Abstract
Assigning function to genes has long been a focus of biomedical research. Even with complete knowledge of the genomic sequences of humans, mice and other experimental organisms, there is still much to be learned about gene function and control. Ablation or overexpression of single genes using knockout or transgenic technologies has provided functional annotation for many genes, but these technologies do not capture the extensive genetic variation present in existing experimental mouse populations. Researchers have only recently begun to truly appreciate naturally occurring genetic variation resulting from single nucleotide substitutions, insertions, deletions, copy number variation, epigenetic changes (DNA methylation, histone modifications, etc.) and gene expression differences and how this variation contributes to complex phenotypes. In this chapter, we will discuss the benefits and limitations of different forward genetic approaches that capture the genetic variation present in inbred mouse strains and present the utility of these approaches for mapping QTL that influence complex behavioral phenotypes.
Keywords: Mouse, Genetics, Behavior, Mapping, QTL, Mutagenesis
Animal models, and rodents in particular, have been used extensively to study behaviors that model human psychiatric diseases. While rats have been particularly well-suited to this research, the mouse has recently emerged as the primary model for studying genetic and genomic aspects of human disease (Cryan and Holmes 2005). This is due, in large part, to the availability of a vast array of genetic and genomic resources for such endeavors in the mouse.
The ability to introduce genes into the mouse (transgenics) and retain tissue-specific expression patterns as well as the ability to disrupt gene function (knockouts) represented a significant advance in a field that has now become known as “functional genomics.” Reverse genetic approaches such as these have been useful for identification and analysis of genes believed to increase risk for schizophrenia (Desbonnet et al. 2009), anxiety (Holmes et al. 2003; Lesch et al. 2003), depression (Savitz et al. 2009; Cryan and Slattery 2010) and autism (Sudhof 2008) but are not without limitations. The types of genetic lesions introduced are often severe, completely knocking out gene function over the course of the lifespan, and may not accurately reflect the genetic basis of many complex human diseases. The ability to make conditional knockouts that limit loss-of-function to specific tissues or developmental timepoints has overcome some of these limitations. However, the reverse genetic approach is generally hypothesis-driven, depending on some prior knowledge regarding a gene’s role in a disease or pathway. Understandably, the genes interrogated are often those in pathways that are already targeted by current drug treatments, making the approach inherently biased. More comprehensive phenotyping might result in identification of gene function not yet attributed to a specific gene. Recently, large-scale reverse genetic projects have been launched in the United States (knockout mouse project (KOMP), Texas A&M Institute for Genomic Medicine (TIGM)), Europe (European Conditional Mouse Mutagenesis (EUCOMM)) and Canada (North American Conditional Mouse Mutagenesis (NorCOMM)) to systematically interrogate the role of every gene by producing targeted knockouts and analyzing the resulting mutants in a variety of phenotypic assays (Collins et al. 2007; Gailus-Durner et al. 2009). These efforts to catalog gene function in a non-hypothesis-driven, unbiased manner will surely provide new insights into gene function. However, studying induced mutants exclusively fails to account for naturally occurring variation, genetic modifiers and gene-gene interactions that may be of primary importance in complex human diseases. In addition, recent research in human populations indicates that loss-of-function variants can occur far more frequently than expected with no discernable effect on health (Macarthur and Tyler-Smith 2010), suggesting that null mutations may not be the best approach for modeling many human diseases.
1. The Importance of Phenotype
In the earliest days of mouse genetics, the phenotype was the primary starting point for genetic analysis. Observations of spontaneous, chemical- or radiation-induced mutations progressed to identification of linkage groups and, eventually, to chromosomal assignment (for excellent reviews, see (Lyon 2002; Paigen 2003)). As molecular tools advanced, starting with the identification of genetic markers and progressing to complete sequencing of the mouse genome, the means for exact gene localization were realized. Knowledge regarding the organization of the genome increased along with an appreciation for the complexities therein, resulting in a renewed interest in the study of complex traits from a systems biology perspective. Recent years have witnessed a resurgence of forward genetic approaches that consider naturally occurring genetic and phenotypic variation and place more emphasis on the genetic complexities that are likely to explain a large portion of human phenotypic variation.
As opposed to reverse genetics, forward genetic studies start with the measurement or observation of a phenotype followed by mapping of the causative loci or genes. A major advantage of this approach is that the role of the gene or genes in a specific biological process is/are proven a priori. In addition, forward genetic approaches are, by definition, unbiased in terms of gene identification since the phenotype is the primary level of measurement. The term “forward genetics” applies to any phenotype-driven mapping approach, including the use of standard inbred strain crosses and haplotype association analysis—both of which result in the identification of quantitative trait loci (QTL).
In this chapter, we will describe forward genetic approaches for gene identification and functional analysis, detailing the advantages, disadvantages, successes and failures of several commonly used forward genetic models.
2. Forward Genetics and Inbred Strains: The QTL Approach
Early mouse geneticists, including pioneers of the field William Castle and Clarence Little, utilized visible characteristics such as coat color and tumor susceptibility to study Mendelian inheritance. These studies eventually led to the development of inbred mouse strains in the early 1900s (reviewed in (Crow 2002)). These strains are the starting point for most forward genetic approaches. Inbred strains are produced by brother-sister mating for 20 or more consecutive generations to fix all loci in a homozygous state. There are now hundreds of inbred strains available, and while mice within a single inbred strain are genetically identical, substantial genetic and phenotypic variation exists across strains. This phenotypic and genetic variation has been used for decades to further our understanding of the genetic basis of human disease. Inbred lines and the crosses derived from them have been instrumental in the search for genes and genomic loci that contribute to the phenotypic variability for complex behavioral traits. These loci, termed quantitative trait loci, or QTL, are regions of the genome that are associated with the phenotypic expression of complex traits, and the genomic locations of these regions can be identified using a process known as QTL mapping.
3. QTL Mapping: The Basics
In its simplest form, QTL mapping can be described as a statistical association analysis between phenotype and genotype at specific locations across the genome. In order to achieve this, two things are necessary—phenotypic and genetic variation. Phenotypic variation is easily observed among inbred strains (Hamilton and Frankel 2001), and experimental crosses between inbred strains provide both phenotypic and genetic variation. Genetic variation across the genome is measured using a set of markers that can distinguish between the two or more strains used in an experimental cross. The source of these markers has changed over the years with advances in technology and knowledge of genomic structure. Genetic markers initially took the form of linked visible characteristics such as coat color and progressed to restriction fragment length polymorphisms (RFLPs). With the advent of the polymerase chain reaction (PCR), scientists had access to a trove of genetic markers called simple sequence length polymorphisms (SSLPs) that were more numerous and provided dense genetic maps. Single nucleotide polymorphisms (SNPs) are single nucleotide changes that are also detectable by PCR, and millions of them exist between inbred strains, providing even greater resolution for genetic mapping. SNPs are now the primary source of genetic variation measured in mapping studies. Genetic markers need not be causative alleles and simply serve as a means of ascertaining allelic state at a genomic location.
Once genotypes and phenotypes are collected, association of genotype with phenotype can be performed. This aim can be accomplished by single-marker analysis using basic statistical techniques (ANOVA, marker regression) and comparing marker status at each genomic location with phenotype. In the late 1980s a technique for analyzing the interval between two genotyped markers was developed by Lander and Botstein (1989), called interval mapping (IM). This technique involves calculating the likelihood of a particular genotype based on the recombination frequency and genetic distance between two markers. A LOD score is calculated to indicate the strength of the association between the phenotype and genotype. This technique has been refined over the years—Jansen (1993) and Zeng (1993) developed an algorithm for composite interval mapping (CIM) that allows for interval mapping in combination with multiple-regression analysis to account for background QTL. The specifics of QTL mapping algorithms are beyond the scope of this chapter, but several books are available that provide a thorough discussion (Broman and Sen 2009; Wu et al. 2010). Linkage and mapping programs are freely available, including mapmaker QTL (Lander et al. 1987), QTL Cartographer (Basten et al. 1994, 2002), map manager QTX (Manly et al. 2001) and R/qtl (Broman et al. 2003) or its graphical user interface, J/qtl (Smith et al. 2009). Most mapping programs were designed to analyze the basic mapping populations described below, but special attention must be given to populations frequently used for fine mapping QTL.
4. Initial QTL Identification: Standard Mapping Populations
4.1. Recombinant Inbred Lines
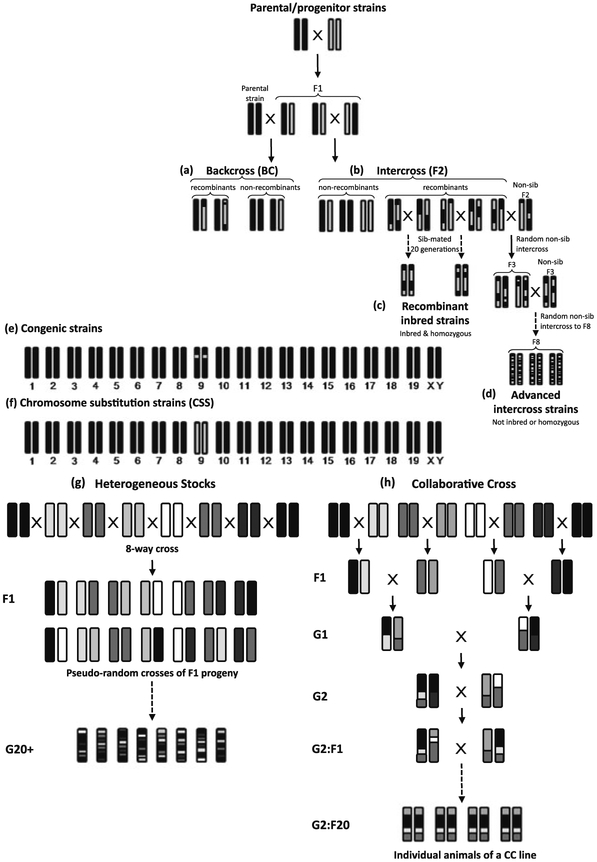
Donald Bailey and Benjamin Taylor developed RI lines at the Jackson Laboratory with the goal of creating a recombinant population for linkage mapping. RI lines are created by intercrossing the F1 hybrids of two inbred progenitor strains to generate F2 progeny, which are then sib-mated for at least 20 generations, resulting in homozygosity at each locus and a mixture of the parental alleles providing genetic variation necessary for QTL mapping. Each RI line consists of animals that are genetically identical but distinct from other RI lines in the set (Fig. 1c) (Bailey 1971).
Fig. 1.
a–h Breeding schemes of various genetic mapping populations. Backcross mice (a) are generated by crossing two inbred strains to recover F1 mice and then backcrossing the F1 mice to one of the parental strains, resulting in one copy of a chromosome that has undergone meiotic recombination and another copy from the inbred strain to which the F1 was backcrossed. Intercross mice (b) result from crossing F1 mice, resulting in two recombined chromosomes. Recombinant inbred strains (c) are produced by continuous brother-sister mating among F2s, resulting in homozygosity at each locus but with a reassortment of alleles from each parental strain. Advanced intercross lines (d) are also produced by crossing F2s, but inbreeding is avoided by restricting breeding partners to exclude common parents or grandparents and a large breeding population is maintained to avoid allelic drift. Congenic strains (e) have a small portion of one chromosome from a donor strain introgressed onto a recipient background strain, while chromosome substitution strains (f) have an entire donor chromosome introgressed onto a recipient background strain (examples of a congenic interval on Chr 9 (e) and a Chr 9 CSS (f) are shown). Heterogeneous stocks (g) are similar to advanced intercross lines but are derived from eight different inbred strains to increase genetic diversity. The collaborative cross (h) also has eight parental strains, but the breeding scheme is similar to recombinant inbred lines to maximize recombination while breeding each CC line to homozygosity
The first murine RI panel, the CXB line, was constructed by Bailey from a BALB/cBy × C57BL/6By cross. Bailey used the seven lines resulting from the initial crosses to identify loci associated with histocompatibility factors and coat color (Taylor 1978). Twelve CXB lines now exist and have been used to map QTL that influence exploratory activity (Blizard and Bailey 1979; Crabbe et al. 1982; Neiderhiser et al. 1992), circadian rhythms of locomotion (Schwartz and Zimmerman 1990), avoidance (Neiderhiser et al. 1992) and sleep behaviors (Tafti et al. 1997). Taylor developed more RI panels, such as the BXH (C57BL/6J × C3H/HeJ) and AKXL (AKR/J × C57L/J) RI sets, which were used in the study of murine leukemia virus (Taylor et al. 1971), drug-induced seizures (Taylor 1976) and lipopolysaccharide response (Watson et al. 1977). There are now RI panels for crosses between A/J and C57BL/6J (AXB and BXA), B6(Cg)-Tyrc-2 J/J and C3H/HeJ (BXH) and C57BL/6J and DBA/2J (BXD), as well as RI lines derived from mice selectively bred for righting response to ethanol—the Long Sleep and Short Sleep mice, ILS/IbgTejJ and ISS/IbgTejJ (ILSXISS) (http://jaxmice.jax.org).
The BXD RI panel is by far the most frequently used, beginning with an initial set of 25 lines (Taylor et al. 1977) but now consisting of 79 strains (Peirce et al. 2004) (http://www.genenetwork.org). QTL for alcohol (Cunningham 1995; Phillips et al. 1994, 1995; Rodriguez et al. 1994, 1995) and drug-related (Alexander et al. 1996; Grisel et al. 1997; Jones et al. 1999; Mogil et al. 1997; Phillips et al. 1998) behaviors were among the first to be dissected using BXD lines. More recently, concerted efforts have been made to characterize the full BXD panel for neurobehavioral phenotypes, particularly those relating to pain, drug addiction, anxiety, stress and locomotor activity (Philip et al. 2010). One innovative approach uses brain-specific gene expression data from a microarray analysis of the BXD panel to map neurobehavioral QTL (Bao et al. 2007).
RI lines offer several advantages for QTL mapping. RI lines are genetic reference populations—genetically stable over time and infinitely reproducible. Therefore, each line must be genotyped only once, and that data becomes available to anyone using the line in the future (Taylor 1978). Comparisons of phenotypic data can be made across laboratories enabling genetic correlations and investigation of similarities among phenotypes. Phenotypic data can be examined retrospectively as marker density increases and more phenotypes are collected. This process has been automated by Williams and colleagues, who established the GeneNetwork database that provides tools for RI-QTL mapping and correlations among more than 1,200 phenotype datasets, including gene expression (Wang et al. 2003). RI lines are also particularly well-suited for mapping QTL for phenotypes for which genetic variance is small compared to environmental variance. This feature of RI lines is derived from the ability to conduct repeated sampling from individuals with the same genetic background, resulting in strain means that reduce environmental noise and more accurately reflect the strain phenotype.
The ability to map QTL for complex traits is dependent upon the availability of a sufficiently large RI panel that provides enough power to detect QTL of small effect size (Gora-Maslak et al. 1991). However, the RI strains were developed to investigate Mendelian traits, and the small number of strains per line has limited their usefulness for identifying QTL for complex phenotypes. Prior to the development of dense sets of genetic markers, the utility of RIs was also limited by the availability of loci for mapping. The expansion of the BXD line, along with the development of a more dense set of polymorphic markers, now allows for mapping of QTL responsible for 10% of the phenotypic variance to approximately 1–2 megabases (Mb) (Peirce et al. 2004).
However, even the largest RI sets have several characteristics that mitigate their usefulness in complex trait analysis. Existing RI panels were produced by crossing only two inbred strains and from only a limited number of inbred strains, resulting in limited genetic variability both within and across panels. Both the advantages and disadvantages of RI lines were considered when planning the collaborative cross (CC) (Churchill et al. 2004), an experimental population akin to RI strains but with added genetic variability. The CC is discussed in more detail below.
4.2. Intercross and Backcross Mapping Populations
The limited power for mapping provided by existing RI lines led to the use of other types of mapping populations—either for replication of RI-identified QTL or as alternative crosses for initial QTL identification. The most commonly used QTL mapping populations are intercross (F2) or backcross (BC) lines produced by breeding two inbred strains known to differ for the complex trait(s) of interest. The resulting F1 hybrid progeny are either intercrossed to produce an F2 generation (Fig. 1b) or backcrossed to one of the parental strains to produce a BC generation (Fig. 1a). At each locus, an F2 animal will have one of three genotypes: homozygous for either of the parental alleles or heterozygous, possessing one allele from each of the parental strains. A BC animal has only two genotype possibilities: heterozygous or homozygous for the allele of the parental strain to which the F1 progeny were backcrossed.
Unlike RI animals, intercross and backcross progeny are genetically heterogeneous; each F2 or BC individual within the population is characterized by a unique pattern of genome-wide recombination. Therefore, each set of animals must be genotyped and the informativeness of these genotypes is limited to the set of phenotypes collected for any particular set of animals. When genotyping costs were prohibitive, selective genotyping of animals from the top and bottom 15% of the phenotypic distribution was frequently performed using DNA pools (Darvasi and Soller 1994; Wang and Paterson 1994). The genetic (DNA) contribution of each individual to the pool was often weighted toward individuals at the phenotypic extremes (Taylor et al. 1999, 2001). Individuals in each pool were genotyped only at loci for which a significant or suggestive QTL were identified, and the entire mapping population was only genotyped for those loci that proved significant. As genotyping costs have decreased, the use of pooling has waned.
The number of markers required to genotype either an F2 or BC is relatively low based on the limited number of recombinations in these populations. A polymorphic marker every 40 Mb provides adequate coverage, as each marker will sweep 20 Mb in each direction. The choice of which type of cross to use is dependent on the genetic architecture of the phenotype of interest. Using an F2 mapping population is recommended to obtain an overview of the number, location and estimated effect sizes of both additive and dominant QTL segregating in the population, as well as QTL interactions (Darvasi 1998). Approximately 30% fewer F2 animals are required to detect additive QTL in comparison with a BC population due to the increased number of recombinations in the F2. However, BC mapping populations are generally considered more efficient for QTL identification, specifically for QTL with dominant effects. The decreased genetic variance in a BC—a population with a 1:1 genotypic ratio and only two prospective genotypes at any given locus—results in fewer genetic interactions than in an F2. The LOD threshold for significance is therefore lower for a BC, and gene effects are more prominent because the decreased genetic variance results in fewer genetic interactions in the BC (Darvasi 1998; Silver 1995).
Regardless of the cross type, QTL mapping has been successful in identifying thousands of loci. Conventional F2 populations derived from various inbred parental strains have been used to identify QTL for behaviors such as contextual fear conditioning (Wehner et al. 1997), spatial learning (Steinberger et al. 2003), alcohol preference (Fernandez et al. 2000) and consumption (Boyle and Gill 2008), anxiety (Bailey et al. 2008; Eisener-Dorman et al. 2010; Henderson et al. 2004; Turri et al. 2001a; Turri et al. 2004), home cage activity (Turri et al. 2001b; Umemori et al. 2009), ethanol-induced (Hitzemann et al. 1998) and cocaine-induced (Boyle and Gill 2009) locomotor activation and locomotor activity in the open field (Bailey et al. 2008; Eisener-Dorman et al. 2010; Gershenfeld et al. 1997; Kelly et al. 2003; Koyner et al. 2000). QTL for locomotor activity in the open field (Eisener-Dorman et al. 2010), alcohol preference (Melo et al. 1996) and body weight regulation (Zhang and Gershenfeld 2003) have been identified using BC populations.
The deluge of QTL for various phenotypes caused concerns regarding the possibility of numerous false positives (Type I Error). A paper published in 1995 proposed thresholds for reporting significant or suggestive QTL loci (Lander and Kruglyak 1995). Some researchers worried that the thresholds were too restrictive and that it would be cost prohibitive to phenotype and genotype populations of the size necessary to result in the levels of significance proposed (Yoon 1996). Others recommended the use of multiple strategies, including provisional mapping and replication of suggestive and significant QTL in independent populations and pooling results (Atkins 2001). At the time, however, the significance levels proposed by Lander and Kruglyak were almost universally adopted when reporting QTL. Advances in computational power now make it more common to generate significance thresholds based on individual experimental populations by running permutation tests (Churchill and Doerge 1994), and this function is present in most QTL mapping software packages.
Large F2 or BC populations have substantial power for detecting QTL, but they also have limitations. Neither F2 nor BC mapping panels are capable of providing high-resolution QTL localization due to the relatively low number of recombinations present in these populations. This results in coarsely mapped QTL with broad peaks that can encompass half of a chromosome and contain hundreds of candidate genes. Because the QTL regions identified are so large, the individual effects of closely-linked QTL are difficult to dissect. QTL comprised of multiple closely-linked, small-effect QTL may fail to be detected if the loci have opposing phenotypic effects. Consequently, the fine mapping necessary to narrow QTL intervals and, ultimately, identify quantitative trait genes (QTG) must be carried out in specialized populations.
4.3. Consomic Lines or Chromosome Substitution Strains
Consomic lines, or chromosome substitution strains (CSS), are produced by introgressing a single chromosome from a donor strain onto a recipient strain background by repeated backcrossing for at least 10 generations (Fig. 1f). A full CSS panel consists of 21 lines—one for each of the 19 autosomes and 2 sex chromosomes. Established CSS lines are genetic reference populations that allow for relatively fast localization of a QTL or group of QTL to a single chromosome and have several advantages over standard mapping populations. Because statistical comparisons are made across 21 CSS lines and the background strain rather than across hundreds of markers in hundreds of F2 or BC animals, the P-value required for significance (corrected for multiple comparisons) in a CSS experiment will be lower. The total number of mice necessary for mapping QTL to a single chromosome in a CSS panel is only slightly lower than in an F2 cross based on sample size estimates required to detect a QTL that accounts for a similar amount of the variance in either population (Belknap 2003). However, no genotyping is required in the initial scan, making the CSS more economical. In addition, the effect size of a QTL will be amplified in the CSS due to the presence of only homozygotes.
These advantages make the CSS an attractive option for initial identification of QTL. However, the end result is an entire chromosome, or multiple chromosomes, on which reside QTL that must be localized and fine-mapped. CSS provide a starting population that requires fewer generations of breeding to produce interval-specific congenic strains or mice for recombinant progeny testing. Since QTL were identified and present on a homogeneous genetic background in the CSS, their effect size should be sufficient for recovery in congenic lines. However, if a significant result on a single chromosome represents a cluster of two or more QTL, the effect might be lost upon construction of congenic or subcongenic lines, as described below.
Constructing a CSS panel is costly and takes years of breeding. Realistically, therefore, most researchers are limited to existing CSS panels and the genetic and phenotypic variability that exists therein. Three CSS panels have been reported in the literature, beginning with the B6. A panel (A/J introgressed on to C57BL/6J) (Singer et al. 2004). This panel has been the most extensively used for mapping QTL for behavioral traits such as prepulse inhibition (Leussis et al. 2009; Petryshen et al. 2005), anxiety (Singer et al. 2005) and sleep-related epilepsy (Strohl et al. 2007) and other complex phenotypes, such as onset of puberty (Nathan et al. 2006) and resistance to diet-induced obesity (Buchner et al. 2008). Takada et al. produced a CSS panel with MSM/Ms introgressed onto C57BL/6J (Takada et al. 2008) that has been used to identify QTL for emotionality (Takahashi et al. 2008), social interactions (Takahashi et al. 2010) and home cage activity (Nishi et al. 2010). Finally, Gregorova et al. (Gregorova et al. 2008) have introgressed PWD/Ph onto C57BL/6J and have characterized the lines for blood chemistry-related phenotypes.
5. QTL Fine Mapping
The identification of QTL is fairly straightforward, as evidenced by the thousands that now exist in online databases and in the literature (Flint et al. 2005). However, identifying the gene, or genes, underlying a QTL peak has been a persistent limitation of commonly used QTL methods. The limited resolution of standard QTL crosses (RIs, F2s, BCs) results in identification of large genomic regions, usually > 20 Mb, containing hundreds of genes. To identify the quantitative trait gene (QTG), the QTL region must be narrowed considerably before a manageable list of candidate genes can be interrogated. Fine mapping of QTL has been the focus of a great deal of research in recent years (Flint et al. 2005). Initially, congenic and subcongenic strains were pursued and have resulted in some success. However, partitioning QTL in this way is not always possible, and efforts toward the use of primary crosses have resulted in the development of advanced intercross lines (AILs), heterogeneous stocks (HS), outbred mice and the CC.
5.1. Congenic Strains
Congenic strains are derived from repeated backcrossing of a donor strain to a recipient strain until only a small region of the QTL-containing donor strain chromosome is introgressed onto the recipient strain background (Fig. 1e). This process, aided by genotyping the QTL region of interest, takes 13 generations (12 backcrosses followed by an intercross to obtain founders), but the process has been accelerated by marker-assisted selection of both the introgressed region and the remaining recipient background in a strategy called “speed congenics” (Markel et al. 1997; Wakeland et al. 1997). Consequently, it is now possible to produce congenics in less than half the number of generations. QTL intervals in congenic strains can be further narrowed by backcrossing the congenic founders to break up the introgressed region, thereby producing subcongenics.
There have been some successes in using congenic and subcongenic mice to narrow QTL regions. Berrettini and colleagues identified a morphine preference QTL (Mop2) in a C57BL/6J (B6) × DBA/2J (D2) F2 (Berrettini et al. 1994). The 20-Mb QTL was confirmed using a marker-assisted breeding strategy to generate a pair of reciprocal congenic mouse strains—the B6 QTL interval was introgressed onto the D2 recipient background (D2.B6-Mop2), and the D2 QTL interval was introgressed onto the B6 recipient background (B6.D2-Mop2) (Ferraro et al. 2005). Heterozygous D2.B6-Mop2 congenic mice were backcrossed to D2 mice, thereby fragmenting the 28.8-Mb QTL interval of the congenic into smaller, discrete intervals of 9.5 and 17.2 Mb in the resulting subcongenic strains (Doyle et al. 2008). Mice from one of these two subcongenic strains exhibited morphine preferences similar to both the D2.B6-Mop2 congenic and the B6 parental strain mice, thereby narrowing the Chr 10 QTL interval to a 9.5-Mb region containing 39 genes (Doyle et al. 2008). In another study, the obesity-related QTL Fob3 on Chr 15 was identified in an F2 cross originating from two outbred mouse lines selected for either high or low body fat (fat and lean lines) (Horvat et al. 2000). One mouse, selected because it was recombinant within the Fob3 interval, was backcrossed for 10 generations to the Fat line to create two subcongenic strains with the Lean line Fob3 QTL interval introgressed onto the Fat line recipient background (Stylianou et al. 2004). Further backcrossing of the Fob3 subcongenic strains to the Fat line yielded additional recombinants, allowing the isolation of two smaller QTL (Fob3a and Fob3b) within the original Fob3 QTL interval (Stylianou et al. 2004). Continued backcrossing of select recombinants within the 22.39-Mb Fob3b interval generated six additional subcongenic strains with overlapping donor intervals. Four subcongenic strains were intercrossed with the Fat line to generate four F2 mapping populations, which identified two closely-linked QTL, Fob3b1 (4.98 Mb) and Fob3b2 (7.68 Mb), within the Fob3b QTL interval (Prevorsek et al. 2010).
However, QTL effects often disappear during the construction of congenics or subcongenics. Reasons for this loss of QTL effect could be the removal of the QTL from a heterogeneous background, thereby eliminating epistatic interactions that increase QTL effect size. It is also likely that QTL that disappear during congenic production might actually be groups of two or more smaller effect size QTL that appear as a single peak in an F2 or BC but do not have sufficient power alone to result in a statistically significant change in phenotype (Legare et al. 2000; Legare and Frankel 2000).
5.2. Advanced Intercross Lines
Although the ability to restrict genetic heterogeneity to only two strains has advantages (Cheng et al. 2010), the limited number of recombinations present in F2 and BC mice also limits the resolution of these mapping resources. AILs are produced by intercrossing two parental strains to obtain an F2 and then intercrossing each successive generation (Fig. 1d). AILs have the advantage of expanding the genetic map by increasing the number of breakpoints, thereby increasing the recombination fraction between markers. The increased number of recombinations in an F10 can reduce the size of a QTL interval by five-fold in comparison with a similarly-sized population of F2 animals (Darvasi and Soller 1995). However, the increase in resolution comes at the cost of power to detect QTL. This loss in power is due to the increased number of markers necessary to account for the reduction in linkage disequilibrium between markers and genetic drift that results in altered allele frequencies. The latter can be controlled somewhat by choosing an appropriate breeding strategy (Rockman and Kruglyak 2008). In general, maintaining large population sizes and controlling for inbreeding can mitigate concerns about power in an AIL.
One of the largest AILs is the LG/J × SM/J (LG, SM) AIL produced by Cheverud and colleagues at Washington University in St. Louis, Missouri (Norgard et al. 2008). The F9–F10 LG, SM AILs have been used successfully to identify dozens of QTL for bone-length (Kenney-Hunt et al. 2008; Norgard et al. 2008, 2009). More recently, F34 LG, SM AILs were used to replicate and refine QTL from previous studies. Norgard et al. were able to replicate almost 80% of QTL identified in both F2–F3 and F9–F10 populations. However, QTL interval reduction in the F34 was less than expected compared to previous studies. QTL intervals ranged from 0.6 to 14 Mb with half of the QTL having confidence intervals from 2 to 5 Mb. The authors postulated that family structure bias inflated the QTL peaks and decreased the confidence interval sizes reported in earlier studies. An alternative explanation is the presence of multiple linked loci at the QTL peaks—a problem that is encountered with any fine mapping population.
Cheng et al. (2010) took advantage of the power of an F2 and the mapping resolution of the LG, SM AIL to identify QTL for methamphetamine sensitivity. QTL were identified for both methamphetamine- and saline-induced locomotor behavior in a large LG × SM F2 and the F34 LG, SM AIL. Both populations were also combined and analyzed as one large intercross. Cheng et al. found that the population structure present in AILs must be considered when mapping to reduce Type I errors. By using a mixed model that considered relatedness of individuals in the AIL for both mapping and significance threshold determination, the authors were able to identify several QTL intervals at sub-centimorgan resolution—one of which contains only a single gene that is now being assessed for its role in methamphetamine sensitivity. Samocha et al. (2010) used a similar approach to map QTL for acoustic startle response, habituation and prepulse inhibition of the startle response. A large set of F2 identified QTL for multiple behaviors. The population size of the AIL used in this study did not provide enough power to detect significant QTL on its own, but combined analysis of both the F2 and AIL populations narrowed QTL intervals identified initially in the F2. Taken together, these studies indicate that the approach of using both an F2 and AILs, along with taking into account population structure, is an effective method for identification and fine-mapping of QTL.
5.3. Heterogeneous Stocks
Heterogeneous Stocks (HS) lines are produced by pseudo-random breeding over multiple generations with the aim of increasing the number of meiotic crossovers and, thus, the genetic resolution (Fig. 1g). The term “heterogeneous stock” is commonly used to describe two independently established eight-way inbred mouse crosses. The Boulder HS was established in 1970 from an eight-way cross between C57BL/6, BALB/c, RIII, AKR, DBA/2, I, A/J and C3H strains and has been breeding for more than 60 generations (McClearn et al. 1970). Initially, the Boulder HS mice were conceived as a normative population of mice with increased genetic variation in comparison to individual inbred strains (McClearn et al. 1970). The Northport HS, derived from A/J, AKR/J, BALB/cJ, C3H/HeJ, C57BL/6J, CBA/J, DBA/2J and LP/J strains, was established in 1994 as a stock for selective breeding (Hitzemann et al. 1991) and has been breeding for more than 50 generations (Demarest et al. 2001; Hitzemann et al. 1994). More recently, two additional HS lines have been reported. A collaborative cross HS (CC-HS), derived from the same eight founder strains that were used to establish the CC, was recently reported by the Hitzemann laboratory and is currently at the 12th generation of outbreeding (Iancu et al. 2010). In addition, a cross between four inbred strains, C57BL/6J, DBA/2J, BALB/cJ and LP/J, called the HS4 has also been produced and maintained in the Hitzemann laboratory for 19 generations (Malmanger et al. 2006).
It is estimated that HS mice at 60 generations of random breeding could increase mapping resolution by 30-fold over an F2 or BC. Talbot et al. (1999) used the Boulder HS to fine map a QTL for open field behavior identified on Chr 1 in previous studies (Caldarone et al. 1997; Flint et al. 1995; Gershenfeld et al. 1997; Wehner et al. 1997) and were able to refine the chromosomal location to a 1.6-Mb region. Using single-marker association analysis, they also replicated a QTL on Chr 12 but failed to replicate three additional QTL on Chrs 1, 10 and 15. The inability to replicate QTL in the HS was attributed to the inability to distinguish between identical alleles contributed by different progenitor strains. This shortcoming was overcome by the development of a multipoint mapping model that takes into account information from flanking markers and progenitor haplotypes. This mapping algorithm, called HAPPY, is available for download or use as a web-based program (http://www.well.ox.ac.uk/~rmott/happy.html). Using the multipoint mapping method, all three previously undetected loci were mapped.
The Northport HS has been used to fine map a QTL on Chr 2 for ethanol-induced locomotor activity. The QTL was initially identified in the BXD RIs and replicated in a B6XD2 F2. Using a G32–35 HS, Demarest et al. (2001) were able to replicate the Chr 2 QTL and resolve the region into three separate peaks.
Mott and Flint (2002) also developed a technique, called the inbred-outbred cross, for using HS mice to both detect and fine map QTL. The technique involves generating an F2 cross using HS and a genetically-distinct line (inbred strain, knockout, transgenic), detecting QTL using standard methods (low marker resolution) and then dense genotyping in candidate QTL regions to take advantage of the increased resolution of the HS-contributed genetic material. Through simulations, Mott and Flint showed that a 5% effect size QTL could be detected and fine mapped with 50% probability to within 6 Mb by genotyping 1,500 animals.
HS mice have been used primarily to fine map previously identified QTL (Demarest et al. 2001; Malmanger et al. 2006; Talbot et al. 1999; Turri et al. 1999), although their increased genetic variability and expanded genetic map also make them suitable for genome-wide association studies. However, several procedural and analytical hurdles had to be addressed before genome-wide QTL identification in the HS could be realized. First, because of the expansion of the genetic map, 100 times more markers and 10 times more animals are required to have the power to detect QTL with 5% effect or less (Flint et al. 2005; Valdar et al. 2006b), making genome-wide mapping in the HS an expensive proposition. Also, unknown selective pressures and random fluctuations in allele frequencies during production and/or maintenance of the stock may affect the resolving power of the HS. These issues are detailed in the first publication of genome-wide association in the Northport HS (Valdar et al. 2006b). Costs for line production and genotyping were reduced by collecting over 100 phenotypes in parallel for each mouse (Solberg et al. 2006), and model-averaging techniques were developed to analyze multiple QTL models and overcome genotype correlations (linkage disequilibrium) and family structure. Hundreds of QTL for dozens of phenotypes were detected with confidence intervals averaging 2.8 Mb—a substantial improvement over standard F2 crosses.
5.4. Outbred Mice
Outbred stocks are closed populations of genetically variable animals that are bred to maintain maximum heterozygosity (Chia et al. 2005). Dozens of outbred stocks exist, and like HS mice, they offer the advantage of increased mapping resolution but suffer some of the same drawbacks—increased sample size and marker density are required for QTL detection. In addition, outbred stocks (with the exception of HS lines) do not have the advantage of progenitor allele information to derive the origin of alleles in the offspring. This limitation introduces difficulties in the use of these stocks for QTL mapping. Only one example of a behavioral QTL has been published thus far using outbred stocks. The Rgs2 gene that influences anxiety in mice was identified using outbred MF1 mice (Yalcin et al. 2004). Yalcin et al. determined that the haplotype patterns in MF1 mice were very similar to those found in standard inbred strains. Thus, they were able to use similar techniques for mapping as those used with HS mice.
Aldinger et al. (2009) recently examined genetic variation and population structure in CD-1 outbred stocks and determined that CD-1 mice are reasonably outbred, polymorphic at a significant number of loci and resemble human populations in terms of complex genetic history. Although CD-1 mice have population substructure that may affect mapping results, the authors believe this structure could be exploited and that the CD-1 mice represent a valuable genetic mapping resource. Williams et al. (2009) used CD-1 mice to confirm the effect of a duplication of the Glo1 gene on anxiety-related behavior in mice. However, outbred mice have not yet been used for genome-wide association mapping of behavioral traits. Perhaps as genomic and analytical resources are developed, these untapped resources of genetic variability will become utilized more frequently in the identification and fine mapping of QTL.
5.5. The Collaborative Cross
In the late 1990s and early 2000s, the discussion in the mouse genetics community started to focus on the problem of QTG identification; it was determined that current mouse resources were not suitable for tackling such a difficult problem. Various standard mapping populations had the features necessary for identifying complex trait loci, but not one mapping resource had all of the features that were necessary if such an endeavor was to be successful down to the point of gene identification. Mouse geneticists began to envision a reference population of mice, the CC, that would capitalize on the advantages of existing resources by offering genetic diversity, mapping power and high-resolution. The CC would also provide a platform for systems genetic studies and modeling of complex networks, including gene by gene and gene by environment interactions. The CC was to be derived from eight divergent inbred strains (A/J, C57BL/6J, 129S1/SvImJ, NOD/LtJ, NZO/HlLt, CAST/EiJ, PWK/PhJ and WSB/EiJ) to maximize genetic diversity and ensure that the resulting population would provide the phenotypic diversity necessary to study any trait of interest. Like the AIL and HS lines, the CC would be subjected to multiple generations of breeding to increase meiotic recombination and provide the necessary mapping resolution for fine mapping QTL (Fig. 1h). However, the CC would be inbred and, upon completion, would provide a fully-genotyped reference population that could be provided to individual investigators. Finally, the CC would be sufficiently large (~1,000 lines) to provide the power necessary to map QTL of relatively small-effect size.
The initial and largest crosses to produce the CC were begun in 2005 at the Oak Ridge National Laboratory (ORNL). Separate, smaller breeding populations were located in Tel Aviv, Israel and Western Australia. In late 2008, the Department of Energy announced that the ORNL mouse genetics program was being phased out, and the ORNL CC population was transferred to the University of North Carolina (UNC). The Tel Aviv population has recently joined the ORNL lines at UNC. Several CC lines have already reached the inbred state due to an acceleration of the process using marker-assisted inbreeding. At least 100 lines will be completed by the end of 2012, and additional lines will follow over the next several years (personal communication, Darla Miller).
It is likely that the final population of the CC will be closer to 500 rather than 1,000 lines, as first proposed. Simulation studies have shown that 500 lines provide both adequate power and fine resolution for QTL mapping (Broman 2005; Valdar et al. 2006a). A multi-stage strategy has also been proposed using an initial mapping panel of 100 strains followed by a second set of 100 strains that have informative allele combinations and recombination events based on QTL localization from the first stage (Churchill et al. 2004). This strategy will make the CC more accessible for smaller laboratories. The completion of fully-genotyped CC-RI lines would also enable the production of many more CC recombinant inbred intercrosses (CC-RIX) produced by crossing different CC lines. Each CC-RIX mouse will inherit a chromosome from each CC parent and, thus, genotype information can be extrapolated. Heterozygosity will be recaptured in the CC-RIX lines, making them a more realistic model for human populations and allowing for more accurate modeling of complex human diseases.
6. Haplotype Association Mapping
Inbred strain surveys are an important initial step in understanding the genetic and phenotypic architecture of complex traits. Until recently, however, strain surveys had become less frequently used, meaning that researchers had a limited set of data on which to base experiments. In 2000, the Mouse Phenome Project was started as an effort to collect phenotypic data from a core set of inbred strains. These data would be stored in a central location, be publicly available on the Mouse Phenome Database (MPD; http://phenome.jax.org/ and Bogue et al. (2007)) and would offer investigators a wide range of phenotypes with which to inform research decisions for complex trait analysis (Bogue 2003; Paigen and Eppig 2000).
The effort to collect phenotype data for inbred strains coincided with efforts to increase the number of single nucleotide polymorphisms (SNPs) identified in inbred mouse lines and construct a SNP haplotype map of the mouse similar to the HapMap being constructed in humans (2003) (http://hapmap.ncbi.nlm.nih.gov/). This convergence of phenotype data and SNP genotype data presented an obvious opportunity to take advantage of both the phenotypic and genotypic diversity among inbred strains. In the context of genetic mapping, inbred strains offer several advantages over existing mapping populations. Inbred strains are, of course, a reference population of mice enabling collection and storage of phenotypic and genetic data that can be utilized repeatedly. In addition, inbred strains, like RIs, allow for repeated sampling within a strain to reduce environmental variance. Inbred strains also have increased genetic and phenotypic diversity in comparison to RI or standard F2 or BC populations, as well as a dense genetic map due to increased recombination.
In 2001, Grupe et al. used existing inbred strain phenotype data from the MPD and over 3,000 SNPs identified in their laboratory and at the Whitehead Institute (Lindblad-Toh et al. 2000) to conduct “in silico QTL mapping” in mouse. The basic approach is straightforward—as described previously, inbred strains are genetically identical within a strain but genetically diverse across strains. Therefore, associating SNP genotypes across the genome with strain phenotypes, similar to an RI-QTL study, can be expected to yield chromosomal regions that are associated with the trait of interest. The in silico mapping technique was successful in identifying QTL that overlapped with previously published reports using standard crosses (Grupe et al. 2001). Nevertheless, subsequent commentary on the method exposed several weaknesses, including lack of power, insufficient genetic variability and failure to control for both Type I and II errors (Chesler et al. 2001; Darvasi 2001). Darvasi (2001) estimated that between 40 and 150 strains would be necessary to identify a QTL affecting a quantitative trait with a heritability of 50%, and the number of strains used in the Grupe study ranged from 4 to 8. However, Grupe et al. argued that this estimate was based on power calculations using Lander and Schork significance levels based on an infinite density of genetic markers and was not applicable to their study.
Regardless, it was generally agreed that in silico mapping, more recently termed haplotype association mapping (HAM), might be a useful tool for identification of QTL. Three factors were necessary for its successful implementation: dense SNP coverage across the genome for more than just a few strains, phenotype data for multiple strains and appropriate analysis tools for genotype/phenotype associations. Pletcher et al. (2004) addressed two of these problems by identifying over 10,000 SNPs in 48 strains and developing a HAM algorithm called SNPster (http://snpster.gnf.org) to perform genotype/phenotype associations across the genome using a three-SNP sliding haplotype window. Using this method, the authors were able to identify QTL peaks for several Mendelian traits, including coat color and retinal degeneration. They were also able to replicate previously identified QTL for more complex traits such as saccharin preference, high-density lipoprotein (HDL) levels and gallstone formation. In addition, most map locations spanned intervals of 1 Mb or less—a significant improvement over standard F2 or BC mapping.
However, several hurdles still remained for HAM. As more dense SNP panels were genotyped and more was known about the genetic structure of the inbred strains, it became clear that an extensive family structure existed that could lead to spurious associations. In addition, only a limited number of strains existed for which there was both phenotype and genotype data, resulting in limited power to detect QTL for complex phenotypes. McClurg et al. (2007) attempted to address family structure by calculating a genetic similarity matrix along with a weighted bootstrap method to decrease the significance of nonspecific associations. In addition, gene expression data were used to optimize the power of the SNPster algorithm to identify cis-acting expression QTL that are considered, by some, to be a highly-enriched set of true positives with a low false-positive rate (Chesler et al. 2006). Additional analysis programs have been developed to perform association mapping in inbred strains, including Efficient Mixed-Model Association (EMMA; (Kang et al. 2008)) and hmmSNP, which uses a hidden Markov model (HMM)-based algorithm (Tsaih and Korstanje 2009). Each program uses a different algorithm and has a slightly different way of dealing with population structure. However, most programs give qualitatively similar results (unpublished data, Tim Wiltshire).
The full power of HAM has not yet been realized, as the resources for performing such analyzes are still being compiled. The quantity of SNPs in mice now number in the millions across more than 100 inbred strains. The number of phenotypes in the mouse phenome database continue to grow, with over 2,000 phenotypes stored and an average of 19 mouse strains tested per phenotype. However, the HAM technique has been used successfully to narrow down previously identified QTL intervals (Burgess-Herbert et al. 2009; Cervino et al. 2005; Harrill et al. 2009; Park et al. 2003; Wang et al. 2005) and to identify new QTL (Bopp et al. 2010; Liao et al. 2004; Liu et al. 2006; Pletcher et al. 2004).
Few HAM studies have been published for behavior. Webb et al. conducted a whole genome association study for prepulse inhibition and identified QTL on Chrs 1 and 13 (Webb et al. 2009) that overlap with genes that alter PPI when knocked out or regions of the human genome that contain genes that have been implicated in schizophrenia. Miller et al. (2010) measured tail suspension, an animal model of behavioral despair or depression, in 33 inbred strains of mice and identified four QTLs, including one that overlapped a human region that contains a locus associated with major depressive disorder and bipolar disorder.
Initial reports of QTL mapping using HAM resulted in concern about the fate of standard QTL studies (Chesler et al. 2001; Darvasi 2001). However, these concerns have not been realized. In general, HAM is viewed as one step in QTL identification that can be used for initial identification of QTL regions that must then be replicated using standard approaches. More commonly, HAM can be used to both replicate and narrow QTL that were identified using standard approaches (Burgess-Herbert et al. 2008, 2009; DiPetrillo et al. 2005). This technique is particularly effective when using strains that are genetically similar. Although genetic similarity may result in phenotypic similarity, QTL can be detected even when using parental strains that are phenotypically similar (Bailey et al. 2008; Eisener-Dorman et al. 2010). This is due to transgressive segregation, the reshuffling of alleles that occurs when two strains are crossed (Rieseberg et al. 1999). QTL identified in these crosses may be fine mapped to relatively small genomic regions using haplotype comparisons since regions of shared haplotype can be eliminated from consideration (Bailey et al. 2008; Eisener-Dorman et al. 2010).
As more phenotype data are collected for larger numbers of inbred strains and more information is gathered on the genomic structure of the laboratory mouse, HAM analysis will become an even more vital tool with which to identify complex disease genes.
7. Mutagenesis
In the late 1990s, with QTL accumulating in the literature and very few identified genes, the scientific community was looking for alternative approaches to identification of complex disease genes. Mutagenesis, the induction of mutations by chemicals or radiation, was particularly attractive. Mutagenesis in the mouse was not new—radiation-induced mutagenesis experiments to determine the genetic effects of radiation on mammals had been ongoing at the Department of Energy’s Oak Ridge National Laboratory (ORNL) since just after World War II. The use of N-ethyl-N-nitrosourea (ENU) as a chemical mutagen in mice began in 1978 when it was shown to have a mutation rate 12 times higher in male spermatogonia than that of X-rays (Justice 2004).
ENU is an alkylating agent that causes single base pair mutations—usually A–T to T–A transversions or A–T to G–C transitions. ENU induces random heritable mutations at a rate of approximately 1.4 × 10−6 per nucleotide site (Takahasi et al. 2007), although this rate varies depending on the protocol and the species or inbred strain background. Because of the random nature of mutations resulting from ENU, it was proposed as an unbiased way to induce a single mutation that would affect a complex phenotype. Thus, unlike transgenic and knockout mouse technologies, which target a specific gene of interest, a strength of the ENU mutagenesis approach is that it is not hypothesis-driven and is not limited to the study of known genes previously associated with specific biological pathways. Downstream analysis for ENU studies included mapping of the causative mutation followed by identification of the genetic lesion. The presence of only one causative mutation was anticipated to render the gene easier to identify.
In 1997, several large-scale ENU mutagenesis centers were set up in the UK and Germany to produce and screen ENU-generated mutants using batteries of phenotypic tests, including dysmorphologies, behavior, neurological abnormalities, immunology, clinical chemistry and hearing (Hrabe de Angelis et al. 2000; Nolan et al. 2000). In 2000, the NIH awarded grant funding to three large-scale mutagenesis centers in the United States at the University of Tennessee at Memphis, Northwestern University and the Jackson Laboratory with a mandate on production, identification and dissemination of ENU-induced mutants displaying alterations in nervous system function and behavior (collectively called the Neuromice.org Consortium). Additional ENU mutagenesis centers were formed outside the auspices of NIH, both small and large-scale, in academia and industry (for a complete list see (Cordes 2005)).
The mutagenesis centers quickly began producing hundreds of mutants for dozens of disease-related domains. However, it quickly became obvious that identification of the causative mutation would not be as straightforward as hoped. There have been many successes in gene identification for ENU-induced mutants in the areas of immunology (Hoebe and Beutler 2008; Sandberg et al. 2005; Tabeta et al. 2006; Theodoratos et al. 2010), development (Herron et al. 2002; Garcia–Garcia et al. 2005; Stottmann et al. 2009) and metabolism (Lloyd et al. 2005, 2006, 2010; Wilkes et al. 2009). ENU-disrupted genes responsible for neurological traits that might be considered less susceptible to environmental fluctuations have also been successfully identified in the areas of deafness (Grillet et al. 2009; Mackenzie et al. 2009; Parker et al. 2010; Schwander et al. 2009a, b), ataxia (Sharkey et al. 2009; Swanson et al. 2010; Xie et al. 2010) and epilepsy (Frankel et al. 2009; Tokuda et al. 2011). Mapping mutants for more complex behavioral phenotypes, on the other hand, has proven problematic.
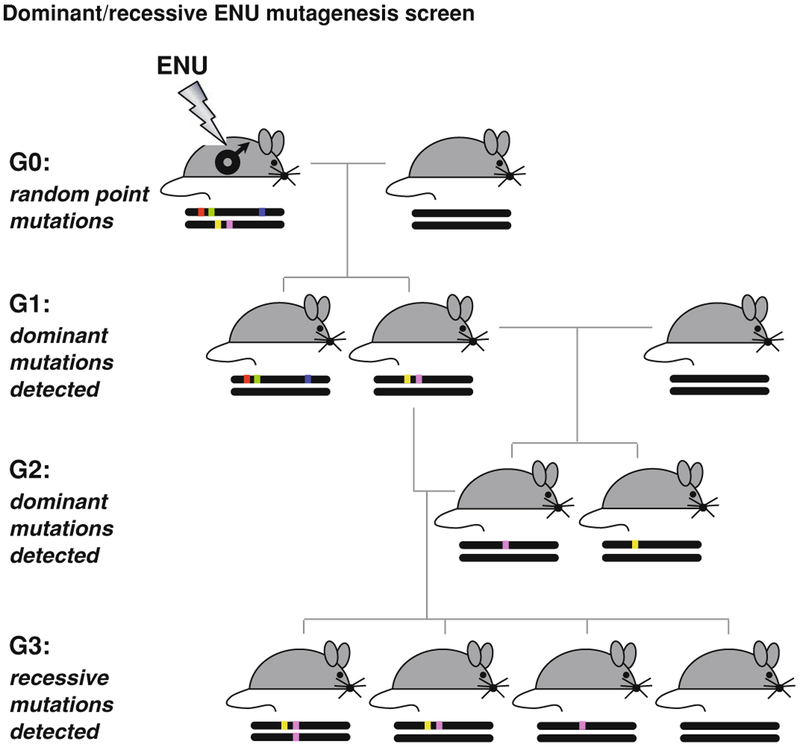
The basic steps in ENU mutagenesis (Fig. 2) include induction of mutations by injection of the mutagen in male mice. These G0 mice are then bred with wildtype females, and the offspring of that cross, the G1, can be screened for dominant mutations. Further breeding of the G1 males to wildtype females produces G2 females that can be bred back to the G1 male. The resulting G3 animals will carry recessive mutations at a rate of one per eight G3s. Once an outlier is identified, the standard course of action is to prove heritability by crossing the animal back to a wildtype, produce F1 mice and then intercross to recover the mutation in the F2 progeny—one-quarter of which should exhibit a fully-penetrant, recessive phenotype. If no F2 mice exhibit the expected phenotype, it is assumed that the original outlier had multiple alleles contributing to the phenotype (ENU produces one mutation approximately every 1–2 Mb; Kile and Hilton 2005) that were dissociated by outcrossing OR that the original outlier displayed an abnormal phenotype due to non-genetic reasons (environmental variability, etc.). If the ratio of affected mice in the F2 is lower than expected, reduced penetrance may be an issue that needs to be considered during the mapping process. If, however, the heritability cross proves successful, the mutants enter into a mapping funnel that starts with outcrossing a proven mutant to a different inbred mapping strain and F2 or BC animals are produced and phenotyped. Here we start to enter into familiar “QTL” territory—if the phenotype produced by the mutation is too weak to be observed on a mixed background or interacts with QTL in the mapping cross, the phenotype may disappear and mapping attempts will be unsuccessful.
Fig. 2.
Standard ENU mutagenesis. The standard ENU mutagenesis scheme includes injecting male mice (G0) with ENU. Doses vary depending upon strain, but generally, three weekly injections of 85–100 mg/kg ENU works well in B6 mice. ENU-treated males become infertile following treatment but many will regain fertility after 10–12 weeks. Loss of fertility is often used as an indicator that the ENU was effective. After regaining fertility, ENU-treated males are bred to wildtype females of the same strain or a different strain. G1 animals produced from this cross can be tested for dominant mutations. G1 mice will inherit a mutagenized genome from the father and the mutations will differ between G1s. To recover recessive mutations, the G1 females are crossed back to the G1 father to generate G2s. Two copies of any mutations that the father and daughter share will be recovered in the G3 at a rate of one mutation per eight G3s. For this reason, G3 pedigrees are often screened for phenotypes in order to recover at least one or two affected animals with which to propagate the new mutant colony
One way to deal with a non-robust phenotype is to cross the mutant to a number of inbred strains in order to find a heterogeneous background that allows for the recovery of the mutant phenotype in the mapping cross, but this increases mapping costs. Alternatively, sufficient SNP data are now available that allow mapping to strains that are closely related to the background strain and might reduce the number of interacting QTL (Bailey et al. 2008; Eisener-Dorman et al. 2010; Xia et al. 2010). The mutant phenotype might also be preserved by limiting the amount of the mapping strain background by backcrossing the F1 animals to a mutant. This strategy also increases the number of animals in the mapping cross that carry the causative mutation. Finally, a contemporaneous population of mice not carrying the mutation can be produced, phenotyped and genotyped to provide a control for both choosing outliers in the mutant cross and identifying background QTL.
Even with these strategies, however, identification of genes in ENU mutants that exhibit behavioral anomalies has been painfully slow, even when the phenotype appears robust. Many mutants with abnormal behavioral profiles have been reported (Rastan et al. 2004; Reijmers et al. 2006; Hamre et al. 2007; Mathews et al. 2009), yet gene identification for ENU behavioral mutants has been slow to materialize.
Recent publications, however, indicate that ENU-induced behavioral mutants are beginning to yield genes. In 2007, Keays et al. identified an ENU-induced mutation in the guanosine triphosphate (GTP) binding pocket of alpha-1 tubulin (Tuba1). Tuba1 mice exhibited hyperactive behavior but also had a secondary correlated phenotype—body weight. The presence of a secondary phenotype that was more stable and reproducible than the hyperactivity phenotype aided in fine mapping and identifying the gene. Speca et al. (2010) recently reported on the identification of an ENU-induced nonsense mutation in the Unc-79 gene in a mouse mutant (Lightweight) initially identified in a screen for animals with enhanced locomotor activity. The screen was conducted on a sensitized genetic background (mice heterozygous null for dopamine transporter), but the Lightweight mutation acted independently of this background. As the name implies, this mutant also had a correlated phenotype of low body weight. Peaks for both weight and locomotor activity overlapped, and the use of both phenotypes contributed to the identification of the causative ENU-induced mutation. Finally, an ENU-generated missense mutation in the Grin1 gene was identified in a mouse mutant that displayed increased spontaneous locomotor activity (Furuse et al. 2010). It should be noted that all three of these mutants were identified in screens for dominant ENU mutations, and all were initially generated on a mixed genetic background (i.e. ENU mutagenized males were crossed to females of a different inbred strain). These results suggest that dominant mutations are ultimately easier to identify than recessive mutations. Furthermore, using a mixed genetic background in the primary ENU screen may increase the chance of recovering mutant mice in the mapping cross, thereby hastening identification of the mutated gene.
The goals of the NIH-funded ENU centers included generation and distribution of mutagenized mice with the idea that individual researchers could follow up on phenotypes of interest and map the mutated genes. Although gene identification of ENU behavioral mutants has been slow, improvements in next-generation sequencing technologies and decreased costs may accelerate the process. Deep sequencing of ENU-generated mutants, in combination with or independent of mapping information, could yield the causative mutations. Several large libraries of cryopreserved sperm and/or embryos from G1 mice are also available at RIKEN (Yoshiki et al. 2009), Harwell (Glenister and Thornton 2000), the Australian Phenomics Network and the German ENU mutagenesis center. As sequencing prices decrease, these libraries could be screened to identify mutations genome-wide. Cryo-recovery and phenotyping of mice with mutations in specific genes would complement efforts such as the KOMP.
8. Beyond QTL Analysis: Finding the Quantitative Trait Gene
The ultimate goal of QTL mapping is identification of the underlying polymorphism that can provide insight into the biology of the phenotype/disease. Regardless of how a QTL is identified, members of the complex trait community have determined that several methods are appropriate and necessary for QTL validation (Abiola et al. 2003). These criteria include relating the gene function to the QTL phenotype, identifying allelic polymorphisms or assessing gene homology to determine if the sequence is evolutionarily conserved across species. Manipulation of the gene of interest is also advantageous for QTL validation and includes the study of knockout, knockin, transgenic or otherwise genetically-altered mouse models. Genetic or functional complementation of different inbred strains or a genetically-deficient mouse model serves as an additional confirmation of the QTL (Kono et al. 2003; Yalcin et al. 2004).
Identification of the QTG represents the beginning of a new phase of analysis that explores the mechanisms underlying alterations in gene expression and biological pathways (Flint 2003). For example, Rgs2 was mapped as an anxiety QTL and was later genetically dissected and confirmed using quantitative complementation (Yalcin et al. 2004). These findings have since been translated to human anxiety research, with the human RGS2 ortholog correlating with introversion and social anxiety, thereby becoming a potential target for the treatment of social anxiety disorders (Smoller et al. 2008).
9. The Future of Forward Genetics
As knowledge of the genomic sequence and organization of both the human and mouse genomes increases, an appreciation for the complexities that will likely explain the genetic components of complex behaviors has become more apparent. Single-gene, reverse genetic approaches like knockouts and transgenics still have much to offer toward functional annotation of genes. However, it is now widely accepted that naturally occurring genetic variation, along with epigenetic effects and environmental variation, may act synergistically to increase risk for complex neuropsychiatric diseases. Although the complex trait community has made consistent advances in the search for QTL that influence behavior, the progress from QTL to QTG has been painstakingly slow. However, genomic tools in the mouse are being developed at a rapid pace and promise to transform complex trait analysis. For example, dense SNP maps now available make it possible to narrow QTL regions using haplotype comparisons—the substantial reduction of a QTL interval that used to take years can now be accomplished in hours depending on the parental strains utilized (Eisener-Dorman et al. 2010).
New sequencing technologies are driving down costs and allowing for more accurate sequencing of entire genomes. As acquiring sequence becomes less cost prohibitive, the ability to sequence individual mice in a specific genomic interval or genome-wide will become a real option even for smaller laboratories. This will be especially useful for gene identification in ENU mutagenized lines.
Using new sequencing technologies, the Sanger Institute is sequencing the entire genome from 17 different inbred mouse strains, including the CC parental strains, as part of its Mouse Genomes Project (http://www.sanger.ac.uk/resources/mouse/genomes/). These data will provide an invaluable resource for mapping and haplotype analysis in the CC as it comes online. These sequence data, along with the increased genetic variation and mapping resolution offered by the CC, will provide access to QTL that have been unrepresented in current mapping populations.
This is a particularly exciting time for mouse genetics as resources and technology advance at a rapid pace. Behavioral scientists are poised to gain new insights into the biology and genetic control of complex behaviors with the ultimate goal of translational studies in humans.
References
- Abiola O, Angel JM, Avner P, Bachmanov AA, Belknap JK, Bennett B, Blankenhorn EP, Blizard DA, Bolivar V, Brockmann GA, Buck KJ, Bureau JF, Casley WL, Chesler EJ, Cheverud JM, Churchill GA, Cook M, Crabbe JC, Crusio WE, Darvasi A, de Haan G, Dermant P, Doerge RW, Elliot RW, Farber CR, Flaherty L, Flint J, Gershenfeld H, Gibson JP, Gu J, Gu W, Himmelbauer H, Hitzemann R, Hsu HC, Hunter K, Iraqi FF, Jansen RC, Johnson TE, Jones BC, Kempermann G, Lammert F, Lu L, Manly KF, Matthews DB, Medrano JF, Mehrabian M, Mittlemann G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Mountz JD, Nagase H, Nowakowski RS, O’Hara BF, Osadchuk AV, Paigen B, Palmer AA, Peirce JL, Pomp D, Rosemann M, Rosen GD, Schalkwyk LC, Seltzer Z, Settle S, Shimomura K, Shou S, Sikela JM, Siracusa LD, Spearow JL, Teuscher C, Threadgill DW, Toth LA, Toye AA, Vadasz C, Van Zant G, Wakeland E, Williams RW, Zhang HG, Zou F (2003) The nature and identification of quantitative trait loci: a community’s view. Nat Rev Genet 4:911–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldinger KA, Sokoloff G, Rosenberg DM, Palmer AA, Millen KJ (2009) Genetic variation and population substructure in outbred CD-1 mice: implications for genome-wide association studies. PLoS One 4:e4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RC, Wright R, Freed W (1996) Quantitative trait loci contributing to phencyclidine-induced and amphetamine-induced locomotor behavior in inbred mice. Neuropsychopharmacology 15:484–490 [DOI] [PubMed] [Google Scholar]
- Atkins JKBAL (2001) The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mamm Genome 12: 893–899 [DOI] [PubMed] [Google Scholar]
- Bailey DW (1971) Recombinant-inbred strains. An aid to finding identity, linkage, and function of histocompatibility and other genes. Transplantation 11:325–327 [DOI] [PubMed] [Google Scholar]
- Bailey JS, Grabowski-Boase L, Steffy BM, Wiltshire T, Churchill GA, Tarantino LM (2008) Identification of QTL for locomotor activation and anxiety using closely-related inbred strains. Genes Brain Behav 7:761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Peirce JL, Zhou M, Li H, Goldowitz D, Williams RW, Lu L, Cui Y (2007) An integrative genomics strategy for systematic characterization of genetic loci modulating phenotypes. Hum Mol Genet 16:1381–1390 [DOI] [PubMed] [Google Scholar]
- Basten CJ, Weir BS, Zeng Z-B (1994) Zmap—a QTL cartographer In: Smith C, Gavora JS, Benkel B, Chesnais J, Fairfull W, Gibson JP, Kennedy BW, Burnside EB (eds) 5th world congress on genetics applied to livestock production: computing strategies and software. Organizing Committee, 5th world congress on genetics applied to livestock production, Guelph, Ontario, Canada, pp 65–66 [Google Scholar]
- Basten CJ, Weir BS, Zeng Z-B (2002) QTL Cartographer, version 1.16 Department of Statistics, North Carolina State University, Raleigh, NC [Google Scholar]
- Belknap JK (2003) Chromosome substitution strains: some quantitative considerations for genome scans and fine mapping. Mamm Genome 14:723–732 [DOI] [PubMed] [Google Scholar]
- Berrettini WH, Ferraro TN, Alexander RC, Buchberg AM, Vogel WH (1994) Quantitative trait loci mapping of three loci controlling morphine preference using inbred mouse strains. Nat Genet 7:54–58 [DOI] [PubMed] [Google Scholar]
- Blizard DA, Bailey DW (1979) Genetic correlation between open-field activity and defecation: analysis with the CXB recombinant-inbred strains. Behav Genet 9:349–357 [DOI] [PubMed] [Google Scholar]
- Bogue M (2003) Mouse phenome project: understanding human biology through mouse genetics and genomics. J Appl Physiol 95:1335–1337 [DOI] [PubMed] [Google Scholar]
- Bogue MA, Grubb SC, Maddatu TP, Bult CJ (2007) Mouse phenome database (MPD). Nucleic Acids Res 35:D643–D649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp SE, Ramachandran V, Henson K, Luzader A, Lindstrom M, Spooner M, Steffy BM, Suzuki O, Janse C, Waters AP, Zhou Y, Wiltshire T, Winzeler EA (2010) Genome wide analysis of inbred mouse lines identifies a locus containing Ppar-gamma as contributing to enhanced malaria survival. PLoS One 5: e10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AE, Gill KJ (2008) Confirmation of provisional quantitative trait loci for voluntary alcohol consumption: genetic analysis in chromosome substitution strains and F2 crosses derived from A/J and C57BL/6J progenitors. Pharmacogenet Genomics 18:1071–1082 [DOI] [PubMed] [Google Scholar]
- Boyle AE, Gill KJ (2009) A verification of previously identified QTLs for cocaine-induced activation using a panel of B6. A chromosome substitution strains (CSS) and A/J × C57Bl/6J F2 mice. Psychopharmacology (Berl) 207(2):325–334 [DOI] [PubMed] [Google Scholar]
- Broman KW (2005) The genomes of recombinant inbred lines. Genetics 169:1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Sen S (2009) A guide to QTL mapping with R/qtl. Springer, New York [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890 [DOI] [PubMed] [Google Scholar]
- Buchner DA, Burrage LC, Hill AE, Yazbek SN, O’Brien WE, Croniger CM, Nadeau JH (2008) Resistance to diet-induced obesity in mice with a single substituted chromosome. Physiol Genomics 35:116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess-Herbert SL, Cox A, Tsaih SW, Paigen B (2008) Practical applications of the bioinformatics toolbox for narrowing quantitative trait loci. Genetics 180:2227–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess-Herbert SL, Tsaih SW, Stylianou IM, Walsh K, Cox AJ, Paigen B (2009) An experimental assessment of in silico haplotype association mapping in laboratory mice. BMC Genet 10:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldarone B, Saavedra C, Tartaglia K, Wehner JM, Dudek BC, Flaherty L (1997) Quantitative trait loci analysis affecting contextual conditioning in mice. Nat Genet 17:335–337 [DOI] [PubMed] [Google Scholar]
- Cervino AC, Li G, Edwards S, Zhu J, Laurie C, Tokiwa G, Lum PY, Wang S, Castellani LW, Lusis AJ, Carlson S, Sachs AB, Schadt EE (2005) Integrating QTL and high-density SNP analyses in mice to identify Insig2 as a susceptibility gene for plasma cholesterol levels. Genomics 86:505–517 [DOI] [PubMed] [Google Scholar]
- Cheng R, Lim JE, Samocha KE, Sokoloff G, Abney M, Skol AD, Palmer AA (2010) Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics 185:1033–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Bystrykh L, De Haan G, Cooke MP, Su AI, Manly KF, Williams RW (2006) Reply to normalization procedures and detection of linkage signal in genetical-genomics experiments. Nat Genet 38:856–858 [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Rodriguez-Zas SL, Mogil JS (2001) In silico mapping of mouse quantitative trait loci. Science 294:2423. [DOI] [PubMed] [Google Scholar]
- Chia R, Achilli F, Festing MF, Fisher EM (2005) The origins and uses of mouse outbred stocks. Nat Genet 37:1181–1186 [DOI] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu HC, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, Kendziorski C, Kotb M, Kooy RF, Llamas B, Lammert F, Lassalle JM, Lowenstein PR, Lu L, Lusis A, Manly KF, Marcucio R, Matthews D, Medrano JF, Miller DR, Mittleman G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Morris DG, Mott R, Nadeau JH, Nagase H, Nowakowski RS, O’Hara BF, Osadchuk AV, Page GP, Paigen B, Paigen K, Palmer AA, Pan HJ, Peltonen-Palotie L, Peirce J, Pomp D, Pravenec M, Prows DR, Qi Z, Reeves RH, Roder J, Rosen GD, Schadt EE, Schalkwyk LC, Seltzer Z, Shimomura K, Shou S, Sillanpaa MJ, Siracusa LD, Snoeck HW, Spearow JL, Svenson K, Tarantino LM, Threadgill D, Toth LA, Valdar W, de Villena FP, Warden C, Whatley S, Williams RW, Wiltshire T, Yi N, Zhang D, Zhang M, Zou F (2004) The collaborative cross, a community resource for the genetic analysis of complex traits. Nat Genet 36:1133–1137 [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Rossant J, Wurst W (2007) A mouse for all reasons. Cell 128:9–13 [DOI] [PubMed] [Google Scholar]
- Cordes SP (2005) N-ethyl-N-nitrosourea mutagenesis: boarding the mouse mutant express. Microbiol Mol Biol Rev 69:426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Rigter H, Kerbusch S (1982) Analysis of behavioural responses to an ACTH analog in CXB/By recombinant inbred mice. Behav Brain Res 4:289–314 [DOI] [PubMed] [Google Scholar]
- Crow JF (2002) C. C. Little, cancer and inbred mice. Genetics 161:1357–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Holmes A (2005) The ascent of mouse: advances in modelling human depression and anxiety. Nat Rev Drug Discov 4:775–790 [DOI] [PubMed] [Google Scholar]
- Cryan JF, Slattery DA (2010) GABAB receptors and depression. Current status. Adv Pharmacol 58:427–451 [DOI] [PubMed] [Google Scholar]
- Cunningham CL (1995) Localization of genes influencing ethanol-induced conditioned place preference and locomotor activity in BXD recombinant inbred mice. Psychopharmacology (Berl) 120:28–41 [DOI] [PubMed] [Google Scholar]
- Darvasi A (1998) Experimental strategies for the genetic dissection of complex traits in animal models. Nat Genet 18:19–24 [DOI] [PubMed] [Google Scholar]
- Darvasi A (2001) In silico mapping of mouse quantitative trait loci. Science 294:2423. [PubMed] [Google Scholar]
- Darvasi A, Soller M (1994) Selective DNA pooling for determination of linkage between a molecular marker and a quantitative trait locus. Genetics 138:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A, Soller M (1995) Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141:1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarest K, Koyner J, McCaughran J Jr, Cipp L, Hitzemann R (2001) Further characterization and high-resolution mapping of quantitative trait loci for ethanol-induced locomotor activity. Behav Genet 31:79–91 [DOI] [PubMed] [Google Scholar]
- Desbonnet L, Waddington JL, O’Tuathaigh CM (2009) Mutant models for genes associated with schizophrenia. Biochem Soc Trans 37:308–312 [DOI] [PubMed] [Google Scholar]
- DiPetrillo K, Wang X, Stylianou IM, Paigen B (2005) Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet 21:683–692 [DOI] [PubMed] [Google Scholar]
- Doyle GA, Furlong PJ, Schwebel CL, Smith GG, Lohoff FW, Buono RJ, Berrettini WH, Ferraro TN (2008) Fine mapping of a major QTL influencing morphine preference in C57BL/6 and DBA/2 mice using congenic strains. Neuropsychopharmacology 33:2801–2809 [DOI] [PubMed] [Google Scholar]
- Eisener-Dorman AF, Grabowski-Boase L, Steffy BM, Wiltshire T, Tarantino LM (2010) Quantitative trait locus and haplotype mapping in closely related inbred strains identifies a locus for open field behavior. Mamm Genome 21:231–246 [DOI] [PubMed] [Google Scholar]
- Fernandez JR, Tarantino LM, Hofer SM, Vogler GP, McClearn GE (2000) Epistatic quantitative trait loci for alcohol preference in mice. Behav Genet 30:431–437 [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Martin JF, Schwebel CL, Doyle GA, Buono RJ, Berrettini WH (2005) Confirmation of a major QTL influencing oral morphine intake in C57 and DBA mice using reciprocal congenic strains. Neuropsychopharmacology 30:742–746 [DOI] [PubMed] [Google Scholar]
- Flint J (2003) Analysis of quantitative trait loci that influence animal behavior. J Neurobiol 54:46–77 [DOI] [PubMed] [Google Scholar]
- Flint J, Corley R, DeFries JC, Fulker DW, Gray JA, Miller S, Collins AC (1995) A simple genetic basis for a complex psychological trait in laboratory mice. Science 269:1432–1435 [DOI] [PubMed] [Google Scholar]
- Flint J, Valdar W, Shifman S, Mott R (2005) Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet 6:271–286 [DOI] [PubMed] [Google Scholar]
- Frankel WN, Yang Y, Mahaffey CL, Beyer BJ, O’Brien TP (2009) Szt2, a novel gene for seizure threshold in mice. Genes Brain Behav 8:568–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse T, Wada Y, Hattori K, Yamada I, Kushida T, Shibukawa Y, Masuya H, Kaneda H, Miura I, Seno N, Kanda T, Hirose R, Toki S, Nakanishi K, Kobayashi K, Sezutsu H, Gondo Y, Noda T, Yuasa S, Wakana S (2010) Phenotypic characterization of a new Grin1 mutant mouse generated by ENU mutagenesis. Eur J Neurosci 31:1281–1291 [DOI] [PubMed] [Google Scholar]
- Gailus-Durner V, Fuchs H, Adler T, Aguilar Pimentel A, Becker L, Bolle I, Calzada-Wack J, Dalke C, Ehrhardt N, Ferwagner B, Hans W, Holter SM, Holzlwimmer G, Horsch M, Javaheri A, Kallnik M, Kling E, Lengger C, Morth C, Mossbrugger I, Naton B, Prehn C, Puk O, Rathkolb B, Rozman J, Schrewe A, Thiele F, Adamski J, Aigner B, Behrendt H, Busch DH, Favor J, Graw J, Heldmaier G, Ivandic B, Katus H, Klingenspor M, Klopstock T, Kremmer E, Ollert M, Quintanilla-Martinez L, Schulz H, Wolf E, Wurst W, de Angelis MH (2009) Systemic first-line phenotyping. Methods Mol Biol 530:463–509 [DOI] [PubMed] [Google Scholar]
- Garcia–Garcia MJ, Eggenschwiler JT, Caspary T, Alcorn HL, Wyler MR, Huangfu D, Rakeman AS, Lee JD, Feinberg EH, Timmer JR, Anderson KV (2005) Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc Natl Acad Sci USA 102:5913–5919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenfeld HK, Neumann PE, Mathis C, Crawley JN, Li X, Paul SM (1997) Mapping quantitative trait loci for open-field behavior in mice. Behav Genet 27:201–210 [DOI] [PubMed] [Google Scholar]
- Glenister PH, Thornton CE (2000) Cryoconservation—archiving for the future. Mamm Genome 11:565–571 [DOI] [PubMed] [Google Scholar]
- Gora-Maslak G, McClearn GE, Crabbe JC, Phillips TJ, Belknap JK, Plomin R (1991) Use of recombinant inbred strains to identify quantitative trait loci in psychopharmacology. Psychopharmacology (Berl) 104:413–424 [DOI] [PubMed] [Google Scholar]
- Gregorova S, Divina P, Storchova R, Trachtulec Z, Fotopulosova V, Svenson KL, Donahue LR, Paigen B, Forejt J (2008) Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res 18:509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet N, Schwander M, Hildebrand MS, Sczaniecka A, Kolatkar A, Velasco J, Webster JA, Kahrizi K, Najmabadi H, Kimberling WJ, Stephan D, Bahlo M, Wiltshire T, Tarantino LM, Kuhn P, Smith RJ, Muller U (2009) Mutations in LOXHD1, an evolutionarily conserved stereociliary protein, disrupt hair cell function in mice and cause progressive hearing loss in humans. Am J Hum Genet 85:328–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisel JE, Belknap JK, O’Toole LA, Helms ML, Wenger CD, Crabbe JC (1997) Quantitative trait loci affecting methamphetamine responses in BXD recombinant inbred mouse strains. J Neurosci 17:745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupe A, Germer S, Usuka J, Aud D, Belknap JK, Klein RF, Ahluwalia MK, Higuchi R, Peltz G (2001) In silico mapping of complex disease-related traits in mice. Science 292:1915–1918 [DOI] [PubMed] [Google Scholar]
- Hamilton BA, Frankel WN (2001) Of mice and genome sequence. Cell 107:13–16 [DOI] [PubMed] [Google Scholar]
- Hamre KM, Goldowitz D, Wilkinson S, Matthews DB (2007) Screening for ENU-induced mutations in mice that result in aberrant ethanol-related phenotypes. Behav Neurosci 121:665–678 [DOI] [PubMed] [Google Scholar]
- Harrill AH, Watkins PB, Su S, Ross PK, Harbourt DE, Stylianou IM, Boorman GA, Russo MW, Sackler RS, Harris SC, Smith PC, Tennant R, Bogue M, Paigen K, Harris C, Contractor T, Wiltshire T, Rusyn I, Threadgill DW (2009) Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res 19:1507–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ND, Turri MG, DeFries JC, Flint J (2004) QTL analysis of multiple behavioral measures of anxiety in mice. Behav Genet 34:267–293 [DOI] [PubMed] [Google Scholar]
- Herron BJ, Lu W, Rao C, Liu S, Peters H, Bronson RT, Justice MJ, McDonald JD, Beier DR (2002) Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat Genet 30:185–189 [DOI] [PubMed] [Google Scholar]
- Hitzemann B, Dains K, Kanes S, Hitzemann R (1994) Further studies on the relationship between dopamine cell density and haloperidol-induced catalepsy. J Pharmacol Exp Ther 271:969–976 [PubMed] [Google Scholar]
- Hitzemann R, Cipp L, Demarest K, Mahjubi E, McCaughran J Jr (1998) Genetics of ethanol-induced locomotor activation: detection of QTLs in a C57BL/6J × DBA/2J F2 intercross. Mamm Genome 9:956–962 [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Dains K, Bier-Langing CM, Zahniser NR (1991) On the selection of mice for haloperidol response and non-response. Psychopharmacology (Berl) 103:244–250 [DOI] [PubMed] [Google Scholar]
- Hoebe K, Beutler B (2008) Forward genetic analysis of TLR-signaling pathways: an evaluation. Adv Drug Deliv Rev 60:824–829 [DOI] [PubMed] [Google Scholar]
- Holmes A, Murphy DL, Crawley JN (2003) Abnormal behavioral phenotypes of serotonin transporter knockout mice: parallels with human anxiety and depression. Biol Psychiatry 54:953–959 [DOI] [PubMed] [Google Scholar]
- Horvat S, Bunger L, Falconer VM, Mackay P, Law A, Bulfield G, Keightley PD (2000) Mapping of obesity QTLs in a cross between mouse lines divergently selected on fat content. Mamm Genome 11:2–7 [DOI] [PubMed] [Google Scholar]
- Hrabe de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, Reis A, Richter T, Alessandrini F, Jakob T, Fuchs E, Kolb H, Kremmer E, Schaeble K, Rollinski B, Roscher A, Peters C, Meitinger T, Strom T, Steckler T, Holsboer F, Klopstock T, Gekeler F, Schindewolf C, Jung T, Avraham K, Behrendt H, Ring J, Zimmer A, Schughart K, Pfeffer K, Wolf E, Balling R (2000) Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet 25:444–447 [DOI] [PubMed] [Google Scholar]
- Iancu OD, Darakjian P, Walter NA, Malmanger B, Oberbeck D, Belknap J, McWeeney S, Hitzemann R (2010) Genetic diversity and striatal gene networks: focus on the heterogeneous stock-collaborative cross (HS-CC) mouse. BMC Genomics 11:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium (2003) The international HapMap project. Nature 426:789–796 [DOI] [PubMed] [Google Scholar]
- Jansen RC (1993) Interval mapping of multiple quantitative trait loci. Genetics 135:205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BC, Tarantino LM, Rodriguez LA, Reed CL, McClearn GE, Plomin R, Erwin VG (1999) Quantitative-trait loci analysis of cocaine-related behaviours and neurochemistry. Pharmacogenetics 9:607–617 [PubMed] [Google Scholar]
- Justice MJ (2004) From the atomic age to the genome project. Genetica 122:3–7 [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E (2008) Efficient control of population structure in model organism association mapping. Genetics 178:1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keays DA, Tian G, Poirier K, Huang GJ, Siebold C, Cleak J, Oliver PL, Fray M, Harvey RJ, Molnar Z, Pinon MC, Dear N, Valdar W, Brown SD, Davies KE, Rawlins JN, Cowan NJ, Nolan P, Chelly J, Flint J (2007) Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell 128:45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MA, Low MJ, Phillips TJ, Wakeland EK, Yanagisawa M (2003) The mapping of quantitative trait loci underlying strain differences in locomotor activity between 129S6 and C57BL/6J mice. Mamm Genome 14:692–702 [DOI] [PubMed] [Google Scholar]
- Kenney-Hunt JP, Wang B, Norgard EA, Fawcett G, Falk D, Pletscher LS, Jarvis JP, Roseman C, Wolf J, Cheverud JM (2008) Pleiotropic patterns of quantitative trait loci for 70 murine skeletal traits. Genetics 178:2275–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile BT, Hilton DJ (2005) The art and design of genetic screens: mouse. Nat Rev Genet 6:557–567 [DOI] [PubMed] [Google Scholar]
- Kono DH, Park MS, Theofilopoulos AN (2003) Genetic complementation in female (BXSB × NZW) F2 mice. J Immunol 171:6442–6447 [DOI] [PubMed] [Google Scholar]
- Koyner J, Demarest K, McCaughran J Jr, Cipp L, Hitzemann R (2000) Identification and time dependence of quantitative trait loci for basal locomotor activity in the BXD recombinant inbred series and a B6D2 F2 intercross. Behav Genet 30:159–170 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181 [DOI] [PubMed] [Google Scholar]
- Legare ME, Bartlett FS 2nd, Frankel WN (2000) A major effect QTL determined by multiple genes in epileptic EL mice. Genome Res 10:42–48 [PMC free article] [PubMed] [Google Scholar]
- Legare ME, Frankel WN (2000) Multiple seizure susceptibility genes on chromosome 7 in SWXL-4 congenic mouse strains. Genomics 70:62–65 [DOI] [PubMed] [Google Scholar]
- Lesch KP, Zeng Y, Reif A, Gutknecht L (2003) Anxiety-related traits in mice with modified genes of the serotonergic pathway. Eur J Pharmacol 480:185–204 [DOI] [PubMed] [Google Scholar]
- Leussis MP, Frayne ML, Saito M, Berry EM, Aldinger KA, Rockwell GN, Hammer RP Jr, Baskin-Hill AE, Singer JB, Nadeau JH, Sklar P, Petryshen TL (2009) Genomic survey of prepulse inhibition in mouse chromosome substitution strains. Genes Brain Behav 8:806–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Wang J, Guo J, Allard J, Cheng J, Ng A, Shafer S, Puech A, McPherson JD, Foernzler D, Peltz G, Usuka J (2004) In silico genetics: identification of a functional element regulating H2-Ealpha gene expression. Science 306:690–695 [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Winchester E, Daly MJ, Wang DG, Hirschhorn JN, Laviolette JP, Ardlie K, Reich DE, Robinson E, Sklar P, Shah N, Thomas D, Fan JB, Gingeras T, Warrington J, Patil N, Hudson TJ, Lander ES (2000) Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat Genet 24:381–386 [DOI] [PubMed] [Google Scholar]
- Liu P, Wang Y, Vikis H, Maciag A, Wang D, Lu Y, Liu Y, You M (2006) Candidate lung tumor susceptibility genes identified through whole-genome association analyses in inbred mice. Nat Genet 38:888–895 [DOI] [PubMed] [Google Scholar]
- Lloyd DJ, Bohan S, Gekakis N (2006) Obesity, hyperphagia and increased metabolic efficiency in Pc1 mutant mice. Hum Mol Genet 15:1884–1893 [DOI] [PubMed] [Google Scholar]
- Lloyd DJ, Hall FW, Tarantino LM, Gekakis N (2005) Diabetes insipidus in mice with a mutation in Aquaporin-2. PLoS Genet 1:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd DJ, Wheeler MC, Gekakis N (2010) A point mutation in Sec61alpha1 leads to diabetes and hepatosteatosis in mice. Diabetes 59:460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF (2002) A personal history of the mouse genome. Annu Rev Genomics Hum Genet 3:1–16 [DOI] [PubMed] [Google Scholar]
- Macarthur DG, Tyler-Smith C (2010) Loss-of-function variants in the genomes of healthy humans. Hum Mol Genet 19:R125–R130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie FE, Parker A, Parkinson NJ, Oliver PL, Brooker D, Underhill P, Lukashkina VA, Lukashkin AN, Holmes C, Brown SD (2009) Analysis of the mouse mutant cloth-ears shows a role for the voltage-gated sodium channel Scn8a in peripheral neural hearing loss. Genes Brain Behav 8:699–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmanger B, Lawler M, Coulombe S, Murray R, Cooper S, Polyakov Y, Belknap J, Hitzemann R (2006) Further studies on using multiple-cross mapping (MCM) to map quantitative trait loci. Mamm Genome 17:1193–1204 [DOI] [PubMed] [Google Scholar]
- Manly KF, Cudmore RH Jr, Meer JM (2001) Map manager QTX, cross-platform software for genetic mapping. Mamm Genome 12:930–932 [DOI] [PubMed] [Google Scholar]
- Markel P, Shu P, Ebeling C, Carlson GA, Nagle DL, Smutko JS, Moore KJ (1997) Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat Genet 17:280–284 [DOI] [PubMed] [Google Scholar]
- Mathews TA, Brookshire BR, Budygin EA, Hamre K, Goldowitz D, Jones SR (2009) Ethanol-induced hyperactivity is associated with hypodopaminergia in the 22-TNJ ENU-mutated mouse. Alcohol 43:421–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClearn GE, Wilson J, Meredith W (1970) The use of isogenic and heterogenic mouse stock in behavioral research In: Lindzey G, Thiessen D (eds) Contributions to behavioral-genetic analysis: the mouse as a prototype. Appleton-Century-Crofts, New York, pp 3–22 [Google Scholar]
- McClurg P, Janes J, Wu C, Delano DL, Walker JR, Batalov S, Takahashi JS, Shimomura K, Kohsaka A, Bass J, Wiltshire T, Su AI (2007) Genomewide association analysis in diverse inbred mice: power and population structure. Genetics 176:675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, Shendure J, Pociask K, Silver LM (1996) Identification of sex-specific quantitative trait loci controlling alcohol preference in C57BL/6 mice. Nat Genet 13:147–153 [DOI] [PubMed] [Google Scholar]
- Miller BH, Schultz LE, Gulati A, Su AI, Pletcher MT (2010) Phenotypic characterization of a genetically diverse panel of mice for behavioral despair and anxiety. PLoS One 5:e14458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogil JS, Richards SP, O’Toole LA, Helms ML, Mitchell SR, Kest B, Belknap JK (1997) Identification of a sex-specific quantitative trait locus mediating nonopioid stress-induced analgesia in female mice. J Neurosci 17:7995–8002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R, Flint J (2002) Simultaneous detection and fine mapping of quantitative trait loci in mice using heterogeneous stocks. Genetics 160:1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan BM, Hodges CA, Palmert MR (2006) The use of mouse chromosome substitution strains to investigate the genetic regulation of pubertal timing. Mol Cell Endocrinol 254–255:103–108 [DOI] [PubMed] [Google Scholar]
- Neiderhiser JM, Plomin R, McClearn GE (1992) The use of CXB recombinant inbred mice to detect quantitative trait loci in behavior. Physiol Behav 52:429–439 [DOI] [PubMed] [Google Scholar]
- Nishi A, Ishii A, Takahashi A, Shiroishi T, Koide T (2010) QTL analysis of measures of mouse home-cage activity using B6/MSM consomic strains. Mamm Genome 21:477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray IC, Vizor L, Brooker D, Whitehill E, Washbourne R, Hough T, Greenaway S, Hewitt M, Liu X, McCormack S, Pickford K, Selley R, Wells C, Tymowska-Lalanne Z, Roby P, Glenister P, Thornton C, Thaung C, Stevenson JA, Arkell R, Mburu P, Hardisty R, Kiernan A, Erven A, Steel KP, Voegeling S, Guenet JL, Nickols C, Sadri R, Nasse M, Isaacs A, Davies K, Browne M, Fisher EM, Martin J, Rastan S, Brown SD, Hunter J (2000) A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat Genet 25:440–443 [DOI] [PubMed] [Google Scholar]
- Norgard EA, Jarvis JP, Roseman CC, Maxwell TJ, Kenney-Hunt JP, Samocha KE, Pletscher LS, Wang B, Fawcett GL, Leatherwood CJ, Wolf JB, Cheverud JM (2009) Replication of long-bone length QTL in the F9–F10 LG, SM advanced intercross. Mamm Genome 20:224–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgard EA, Roseman CC, Fawcett GL, Pavlicev M, Morgan CD, Pletscher LS, Wang B, Cheverud JM (2008) Identification of quantitative trait loci affecting murine long bone length in a two-generation intercross of LG/J and SM/J Mice. J Bone Miner Res 23:887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K (2003) One hundred years of mouse genetics: an intellectual history. I. The classical period (1902–1980). Genetics 163:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Eppig JT (2000) A mouse phenome project. Mamm Genome 11:715–717 [DOI] [PubMed] [Google Scholar]
- Park YG, Clifford R, Buetow KH, Hunter KW (2003) Multiple cross and inbred strain haplotype mapping of complex-trait candidate genes. Genome Res 13:118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker A, Hardisty-Hughes RE, Wisby L, Joyce S, Brown SD (2010) Melody, an ENU mutation in Caspase 3, alters the catalytic cysteine residue and causes sensorineural hearing loss in mice. Mamm Genome 21:565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW (2004) A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryshen TL, Kirby A, Hammer RP Jr, Purcell S, O’Leary SB, Singer JB, Hill AE, Nadeau JH, Daly MJ, Sklar P (2005) Two quantitative trait Loci for prepulse inhibition of startle identified on mouse chromosome 16 using chromosome substitution strains. Genetics 171:1895–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, Hamre KM, Lariviere WR, Matthews DB, Mittleman G, Goldowitz D, Chesler EJ (2010) High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav 9:129–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Crabbe JC, Metten P, Belknap JK (1994) Localization of genes affecting alcohol drinking in mice. Alcohol Clin Exp Res 18:931–941 [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Huson M, Gwiazdon C, Burkhart-Kasch S, Shen EH (1995) Effects of acute and repeated ethanol exposures on the locomotor activity of BXD recombinant inbred mice. Alcohol Clin Exp Res 19:269–278 [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Huson MG, McKinnon CS (1998) Localization of genes mediating acute and sensitized locomotor responses to cocaine in BXD/Ty recombinant inbred mice. J Neurosci 18:3023–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, Lagler E, Korstanje R, Wang X, Nusskern D, Bogue MA, Mural RJ, Paigen B, Wiltshire T (2004) Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol 2:e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevorsek Z, Gorjanc G, Paigen B, Horvat S (2010) Congenic and bioinformatics analyses resolved a major-effect Fob3b QTL on mouse Chr 15 into two closely linked loci. Mamm Genome 21:172–185 [DOI] [PubMed] [Google Scholar]
- Rastan S, Hough T, Kierman A, Hardisty R, Erven A, Gray IC, Voeling S, Isaacs A, Tsai H, Strivens M, Washbourne R, Thornton C, Greenaway S, Hewitt M, McCormick S, Selley R, Wells C, Tymowska-Lalanne Z, Roby P, Mburu P, Rogers D, Hagan J, Reavill C, Davies K, Glenister P, Fisher EM, Martin J, Vizor L, Bouzyk M, Kelsell D, Guenet JL, Steel KP, Sheardown S, Spurr N, Gray I, Peters J, Nolan PM, Hunter AJ, Brown SD (2004) Towards a mutant map of the mouse–new models of neurological, behavioural, deafness, bone, renal and blood disorders. Genetica 122:47–49 [DOI] [PubMed] [Google Scholar]
- Reijmers LG, Coats JK, Pletcher MT, Wiltshire T, Tarantino LM, Mayford M (2006) A mutant mouse with a highly specific contextual fear-conditioning deficit found in an N-ethyl-N-nitrosourea (ENU) mutagenesis screen. Learn Mem 13:143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK (1999) Transgressive segregation, adaptation and speciation. Heredity 83(Pt 4):363–372 [DOI] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L (2008) Breeding designs for recombinant inbred advanced intercross lines. Genetics 179:1069–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez LA, Plomin R, Blizard DA, Jones BC, McClearn GE (1994) Alcohol acceptance, preference, and sensitivity in mice. I. Quantitative genetic analysis using BXD recombinant inbred strains. Alcohol Clin Exp Res 18:1416–1422 [DOI] [PubMed] [Google Scholar]
- Rodriguez LA, Plomin R, Blizard DA, Jones BC, McClearn GE (1995) Alcohol acceptance, preference, and sensitivity in mice. II. Quantitative trait loci mapping analysis using BXD recombinant inbred strains. Alcohol Clin Exp Res 19:367–373 [DOI] [PubMed] [Google Scholar]
- Samocha KE, Lim JE, Cheng R, Sokoloff G, Palmer AA (2010) Fine mapping of QTL for prepulse inhibition in LG/J and SM/J mice using F(2) and advanced intercross lines. Genes Brain Behav 9:759–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, Cooke MP (2005) c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell 8:153–166 [DOI] [PubMed] [Google Scholar]
- Savitz J, Lucki I, Drevets WC (2009) 5-HT(1A) receptor function in major depressive disorder. Prog Neurobiol 88:17–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander M, Lopes V, Sczaniecka A, Gibbs D, Lillo C, Delano D, Tarantino LM, Wiltshire T, Williams DS, Muller U (2009a) A novel allele of myosin VIIa reveals a critical function for the C-terminal FERM domain for melanosome transport in retinal pigment epithelial cells. J Neurosci 29:15810–15818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander M, Xiong W, Tokita J, Lelli A, Elledge HM, Kazmierczak P, Sczaniecka A, Kolatkar A, Wiltshire T, Kuhn P, Holt JR, Kachar B, Tarantino L, Muller U (2009b) A mouse model for nonsyndromic deafness (DFNB12) links hearing loss to defects in tip links of mechanosensory hair cells. Proc Natl Acad Sci USA 106:5252–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz WJ, Zimmerman P (1990) Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J Neurosci 10:3685–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey LM, Cheng X, Drews V, Buchner DA, Jones JM, Justice MJ, Waxman SG, Dib-Hajj SD, Meisler MH (2009) The ataxia3 mutation in the N-terminal cytoplasmic domain of sodium channel Na(v)1.6 disrupts intracellular trafficking. J Neurosci 29:2733–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver LM (1995) Mouse genetics concepts and applications. Oxford University Press, Oxford [Google Scholar]
- Singer JB, Hill AE, Burrage LC, Olszens KR, Song J, Justice M, O’Brien WE, Conti DV, Witte JS, Lander ES, Nadeau JH (2004) Genetic dissection of complex traits with chromosome substitution strains of mice. Science 304:445–448 [DOI] [PubMed] [Google Scholar]
- Singer JB, Hill AE, Nadeau JH, Lander ES (2005) Mapping quantitative trait loci for anxiety in chromosome substitution strains of mice. Genetics 169:855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Sheppard K, DiPetrillo K, Churchill G (2009) Quantitative trait locus analysis using J/qtl. Methods Mol Biol 573:175–188 [DOI] [PubMed] [Google Scholar]
- Smoller JW, Paulus MP, Fagerness JA, Purcell S, Yamaki LH, Hirshfeld-Becker D, Biederman J, Rosenbaum JF, Gelernter J, Stein MB (2008) Influence of RGS2 on anxiety-related temperament, personality, and brain function. Arch Gen Psychiatry 65:298–308 [DOI] [PubMed] [Google Scholar]
- Solberg LC, Valdar W, Gauguier D, Nunez G, Taylor A, Burnett S, Arboledas-Hita C, Hernandez-Pliego P, Davidson S, Burns P, Bhattacharya S, Hough T, Higgs D, Klenerman P, Cookson WO, Zhang Y, Deacon RM, Rawlins JN, Mott R, Flint J (2006) A protocol for high-throughput phenotyping, suitable for quantitative trait analysis in mice. Mamm Genome 17:129–146 [DOI] [PubMed] [Google Scholar]
- Speca DJ, Chihara D, Ashique AM, Bowers MS, Pierce-Shimomura JT, Lee J, Rabbee N, Speed TP, Gularte RJ, Chitwood J, Medrano JF, Liao M, Sonner JM, Eger EI 2nd, Peterson AS, McIntire SL (2010) Conserved role of unc-79 in ethanol responses in lightweight mutant mice. PLoS Genet 6(8):e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberger D, Reynolds DS, Ferris P, Lincoln R, Datta S, Stanley J, Paterson A, Dawson GR, Flint J (2003) Genetic mapping of variation in spatial learning in the mouse. J Neurosci 23:2426–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stottmann RW, Tran PV, Turbe-Doan A, Beier DR (2009) Ttc21b is required to restrict sonic hedgehog activity in the developing mouse forebrain. Dev Biol 335:166–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohl KP, Gallaugher L, Lynn A, Friedman L, Hill A, Singer JB, Lander ES, Nadeau J (2007) Sleep-related epilepsy in the A/J mouse. Sleep 30:169–176 [DOI] [PubMed] [Google Scholar]
- Stylianou IM, Christians JK, Keightley PD, Bunger L, Clinton M, Bulfield G, Horvat S (2004) Genetic complexity of an obesity QTL (Fob3) revealed by detailed genetic mapping. Mamm Genome 15:472–481 [DOI] [PubMed] [Google Scholar]
- Sudhof TC (2008) Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455:903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson DJ, Steshina EY, Wakenight P, Aldinger KA, Goldowitz D, Millen KJ, Chizhikov VV (2010) Phenotypic and genetic analysis of the cerebellar mutant tmgc26, a new ENU-induced ROR-alpha allele. Eur J Neurosci 32:707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, Shamel L, Herskovits AA, Portnoy DA, Cooke M, Tarantino LM, Wiltshire T, Steinberg BE, Grinstein S, Beutler B (2006) The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol 7:156–164 [DOI] [PubMed] [Google Scholar]
- Tafti M, Franken P, Kitahama K, Malafosse A, Jouvet M, Valatx JL (1997) Localization of candidate genomic regions influencing paradoxical sleep in mice. Neuroreport 8:3755–3758 [DOI] [PubMed] [Google Scholar]
- Takada T, Mita A, Maeno A, Sakai T, Shitara H, Kikkawa Y, Moriwaki K, Yonekawa H, Shiroishi T (2008) Mouse inter-subspecific consomic strains for genetic dissection of quantitative complex traits. Genome Res 18:500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Nishi A, Ishii A, Shiroishi T, Koide T (2008) Systematic analysis of emotionality in consomic mouse strains established from C57BL/6J and wild-derived MSM/Ms. Genes Brain Behav 7:849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Tomihara K, Shiroishi T, Koide T (2010) Genetic mapping of social interaction behavior in B6/MSM consomic mouse strains. Behav Genet 40:366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi KR, Sakuraba Y, Gondo Y (2007) Mutational pattern and frequency of induced nucleotide changes in mouse ENU mutagenesis. BMC Mol Biol 8:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot CJ, Nicod A, Cherny SS, Fulker DW, Collins AC, Flint J (1999) High-resolution mapping of quantitative trait loci in outbred mice. Nat Genet 21:305–308 [DOI] [PubMed] [Google Scholar]
- Taylor BA (1976) Genetic analysis of susceptibility to isoniazid-induced seizures in mice. Genetics 83:373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BA (1978) Recombinant inbred strains: use in gene mapping In: Morse HC (ed) Origins of inbred Mice: proceedings of a workshop, Bethesda, Maryland, 14–16 February. Academic Press, New York, pp 423–438 [Google Scholar]
- Taylor BA, Bedigian HG, Meier H (1977) Genetic studies of the Fv-1 locus of mice: linkage with Gpd-1 in recombinant inbred lines. J Virol 23:106–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BA, Meier H, Myers DD (1971) Host-gene control of C-type RNA tumor virus: inheritance of the group-specific antigen of murine leukemia virus. Proc Natl Acad Sci USA 68:3190–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BA, Tarantino LM, Phillips SJ (1999) Gender-influenced obesity QTLs identified in a cross involving the KK type II diabetes-prone mouse strain. Mamm Genome 10:963–968 [DOI] [PubMed] [Google Scholar]
- Taylor BA, Wnek C, Schroeder D, Phillips SJ (2001) Multiple obesity QTLs identified in an intercross between the NZO (New Zealand obese) and the SM (small) mouse strains. Mamm Genome 12:95–103 [DOI] [PubMed] [Google Scholar]
- Theodoratos A, Whittle B, Enders A, Tscharke DC, Roots CM, Goodnow CC, Fahrer AM (2010) Mouse strains with point mutations in TAP1 and TAP2. Immunol Cell Biol 88:72–78 [DOI] [PubMed] [Google Scholar]
- Tokuda S, Mahaffey CL, Monks B, Faulkner CR, Birnbaum MJ, Danzer SC, Frankel WN (2011) A novel Akt3 mutation associated with enhanced kinase activity and seizure susceptibility in mice. Hum Mol Genet 20(5):988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaih SW, Korstanje R (2009) Haplotype association mapping in mice. Methods Mol Biol 573:213–222 [DOI] [PubMed] [Google Scholar]
- Turri MG, Datta SR, DeFries J, Henderson ND, Flint J (2001a) QTL analysis identifies multiple behavioral dimensions in ethological tests of anxiety in laboratory mice. Curr Biol 11:725–734 [DOI] [PubMed] [Google Scholar]
- Turri MG, DeFries JC, Henderson ND, Flint J (2004) Multivariate analysis of quantitative trait loci influencing variation in anxiety-related behavior in laboratory mice. Mamm Genome 15:69–76 [DOI] [PubMed] [Google Scholar]
- Turri MG, Henderson ND, DeFries JC, Flint J (2001b) Quantitative trait locus mapping in laboratory mice derived from a replicated selection experiment for open-field activity. Genetics 158:1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turri MG, Talbot CJ, Radcliffe RA, Wehner JM, Flint J (1999) High-resolution mapping of quantitative trait loci for emotionality in selected strains of mice. Mamm Genome 10:1098–1101 [DOI] [PubMed] [Google Scholar]
- Umemori J, Nishi A, Lionikas A, Sakaguchi T, Kuriki S, Blizard DA, Koide T (2009) QTL analyses of temporal and intensity components of home-cage activity in KJR and C57BL/6J strains. BMC Genet 10:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Flint J, Mott R (2006a) Simulating the collaborative cross: power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics 172:1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J (2006b) Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet 38:879–887 [DOI] [PubMed] [Google Scholar]
- Wakeland E, Morel L, Achey K, Yui M, Longmate J (1997) Speed congenics: a classic technique in the fast lane (relatively speaking). Immunol Today 18:472–477 [DOI] [PubMed] [Google Scholar]
- Wang GL, Paterson AH (1994) Assessment of DNA pooling strategies for mapping of QTLs. Theor Appl Genet 88:355–361 [DOI] [PubMed] [Google Scholar]
- Wang J, Liao G, Usuka J, Peltz G (2005) Computational genetics: from mouse to human? Trends Genet 21:526–532 [DOI] [PubMed] [Google Scholar]
- Wang J, Williams RW, Manly KF (2003) WebQTL: web-based complex trait analysis. Neuroinformatics 1:299–308 [DOI] [PubMed] [Google Scholar]
- Watson J, Riblet R, Taylor BA (1977) The response of recombinant inbred strains of mice to bacterial lipopolysaccharides. J Immunol 118:2088–2093 [PubMed] [Google Scholar]
- Webb BT, McClay JL, Vargas-Irwin C, York TP, van den Oord EJ (2009) In silico whole genome association scan for murine prepulse inhibition. PLoS One 4:e5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner JM, Radcliffe RA, Rosmann ST, Christensen SC, Rasmussen DL, Fulker DW, Wiles M (1997) Quantitative trait locus analysis of contextual fear conditioning in mice. Nat Genet 17:331–334 [DOI] [PubMed] [Google Scholar]
- Wilkes JJ, Lloyd DJ, Gekakis N (2009) Loss-of-function mutation in myostatin reduces tumor necrosis factor alpha production and protects liver against obesity-induced insulin resistance. Diabetes 58:1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RT, Lim JE, Harr B, Wing C, Walters R, Distler MG, Teschke M, Wu C, Wiltshire T, Su AI, Sokoloff G, Tarantino LM, Borevitz JO, Palmer AA (2009) A common and unstable copy number variant is associated with differences in Glo1 expression and anxiety-like behavior. PLoS One 4:e4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Ma C, Casella G (2010) Statistical genetics of quantitative traits: linkage, maps and QTL. Springer, New York [Google Scholar]
- Xia Y, Won S, Du X, Lin P, Ross C, La Vine D, Wiltshire S, Leiva G, Vidal SM, Whittle B, Goodnow CC, Koziol J, EM YM, Beutler B (2010) Bulk segregation mapping of mutations in closely related strains of mice. Genetics 186:1139–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G, Harrison J, Clapcote SJ, Huang Y, Zhang JY, Wang LY, Roder JC (2010) A new Kv1.2 channelopathy underlying cerebellar ataxia. J Biol Chem 285:32160–32173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B, Willis-Owen SA, Fullerton J, Meesaq A, Deacon RM, Rawlins JN, Copley RR, Morris AP, Flint J, Mott R (2004) Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat Genet 36:1197–1202 [DOI] [PubMed] [Google Scholar]
- Yoon CK (1996) J. NIH Res. 8:23–24 [Google Scholar]
- Yoshiki A, Ike F, Mekada K, Kitaura Y, Nakata H, Hiraiwa N, Mochida K, Ijuin M, Kadota M, Murakami A, Ogura A, Abe K, Moriwaki K, Obata Y (2009) The mouse resources at the RIKEN BioResource center. Exp Anim 58:85–96 [DOI] [PubMed] [Google Scholar]
- Zeng ZB (1993) Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci USA 90:10972–10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Gershenfeld HK (2003) Genetic contributions to body weight in mice: relationship of exploratory behavior to weight. Obes Res 11:828–838 [DOI] [PubMed] [Google Scholar]