CD82 traffics to bacterial and fungal-containing phagosomes prior to acidification (15). Herein, we provide further insight on the differential regulation of CD82 interaction with full-length and cleaved TLR9. Although a majority of CD82 is EndoH resistant, the minority amount of EndoH-sensitive CD82 can interact with full-length TLR9. Thus, the interaction between CD82 and full-length TLR9 in the ER, as well as cleaved TLR9 interaction with CD82 post-ER, adds significance to the role of CD82 on TLR9 signaling. Our data demonstrate that this interaction is a tightly regulated process. Additionally, we demonstrate that CD82 is required for the interaction between TLR9 and VAMP3 in response to CpG. VAMP3 is found in CpG-containing endosomal compartments and dictates the fate of intracellular pathways (i.e., autophagy) (26, 52, 57). We found that only cleaved TLR9 interacts with VAMP3, suggesting that this interaction occurs in acidified endosomes. CD82 interacts with both full-length and cleaved TLR9, supporting the notion that CD82 directs TLR9 to VAMP3-positive acidified endosomes. Overall, these findings signify the role of CD82 as a key regulator of TLR9 trafficking.
The cross-regulation of cell surface and endosomal TLRs indicates a role for tetraspanins to coordinate TLR signaling with specific sorting adaptor proteins. Despite distinct subcellular localization, TLRs can activate a common signal transduction pathway via adaptor proteins [e.g., MyD88, Toll/interleukin-1 receptor domain-containing adapter protein (TIRAP)] that are shared among cell surface and endosomal TLRs (22). Macrophages lacking Ly49Q, an immunoreceptor tyrosine-based inhibitory motif–bearing inhibitory receptor, demonstrate altered distribution of TLR9, and cells lacking Ly49Q show impaired cytokine production in response to CpG (27). Therefore, it becomes critical to dissect specific regulators of TLRs. The activation of TLR9 signaling relies on recruitment of signal transduction machinery. Cell surface and endosomal TLRs interact with MyD88, promoting recruitment of IRAK-2 and IRAK-4 to form the myddosome, which is essential for NF-κB activation and proinflammatory cytokine production (21–23). Recently, supramolecular organizing centers are described in the context of myddosome formation, which serves as a multifunctional signaling complex (58). The purpose of these supramolecular organizing centers is to provide diverse effector responses that are regulated by oligomeric complexes such as myddosomes (59). Here, we demonstrate that CD82 specifically controls TLR9-induced myddosome formation but is dispensable for TLR1/2-, TLR4-, and TLR7-dependent myddosome assembly. These findings, coupled with our data, emphasize the importance of the precise coordination in assembling signaling complexes, which strongly indicates a key role of tetraspanins.
Following myddosome assembly, NF-κB translocates to the nucleus to induce cytokine production (23). We demonstrate that CD82 controls TLR9-mediated NF-κB activation. In the absence of CD82, TNF-α production in response to CpG was significantly reduced but not completely absent, suggesting that other pathways can induce TLR9-dependent TNF-α production. In response to Aspergillus fumigatus infection, TLR9 activates nuclear factor of activated T cells (NFAT) translocation to the nucleus, which collaborates with NF-κB to induce TNF-α production in macrophages (60). NFAT activation is associated with intracellular calcium influx. Engagement of CD82 has been associated with intracellular calcium influx (61). Although it is tempting to speculate that upon CpG stimulation of macrophages, CD82 induces crosstalk between NFAT and NF-κB, further studies are warranted.
The regulation of TLR signaling may have implications in human disease. For example, TLR9 recognition of self-DNA triggers a chronic inflammatory state that contributes to the pathogenicity of autoimmunity (62). We speculate that therapeutically targeting CD82 to tune the inflammatory response may attenuate autoimmunity. Our results demonstrate the requirement of CD82 in mice for TNF-α production in response to CpG. Previous findings in humans have identified that tetraspanin deficiencies attenuate cellular immunity, resulting in immunodeficiency and increased susceptibility to bacterial, viral, and fungal infections. For example, deficiency of the tetraspanin CD53 in humans has implications in recurrent bacterial and fungal infections (63). In addition, the absence of CD81 in humans has been demonstrated to result in defective B-cell receptor signaling, which has implications in immunodeficiency (64). Recently, we identified 3 CD82 single nucleotide polymorphisms in human blood associated with increased candidemia susceptibility and cytokine changes (34), yet a deficiency in CD82 has not been reported in humans. We predict CD82 regulation of TLR9 signaling is a generalizable mechanism of control; however, we have not formally investigated these findings in human cells. Based on our data, it is likely that CD82 synchronizes host innate immune response during bacterial, fungal, and viral infections.
CpG is a promising candidate as a vaccine adjuvant because of its immunostimulatory properties (65–67). TLR9 activation by CpG enhances the anti-tumor activity of cancer therapies including chemotherapy by inducing activation of APCs and T-helper cells that target cancer cells (66, 67). CpG uptake and antigen presentation by immune cells is an actin-dependent process. In cancer cells, CD82 plays a critical role in regulating actin polymerization and attenuating cellular protrusion (68). In APCs, CD82 is found on the endolysosomal tubules with class II MHC that shuttle antigens to the cell surface for presentation (17). Here, we demonstrate that CD82 controls TLR9 trafficking to acidified CpG-containing compartments. It is possible that in response to CpG, CD82 facilitates TLR9 trafficking to class II MHC–positive antigen-processing compartments for enhanced cell surface presentation of tumor antigens. All experiments used class B CpG; the effects of class A and C CpG remains unknown. In addition to the important role of CD82 in suppressing tumor metastasis (69), our findings suggest that CD82 could have important implications for tuning the TLR9-dependent response to CpG during cancer immunotherapy.
We revealed a critical role of the tetraspanin CD82 in innate immune signaling. Despite the existing challenges in understanding tetraspanin biology, we define the specific role of CD82 in TLR9 signaling in macrophages. We show a functional interaction between TLR9 and CD82, which advances our understanding of tetraspanin interactions in immune cells. Our results provide novel insights on the role of CD82 in coordinating trafficking and organization of TLR9. We demonstrate that CD82 has specificity in forming molecular interactions with PRRs and costimulatory molecules. Furthermore, we elucidate the mechanism of selective TLR9 signaling in response to CpG. The increasing interest in the uses of TLR9 agonists including CpG for therapeutic vaccine adjuvants has promising potential in immunotherapies. These results highlight the critical role of the tetraspanin CD82 in shaping innate immune signaling.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank Nicole Wolf for the artwork displayed in the graphical abstract (Fig. 7), Shizuo Akira (Osaka University, Osaka, Japan) for the TLR9 knockout (TLR9KO) mice, and Douglas Golenbock (University of Massachusetts Medical Center, Worcester, MA, USA) for the TLR9KO macrophage cell line. The authors also thank Kensuyke Miyake and Ryutaro Fukui (University of Tokyo, Tokyo, Japan) for the TLR9 monoclonal and polyclonal antibodies (41), and Gregory Barton and Bo Liu (University of California–Berkeley, Berkeley, CA, USA) for the TLR7-FLAG construct. This work was supported by U.S. National Institutes of Health, National Institute of Allergy and Infectious Diseases Grants R01 AI092084 and R01 AI097519 (to J.M.V.) and R01 AI025780 and R01 AI139615 (to S.M.L.). This work was also supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001076), the UK Medical Research Council (FC001076), and the Wellcome Trust (FC001076). The authors declare no conflicts of interest.
Glossary
- APC
antigen presenting cell
- BMDM
bone marrow–derived macrophage
- CpG
cytosine-phosphate-guanine
- D-GalN
d-galactosamine
- EndoH
endoglycosidase H
- ER
endoplasmic reticulum
- FACS
fluorescence-activated cell sorting
- GFP
green fluorescent protein
- HA
hemagglutinin
- HRP
horseradish peroxidase
- IRAK
interleukin-1 receptor-associated kinase
- MHC
major histocompatibility complex
- NFAT
nuclear factor of activated T cells
- NP-40
Nonidet P-40
- KO
knockout
- MGH
Massachusetts General Hospital
- MyD88
myeloid differentiation primary response 88
- PBS-T
PBS-0.01% Tween 20
- PCC
Pearson correlation coefficient
- PNGaseF
peptide-N-glycosidase F
- PRR
pattern recognition receptor
- RAW
RAW264.7
- TEM
tetraspanin-enriched microdomain
- UNC93B1
Unc-93 homolog B1
- VAMP3
vesicle associated membrane protein 3
- WT
wild type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
N. S. Khan, Z. G. Ramirez-Ortiz, P. V. Kasperkovitz, E.-M. Frickel, Y.-M. Kim, H. L. Ploegh, and J. M. Vyas designed the research; N. S. Khan, D. P. Lukason, M. Feliu, Z. G. Ramirez-Ortiz, J. M. Tam, P. E. Negoro, M. Acharaya, E.-M. Frickel, C. E. Becker, and Y.-M. Kim performed research; P. V. Kasperkovitz, M. M. Brinkmann, K. Artavanis-Tsakonas, E. Latz, H. L. Ploegh, and C. K. Miranti contributed new reagents; N. S. Khan, D. P. Lukason, M. Feliu, R. A. Ward, A. K. Lord, J. L. Reedy, J. M. Tam, T. D. Vyas, S. Xu, M. M. Brinkmann, K. Artavanis-Tsakonas, E.-M. Frickel, Z. Dagher, Y.-M. Kim, E. Latz, H. L. Ploegh, M. K. Mansour, C. K. Miranti, S. M. Levitz, and J. M. Vyas analyzed data; and N. S. Khan, R. A. Ward, A. K. Lord, and J. M. Vyas wrote the manuscript.
REFERENCES
- 1.Hemler M. E. (2005) Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 [DOI] [PubMed] [Google Scholar]
- 2.Van Spriel A. B., Figdor C. G. (2010) The role of tetraspanins in the pathogenesis of infectious diseases. Microbes Infect. 12, 106–112 [DOI] [PubMed] [Google Scholar]
- 3.Hemler M. E. (2014) Tetraspanin proteins promote multiple cancer stages. Nat. Rev. Cancer 14, 49–60 [DOI] [PubMed] [Google Scholar]
- 4.Rocha-Perugini V., Sánchez-Madrid F., Martínez Del Hoyo G. (2016) Function and dynamics of tetraspanins during antigen recognition and immunological synapse formation. Front. Immunol. 6, 653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., Sánchez-Madrid F. (2009) Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446 [DOI] [PubMed] [Google Scholar]
- 6.Delos Santos R. C., Garay C., Antonescu C. N. (2015) Charming neighborhoods on the cell surface: plasma membrane microdomains regulate receptor tyrosine kinase signaling. Cell. Signal. 27, 1963–1976 [DOI] [PubMed] [Google Scholar]
- 7.Hur J., Choi J. I., Lee H., Nham P., Kim T. W., Chae C. W., Yun J. Y., Kang J. A., Kang J., Lee S. E., Yoon C. H., Boo K., Ham S., Roh T. Y., Jun J. K., Lee H., Baek S. H., Kim H. S. (2016) CD82/KAI1 maintains the dormancy of long-term hematopoietic stem cells through interaction with DARC-expressing macrophages. Cell Stem Cell 18, 508–521 [DOI] [PubMed] [Google Scholar]
- 8.Marjon K. D., Termini C. M., Karlen K. L., Saito-Reis C., Soria C. E., Lidke K. A., Gillette J. M. (2016) Tetraspanin CD82 regulates bone marrow homing of acute myeloid leukemia by modulating the molecular organization of N-cadherin. Oncogene 35, 4132–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uchtmann K., Park E. R., Bergsma A., Segula J., Edick M. J., Miranti C. K. (2015) Homozygous loss of mouse tetraspanin CD82 enhances integrin αIIbβ3 expression and clot retraction in platelets. Exp. Cell Res. 339, 261–269 [DOI] [PubMed] [Google Scholar]
- 10.Tonoli H., Barrett J. C. (2005) CD82 metastasis suppressor gene: a potential target for new therapeutics? Trends Mol. Med. 11, 563–570 [DOI] [PubMed] [Google Scholar]
- 11.Tsai Y. C., Weissman A. M. (2011) Dissecting the diverse functions of the metastasis suppressor CD82/KAI1. FEBS Lett. 585, 3166–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemler M. E. (2008) Targeting of tetraspanin proteins--potential benefits and strategies. Nat. Rev. Drug Discov. 7, 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kropshofer H., Spindeldreher S., Röhn T. A., Platania N., Grygar C., Daniel N., Wölpl A., Langen H., Horejsi V., Vogt A. B. (2002) Tetraspan microdomains distinct from lipid rafts enrich select peptide-MHC class II complexes. Nat. Immunol. 3, 61–68 [DOI] [PubMed] [Google Scholar]
- 14.Vyas J. M., Kim Y. M., Artavanis-Tsakonas K., Love J. C., Van der Veen A. G., Ploegh H. L. (2007) Tubulation of class II MHC compartments is microtubule dependent and involves multiple endolysosomal membrane proteins in primary dendritic cells. J. Immunol. 178, 7199–7210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artavanis-Tsakonas K., Kasperkovitz P. V., Papa E., Cardenas M. L., Khan N. S., Van der Veen A. G., Ploegh H. L., Vyas J. M. (2011) The tetraspanin CD82 is specifically recruited to fungal and bacterial phagosomes prior to acidification. Infect. Immun. 79, 1098–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammond C., Denzin L. K., Pan M., Griffith J. M., Geuze H. J., Cresswell P. (1998) The tetraspan protein CD82 is a resident of MHC class II compartments where it associates with HLA-DR, -DM, and -DO molecules. J. Immunol. 161, 3282–3291 [PubMed] [Google Scholar]
- 17.Jones E. L., Wee J. L., Demaria M. C., Blakeley J., Ho P. K., Vega-Ramos J., Villadangos J. A., van Spriel A. B., Hickey M. J., Hämmerling G. J., Wright M. D. (2016) Dendritic cell migration and antigen presentation are coordinated by the opposing functions of the tetraspanins CD82 and CD37. J. Immunol. 196, 978–987 [DOI] [PubMed] [Google Scholar]
- 18.Barton G. M., Kagan J. C. (2009) A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat. Rev. Immunol. 9, 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai T., Akira S. (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 20.Latz E., Verma A., Visintin A., Gong M., Sirois C. M., Klein D. C., Monks B. G., McKnight C. J., Lamphier M. S., Duprex W. P., Espevik T., Golenbock D. T. (2007) Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat. Immunol. 8, 772–779 [DOI] [PubMed] [Google Scholar]
- 21.Motshwene P. G., Moncrieffe M. C., Grossmann J. G., Kao C., Ayaluru M., Sandercock A. M., Robinson C. V., Latz E., Gay N. J. (2009) An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J. Biol. Chem. 284, 25404–25411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonham K. S., Orzalli M. H., Hayashi K., Wolf A. I., Glanemann C., Weninger W., Iwasaki A., Knipe D. M., Kagan J. C. (2014) A promiscuous lipid-binding protein diversifies the subcellular sites of toll-like receptor signal transduction. Cell 156, 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin S. C., Lo Y. C., Wu H. (2010) Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 465, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brinkmann M. M., Spooner E., Hoebe K., Beutler B., Ploegh H. L., Kim Y. M. (2007) The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J. Cell Biol. 177, 265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y. M., Brinkmann M. M., Paquet M. E., Ploegh H. L. (2008) UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 452, 234–238 [DOI] [PubMed] [Google Scholar]
- 26.Sasai M., Linehan M. M., Iwasaki A. (2010) Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science 329, 1530–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshizaki M., Tazawa A., Kasumi E., Sasawatari S., Itoh K., Dohi T., Sasazuki T., Inaba K., Makrigiannis A. P., Toyama-Sorimachi N. (2009) Spatiotemporal regulation of intracellular trafficking of Toll-like receptor 9 by an inhibitory receptor, Ly49Q. Blood 114, 1518–1527 [DOI] [PubMed] [Google Scholar]
- 28.Ewald S. E., Lee B. L., Lau L., Wickliffe K. E., Shi G. P., Chapman H. A., Barton G. M. (2008) The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456, 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park B., Brinkmann M. M., Spooner E., Lee C. C., Kim Y. M., Ploegh H. L. (2008) Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat. Immunol. 9, 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avalos A. M., Kirak O., Oelkers J. M., Pils M. C., Kim Y. M., Ottinger M., Jaenisch R., Ploegh H. L., Brinkmann M. M. (2013) Cell-specific TLR9 trafficking in primary APCs of transgenic TLR9-GFP mice. J. Immunol. 190, 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., Akira S. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 [DOI] [PubMed] [Google Scholar]
- 32.Latz E., Schoenemeyer A., Visintin A., Fitzgerald K. A., Monks B. G., Knetter C. F., Lien E., Nilsen N. J., Espevik T., Golenbock D. T. (2004) TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5, 190–198 [DOI] [PubMed] [Google Scholar]
- 33.Khan N. S., Kasperkovitz P. V., Timmons A. K., Mansour M. K., Tam J. M., Seward M. W., Reedy J. L., Puranam S., Feliu M., Vyas J. M. (2016) Dectin-1 controls TLR9 trafficking to phagosomes containing β-1,3 glucan. J. Immunol. 196, 2249–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tam J. M., Reedy J. L., Lukason D. P., Kuna S. G., Acharya M., Khan N. S., Negoro P. E., Xu S., Ward R. A., Feldman M. B., Dutko R. A., Jeffery J. B., Sokolovska A., Wivagg C. N., Lassen K. G., Le Naour F., Matzaraki V., Garner E. C., Xavier R. J., Kumar V., van de Veerdonk F. L., Netea M. G., Miranti C. K., Mansour M. K., Vyas J. M. (2019) Tetraspanin CD82 organizes dectin-1 into signaling domains to mediate cellular responses to Candida albicans. J. Immunol. 202, 3256–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babdor J., Descamps D., Adiko A. C., Tohmé M., Maschalidi S., Evnouchidou I., Vasconcellos L. R., De Luca M., Mauvais F. X., Garfa-Traore M., Brinkmann M. M., Chignard M., Manoury B., Saveanu L. (2017) IRAP+ endosomes restrict TLR9 activation and signaling. Nat. Immunol. 18, 509–518 [DOI] [PubMed] [Google Scholar]
- 36.Kasperkovitz P. V., Cardenas M. L., Vyas J. M. (2010) TLR9 is actively recruited to Aspergillus fumigatus phagosomes and requires the N-terminal proteolytic cleavage domain for proper intracellular trafficking. J. Immunol. 185, 7614–7622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halle A., Hornung V., Petzold G. C., Stewart C. R., Monks B. G., Reinheckel T., Fitzgerald K. A., Latz E., Moore K. J., Golenbock D. T. (2008) The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat. Immunol. 9, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman Z. R., Young J. M., Ingolia N. T., Barton G. M. (2016) Differences in codon bias and GC content contribute to the balanced expression of TLR7 and TLR9. Proc. Natl. Acad. Sci. USA 113, E1362–E1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanqué R., Meakin C., Millet S., Gardner C. R. (1998) Selective enhancement of LPS-induced serum TNF-alpha production by carrageenan pretreatment in mice. Gen. Pharmacol. 31, 301–306 [DOI] [PubMed] [Google Scholar]
- 40.Murakami Y., Fukui R., Motoi Y., Shibata T., Saitoh S. I., Sato R., Miyake K. (2017) The protective effect of the anti-Toll-like receptor 9 antibody against acute cytokine storm caused by immunostimulatory DNA. Sci. Rep. 7, 44042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukui R., Yamamoto C., Matsumoto F., Onji M., Shibata T., Murakami Y., Kanno A., Hayashi T., Tanimura N., Yoshida N., Miyake K. (2018) Cleavage of toll-like receptor 9 ectodomain is required for in vivo responses to single strand DNA. Front. Immunol. 9, 1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoorn T. v., Paul P., Janssen L., Janssen H., Neefjes J. (2012) Dynamics within tetraspanin pairs affect MHC class II expression. J. Cell Sci. 125, 328–339 [DOI] [PubMed] [Google Scholar]
- 43.Kawai T., Akira S. (2007) Signaling to NF-kappaB by Toll-like receptors. Trends Mol. Med. 13, 460–469 [DOI] [PubMed] [Google Scholar]
- 44.Levy S., Shoham T. (2005) The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5, 136–148 [DOI] [PubMed] [Google Scholar]
- 45.Suzuki M., Tachibana I., Takeda Y., He P., Minami S., Iwasaki T., Kida H., Goya S., Kijima T., Yoshida M., Kumagai T., Osaki T., Kawase I. (2009) Tetraspanin CD9 negatively regulates lipopolysaccharide-induced macrophage activation and lung inflammation. J. Immunol. 182, 6485–6493 [DOI] [PubMed] [Google Scholar]
- 46.Sparwasser T., Miethke T., Lipford G., Erdmann A., Häcker H., Heeg K., Wagner H. (1997) Macrophages sense pathogens via DNA motifs: induction of tumor necrosis factor-alpha-mediated shock. Eur. J. Immunol. 27, 1671–1679 [DOI] [PubMed] [Google Scholar]
- 47.Yi A. K., Yoon H., Park J. E., Kim B. S., Kim H. J., Martinez-Hernandez A. (2006) CpG DNA-mediated induction of acute liver injury in D-galactosamine-sensitized mice: the mitochondrial apoptotic pathway-dependent death of hepatocytes. J. Biol. Chem. 281, 15001–15012 [DOI] [PubMed] [Google Scholar]
- 48.Meyer-Wentrup F., Figdor C. G., Ansems M., Brossart P., Wright M. D., Adema G. J., van Spriel A. B. (2007) Dectin-1 interaction with tetraspanin CD37 inhibits IL-6 production. J. Immunol. 178, 154–162 [DOI] [PubMed] [Google Scholar]
- 49.Koh H. J., Kim Y. R., Kim J. S., Yun J. S., Kim S., Kim S. Y., Jang K., Yang C. S. (2018) CD82 hypomethylation is essential for tuberculosis pathogenesis via regulation of RUNX1-Rab5/22. Exp. Mol. Med. 50, 62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tabeta K., Hoebe K., Janssen E. M., Du X., Georgel P., Crozat K., Mudd S., Mann N., Sovath S., Goode J., Shamel L., Herskovits A. A., Portnoy D. A., Cooke M., Tarantino L. M., Wiltshire T., Steinberg B. E., Grinstein S., Beutler B. (2006) The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat. Immunol. 7, 156–164 [DOI] [PubMed] [Google Scholar]
- 51.Sepulveda F. E., Maschalidi S., Colisson R., Heslop L., Ghirelli C., Sakka E., Lennon-Duménil A. M., Amigorena S., Cabanie L., Manoury B. (2009) Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity 31, 737–748 [DOI] [PubMed] [Google Scholar]
- 52.Danglot L., Chaineau M., Dahan M., Gendron M. C., Boggetto N., Perez F., Galli T. (2010) Role of TI-VAMP and CD82 in EGFR cell-surface dynamics and signaling. J. Cell Sci. 123, 723–735 [DOI] [PubMed] [Google Scholar]
- 53.Inoue M., Shinohara M. L. (2014) Clustering of pattern recognition receptors for fungal detection. PLoS Pathog. 10, e1003873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodridge H. S., Reyes C. N., Becker C. A., Katsumoto T. R., Ma J., Wolf A. J., Bose N., Chan A. S., Magee A. S., Danielson M. E., Weiss A., Vasilakos J. P., Underhill D. M. (2011) Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature 472, 471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown G. D., Herre J., Williams D. L., Willment J. A., Marshall A. S., Gordon S. (2003) Dectin-1 mediates the biological effects of beta-glucans. J. Exp. Med. 197, 1119–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gantner B. N., Simmons R. M., Canavera S. J., Akira S., Underhill D. M. (2003) Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 197, 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ligeon L. A., Moreau K., Barois N., Bongiovanni A., Lacorre D. A., Werkmeister E., Proux-Gillardeaux V., Galli T., Lafont F. (2014) Role of VAMP3 and VAMP7 in the commitment of Yersinia pseudotuberculosis to LC3-associated pathways involving single- or double-membrane vacuoles. Autophagy 10, 1588–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan Y., Kagan J. C. (2019) Innate immune signaling organelles display natural and programmable signaling flexibility. Cell 177, 384–398.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kagan J. C., Magupalli V. G., Wu H. (2014) SMOCs: supramolecular organizing centres that control innate immunity. Nat. Rev. Immunol. 14, 821–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herbst S., Shah A., Mazon Moya M., Marzola V., Jensen B., Reed A., Birrell M. A., Saijo S., Mostowy S., Shaunak S., Armstrong-James D. (2015) Phagocytosis-dependent activation of a TLR9-BTK-calcineurin-NFAT pathway co-ordinates innate immunity to Aspergillus fumigatus. EMBO Mol. Med. 7, 240–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lebel-Binay S., Lagaudrière C., Fradelizi D., Conjeaud H. (1995) CD82, tetra-span-transmembrane protein, is a regulated transducing molecule on U937 monocytic cell line. J. Leukoc. Biol. 57, 956–963 [DOI] [PubMed] [Google Scholar]
- 62.Zhou Y., Fang L., Peng L., Qiu W. (2017) TLR9 and its signaling pathway in multiple sclerosis. J. Neurol. Sci. 373, 95–99 [DOI] [PubMed] [Google Scholar]
- 63.Mollinedo F., Fontán G., Barasoain I., Lazo P. A. (1997) Recurrent infectious diseases in human CD53 deficiency. Clin. Diagn. Lab. Immunol. 4, 229–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Zelm M. C., Smet J., Adams B., Mascart F., Schandené L., Janssen F., Ferster A., Kuo C. C., Levy S., van Dongen J. J., van der Burg M. (2010) CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. J. Clin. Invest. 120, 1265–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Den Brok M. H., Sutmuller R. P., Nierkens S., Bennink E. J., Toonen L. W., Figdor C. G., Ruers T. J., Adema G. J. (2006) Synergy between in situ cryoablation and TLR9 stimulation results in a highly effective in vivo dendritic cell vaccine. Cancer Res. 66, 7285–7292 [DOI] [PubMed] [Google Scholar]
- 66.Jahrsdörfer B., Weiner G. J. (2008) CpG oligodeoxynucleotides as immunotherapy in cancer. Update Cancer Ther. 3, 27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shirota H., Sano K., Hirasawa N., Terui T., Ohuchi K., Hattori T., Shirato K., Tamura G. (2001) Novel roles of CpG oligodeoxynucleotides as a leader for the sampling and presentation of CpG-tagged antigen by dendritic cells. J. Immunol. 167, 66–74 [DOI] [PubMed] [Google Scholar]
- 68.Liu W. M., Zhang F., Moshiach S., Zhou B., Huang C., Srinivasan K., Khurana S., Zheng Y., Lahti J. M., Zhang X. A. (2012) Tetraspanin CD82 inhibits protrusion and retraction in cell movement by attenuating the plasma membrane-dependent actin organization. PLoS One 7, e51797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bandyopadhyay S., Zhan R., Chaudhuri A., Watabe M., Pai S. K., Hirota S., Hosobe S., Tsukada T., Miura K., Takano Y., Saito K., Pauza M. E., Hayashi S., Wang Y., Mohinta S., Mashimo T., Iiizumi M., Furuta E., Watabe K. (2006) Interaction of KAI1 on tumor cells with DARC on vascular endothelium leads to metastasis suppression. Nat. Med. 12, 933–938 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.