ABSTRACT
Duplicated ribosomal protein (RP) genes in the Drosophila melanogaster eRpL22 family encode structurally-divergent and differentially-expressed rRNA-binding RPs. eRpL22 is expressed ubiquitously and eRpL22-like expression is tissue-restricted with highest levels in the adult male germline. We explored paralogue functional equivalence using the GAL4-UAS system for paralogue knockdown or overexpression and a conditional eRpL22-like knockout in a heat- shock flippase/FRT line. Ubiquitous eRpL22 knockdown with Actin-GAL4 resulted in embryonic lethality, confirming eRpL22 essentiality. eRpL22-like knockdown (60%) was insufficient to cause lethality; yet, conditional eRpL22-like knockout at one hour following egg deposition caused lethality within each developmental stage. Therefore, each paralogue is essential. Variation in timing of heat-shock-induced eRpL22-like knockout highlighted early embryogenesis as the critical period where eRpL22-like expression (not compensated for by eRpL22) is required for normal development of several organ systems, including testis development and subsequent sperm production. To determine if eRpL22-like can substitute for eRpL22, we used Actin-GAL4 for ubiquitous eRpL22 knockdown and eRpL22-like-FLAG (or FLAG-eRpL22: control) overexpression. Emergence of adults demonstrated that ubiquitous eRpL22-like-FLAG or FLAG-eRpL22 expression eliminates embryonic lethality resulting from eRpL22 depletion. Adults rescued by eRpL22-like-FLAG (but not by FLAG-eRpL22) overexpression had reduced fertility and longevity. We conclude that eRpL22 paralogue roles are not completely interchangeable and include functionally-diverse roles in development and spermatogenesis. Testis-specific paralogue knockdown revealed molecular phenotypes, including increases in eRpL22 protein and mRNA levels following eRpL22-like depletion, implicating a negative crosstalk mechanism regulating eRpL22 expression. Paralogue depletion unmasked mechanisms, yet to be defined that impact paralogue co-expression within germ cells.
KEYWORDS: Cell biology, fertilization, gene regulation, molecular genetics, reproduction, RNAi
Introduction
In several ribosomal protein (RP) families, duplicated genes exist as a result of whole genome or gene duplication events in the evolutionary history of the gene family, giving rise to protein paralogues that may display redundant or specialized roles in cellular processes. Once thought only to occupy structural roles within the ribosome, several RPs are now known to moonlight in regulatory roles in other cellular pathways unrelated to ribosome biogenesis or ribosome function (reviewed by [1]). Further, the existence of RP paralogues in many organisms broadens the possibility of discovering that paralogue functions have diverged over time and that novel extraribosomal protein roles may be uncovered. With emerging interest in the role of the ribosome as a dynamic macromolecular complex involved in specifying cell and tissue-specific translation in developmental processes [2,3] (reviewed by [4]), assembly of RP paralogues into ribosomes during biogenesis contributes an additional level of ribosome heterogeneity, possibly affecting differential translation or the scope of regulatory mechanisms affecting protein expression.
The eukaryotic-specific eRpL22 ribosomal protein gene family is an excellent model system for exploring functional divergence and extraribosomal roles among RP paralogues (note the adoption of recommended nomenclature for eukaryotic-specific RPs by [5]). Recent studies of highly conserved paralogues Rpl22 and Rpl22-Like 1 (Rpl22l1) in zebrafish and mice models demonstrated functionally distinct and antagonistic regulatory roles for these paralogues as modulators of alternative splicing patterns for smad mRNA during development of hematopoietic cells [6]. Importantly, regulatory roles for these paralogues are dependent on nuclear localization apart from a canonical role in translation within the cytoplasmic compartment. Several studies show that Rpl22 paralogue expression is autoregulated through paralogue binding to pre-mRNA to inhibit splicing (in yeast [7]) or through unknown mechanisms (in zebrafish and mice [8]). In zebrafish and mice, Rpl22l1 levels are negatively regulated by Rpl22. In yeast, additional extraribosomal Rpl22 paralogue functions show that Rpl22A and Rpl22B are required for selective translation of IME1 mRNA that governs the switch from mitosis to meiosis and the transition to gametogenesis [9].
We have focused on the Drosophila melanogaster eRpL22 gene family because of paralogue expression differences and striking divergence in paralogue primary structure compared to highly homologous orthologues found in other systems (see www.ncbi.nlm.nih.gov/homologene/37378 for genes identified as putative eRpL22 orthologues). In Drosophila melanogaster, eRpL22 and eRpL22-like are differentially expressed throughout development. eRpL22 is ubiquitously expressed [10–13]. Cell- or tissue-specific expression of eRpL22-like has been confirmed by in situ hybridization in pole cells, embryonic gonads, and the stomatogastric nervous system of developing embryos [10,11], by Western blot analysis predominately in the adult male germline and in the eye of both males and females [13], and by mass spectrometry in adult heads [14].
In Drosophila, eRpL22 is an essential gene, shown in vivo [15] and in S2 tissue culture cells [16]. Whether or not eRpL22-like is also essential throughout fly development is unclear although it is significant that P-element insertion upstream of the eRpL22-like transcription start site is homozygous lethal, perhaps reflecting disruption of eRpL22-like regulatory elements or other cis-elements not as yet annotated (Flybase.org: FB2017_03).
Drosophila eRpL22 and eRpL22-like are highly conserved in the C-terminal rRNA binding domain (as are other eukaryotic eRpl22 orthologues), but are highly divergent in the unique N-terminal domain extension, resulting in an exceptionally low degree of amino acid conservation overall (only 38% [17]) compared to a high degree of amino acid identity (64–100%) for most Drosophila RP paralogues [17]. The unique N-terminal domain of both fly eRpL22 and eRpL22-like has homology to the C-terminal portion of histone H1 [13,18]. Whether or not this domain contributes to a non-ribosomal role for Drosophila eRpL22 in chromatin binding and suppression of global gene expression [19] is unknown and awaits exploration.
Recently we determined that fly eRpL22 undergoes post-translation modification (PTM) by SUMOylation in a variety of cell types, with additional unique phosphorylation and SUMOylation modifications occurring in the male germline during spermatogenesis [20]. While testis eRpL22-like is primarily cytoplasmic in all germ cell stages [13,20], the subcellular distribution of eRpL22 changes during the course of sperm maturation. In mitotic stages, eRpL22 is found within nucleoli and the cytoplasm, consistent with a ribosomal function for eRpL22 in spermatogonia during the transit amplification stage of spermatogenesis. In meiotic spermatocytes, however, eRpL22 is found in the nucleoplasm of germ cells, suggesting a transition from a ribosomal role for eRpL22 to a nuclear role. Notably, SUMOylated eRpL22 is not a component of translationally active ribosomes in S2 cells, suggesting that SUMOylation may regulate eRpL22 incorporation into the ribosome [20]. We have proposed that eRpL22 and eRpL22-like have overlapping ribosomal-type functions, particularly in mitotic stages of spermatogenesis, but a unique set of nuclear, non-ribosomal functions may be defined for SUMOylated eRpL22, where it is highly enriched in the nucleoplasm of meiotic spermatocytes.
Here, we investigate the extent of redundancy or divergence in eRpL22 paralogue function in the testis or in development using the GAL4-UAS system for RNAi-mediated depletion or overexpression of eRpL22 or eRpL22-like and a conditional knockout of eRpL22-like in a heat shock flippase/FRT line. Ubiquitous knockdown of eRpL22 using an Actin-GAL4 driver and conditional heat shock-induced knockout of eRpL22-like within an hour of egg laying generate lethal phenotypes, demonstrating that both paralogues are essential for development. eRpL22-like knockout during embryogenesis (within several hours of egg laying) disrupted development of several organ systems, including testis development, thereby revealing a critical time in embryogenesis where eRpL22-like function is essential. Conditional eRpL22-like knockout experiments, in addition to rescue experiments (where eRpL22-like is overexpressed when eRpL22 is depleted), reveal overlapping paralogue functions in development and spermatogenesis, but also uncover a unique set of functions for eRpL22 paralogues as well. While testis-specific paralogue knockdown by RNAi revealed no remarkable development or fertility phenotypes, we show that at the molecular level, eRpL22 mRNA and protein levels increase resulting from eRpL22-like depletion. A negative crosstalk mechanism is proposed that affects eRpL22 expression in germ cells.
Materials and methods
Plasmid construction and injection
We thank the TRiP at Harvard Medical School (https://fgr.hms.harvard.edu/fly-in-vivo-rnai) for providing transgenic RNAi fly stocks and/or plasmid vectors used in this study. For RNAi, we utilized the pVALIUM10 strategy developed by TRiP. Codons 1–100 were used to target eRpL22 (FBid: FBgn0015288) and eRpL22-like (FBid: FBgn0034837) mRNAs separately because of lower amino acid similarity at the N-terminus. Off target hits were ruled out of the selected targeted region by analysis with the SnapDragon dsRNA design tool provided by the TRiP. Construction followed a two-stage gateway cloning procedure provided by the TRiP. Briefly, the targeted regions were subcloned from cDNA [13] into the directional entry vector pENTR/D-TOPO (Invitrogen). Sequences of selected clones were confirmed by Sanger sequencing and cloned into the designation vector pVALIUM10 (TRiP) using LR clonase (Invitrogen). Clones were selected and sequenced to confirm proper orientation and sequence. pENTR/D-TOPO and pVALIUM10 were propagated in TOP10 E. coli cells (Invitrogen) and ccdB Survival T1R E. coli cells (Invitrogen), respectively. TOP10 E. coli cells were used for all cloning steps.
Plasmid DNA was purified using the QIAGEN plasmid maxiprep kit and resuspended in sterile dH20 for phiC3-integrase-mediated site-specific transgenesis. Plasmid DNA was injected into y, v, nanos-integrase; attP2 embryos for integration into chromosome 3L. Transgenics were selected, backcrossed, and balanced to homozygosity with y v; Sb/TM3, Ser. Injection and balancing was performed by Genetic Services.
To determine if endogenous levels of the eRpL22 paralogues are sensitive to the expression levels of their paralogues, we generated overexpression lines for FLAG-eRpL22 and eRpL22-like-FLAG. For each paralogue, previously cloned cDNAs [13,20] were used as templates for addition of a FLAG tag by PCR with High Fidelity Platinum Taq Master Mix (Invitrogen) (see Table S3 for primers). Amplicons were cloned into pENTR/D-TOPO (Invitrogen), verified by Sanger sequencing, subsequently gateway-cloned into destination vector pVALIUM10-roe (provided by the Drosophila RNAi Screening Center) with LR clonase (Invitrogen), and verified by Sanger sequencing. Injection into y, v, nanos-integrase; attp2 embryos for site-directed insertion on chromosome 3 at the attP2 locus and balancing to homozygosity was performed by Genetic Services.
To determine if eRpL22-like is essential during development, the eRpL22-like coding sequence was excised upon heat shock-induced flippase expression. A conditional knockout allele was engineered (by Genetivision, Inc.) to contain a single allele of dsRED (controlled by a truncated tubulin promoter [21]) downstream of the eRpL22-like 3ʹUTR. The entire insert is flanked by parallel FRT sites on either side of the cassette.
Fly stocks
All stocks were kept at room temperature on standard cornmeal media. The Actin-GAL4 driver (y[1] w[*]; P{Act5C-GAL4-w}E1/CyO), nos-GAL4 driver (P{w[+mC] = UAS-Dcr-2.D}1, w[1118]; P{w[+mC] = GAL4-nos.NGT}40), UAS-VAL10-GFP stock (y[1] v[1]; P{y[+t7.7] v[+t1.8] = UAS-GFP.VALIUM10}attP2), 7198 (w[*]; Kr[If-1]/CyO; D[1]/TM3, Ser[1]), and 4558 (FM7a, l(1)TW24[1]/oc[1] ptg[3] l(1)TW1[cs]; CyO/l(2)DTS91[1]), and HS-FLP (w[1118]; MKRS, P{ry[+t7.2] = hsFLP}86E/TM6B) were obtained from the Bloomington Stock Center. The bam-GAL4-VP16, UAS-Dicer2 (y, w; bam-GAL4, UAS-Dicer2) driver was a kind gift from Marina Wolfner (Cornell), but was originally developed by Dennis McKearin [22]. The eRpl22.IR line (y[1], v[1]; pVALIUM10{UAS-eRpL22.IR} attP2/TM3, Ser; Chromosomes:1;3) was kept over the TM3, Ser balancer due to it being male sterile when homozygous. The eRpL22-like.IR line (y[1], v[1]; pVALIUM10{UAS-eRpL22-like.IR} attP2; Chromosomes:1;3), eRpL22-like overexpression line (y,v; P{pVALIUM10-roe-eRpL22-like-FLAG} attP2; Chromosomes:1;3), and eRpL22 overexpression line (y,v; p{pVALIUM10-roe-FLAG-eRpL22}; Chromosomes:1;3) could each be propagated as a homozygous line without any noticeable effect on development or fertility. The eRpL22-like-FRT-conditional knockout allele (CKA) (w[*]; eRpL22-like FRT Δtub-DsRed – Δtubulin – eRpL22-like – FRT; chromosome 1;2) was constructed by Genetivision, Inc., as described above.
Crosses
All RNAi crosses were set up at 27–29°C to achieve maximum GAL4 activity without inducing heat stress. In all cases unless noted (see sperm motility and fertility assays), two 1–5 day males harboring the desired GAL4 driver were mated with four 1–5 day old virgin females harboring the desired UAS element. F1 males (1–3 day old) were used for Western analysis and phase contrast microscopy. Western analysis, including antibody specifications, was completed as previously reported [20]. For phase contrast microscopy, testes were dissected in 1X PBS and gently squashed with a cover slip and imaged with a Nikon Eclipse TE200U.
eRpL22-like knockout (KO) crosses were set up at 25°C. The eRpL22-like KO flies were created through multiple crosses. First, the HS-FLP or CKA flies were independently mated to 7198 balancer flies, and curly and serrated wing progeny were selected. Second, HS-FLP/7198 progeny were mated with CKA/7198 progeny and resulting F1 progeny were genotyped for Flippase and DsRed (primers in Table S3). F1 progeny that contained both Flippase and DsRed (marker for CKA) were mated and raised to homozygosity. eRpL22-like flippase-mediated KO was initiated by heat-shocking the embryos at 38°C for 60 min. Excision of eRpL22-like was confirmed by PCR.
Rescue crosses were set up at 25°C. In the primary eRpL22-like-FLAG rescue cross, four 1–5 day old virgin females harboring Actin-GAL4 were mated to two 1–5 day old males harboring the triple balancer (Bloomington stock no. 7198 – referred to as ‘7198’ in this paper). Four 1–5 day old females harboring the eRpL22-like-FLAG overexpression UAS were mated to two 1–5 day old males harboring the triple balancer 7198. Progeny from the previous two crosses were then mated in reciprocal. This yielded progeny harboring the eRpL22-like-FLAG overexpression UAS and Actin-GAL4; progeny were genotyped for Actin. Four 1–5 day old females containing the eRpL22-like-FLAG overexpression UAS and Actin-GAL4 (determined through genotyping) were mated to two 1–5 day old males harboring eRpL22.IR. Progeny of various ages were collected and used for Western blotting, immunohistochemistry, phase contrast imaging, and fertility assays. In the second rescue cross, four 1–5 day old eRpL22-like-FLAG overexpression UAS virgin females were mated with two 1–5 day old Actin-GAL4 males. Virgin female progeny from this cross expressing the eRpL22-like-FLAG overexpression UAS and Actin-GAL4 were collected. Four 1–5 day old virgin females expressing Act>eRpL22-like-FLAG overexpression were mated with two 1–5 day old 4558 males. Progeny from this cross were genotyped to ensure the presence of Actin-GAL4 and eRpL22-like-FLAG overexpression alleles. Four 1–5 day old virgin females harboring Actin-GAL4 and eRpL22-like-FLAG overexpression elements were mated with two 1–5 day old eRpL22.IR (see Figure S3 for overview of scheme for rescue crosses). Various ages of progeny were collected for Western blots, phase contrast imaging, and fertility assays.
FLAG-eRpL22 rescue crosses were set up using the same scheme as for eRpL22-like-FLAG rescue crosses except that the FLAG-eRpL22 overexpression line was used in place of the eRpL22-like-FLAG overexpression line.
Fertility and longevity assay
Fertility crosses were set up at 25°C for all rescue crosses and 29°C for eRpL22.IR and eRpL22-like.IR crosses. To assess fecundity of each male, a single, 1–5 day old virgin male was mated with a single, age matched virgin wild type female until larva were present, then parents were removed. Larval, pupae and adult stages were monitored for developmental acceleration or delays. Adult progeny were counted each day for five consecutive days.
Longevity was assessed observing multiple isolated males (20 per genotype) for thirty days. Flies were transferred to fresh media every four days.
qRT-PCR
RNA isolation, cDNA preparation and qRT-PCR specifications were completed as previously described [13]. Expression levels were normalized to Act5C (ΔCT). Fold change was determined by first calculating ΔΔCT = ΔCT (Experimental) – ΔCT (Control), and then calculating fold change as 2−ΔΔCT. Primers used can be found in Table S3.
Antibodies
Rabbit polyclonal anti-Drosophila eRpl22 and mouse polyclonal anti-Drosophila eRpl22-like were previously developed by Kearse et al. [13] and were used at 1:1000 for Western analysis and 1:100 for immunohistochemistry (IHC). Anti-β-tubulin, and anti-FLAG were used, as previously described in Kearse et al. [20]. Anti-eRpL23a (Abgent #AP1939b) was used at 1:1000 for Western analysis. HRP-conjugated goat anti-mouse IgG and goat anti-rabbit IgG secondary were obtained from Promega (#W4021 and #Q4011, respectively) and used at 1:50,000 for Western analysis. Goat anti-mouse/Alexa Fluor 488 and goat anti-rabbit/Alexa Fluor 568 were obtained from Invitrogen (#A11029 and #A11036, respectively) and used at 1:200 for IHC.
Fly tissue preparation
Adult Drosophila testes and bodies were dissected in 1x PBS and immediately frozen on dry ice. Approximately seven pairs of testes were homogenized in reducing (βME) sample buffer. Approximately seven bodies were lysed in 30μL of RIPA buffer supplemented with 1mM PMSF for 10 minutes on ice. Lysates were centrifuged at 24,000 x g to clear cell debris, nuclei, and mitochondria. The resulting soluble supernatant was then quantified using the Bio-Rad DC Protein Assay Kit with BSA standards (#500–0112). The resulting insoluble pellet was homogenized in 50μL of reducing (βME) sample buffer. Soluble lysates (20μg) were mixed with reducing (βME) sample buffer.
Western analysis
Proteins were separated by SDS-PAGE and electroblotted onto Westran-S PVDF membrane (Whatman #10413096) for 1 hour in chilled transfer buffer. Following blocking with 5% nonfat dry milk (NFDM) for 1 hour, primary antibodies were incubated overnight in 3% NFDM at 4°C. HRP-conjugated secondary antibodies were incubated for 2 hours at 4°C in 3% NFDM. Chemiluminescence detection was performed with ECL2 (Thermo Scientific) and Kodak Bio-Max film.
Densitometry was performed on scanned Western blot images using ImageJ software [23]. Bands of interest were boxed and the area under the curve was calculated using the gel analysis tool. Experimental areas were normalized by making a ratio of the experimental band to the control band. Fold change was calculated by comparing the experimental ratio to the control ratio.
Genotype analysis
Genotype analysis was performed as previously reported [24]. A wing pair from an anesthetized adult Drosophila was placed in a 1.5mL microcentrifuge tube with 10μL of 400μg/mL protease K in buffer A (10mM Tris-Cl pH 8.2, 1mM EDTA, 25mM NaCl). The tube was incubated at 37°C for 1 hour, then incubated at 95°C to heat inactivate protease K. PCR amplification of genes of interest was performed using the following PCR conditions: Platinum blue PCR supermix (Invitrogen), 3μL Drosophila wing DNA, 10μM forward and reverse primers listed in Table S3. Thermocycler conditions were: 1 cycle of initial denaturation (95°C for 5 min); 35 cycles of denaturation (95°C for 1 min), annealing (primer and product dependent), extension (72°C for 1min); and 1 cycle of final extension (72°C for 10 min). PCR products were analyzed on agarose gels.
For confirmation of eRpL22-like knockout, tissues were dissected in 1x PBS and frozen immediately on dry ice. After thawing on ice, tissues were homogenized in buffer (0.2M sucrose, 0.1M Tris HCl pH 9.2, 50 mM EDTA, 0.5% SDS), vortexed briefly, heated at 65°C for 10 minutes, and DNA isolated by phenol/chlorform extraction and ethanol precipitation. DNAs were resuspended in sterile nuclease-free water and subsequently used for PCR analysis as above. Thermocycler conditions were: 1 cycle of initial denaturation (94°C for 2 min); 35 cycles of denaturation (94°C for 30 sec), annealing (52°C for 30 sec), extension (72°C for 5 min); and 1 cycle of final extension (72°C for 10 min).
Stereoscope imaging
Images of age-matched larvae (5 days after egg deposition) were obtained using a Nikon D5-F12 camera mounted on a Nikon SMZ1500 stereo microscope.
Immunohistochemistry
Testis squashes and IHC were performed as previously described [13]. For Malpighian tubule IHC, age-matched tissue was dissected in cold 1x PBS, fixed for 15 minutes in 3.7% formaldehyde (in 1x PBS), washed three times in 1x PBS, and incubated in 1x PBS for 30 minutes. Tissue was permeabilized with 1x PBS containing 0.1% Triton-X100 (PBX) for 30 minutes, blocked for 20 minutes in PBX containing 5% normal goat serum, and incubated overnight with primary antibody in blocking solution at 4°C. Tissue was washed three times with PBX, incubated in PBX for 4 hours at room temperature, and then incubated with Alexafluor-conjugated secondary antibodies overnight at 4°C. Samples were washed three times and incubated in PBX for 4 hours. Tissue was then incubated with 4ʹ, 6-diamidino-2-phenylindole ([DAPI]; 0.4µg/ml; Life Technologies) for 20 minutes at room temperature. After three washes in PBX, tissue was mounted in Slow Fade Diamond Antifade Mountant (Life Technologies). Malpighian tubule images were acquired using a Zeiss LSM 880 confocal microscope.
Phase contrast imaging
Males of various ages were maintained separately from females for three days before dissected testes were subjected to gentle squashes and phase contrast microscopy to score the presence of motile sperm.
Image analysis
Measurements of the width of apical tips, midpoints of testes, width of Malpighian tubules and length of larva were taken using the ImageJ line tool [23]. A ratio of apical tip: midpoint testes widths was calculated by dividing the ImageJ value for the apical tip by the ImageJ value for the midpoint of the testis. The apical tip: midpoint ratio was averaged for ten individuals for each genotype. The fold change was calculated by dividing the average ratio for rescued males by the average ratio of WT males. Malpighian tubules widths were measured at various points through the length of each tubule to account for width differences within one tubule. eRpL22-like fluorescence differences were measured using the ImageJ freehand tool and were divided by the total number of nuclei within the measured area.
Results
eRpL22 gene expression is essential for development
Using the UAS-GAL4 binary system, we tested the requirement of eRpL22 and eRpL22-like separately in Drosophila development by expressing inverted repeats (IR) forming shRNA to drive RNAi. Nucleotide similarity between eRpL22 paralogous genes is lowest at the 5ʹ end of the coding sequence [13], allowing for separate targeting by RNAi. Hereafter, the eRpL22 RNAi line is referred to eRpL22.IR and the eRpL22-like RNAi line is referred as eRpL22-like.IR. As the most appropriate genetic control available, we obtained a GFP expressing line that harbors the identical expression construct backbone as our developed RNAi lines, all of which have been integrated at the attP2 locus on chromosome 3. To test whether each eRpL22 paralogue is essential for development, we compared the F1 ratios between the genetic control, eRpL22.IR, and eRpL22-like.IR when crossed with the Actin-GAL4 driver for ubiquitous knockdown. F1 progeny carrying both UAS-GAL4 elements (unbalanced) are distinguished from other F1 progeny by the absence of dominant wing markers (CyO and TM3, Ser). We hypothesized that ubiquitous knockdown of eRpL22 is lethal, as it has been reported by P-element gene disruption [15] and by RNAi in S2 cells [16] that eRpL22 is an essential gene. Our data show that ubiquitous knockdown of eRpL22 in Drosophila is lethal, as F1 males or females harboring both the eRpL22.IR and GAL4 elements (unbalanced F1) were not found (Figure 1(a)), confirming that eRpL22 is an essential gene in males and females. We suspect that eRpL22 depletion is embryonic lethal, as an accumulation of non-developing larvae or pupae was not evident.
Figure 1.
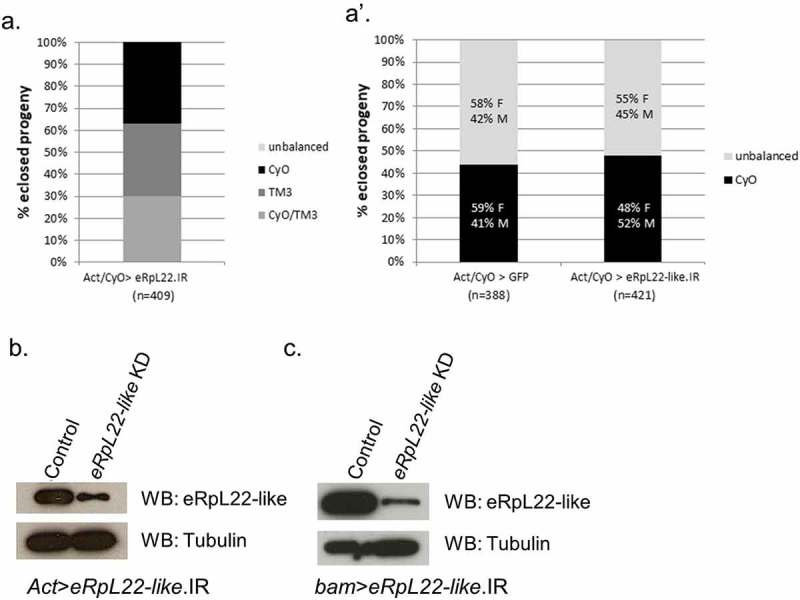
eRpL22 family paralogue knockdown generates differential outcomes in fly development. A & A’) F1 progeny were scored to assess the impact of knockdown (KD) of eRpL22 paralogues on development. The lack of F1 adults harboring Actin-GAL4 and eRpL22.IR, represented as unbalanced progeny (lacking CyO and TM3 balancers), suggests eRpL22 is an essential gene (a). Equal representation of the CyO balancer and Actin-GAL4 driver, with no significant differences in progeny number or sex ratios compared to Act>GFP control (left), suggests eRpL22-like is dispensable for viability (right) (a’). F: Females; M: Males. Numbers of progeny scored are indicated below each genotype. b) Western analysis of testis tissue confirms eRpL22-like knockdown (Act>eRpL22-like.IR). Tubulin was used as a loading control. c) Western analysis of testis tissue confirms an 80% eRpL22-like knockdown (bam>eRpL22-like.IR). Tubulin was used as a loading control.
eRpL22-like is essential early in embryogenesis for development of several organ systems
Whether or not eRpL22-like is also essential has not been thoroughly investigated. FlyBase (FB2017_3) reports that a P-element insertion 156 nucleotides upstream of the eRpL22-like transcription start site is homozygous lethal, suggesting that eRpL22-like is essential. We tested this by ubiquitous knockdown of eRpL22-like using the Actin-GAL4 driver. Act>eRpL22-like.IR adult F1 ratios are comparable to the genetic control (Figure 1(a’)). The CyO balancer and a genotype harboring both elements necessary for knockdown (UAS.IR and GAL4; seen as unbalanced) segregated equally between males and females (Figure 1(a’)) in the expected developmental time frame. To confirm knockdown, we analyzed eRpL22-like protein levels in control and Act>eRpL22-like.IR testis tissue. Western analysis shows a significant reduction (~60% KD) in eRpL22-like protein levels in F1 Act>eRpL22-like.IR testes compared to the control (Figure 1(b)). Therefore, we conclude that 60% knockdown of eRpL22-like in tissues where expressed (e.g., pole cells, developing gonads, stomatogastric nervous system of developing embryos – [10]) is insufficient to disrupt embryonic developmental progression to the adult stage in males and females. The absence of viable progeny precludes the ability to quantify the degree of eRpL22 knockdown with the Actin driver; however, an assumption of comparable knockdown of eRpL22 (at a level of 60%, as demonstrated for eRpL22-like) results in embryonic lethality.
To rule out the possibility that residual eRpL22-like function (remaining after 60% knockdown) was adequate to support developmental progression, we used an alternative strategy to determine if eRpL22-like is essential for development. A conditional knockout (CKO) of eRpL22-like was performed. eRpL22-like was flanked by FRT sites in the genome at the endogenous locus, where upon heat shock treatment, a heat shock-flippase (HS_FLP) allele would be activated to excise eRpL22-like from the genome (Figure 2(a)). Heat shock was performed 1 hour after egg laying to induce eRpL22-like excision at the beginning of embryonic development. Numerous developmental defects were evident when eRpL22-like was deleted at this time point. The most notable phenotype was that only two embryos reached adulthood (2/22); one succumbed within 24 hours after eclosure, while the other had only noticeable eye bristle defects (Table S1). Heat shock treatment of wildtype embryos at 1 hour likewise had a detrimental effect on embryo survivability, with only 16/51 embryos surviving to the adult stage, but no developmental anomalies were observed in surviving animals. Low rates of survival into the adult stage following embryonic heat shock have been reported previously by Bergh and Arking [25], but increase significantly as heat shock is applied at later stages of embryonic development. We also note a significant increase in survivability when heat shock is applied at later times after egg laying (data not shown).
Figure 2.
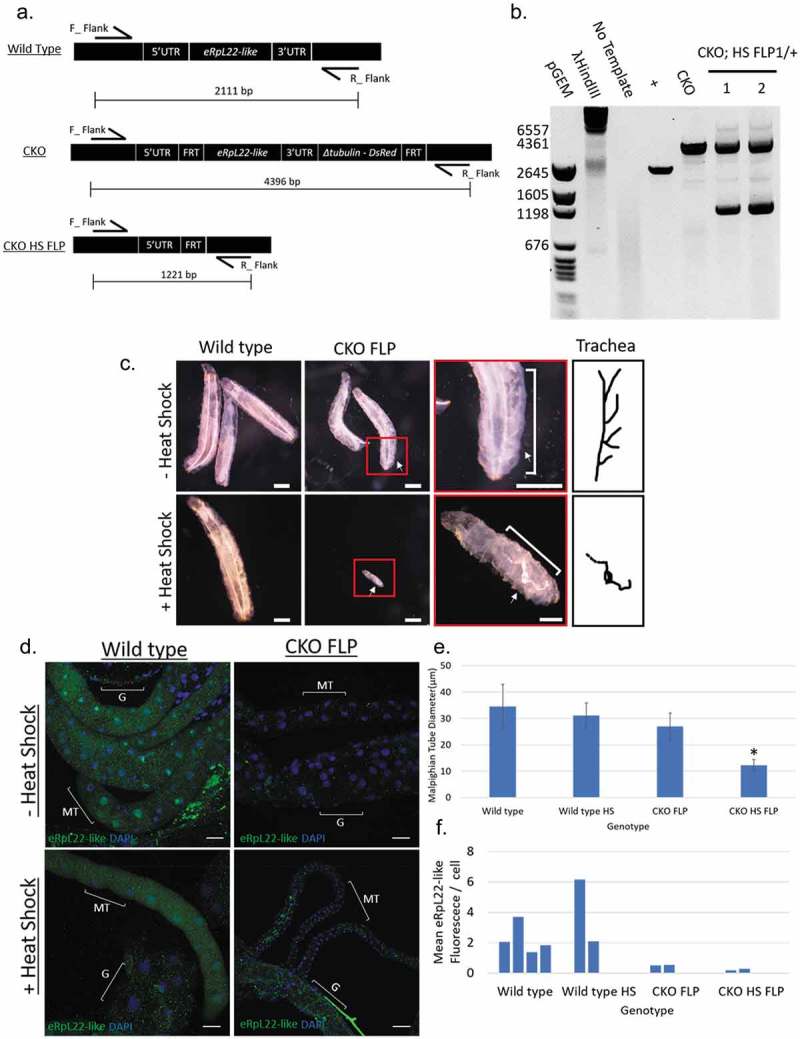
Conditional, heat shock-induced knockout of eRpL22-like. a) Diagram of the eRpL22-like locus (top) and diagram of the FRT flanked eRpL22-like locus before (middle) and after heat shock induced FLP expression (bottom). b) PCR confirmation of eRpL22-like knockout showing an expected shift in PCR products corresponding to FLP-FRT mediated excision of genomic sequence. c) Dissecting scope images show gross morphological consequences of eRpL22-like knockout on larval development. Differences in overall larval growth are apparent. White arrows how differences in trachea development with and without heat shock. Scale bar: 400 μm, red box scale bar: 100 μm. d) IHC analysis of Malpighian tubules (white arrow) at day 5 post embryo deposition. Tissue is stained against eRpL22-like (Green) and DAPI (Blue) is used as a nuclear stain. Scale bar: 20 μm. Heat shock treatment shows no effect on eRpL22-like expression and localization. CKOFLP tissue does not recapitulate wild type eRpL22-like staining, indicating potential differences in eRpL22-like expression between wild type and CKOFLP. Heat shock treatment results in dramatic defects in Malpighian tubule development. e) Diameter of Malpighian tubes was quantified using ImageJ. Malpighian tubules are significantly smaller. Error bars represent one standard deviation, *p < 0.01 according to t-test. f) Overall fluorescence was measured and normalized to the number of cells within the area measured to control for growth differences. The number of cells was determined by counting nuclei of each tube.
To confirm eRpL22-like knockout and to identify what tissues were affected when heat shock was applied at 1 hour post-egg laying, we dissected three of the four individuals (except the adult female with eye bristle defects) that developed beyond the 1st instar larval stage (1 larva, 1 midpupa, and 1 adult; Table S1). PCR analysis confirmed eRpL22-like knockout in the midpupa and adult since the predicted PCR product of 1221bp was observed (Figure 2(b)). In each individual, the presence of PCR products representing the full length eRpL22-like FRT cassette (4396bp) and the deleted version (1221bp) is likely due to mosaicism resulting from variable heat shock effects applied throughout the entire organism to generate gene excision in some cells, but not in others (Figure 2(b)).
Larval phenotypes included defects in tracheal branching and growth (Figure 2(c)) and comparatively smaller and less developed Malpighian tubules (Figure 2(d,e)) with less eRpL22-like expression compared to wildtype (Figure 2(f)). Malpighian tubule diameter is significantly reduced at 10 μm compared to controls (~30 μm). The reduction in eRpL22-like staining without heat shock in the Malpighian tubules may be due to changes in eRpL22-like expression resulting from insertion of the FRT cassette within the eRpL22-like locus. Age-matched wildtype or CKO-FLP larvae were >3X longer than the heat-shocked CKO-FLP larva (Figure 2(c), Table S2), suggesting considerable growth retardation in the absence of eRpL22-like function.
In each case where heat shocked embryos developed beyond embryonic stages, progeny displayed behavioral abnormalities. Larval movements were sluggish, suggesting muscle movement phenotypes. In fact, eRpL22-like and actin staining of wildtype 1st instar larval muscle tissue shows the presence of eRpL22-like, suggesting that eRpL22-like expression may be required for normal muscle development (Figure S1). It is also noteworthy that eRpL22-like and actin staining do not completely overlap in this tissue, suggesting that actin expression at this stage may not be ‘ubiquitous’ in all tissues (Figure S1). A more complete larval eRpL22-like expression profile is currently in progress (Gershman, Pritchard, and Ware, unpublished). Incomplete overlap of actin and eRpL22-like expression in critical tissues may account for insufficient Actin-GAL4 driven RNAi-mediated knockdown, resulting in enough residual eRpL22-like to support development into the adult stage.
Based on previous work of Shigenobu et al. [10] showing eRpL22-like mRNA expression in gonads and the stomatogastric nervous system of developing embryos by in situ hybridization, we reasoned that tissues of the enteric system (innervated by the stomatogastric nervous system) might be impacted by deletion of eRpL22-like. In the heat shocked embryo that survived to the midpupal stage, gut defects were indeed apparent. A well-developed gut was completely absent, with only immature tubes attached to the body wall. Based on the presence of sex combs, this animal was determined to be male, but external genitalia were severely deformed (data not shown).
Internal dissection of the adult that succumbed shortly after eclosure revealed no clearly distinguishable tissues or organs. Tissue death was particularly apparent in bristles, midgut, and the eye. Taken together, we conclude that expression of eRpL22-like in embryonic and larval tissues is necessary for complete animal development. Whether or not eRpL22-like has a special role in tubular organogenesis (e.g., trachea, gut, testis, Malpighian tubules) remains to be determined.
Variation in the timing of heat shock following egg deposition allowed us to probe eRpL22-like knockout effects at different times in development. When heat shock was applied at 24–36 hours after egg deposition, only 5/20 (25%) embryos developed to adulthood whereas 62/97 (~64%) wildtype embryos developed to adulthood. Improvement in survivability with heat shock at later stages of development is consistent with increases in survival rates for wildtype flies after heat shock, previously disclosed by Bergh and Arking [25]. F1 progeny from this heat shock experiment were then allowed to mate to determine if reproductive potential was affected. The female deposited 125 eggs over the course of approximately 21 days, but none of them developed, suggesting that fertility was compromised in the male or female or in both. When the CKO female was mated with two wildtype males, 11 embryos were deposited, eventually yielding five F1 progeny. These data indicate that eggs produced by the 24–36 hour heat shock CKO female could be fertilized and suggest that the original male (from a heat-shocked at 24–36 hours after egg deposition experiment) was sterile. We note that while the CKO female was fertile, there were noticeable anomalies in ovaries, including nurse cell death and follicle cell polarity defects within some egg chambers (data not shown). Expression of eRpL22-like has been documented within germline stem cells in adult ovaries [26], but the impact of eRpL22-like depletion within the ovary remains to be investigated.
Subsequent dissection of the reproductive system from the sterile CKO male revealed several abnormalities compared to wildtype or CKO parental lines (Figure 3(a)). The most obvious defect was incomplete testis development. Testis tube elongation and coiling were noticeably absent in the heat-shocked CKO case. Fully formed seminal vesicles were not apparent. Accessory gland morphology appeared similar to wildtype. PCR confirmed eRpL22-like knockout in the reproductive system from this male (Figure 3(b)). The reproductive systems of three other heat-shocked CKO males (confirmed by PCR and from a separate heat shock experiment in the 24–36 hour window) appeared relatively normal in organization and morphology, with elongated and coiled testes and motile sperm present (data not shown). We postulate that differences in timing of heat shock application within the 24–36 hour window of egg laying most likely account for variation in reproductive system development, as seen for CKO males in this experiment.
Figure 3.
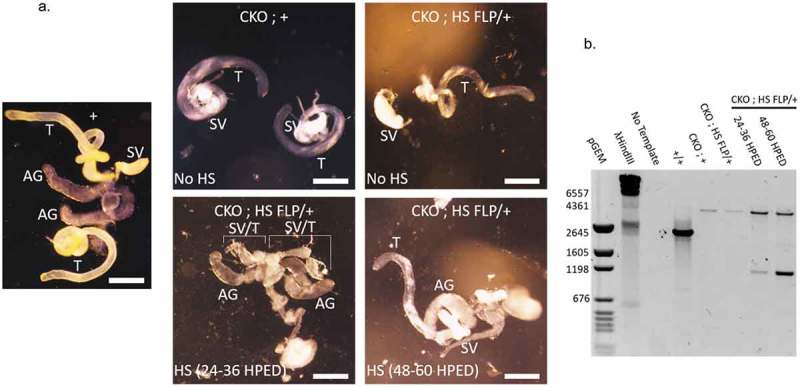
Effect of eRpL22-like knockout on testes development. a) Dissecting microscope images of gross morphological changes to adult testis development resulting from heat shock treatment of embryos at 24–36 hours post-embryo deposition ([HPED]; bottom left) and 48–60 HPED (bottom right). Heat shock 24–36 HPED results in severe defects in both testis and accessory gland growth. Heat shock 48–60 HPED results in overall normal reproductive system. (+, CKO; +, CKO; HS-FLP/+). Scale bar: 300 μm. T- testis, AG- accessory gland, SV- seminal vesicle. SV/T indicates the tissue in the labeled region is either malformed testis, seminal vesicle, or both tissues. b) eRpL22-like knockout was confirmed through PCR analysis of DNA extracted from heat-shock treated CKO; HS-FLP/+ testis tissue compared to untreated control tissue.
When heat shock was applied at 48–60 hours after egg deposition, testis development was not noticeably altered and appeared similar to that of CKO flies (compare Figure 3(a)). Testis tube elongation was apparent and other reproductive organs were more clearly developed than when heat shock was applied earlier in development. F1 males and females derived from the 48–60 hour heat shock time point were fertile, confirmed by the appearance of at least 30 F2 progeny. These experiments establish that eRpL22-like expression early in embryogenesis is essential for proper development of critical organ systems required for viability and for testis development and subsequent germ cell production.
eRpL22-like levels remaining after incomplete knockdown in mitotic cysts are sufficient for normal germline development and fertility
To test the requirements for each paralogue in germline development and fertility in adults, we used the mitotic cyst germline-specific bam-GAL4-VP16 driver [22] for germline-specific RNAi knockdown. Bam is an important differentiation factor required during the transition of mitotic spermatogonia to the spermatocyte stage; yet its downregulation in spermatocytes is necessary for differentiation of mature spermatids [27]. Thus, based on the pattern of bam expression during spermatogenesis, we would expect most efficient depletion of eRpL22 paralogues in the window up to the spermatocyte stage.
Knockdown of eRpL22-like with bam-GAL4-VP16 shows significant reduction (~80% KD) in eRpL22-like protein levels compared to control by Western analysis, confirming knockdown (Figure 1(c)). Incomplete eRpL22-like knockdown with the bam-GAL4 driver (active in mitotic germ cells: late spermatogonia and early spermatocytes; reviewed by [28]) may be a consequence of post-mitotic transcription of eRpL22-like after RNAi expression or the existence of stable eRpL22-like transcripts that await translation in later stages of spermatogenesis, as is the case for many testis-specific transcripts (e.g., see review by [28]). Phase microscopy revealed no obvious phenotypes and motile sperm were evident and capable of fertilization (Figure S2 and Figure 4). No significant levels of knockdown were observed for bam>eRpL22.IR.
Figure 4.
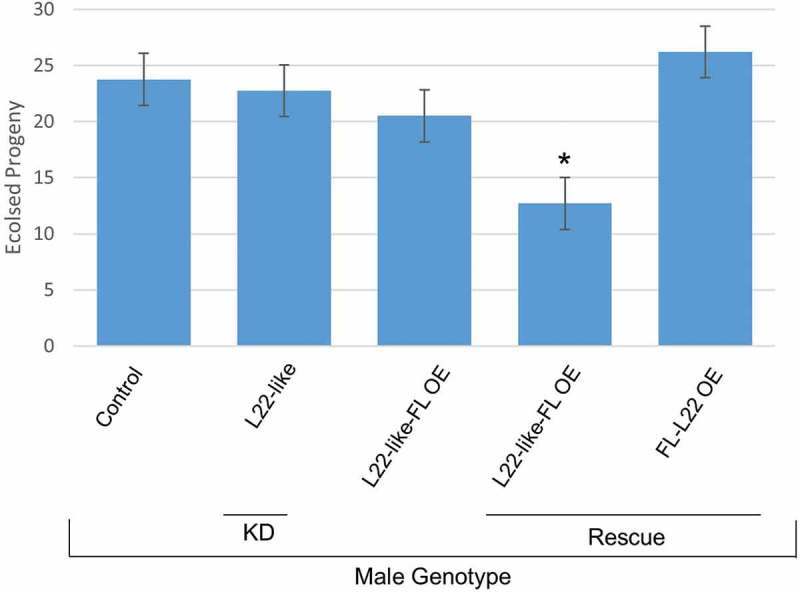
Assessment of fertility in paralogue knockdown and rescue genotypes determined by number of eclosed progeny. F1 male fertility was assessed by number of eclosed progeny per male when mated with a single wildtype female. Compared to all other genotypes, eRpL22-like-FLAG rescue males have a significant decrease in number of progeny per male. eRpL22-like paralogue knockdown was performed with bam-GAL4-VP16. eRpL22-like-FLAG overexpression was driven by Actin-GAL4. The eRpL22-like OE and eRpL22 OE rescues were driven with Actin-GAL4. FLAG (FL), knockdown (KD), overexpression (OE), and rescue are listed below genotypes. Error bars represents one standard deviation, * represents a p < 0.01 according to t-test, graph represents the mean of 10 males. The genotypes of males from each fertility cross are listed from left to right: Wildtype, bam>eRpL22-like.IR, Act>eRpL22-like-FLAG, Act>eRpL22-like-FLAG/eRpL22.IR, Act>FLAG-eRpL22/eRpL22.IR.
Overexpression of eRpL22-like-FLAG in eRpL22-depleted flies rescues embryonic viability but results in both reduced fertility and life span
Despite insufficient buildup of morphological phenotypes that would arrest or impede spermatogenesis progression in germline paralogue knockdown experiments, ubiquitous knockdown of eRpL22 (with an Actin-GAL4 driver) results in embryonic lethality. We therefore asked if eRpL22-like-FLAG overexpression could rescue viability in flies depleted in eRpL22 (cross shown in Figure S3). eRpL22-like overexpression was confirmed by Western blot (Figure 5(a)), showing accumulation of eRpL22-like in adult bodies where it is not normally expressed. Knockdown of eRpL22 (15% for unmodified 33kD and 65% for ≥ 55α) is also observed in bodies (Figure 5(a)). Overexpression of eRpL22-like-FLAG in an eRpL22 knockdown background produced F1 progeny (Figure 4), but specific testis and longevity phenotypes were noticed (Table 1). As a control to determine if eRpL22-like overexpression phenotypes were due to insufficient expression of eRpL22-like from the Actin driver, we overexpressed FLAG-eRpL22 in an eRpL22 knockdown background as well. Viable F1 progeny were also produced and noticeably, no fertility or longevity phenotypes were evident (Figure 4, Table 1). In this case, testis morphology, fertility, and longevity were indistinguishable from wildtype. Therefore, we conclude that phenotypes observed with eRpL22-like-FLAG overexpression are not the consequence of inadequate expression from the Actin driver.
Figure 5.
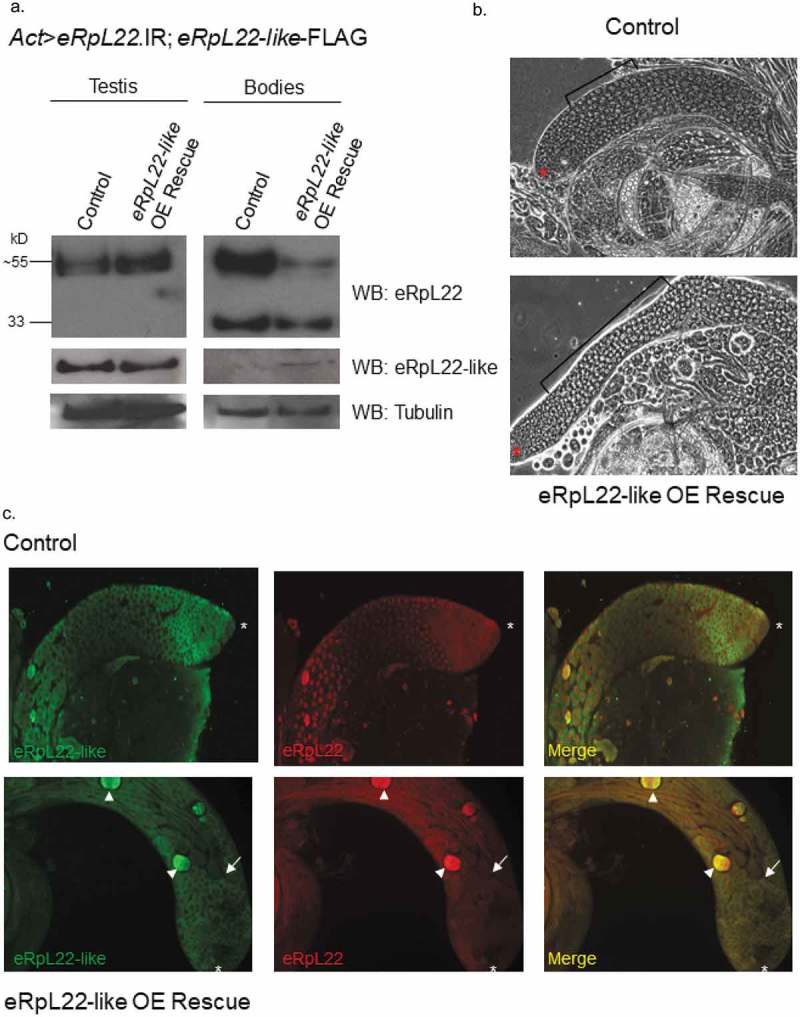
Rescue of embryonic viability is achieved by overexpression of eRpL22-like in flies ubiquitously depleted of eRpL22. (a) Western analysis shows no change in accumulation of eRpL22 or eRpL22-like levels in testes from 3 day old males. In gonadectomized bodies, eRpL22-like is detected due to ubiquitous overexpression (OE) in all tissues (using Act>eRpL22-like-FLAG). Western blot analysis shows a ~ 15% loss of unmodified eRpL22 (33kD) and a ~ 65 % loss of SUMOylated eRpL22 (~≥55kD). Tubulin was used as a loading control. (b) Representative phase contrast microscopy images of wildtype control and eRpL22-like-FLAG OE rescue testes from 2 day old males. All stages of spermatogenesis are evident; however, an increase in immature spermatocytes (brackets) is noticeable within eRpL22-like OE rescued testes. An asterisk denotes the apical tip. (10x magnification). (c) IHC of wildtype control or eRpL22-like OE rescued from testes of 3 day old males. End of spermatid bundles (arrow) are closer to the apical tip (asterisk), and degenerating cysts are present (arrowhead). (10x magnification) Images in B and C are representative of rescued testes. Control is age matched wildtype males.
Table 1.
Overview of outcomes of RNAi or overexpression crosses described. Viability was assessed using longevity assays. Motile sperm was assessed using testes squashes from phase contrast imaging; fertility was assessed in a 1:1 female: male ratio as described in Methods. Effects of paralogue depletion or overexpression were determined by Western analysis and ImageJ quantification.
| Cross | Viable | Motile Sperm | Fertile | Effect on Paralogue |
|---|---|---|---|---|
| Act>eRpL22.IR | No | - | - | - |
| Act>eRpL22-like.IR | Yes (>30 days) | Yes | Yes | Increase eRpL22 |
| bam>eRpL22-like.IR | Yes (>30 days) | Yes | Yes | Increase eRpL22 |
| Act>eRpL22-like-FLAG | Yes (>30 days) | Yes | Yes | Decrease in 33kD eRpL22 |
| Act>eRpL22-like-FLAG/eRpL22.IR | Yes (~14 days) | Yes, Reduced | Yes, Reduced | Bodies: Decrease eRpL22, Increase eRpl22-like Testes: No significant change |
| Act>FLAG-eRpL22/eRpL22.IR | Yes (>30 days) | Yes | Yes | No significant change |
| bam>eRpL22-like-FLAG/eRpL22.IR | Yes (>30 days) | Yes | Yes | No significant change |
Several notable phenotypes were observed in testes from flies depleted of eRpL22 and rescued by eRpL22-like overexpression. Phase contrast microscopy images show testes that are ~20% more narrow than in wildtype, based on ImageJ measurements of 10 testes from each genotype (SD: ± 0.16) (Figure 5(b)). A larger zone of immature spermatocytes is apparent compared to wildtype testes. It is unclear if accumulation of immature early meiotic spermatocytes is due to a delay in normal numbers of spermatocytes (from continuous supply of mitotic cysts from the hub) progressing to the spermatid stage or alternatively, if greater numbers of mitotic cysts are emerging from the stem cell niche at the hub, but the spermatocyte-to-spermatid transition is not delayed. IHC shows that numerous degenerating cysts are apparent in rescued testes (Figure 5(c)). Fertility was significantly reduced (Figure 4) and flies had reduced longevity (Table 1). It is likely that reduced fertility is associated with spermatogenesis phenotypes previously described.
It is noteworthy that longevity phenotypes with the Actin driver are unlikely consequences of testis-specific anomalies, but are likely outcomes of downregulation of eRpL22 in somatic tissues and not consequences of eRpL22-like-FLAG overexpression, since neither longevity nor fertility defects were present in flies that ubiquitously overexpress eRpL22-like-FLAG from the Actin driver (Figure 4, Table 1). eRpL22-like expression is sufficient to rescue complete development in flies ubiquitously depleted of eRpL22; however, some functions required for completion of normal spermatogenesis to produce an abundance of fertile sperm, as well as functions affecting lifespan are not completely restored. Roles for eRpL22 and eRpL22-like in development and spermatogenesis are not completely interchangeable, and must include functionally distinct roles.
Mitotic germline depletion of eRpL22 paralogues alters expression levels of the opposing paralogue
To explore possible compensatory mechanisms from the effect of 80% eRpL22-like knockdown, we assessed eRpL22 expression levels in eRpL22-like RNAi-depleted tissue (knockdown seen in Figure 1(c)) given the increase in eRpL22 staining shown by IHC (Figure S2B). Interestingly, all molecular mass species of eRpL22 increase (although in varying amounts) in eRpL22-like-depleted tissue when compared to the control (Figure 6(a)). Unmodified 33kD (known to be in polysomes in S2 cells [20]) and ≥55kD (SUMOylated and phosphorylated, but not a component of translating ribosomes in S2 cells [20]) eRpL22 species are the most upregulated, with a 42% increase in the amount of unmodified eRpL22 and a ~ 40% increase in the ≥55kD species. RpL23a levels are not altered, suggesting a specific effect relevant to eRpL22 paralogues rather than a global increase in ribosomal protein expression. Molecular confirmation of an increase in eRpL22 in eRpL22-like-depleted testes by Western blot mirrors the increased eRpL22 staining pattern seen in IHC images of eRpL22-like-depleted testes (Figure S2B).
Figure 6.
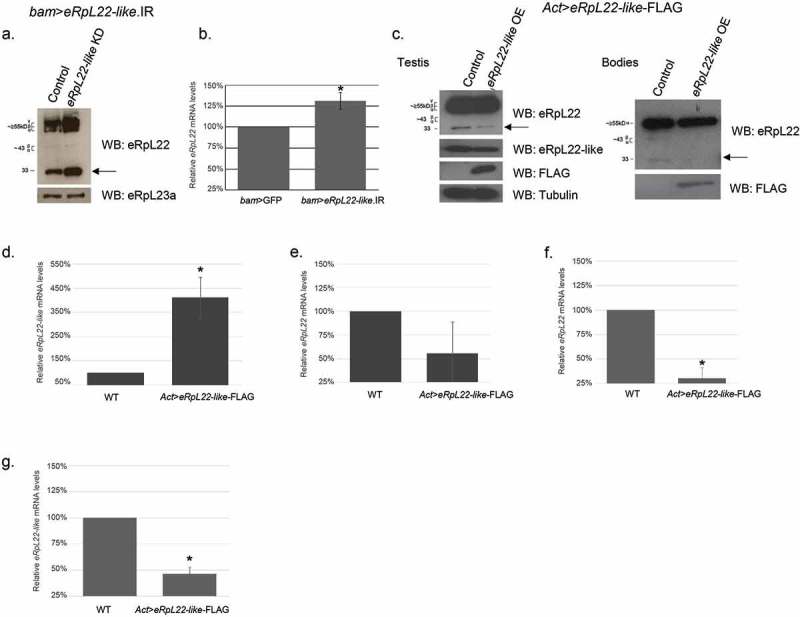
Mitotic germline eRpL22 paralogue depletion or overexpression affects opposing paralogue expression levels. a) An increase in testis eRpL22 protein levels is observed when eRpL22-like is specifically depleted in the male germline (bam-GAL4-VP16) when compared to control (wildtype). The most notable increase occurs in the unmodified (33kD) species (arrow). Testis eRpL23a levels remain constant. b) qRT-PCR shows a statistically significant increase (average: 31% [1.31 fold change]; range: 17–47%) in testis eRpL22 mRNA levels when eRpL22-like is depleted from the germline. Error bars represent standard error, n = 3, *p < 0.01 according to t-test. c) eRpL22-like overexpression (using an Actin-GAL4 driver) results in a decrease of eRpL22 in the Drosophila testis and bodies compared to controls (wildtype). Significant knockdown is seen in the unmodified (33kD) species of eRpL22 (arrow). FLAG signal confirms the presence of the overexpression construct eRpL22-like-FLAG. Tubulin was used as loading control for testes samples and protein assays were used as a loading control for gonadectomized bodies. d) qRT-PCR shows a statistically significant increase (average: 311% [fold-change 4.11]) in gonadectomized bodies eRpL22-like mRNA levels when eRpL22-like is ubiquitously overexpressed. Error bars represent one standard deviation, n = 3, *p < 0.01 according to t-test. e) qRT-PCR shows a reduction (average: 44% [0.56 fold-change]) in eRpL22 mRNA levels in gonadectomized bodies from ubiquitous overexpression of eRpL22-like. Error bars represent one standard deviation, n = 3. f) qRT-PCR shows a significant reduction (average: 69% [0.31 fold-change]) in testis eRpL22 mRNA levels when eRpL22-like is ubiquitously overexpressed. Error bars represent one standard deviation, n = 9, *p < 0.01 according to t-test g) qRT-PCR shows a significant reduction (average: 53% [0.47 fold-change]) in testis eRpL22-like mRNA levels upon ubiquitous eRpL22-like overexpression. Error bars represent one standard deviation, n = 9, *p < 0.01 according to t-test.
To characterize this effect, we analyzed eRpL22 mRNA levels to determine if the increased eRpL22 accumulation correlates with an increase in mRNA level. qRT-PCR shows a statistically significant increase (average 31%; 1.31 fold change) in testis eRpL22 mRNA levels compared to the control when eRpL22-like is depleted in the male germline (Figure 6(b)). The increase may be attributable to an increase in eRpL22 transcription and/or eRpL22 mRNA stability. Together, testis eRpL22 protein and mRNA levels increase when eRpL22-like levels are depleted (Figure 6(a,b), respectively), suggesting a negative regulatory effect of eRpL22-like on eRpL22 levels, mediated through an effect on eRpL22 mRNA. Thus, eRpL22 protein levels may be regulated by a direct or indirect role of eRpL22-like in processes that inhibit eRpL22 transcription and/or promote eRpL22 mRNA turnover in germ cells.
As a further test of the hypothesis that eRpL22-like has an inhibitory effect on eRpL22 levels, we overexpressed eRpL22-like-FLAG with an Actin driver and measured eRpL22 protein accumulation in testes and gonadectomized flies, predicting that eRpL22 levels would be diminished in this case. Indeed, ubiquitous overexpression of eRpL22-like-FLAG had an inverse effect on eRpL22 accumulation in testes and gonadectomized flies: eRpL22 levels were diminished by ~50% in testes and in residual bodies, consistent with a negative regulation model (Figure 6(c)). We used qRT-PCR to confirm paralogue mRNA levels from testes and gonadectomized bodies in these flies. A 4.11-fold increase in total eRpL22-like mRNA was observed, confirming eRpL22-like overexpression in bodies (Figure 6(d)). There was a 0.56-fold reduction in eRpL22 mRNA in gonadectomized bodies when eRpL22-like was overexpressed, confirming a negative regulation model at the mRNA level (Figure 6(e)). Given that the eRpL22-like negative effect on eRpL22 levels could be recapitulated outside of the testis environment, we posit that testis-specific factors are not required for basic features of this regulatory mechanism.
Based on knockdown of eRpL22-like showing an increase in eRpL22 mRNA levels, we would predict a decrease in eRpL22 mRNA levels when eRpL22-like is overexpressed. Depletion of eRpL22 resulted in a decrease in eRpL22-like. qRT-PCR confirmed a 0.31-fold (69%) decrease in eRpL22 mRNA in eRpL22-like overexpression testes, but eRpL22-like mRNA levels were diminished by 0.47 fold (53%) (Figure 6(f,g)). Taken together, resulting eRpL22-like protein levels may be explained by a regulatory feedback loop. We propose that mitotic germline knockdown of eRpL22 paralogues affects accumulation of opposing paralogue levels, but that different mechanisms for each paralogue are likely in play. The increase in eRpL22 upon eRpL22-like depletion is unlikely to be explained as a simple compensatory global increase in ribosome production in response to depleted eRpL22-like levels, since no increase in RpL23a (an early rRNA binding RP required for ribosome biogenesis; e.g. [29],) was noted. Changes in the proportion of ribosomes containing each paralogue might be predicted as an outcome of paralogue knockdown; however, the impact of those changes on progression of spermatogenesis within maturing mitotic spermatogonia or early spermatocytes is unknown. Determination of possible changes in the proportion of ribosomes containing each paralogue after opposing paralogue knockdown must await future experimental analysis.
Discussion
eRpL22 paralogues are partially functionally redundant but specify unique roles in development and in spermatogenesis
A major goal of the work described here was to decipher aspects of the functional complexity of eRpL22 and eRpL22-like by exploring the degree to which roles for these proteins overlap or are unique in development and in spermatogenesis. Our first consideration was to confirm the essential requirement for eRpL22 [15,16] and eRpL22-like (FB2017_3) in fly development using our RNAi-mediated system to deplete ubiquitous expression of each paralogue in development. Interestingly, eRpL22 has not uniformly been characterized as an essential gene: it is essential for viability in the fly and C. elegans but not in other eukaryotic systems studied (fly: [15,16]; C. elegans: [30]; rat: [31]; yeast: [32]; zebrafish and mice: [33,34]. In zebrafish and mice, knockdown of Rpl22 dramatically affects differentiation in the T-cell lineage [33,34]. The absence of a requirement for eRpL22 for viability in other organisms suggests that an additional function(s), critical for developmental progression in flies and worms, may be specified. Our RNAi data demonstrate an essential requirement for eRpL22 in Drosophila development (summarized in Figure 7).
Figure 7.
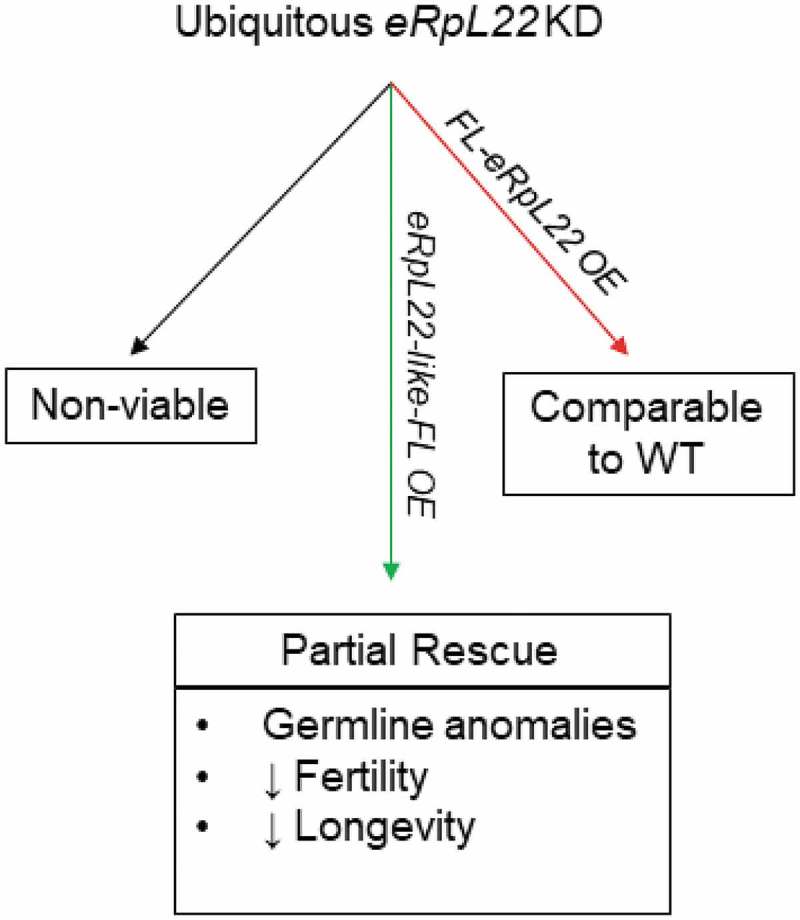
Summary of eRpL22 knockdown and rescue effects. Outcomes from eRpL22-depletion are shown in boxes. Arrows are color-coded (green: eRpL22-like; red: eRpL22) to depict paralogue gene overexpressed for rescue. Ubiquitous expression was driven by Actin-GAL4. KD: knockdown; OE: overexpression; FL: FLAG.
An essential requirement for eRpL22-like had also been proposed based on reports that a P-element insertion 156 nucleotides upstream of the eRpL22-like transcription start site is homozygous lethal (FB2017_3). Our ubiquitous RNAi knockdown experiments using an Actin-GAL4 driver were insufficient to block fly development or negatively impact male fertility compared to wildtype, despite localized disruptions in some cysts. However, our conditional eRpL22-like knockout experiments demonstrate that eRpL22-like expression (and not expression of an upstream regulatory element) during early embryogenesis is essential for normal fly development. Given the degree of eRpL22-like expression in several 1st instar larval tissues, it is not surprising that lethal phenotypes result when eRpL22-like gene knockout occurs in early embryonic development. It is interesting that levels of eRpL22-like remaining (40%) after ubiquitous RNAi depletion must have been sufficient to sustain development overall in critical tissues (and in developing germ cells) and may be explained by partial overlap between eRpL22-like and actin expression (as seen in Figure S1).
Germline knockdown of eRpL22-like was insufficient to block sperm production and fertility compared to wildtype controls, despite the presence of some morphological phenotypes. One explanation that may account for the absence of an overall fertility effect is that eRpL22 (mRNA and protein levels upregulated following eRpL22-like knockdown) replaces functions lost when eRpL22-like is depleted. In the latter case, it would be expected that eRpL22 would act to rescue eRpL22-like ribosomal functions in the germline. What remains unclear is the extent to which eRpL22 and eRpL22-like ribosomes may be functionally equivalent.
Rescue experiments clearly demonstrate that eRpL22 paralogues, at a minimum, retain a set of overlapping ribosomal functions, but are not completely functionally redundant since rescued adults display fertility and longevity deficiencies. It remains to be determined if defects result from functional differences between eRpL22-specific and eRpL22-like-specific ribosomes, unique expression profiles for paralogues in some larval tissues, and/or extraribosomal roles for SUMOylated eRpL22 within meiotic spermatocytes and/or in other cell types (e.g., as shown in S2 cells) that are not replaced by eRpL22-like.
Widespread evidence is accumulating that paralogous RPs are not completely functionally redundant. Early studies showed that overexpression of one RP often rescued the growth defect resulting from genetic or biochemical depletion of its paralogue, thereby supporting the notion of functional redundancy [35]. Further, Deutschbauer et al. [32] reported that duplicated RP genes have less severe haploinsufficiency defects than their non-duplicated partners, indicating that duplication provides fitness to the organism. Yet, many of the 14 duplicated RPs in Drosophila are not equally associated with the haploinsufficiency-related Minute phenotype [17], suggesting non-redundancy.
If paralogues are functionally redundant, one would expect to see identical phenotypes upon deletion or alteration. Komili et al. [36] provided very compelling evidence for non-redundancy as they reported RP deletions in yeast that exhibit paralogue-specific effects on transcription profiles and cause unique phenotypes under different physiological conditions. Whether the differential effects seen between yeast paralogues are due to changes in the translation machinery (and ultimately which mRNAs are translated) or are the result of a secondary effect from extraribosomal functions of RPs remains to be investigated.
Differential expression of RP paralogues may be indicative of specialized functions of ribosomes expressed in particular tissues or cells. Indeed, compelling evidence for ribosome heterogeneity at the level of core RPs affecting mRNA translation has recently been shown (reviewed by [4]). Certainly, extensive structural diversity within eRpL22 paralogues adds structural heterogeneity to the ribosome pool within germ cells (where paralogues are known to be co-expressed) beyond that provided by core RPs. Based on mounting evidence for the existence of specialized ribosomes [e.g., 3, 4,37] (Mageeney and Ware, unpublished), we propose that ribosomes containing eRpL22 or eRpL22-like are not functionally equivalent, and thus the failure of eRpL22-like expression to fully rescue wildtype phenotypes in eRpL22-depleted flies may result from differences in translation capacity of paralogue-specific ribosomes. A complete analysis of mRNAs translated on eRpL22 and eRpL22-like-specific ribosomes (as one of several possible regulatory differences between ribosome types) is currently in progress and should clarify if distinctive and/or overlapping ribosomal roles (at the level of mRNA translational specificity) exist for these paralogues in spermatogenesis (Mageeney and Ware, in preparation).
On the other hand, co-expression of RP paralogues adds an additional consideration beyond a specialized ribosome function: that RP paralogues confer unrelated functions in cells that are exempt from the translation pathway. Over the course of evolution, duplicated RPs may have segregated functions previously ascribed to an ancestral protein or duplicated proteins may acquire novel functions that are extraribosomal in nature. Both ribosomal and extraribosomal roles have been documented for several RPs [1], allowing consideration of eRpL22 paralogue functionality within the ribosome cycle and in extraribosomal pathways in the Drosophila male germline. Is there evidence that eRpL22 paralogues function in extraribosomal pathways in the testis?
Mitotic germline deposition of eRpL22 and eRpL22-like shows both proteins in the nucleolus and cytoplasm, consistent with a ribosomal role [20]. It is not clear if eRpL22 has a predominately ribosomal function in post-meiotic cells undergoing extensive morphological and genomic remodeling to become mature sperm. A shift in eRpL22 function may occur in meiotic spermatocytes that separates predominant roles for eRpL22 paralogues in germ cell differentiation. Polysome profiles confirmed that SUMOylated eRpL22, found in a wide variety of cell types, does not co-sediment with actively translating ribosomes in S2 cells, supporting an extraribosomal role for SUMOylated eRpL22. The presence of additional testis-specific posttranslational modifications correlates with nuclear distribution of eRpL22 [20]. Cytoplasmic distribution of eRpL22-like is consistent with a ribosomal role throughout spermatogenesis, although additional possibilities have not, as yet, been examined rigorously.
A proposed auxiliary function provided by SUMOylated eRpL22 may be required for Drosophila development. Drosophila eRpL22 harbors a fly-specific N-terminal extension of unknown function, but is homologous to the C-terminus of histone H1 [18]. Interestingly, our previous work has shown that deleting this N-terminal domain does not hinder incorporation of the residual, truncated protein into functional ribosomes in S2 cells [20], leading to the interpretation that the fly-specific extension is neither required for assembly into ribosomes nor for ribosome function. Therefore, the eRpL22 N-terminal extension may be necessary for a proposed extraribosomal role of this paralogue in the fly and provides an explanation for why eRpL22 is essential in the fly. Even though SUMOylation motifs are predicted for each paralogue within the structurally diverse N terminal domain at different positions within the domain [20], there is no expectation that eRpL22-like would assume the proposed nuclear function for SUMOylated eRpL22 in somatic cells or in the male germline, possibly contributing to abnormalities in longevity and fertility.
Overall, these studies attribute unique functions to eRpL22 paralogues in the male germline in addition to common roles as ribosomal components. Further explorations are forthcoming to reveal the extent of overlap in ribosomal functions and the role of modified eRpL22.
Gene expression of eRpL22 paralogues is uniquely coordinated in Drosophila
In yeast, ~70% of yeast duplicated RP genes are asymmetrically expressed and are generally regulated to maintain the expression ratio, rather than the dosage of RPs [38]. Whether this regulatory pattern is similar in higher eukaryotes remains to be determined. Parenteau et al. [38] demonstrated that yeast eRpl22 paralogues (84.4% sequence identity and 95.6% sequence similarity [39]) display intergenic intron-dependent regulation (e.g. the intron of eRpl22A regulates the expression of eRpl22B, and vice versa). Furthermore, deleting introns from both genes leads to increased expression of both paralogues at a level of 2–6 fold [38]. Coordinated gene expression between eRpL22 paralogues is evident in yeast, but whether this is similar in Drosophila had yet to be investigated until this point. In Drosophila, expression of testis-specific (or testis-biased) genes with paralogues is often coordinated [40], but specific mechanisms remain to be determined.
Unlike the ubiquitous expression profile for Rpl22 paralogues in zebrafish and mice, Drosophila eRpL22-like has tissue-restricted expression and is co-expressed along with eRpL22 in the adult male germline. As such, some mechanistic differences might be predicted to account for regulation of eRpL22 paralogue expression in Drosophila compared to other model systems. One common regulatory thread is that one paralogue negatively regulates the expression of the opposing paralogue. In Drosophila, eRpL22 expression increases when eRpL22-like is depleted in the male germline. The absence of a change in RpL23a levels appears to eliminate a general global effect on RP synthesis in favor of a specific eRpL22 paralogue effect. This physiological response to a decrease in the level of eRpL22-like is consistent with a crosstalk regulation model based on overall expression ratios of eRpL22 and eRpL22-like.
Notable differences in eRpL22 paralogue crosstalk are evident in Drosophila compared to zebrafish and mice with respect to which paralogue functions as the negative regulator. In zebrafish and mice, RpL22 functions as the negative regulator impacting Rpl22l1 mRNA stability through yet undetermined mechanisms. In Drosophila, it is currently unknown if eRpL22-like binds eRpL22 mRNA and impacts mRNA stability, in a comparable manner as has been reported for mouse Rpl22 effects on Rpl22l1 mRNA stability [8]. Whether or not Drosophila eRpL22 paralogues bind their own mRNA or the opposing paralogue mRNA has not as yet been extensively investigated. In preliminary polysome profiling experiments, eRpL22 mRNA is co-immunoprecipitated from fractions at the top of ribosomal gradients using eRpL22-like-specific antibodies, showing possible interactions between eRpL22 mRNA and eRpL22-like protein (Mageeney and Ware, unpublished). Further investigation of proposed interactions should provide insight into mechanisms by which paralogue expression levels are regulated.
Coordinated changes in gene expression for eRpL22 and eRpL22-like may occur based on shared regulatory elements for the two genes, originally derived from gene duplication. Regulatory elements (genomic and mRNA) in the eRpL22 gene family have not been reported in any model system. RP promoter elements have been studied in Drosophila, but those for eRpL22 paralogues or other RP paralogues have not investigated [41]. Eukaryotic RP mRNAs, particularly in mammals, often contain 5ʹ-TOP (5ʹ terminal oligopyrimidine tract) regulatory elements that respond to cellular growth cues [42] and have recently been identified as targets for miRNA regulation [43]. However, Drosophila RP regulation through 5ʹ-TOP motifs has not been thoroughly investigated [44]. Exploring cis-regulatory elements, shared or unique to each paralogue, may provide insight to the coordinated gene expression of eRpL22 paralogues.
Negative regulation of eRpL22 levels by eRpL22-like may constitute part of a mechanism that regulates ribosome composition in germ cells. Additionally, these studies offer a perspective on mechanisms that may block incorporation of a paralogue into tissue-specific ribosomes, and divert the RP paralogue into new cellular functions that are extraribosomal in nature. Zhang et al. [6] have recently reported antagonistic roles for zebrafish RpL22 and Rpl22l1 acting outside of the ribosome as modulators of splicing of pre-mRNAs involved in gastrulation. Though the mechanism by which zebrafish paralogues are diverted from ribosomal function during gastrulation remains unknown, no doubt a complex interplay of interactions awaits discovery. In Drosophila, further investigation of the precise role of post-translationally modified eRpL22 will be crucial to determine if eRpL22 assembles into ribosomes at specific stages of male germline maturation beyond early mitotic stages of spermatogenesis.
Funding Statement
Lehigh University Faculty Research Grants 607233 and 607277; National Academy of Science Grants-In-Aid of Research G20100315152292 and G2016100191865376, sponsored by Sigma Xi.
Acknowledgments
Funding was provided in part by Faculty Research Grants (607233 and 607277) from Lehigh University to VCW. Work described here fulfilled a part of the requirements for the PhD for MGK and for CMM. Both MGK and CMM were partially supported by Nemes Fellowships. JMC was supported by an undergraduate research grant funded by the College of Arts and Sciences at Lehigh University. We thank TRiP at Harvard University and members of the fly community for fly stocks and plasmids as described in Materials and Methods. Members of MGK’s dissertation committee and CMM’s dissertation committee are acknowledged for critical input about the work described here. Funding to produce transgenics was provided by a National Academy of Science Grants-In-Aid of Research (G20100315152292) administrated by Sigma Xi to MGK and G2016100191865376 to BWG.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- 1.Warner JR, McIntosh KB.. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simsek D, Tiu GC, Flynn RA, et al. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell. 2017;169:1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Z, Fujii K, Kovary KM, et al. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol Cell. 2017;67:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genuth NR, Barna M. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol Cell. 2018;71(3):364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ban N, Beckmann R, Cate JH, et al. A new system for naming ribosomal proteins. Curr Opin in Struct Biol. 2014;24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, O’Leary MN, Peri S, et al. Ribosomal proteins Rpl22 and Rpl22l1 control morphogenesis by regulating pre-mRNA splicing. Cell Rep. 2017;18:545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabunilas J, Chanfreau G. Splicing-mediated autoregulation modulates Rpl22p expression in Saccharomyces cerevisiae. PLoS Genet. 2016;12(4):e1005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’ Leary MN, Schreiber KH, Zhang Y, et al. The ribosomal protein Rpl22 controls ribosome composition by directly repressing the expression of its own paralog, Rpl22l1. PLoS Genet. 2013;9(8):e1003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SJ, Strich R. Rpl22 is required for IME1 mRNA translation and meiotic induction in S. cerevisiae. Cell Div. 2016;11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shigenobu S, Arita K, Kitadate Y, et al. Isolation of germline cells from Drosophila embryos by flow cytometry. Dev Growth Differ. 2006a;48:49–57. [DOI] [PubMed] [Google Scholar]
- 11.Shigenobu S, Kitadate Y, Noda C, et al. Molecular characterization of embryonic gonads by gene expression profiling in Drosophila melanogaster. Proc Natl Acad Sci. 2006b;103:13728–13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crosby MA, Goodman JL, Strelets VB, et al. Flybase: genomes by the dozen. Nucleic Acids Res. 2007;35:486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kearse MG, Chen AS, Ware VC. Expression of ribosomal protein L22e family members in Drosophila melanogaster: rpL22-like is differentially expressed and alternatively spliced. Nucleic Acids Res. 2011;39:2701–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aradska J, Bulat T, Sialana FJ, et al. Gel-free mass spectrometry analysis of Drosophila melanogaster heads. Proteomics. 2015;15(19):3356–3360. [DOI] [PubMed] [Google Scholar]
- 15.Bourbon HM, Gonzy-Treboul G, Peronnet F, et al. A P-insertion screen identifying novel X-linked essential genes in Drosoph. Mech Dev. 2002;110: 71–83. [DOI] [PubMed] [Google Scholar]
- 16.Boutros M, Kiges AA, Armknecht S, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. [DOI] [PubMed] [Google Scholar]
- 17.Marygold SJ, Roote J, Reuter G, et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Bio. 2007;8:R218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koyama Y, Katagiri S, Hanai S, et al. Poly(ADP-ribose) polymerase interacts with novel Drosophila ribosomal protein L22 and L23a, with unique histone-like amino-terminal extensions. Gene. 1999;226:339–345. [DOI] [PubMed] [Google Scholar]
- 19.Ni J, Liu L, Gess D, et al. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 2006;20:1959–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kearse MG, Ireland JA, Prem SM, et al. eRpL22, but not eRpL22-like-PA, is SUMOylated and localized to the nucleoplasm of Drosophila meiotic spermatocytes. Nucleus. 2013;4:241–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Arcia S, Perez B, et al. p∆TubHA4C, a new versatile vector for constitutive expression in Drosophila. Mol Biol Reprod. 2013;40:5407–5415. [DOI] [PubMed] [Google Scholar]
- 22.Chen D, McKearin DM. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development. 2003;130:1159–1170. [DOI] [PubMed] [Google Scholar]
- 23.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carvalho GB, Ja WW, Benzer S. Non-lethal genotyping of a single Drosophila. Biotechniques. 2009;46:312–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergh S, Arking R. Development profile of the heat shock response in early embryos in Drosophila. J Exp Zool. 1984;231(3):379–391. [DOI] [PubMed] [Google Scholar]
- 26.Kai T, Williams D, Spradling AC. The expression profile of purified Drosophila germline stem cells. Dev Bio. 2005;283:486–502. [DOI] [PubMed] [Google Scholar]
- 27.Eun SH, Stoiber PM, Wright HJ, et al. MicroRNAs downregulate Bag of marbles to ensure proper terminal differentiation in the Drosophila male germline. Development. 2013;140:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White-Cooper H. Tissue, cell type and stage-specific ectopic gene expression and RNAi induction in the Drosophila testis. Spermatogenesis. 2012;2(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El-Baradi TT, Raué HA, De Regt CH, et al. Stepwise dissociation of yeast 60S ribosomal subunits by LiCl and identification of L25 as a primary 26S rRNA binding protein. Eur J Biochem. 1984;144:393–400. [DOI] [PubMed] [Google Scholar]
- 30.Kamath RS, Fraser AG, Dong Y, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. [DOI] [PubMed] [Google Scholar]
- 31.Lavergne JP, Conquet F, Reboud JP, et al. Role of acidic phosphoproteins in the partial reconstitution of the active 60 S ribosomal subunit. FEBS Lett. 1987;216:83–88. [DOI] [PubMed] [Google Scholar]
- 32.Deutschbauer AM, Jaramillo DF, Proctor M, et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson SJ, Lauritsen JP, Hartman MG, et al. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–772. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Duc AC, Rao S, et al. Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev Cell. 2013;24:411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotenberg MO, Moritz M, Woolford JL Jr.. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988;2:160–172. [DOI] [PubMed] [Google Scholar]
- 36.Komili S, Farny NG, Roth FP, et al. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13(6):355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parenteau J, Durand M, Morin G, et al. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell. 2011;147:320–331. [DOI] [PubMed] [Google Scholar]
- 39.Nakao A, Yoshihama M, Kenmochi N. RPG: the ribosomal protein gene database. Nucleic Acids Res. 2004;32:D168–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikhaylova LM, Nguyen K, Nurminsky DI. Analysis of the Drosophila melanogaster testes transcriptome reveals coordinate regulation of paralogous genes. Genetics. 2008;179:305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma X, Zhang K, Li X. Evolution of Drosophila ribosomal protein gene core promoters. Gene. 2009;432:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. [DOI] [PubMed] [Google Scholar]
- 43.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5ʹUTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. [DOI] [PubMed] [Google Scholar]
- 44.Qin X, Ahn S, Speed TP, et al. Global analyses of mRNA translational control during early Drosophila embryogenesis. Genome Biol. 2007;8(4):R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


