Abstract
Genetic screens in cultured human cells represent a powerful unbiased strategy to identify cellular pathways that determine drug efficacy, providing critical information for clinical development. We used insertional mutagenesis-based screens in haploid cells to identify genes required for the sensitivity to Lasonolide A (LasA), a macrolide derived from a marine sponge that kills certain types of cancer cells at low-nanomolar concentrations. Our screens converged on a single gene, LDAH, encoding a member of the metabolite serine hydrolase family that is localized on the surface of lipid droplets. Mechanistic studies revealed that LasA accumulates in lipid droplets, where it is cleaved into a toxic metabolite by LDAH. We suggest that selective partitioning of hydrophobic drugs into the oil phase of lipid droplets can influence their activation and eventual toxicity to cells.
INTRODUCTION
Small molecules derived from natural sources have been an invaluable resource for the discovery of new drugs and new tools to study a variety of cellular processes. Natural products have complex and diverse structures with multiple stereocenters, allowing them to interact with and alter the function of cellular proteins with high selectivity. However, understanding the proteins and pathways that mediate cellular responses to natural products can be challenging because of their low abundances and often difficult, multi-step chemical syntheses. Genetic screens in cultured human cells, now made routine by the use of haploid cell lines and CRISPR-mediated gene editing, have emerged as powerful methods to discover both the mechanism of action of drugs and the pathways that mediate drug uptake and metabolism. For anti-cancer applications, this information can identify cellular factors that mediate both de novo and acquired resistance and thus identify predictive biomarkers to select the tumor types and patients that would derive the greatest benefit from the drug.
Lasonolide-A (1) (hereafter LasA, Fig.1a) is a polyketide-derived macrolide isolated from a marine sponge (Forcepia) found in the Gulf of Mexico1–2. LasA has been considered a good candidate for development as an anti-cancer drug because it kills cancer cell lines at low nanomolar concentrations and has a unique cytotoxicity profile when tested against the National Cancer Institute (NCI-60) cell panel1. LasA has an unusual biphasic effect on cells. Within minutes, it causes loss of cell adhesion, cell blebbing, and premature chromosome condensation, effects that are reversible with drug withdrawal3–4. Longer treatment leads to irreversible toxicity. While the cellular target of LasA has not been conclusively identified, it triggers protein hyper-phosphorylation in cells and has been implicated in the activation of multiple kinases, including PKC, MAPKs and c-RAF3–5. Taking advantage of our recent total synthesis of LasA6, we used genetic screens in a haploid human cell line (Hap1)7–9 to uncover cellular mechanisms that mediate sensitivity to LasA. The results of our screen, with subsequent mechanistic follow-up, revealed that LasA accumulates in lipid droplets, where it is hydrolyzed into a toxic metabolite by an orphan serine hydrolase enzyme. We suggest that lipid droplets can influence the efficacy of small molecule drugs by providing both a depot for hydrophobic molecules and a complement of enzymes for drug metabolism. It may be possible to harness such lipid-droplet based mechanisms to increase both drug efficacy and selectivity.
Figure 1. Genetic screens in human haploid cells to identify genes required for Lasonolide-A (LasA) toxicity.
(a) Structure of (-) Lasonolide-A and its analogs, with the macrocyclic ring colored in black and the side chain colored in red. Dotted circles highlight the double bond that is endocyclic in LasA but exocyclic in Ces-73. (b) MTT assays for cell viability were used to determine the IC50 values for LasA and its analogs shown in (a). The IC50 values for LasA and Ces-73 are 22 ± 2 nM and 157 ± 11 nM respectively. (c) Volcano plot depicting the results of a haploid screen (conducted once, n=1) using cell survival as the selection in the presence of a concentration of LasA that killed >99% of cells. Each circle represents a gene, with the diameter scaled according to the number of independent retroviral insertions, plotted based on the p-value for enrichment of insertions in the drug-selected population over the control population (y-axis) and the bias toward inactivating intronic insertions (x-axis). Genes with a False Discovery Rate (FDR)-corrected p-value smaller than 0.01 are colored green, with the exception of the top hit in the screen (LDAH) which is colored pink. The entire dataset for the screen is provided in Supplementary Data 1. p-values were calculated using the one-sided Fisher exact test and corrected for multiple testing using the Benjamini and Hochberg method. (d) LasA sensitivity was assessed in LDAHnull cell lines using an MTT assay. Immunoblot (representative of three independent repeats) showing abundance of LDAH protein in two clonal cell lines (LDAHnull1 and LDAHnull2) carrying CRISPR/Cas9-mediated frameshift mutations in LDAH generated using guide RNAs targeting two different exons (Supplementary Fig.2b; uncropped immunoblot is in Supplementary Fig. 10). IC50 values for WT, LDAHnull1 and LDAHnull2 cell lines were 21 ± 2 nM, 636 ± 23 nM and 437 ± 17 nM, respectively. For panels (b) and (d) the circles denote the mean (+/- S.D. from n=4 independent samples). Error bars are often smaller than the diameter of the circles used to denote the mean.
RESULTS
Screens to identify genes required for LasA toxicity
LasA6 was toxic to Hap1 cells, with an IC50 (the concentration causing a 50% inhibition of viability compared to untreated cells) of ~20 nM (Fig.1b). LasA is composed of a macrocyclic ring and a side chain (Fig.1a). A series of Lasonolides (C-G) have been purified from Forcepia that have the same macrocyclic ring but vary in the structure of the side chain2. The toxicity of LasA was sensitive to its chemical structure, suggesting it interacts with specific protein targets in cells. Movement of a double bond from an endocyclic to an exocyclic position in Ces-73 (2) increased the IC50 to ~160nM and expansion of the macrocycle in Ces-24a (3) and Ces-24b (4) abolished toxicity (Figs.1a and 1b).
We conducted genetic screens in Hap1 cells to identify genes required for cellular sensitivity to LasA and Ces-73, similar to how we previously identified genes required for cellular sensitivity to doxorubicin9. Hap1 cells have a near-haploid genome which enables the generation of null alleles for most genes by insertional mutagenesis10. A library of ~100 million mutagenized Hap1 cells was created by random insertions of a gene-trap (GT) cassette delivered using a retrovirus. The entire population was then treated with either LasA at 80 nM or Ces-73 at 400 nM, concentrations expected to kill >99% of cells (the IC99). Surviving cells, presumably carrying inactivating insertions in genes required for drug sensitivity, were isolated and the positions of retroviral insertions mapped by deep sequencing (Supplementary Fig.1). Both screens converged on the discovery that inactivating insertions in LDAH (Lipid Droplet Associated Hydrolase or C2ORF43) made cells resistant to LasA (Figs. 1c and Supplementary Fig. 2a).
LDAH is required for LasA toxicity
We validated the results of the screen by using two independent guide RNAs (sgRNAs) to introduce inactivating frameshift mutations in LDAH (Supplementary Fig. 2b). Both clonal LDAH−/− cell lines showed markedly reduced sensitivity to LasA: the IC50 of LasA was >20-fold higher in LDAH−/− Hap1 cells compared to wild-type (WT) Hap1 cells (Fig. 1d). Importantly, the sensitivity to LasA was restored by stable re-expression of LDAH, excluding off-target effects of CRISPR editing (Fig. 2). Increasing the abundance of LDAH protein beyond that seen in wild-type cells did not further increase sensitivity to LasA, suggesting either that other cellular factors become limiting for LasA toxicity above a threshold of LDAH activity or that over-expression of LDAH leads to secondary effects that mitigate toxicity (Supplementary Fig. 3).
Figure 2. The serine hydrolase activity of LDAH is required for LasA toxicity.
(a) LDAH contains an α/β hydrolase domain (red) with a typical catalytic triad: S139 within a GxSxG motif, D271 and H300 (circled). A hydrophobic hairpin motif that mediates localization to lipid droplets (colored yellow) is present as an insertion in the α/β fold. Numbers refer to amino acid positions. (b) Abundance of LDAH protein in WT cells, LDAH−/− cells (hereafter called LDAHnull) and LDAHnull cells re-expressing wild-type LDAH (LDAHWT) or a mutant (LDAHS139C) in which the catalytic triad serine (shown in A) was changed to cysteine. LDAHWT and LDAHS139C were expressed either as untagged proteins or fused to GFP (uncropped immunoblots are in Supplementary Fig. 10). Immunoblot was repeated three times with similar results. (c) LasA sensitivity of the indicated cell lines was assessed using an MTT assay. IC50 values are as follows: 21 ± 1 nM for WT, 505 ± 22 nM for LDAHnull, 56 ± 3 nM for LDAHnull;LDAHWT, 60 ± 3 nM for LDAHnull;LDAHWT-GFP, 582 ± 25 nM for LDAHnull;LDAHS139C and 599 ± 24 nM for LDAHnull; hLDAHS139C-GFP. Circles denote the mean value (+/- S.D. from n=3 independent samples).
We confirmed that the requirement of LDAH for LasA toxicity was not restricted to Hap1 cells. CRISPR-mediated elimination of LDAH protein from MCF7 breast cancer cells, A549 lung cancer cells and H1650 and RKO colon cancer cells also resulted in decreased sensitivity to LasA (Supplementary Fig. 4). Across the NCI-60 panel of cancer cell lines, we found a statistically-significant direct correlation between the mRNA levels of LDAH and sensitivity to LasA3 (Supplementary Fig.5).
LDAH belongs to a family of ~117 metabolite serine hydrolases in humans that function as esterases, lipases, peptidases or amidases11–12. Transcriptional profiling by RNAseq9 revealed that the mRNA of 99 of these genes was detectable in Hap1 cells (Reads per kilobase of transcript, per million or RPKM>0), with 74 genes being expressed at RPKM>113 (Supplementary Fig.6). While mRNA expression does not guarantee protein expression (and protein expression does not guarantee biochemical activity), the broad representation of metabolite serine hydrolases in the Hap1 transcriptome highlights the striking specificity of the requirement of LDAH for LasA sensitivity.
The metabolite serine hydrolases are characterized by an α/β-hydrolase fold and a Ser-His-Asp catalytic triad14–16 (Fig. 2a). While the physiological function of LDAH is unknown, its loss has been linked to an increased risk of prostate cancer17. Weak enzymatic activity of LDAH was reported against cholesteroyl esters in vitro, but a Ldah−/− mouse was viable and had no defects in cholesterol ester or triacylglycerol metabolism14, 18. While its endogenous substrate is unknown, LDAH is likely to be enzymatically active because the three catalytic triad residues are conserved in its sequence: a GXSXG motif harboring a putative nucleophilic serine (S139), aspartate (D271) and histidine (H290)15 (Fig. 2a). To test if the catalytic activity of LDAH was required for its role in sensitizing cells to LasA, we mutated the conserved Ser139 in LDAH to cysteine (Cys). LDAH−/− cells stably expressing LDAHS139C (Fig.2b) remained resistant to LasA, showing that LDAH catalytic activity was essential for its ability to confer sensitivity to LasA (Fig. 2c). In an important control, the Ser139Cys mutation does not change the abundance of the LDAH protein (Fig. 2b) and also does not change its localization to the surface of lipid droplets14.
LDAH hydrolyzes LasA to its active metabolite LasF
Why would a serine hydrolase be required for sensitivity of cells to LasA? The fact that LDAH−/− cells and Ldah−/− mice are viable18 without any growth defects implies that LasA does not kill cells by blocking LDAH function. Thus, our loss-of-function screens failed to identify the protein target of LasA, perhaps because this target is redundant or required for cell growth or viability. Instead, we considered the possibility that the hydrolytic activity of LDAH may be required for LasA uptake or LasA activation. LasA has two ester bonds, one in the macrocycle and one in the side chain, that could be substrates for a protein with serine hydrolase activity (Fig. 3a). Since cleavage of the ester bond in the macrocyclic ring is likely to destroy activity (based on structure-activity data presented in Fig.1b), we focused on the ester bond in the side chain (Fig. 3a). Cleavage of this bond would leave a carboxylic acid side chain, which would be charged when unprotonated. In fact, this potential cleavage product was previously isolated from Forcepia and named Lasonolide-F (5) (LasF)2. We tested the hypothesis that LDAH cleaves LasA into LasF in cells. Since we failed to reconstitute the enzymatic activity of purified LDAH in vitro, we developed a quantitative mass spectrometry assay to measure the levels of the parent LasA and its cleavage product LasF in cell extracts (See Methods). In extracts from WT Hap1 cells exposed to LasA, the abundance of cleaved LasF was ~10-fold higher than LasA (Fig. 3b). Conversely, in Hap1 cell lines lacking LDAH, the ratio was reversed— the parent LasA was ten-fold more abundant than its cleavage product LasF (Fig. 3b). The ratio of LasA/LasF, an internally controlled metric that can be compared across cell lines, increased by nearly 100-fold when LDAH was disrupted in Hap1 cells (Fig.3c). Re-expression of wild-type LDAH, but not its catalytically inactive Ser139Cys variant, restored the LasA/LasF ratio to that seen in wild-type cells (Figs. 3b and 3c). These data show that the enzymatic activity of LDAH is required for the metabolism of LasA into LasF. The total abundance of all Lasonolide species (LasA+LasF) did not change much when LDAH was depleted (Fig. 3b), so drug uptake cannot explain the 20-fold greater potency of LasA in WT cells compared to LDAH−/− cells.
Figure 3. LDAH converts LasA to LasF by cleaving its side chain.
(a) LDAH may catalyze the hydrolysis of LasA to LasF by cleaving the ester bond (marked 2) in the side chain. (b) The abundances of LasA and LasF, normalized using the genomic DNA content in each sample, were measured by quantitative mass spectrometry (see Methods). (c) The ratio of the abundance of LasA to LasF for each independent experiment is depicted (on a logarithmic scale) for the four indicated cell lines. (d, e) Sensitivity to LasA and LasF in wildtype (WT) and LDAHnull cells was assessed using an MTT assay. IC50 values for LasA (d) in WT and LDAHnull cell lines are 19 ± 1 nM and 586 ± 34 nM respectively and for LasF (e) are 2.4 ± 0.1 μM and 4.3 ± 0.4 μM respectively. Circles denote the mean value (+/- S.D. from n=3 independent samples). Significance in b and c was tested using a two-tailed, unpaired t-test and is indicated as *** (p<0.001) or ** (p<0.01). The circles in b and c denote data from four different experiments in each of the indicated cell lines.
These results support the hypothesis that LDAH is required for cell sensitivity to LasA because it converts the pro-drug LasA into the active drug LasF, the molecule that kills cells rather than LasA itself. The log of the octanol/water partition coefficient of a drug (called the LogP) is commonly used to evaluate the suitability of small molecules to permeate cell membranes. The predicted LogP of LasA is 5.98, while the LogP of the deprotonated, charged form of LasF is predicted to be much lower at -0.55 (the LogP of the protonated form would be ~2.29). The high hydrophobicity of LasA implies that it would readily partition into the plasma membrane but would likely accumulate in the membrane and other lipophilic compartments rather than crossing into the cytoplasm. In contrast, the charged form of LasF may have difficulty partitioning into the plasma membrane and hence in permeating into cells. Indeed, LasF, when directly applied to Hap1 cells, was much less potent than LasA (Figs. 3d and 3e), a result consistent with published data from cancer cells lines2. Importantly, the IC50 of LasF was minimally affected by the depletion of LDAH, supporting the model that the function of LDAH is to convert LasA to LasF (Figs. 3d and 3e).
LasA toxicity requires membrane localized LDAH
To understand how LDAH may gain access to the hydrophobic LasA for cleavage, we first looked at the subcellular localization of Green Fluorescent Protein (GFP) tagged LDAH. LDAH-GFP was functional because it could restore LasA toxicity to LDAH−/− cells (Figs. 2b and 2c). As reported previously15, LDAH was found localized on the surface of lipid droplets and in the endoplasmic reticulum (ER) (Fig. 4a, Supplementary Fig. 7 and Supplementary movie 1). Like some (Class I) lipid droplet proteins, LDAH is thought to move from the ER to the surface of lipid droplets as they emerge from the cytoplasmic leaflet of the ER15, 19. These observations suggested the possibility that LasA activation by cleavage to LasF occurs at a membrane interface—either at the ER membrane or the surface of lipid droplets. To test the idea that membrane localization of LDAH was required for its ability to activate LasA, we took advantage of the previous observation that LDAH inserts into the phospholipid monolayer surface of lipid droplets using a hydrophobic hairpin motif (Fig. 4b)15, 18. Two different truncating mutations that removed portions of this motif eliminated LDAH localization to lipid droplets while maintaining LDAH protein expression (Figs. 4c–4d). Both mutations completely eliminated the ability of LDAH to sensitize cells to LasA (Fig.4e), suggesting that localization of the enzyme to membranes was required for its ability to cleave LasA to LasF.
Figure 4. Membrane localization of LDAH is required for LasA sensitivity.
(a) LDAH-GFP (green) is localized on the surface of lipid droplets, marked by the LipidTox stain (red). Images at two different magnifications are shown in the top (scale bar 10μm) and bottom panel (scale bar 2μm). Localization in live cells is shown in Supplementary Fig.7. Experiment was repeated three times with similar results. (b) The hydrophobic hairpin motif (yellow) in LDAH-GFP was interrupted by replacing two regions (LDAH176–219 or the shorter LDAH188–203) with a flexible linker sequence. These LDAH-GFP mutant proteins were stably expressed in LDAHnull cells and their abundances were assessed by immunoblotting (c) and their localization to lipid droplets was assessed by immunofluorescence (d, scale bar 10μm). Experiments were repeated three times with similar results. Uncropped immunoblots are in Supplementary Fig.10. (e) LasA sensitivity was measured in the indicated cell lines by an MTT assay. IC50 values are as follows: 440 ± 32 nM for LDAHnull, 10 ± 1 nM for Hap1WT, 19 ± 1 nM for LDAHnull;LDAHWT-GFP, 367 ± 26 nM for LDAHnull;LDAHΔ176–219-GFP, 341 ± 29 nM for LDAHnull;LDAHΔ188–203-GFP. Circles denote the mean (+/- S.D. from n=4 independent samples). (f) The IC50 values of LasA, measured by the MTT assay, in cells exposed to DGAT inhibitors (20μM T-863 and 10 μM PF06424439), Triacsin-C (1 μM) or oleic acid (200 μM, right graph). Circles denote mean IC50 (from n=3 independent samples) and error bars denote the standard error of the curve fit used to calculate the IC50. Significance in (f) was determined by a two-tailed, unpaired t-test and is indicated as ** (p<0.01) or * (p<0.05).
Lipid droplets promote the toxicity of LasA
We next tested the concept that lipid droplets play a role in the activation of LasA by LDAH. First, we used established manipulations to increase or decrease lipid droplet number in cells. Oleic acid was used to enhance the lipid droplet content of cells, while treatment with Diacylglycerol O-Acyltransferase (DGAT) inhibitors (T-863 and PF06424439) or a long chain fatty acid acyl-CoA synthetase inhibitor (Triacsin C) decreased the lipid droplet content of cells (Supplementary Fig. 8). The IC50 of LasA correlated with cellular lipid droplet content—increasing the number of lipid droplets with oleic acid increased the potency of the drug while decreasing the lipid droplet content by inhibiting triglyceride and cholesterol ester synthesis reduced the potency of the drug by 2–3-fold (Fig. 4f).
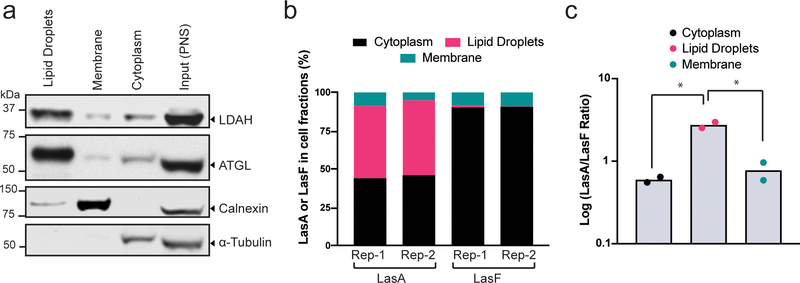
We also analyzed the subcellular distribution of LasA and LasF by cell fractionation. Post-nuclear supernatants derived from LasA treated cells were separated by discontinuous density gradient centrifugation into three fractions20: lipid droplets, which float due to their uniquely lower density in comparison to other cellular compartments, cytoplasm, and cellular membranes, including the ER (Supplementary Fig. 9). The quality of the fractionation was established by immunoblotting using antibodies against markers for each compartment (Fig. 5a): ATGL, a commonly used marker of lipid droplets, and LDAH were predominantly found in the lipid droplet fraction, while Calnexin, an ER marker, was found in the membrane fraction. LasA and LasF were measured in each fraction by our mass spectrometry assay (Fig.3). LasA was equally distributed between the cytoplasm and the lipid droplet fraction, while LasF was mostly found in the cytoplasm (Fig.5b). The abundances of both LasA and LasF were similar in the membrane fractions. As a result, the LasA/LasF ratio is the highest in lipid droplets and lowest in the cytoplasm (Fig.5c). Thus, amongst membrane bound compartments in cells, LasA showed a striking propensity to accumulate in lipid droplets.
Figure 5. Sub-cellular distribution of LasA and LasF.
(a) An equal proportion of each sub-cellular fraction (see Supplementary Fig. 9) was used to assess the abundance of LDAH and established protein markers of the cytoplasm (α-Tubulin), ER (Calnexin), or lipid droplets (ATGL) by immunoblotting (uncropped immunoblots are in Supplementary Fig. 11). Experiment was repeated twice with similar results. (b) The distribution of total LasA and LasF in each of the three fractions as measured by quantitative mass spectrometry. Rep-1 and Rep-2 show results from two different experiments. (c) The ratio of the abundance of LasA to LasF in the lipid droplet, cytoplasm and membrane fraction is shown on a logarithmic scale. Each dot represents a different experiment (n=2). Significance in (c) was determined by a two-tailed, unpaired t-test and is indicated by * (p<0.05).
DISCUSSION
Our results uncovered an unexpected cellular route for the activation of the potent cytotoxic macrolide LasA (depicted in Fig. 6). We propose that LasA partitions into the plasma membrane and then gains access to lipid droplets, either directly or via the membrane of the ER, since the phospholipid monolayer that invests lipid droplets is derived from the cytoplasmic leaflet of the ER. How LasA moves from the plasma membrane, the first barrier for any drug, to the ER or to lipid droplets remains unknown but we speculate that this may occur at inter-organelle contact sites. Importantly, LasA seems to preferentially accumulate in lipid droplets over other membrane compartments (Fig. 5b) while LasF, the cleavage product of LasA and likely the active metabolite, was mostly localized in the cytoplasm. Given the highly hydrophobic nature of LasA (calculated LogP~6), the presence of LasA in the cytoplasm (Fig. 5b) was unexpected, though we cannot exclude artefacts caused by organelle damage and consequent drug leakage during the inherently disruptive process of cell fractionation. Alternatively, LasA may bind to cytoplasmic proteins that shuttle it to lipid droplets. The mechanism for why LasA accumulates in lipid droplets is unknown, but its highly hydrophobic nature may favor selective partitioning into the oil-like hydrophobic core composed of neutral lipids like triglycerides and cholesterol esters. Cleavage of LasA to the more polar LasF (Fig. 3a) would undoubtedly reduce its propensity to partition in the oil-phase core of lipid droplets and hence promote its release into the cytoplasm.
Figure 6. A proposed mechanism for the uptake of LasA and its activation by LDAH-mediated cleavage.
LasA traverses the plasma membrane and accumulates in lipid droplets, likely by selective partitioning into its oil-like core (light yellow). LDAH is also found in the ER, but it accumulates to the highest levels on the surface of lipid droplets proximal to the reservoir of LasA in the lipid droplet core. LDAH can likely cleave LasA to LasF either at the ER or lipid droplet surface.
The accumulation of both LasA and LDAH in lipid droplets (Figs. 4a and 5b) may accelerate the LasA to LasF cleavage reaction by concentrating the enzyme and its substrate in close proximity. This property of concentrating reactants by selective partitioning is emerging as one of the key biochemical functions of phase-separated compartments in cells14, 21. More generally, lipid droplets and other phase separated organelles may function as reservoirs for drug-like small molecules in cells. For example, a recent study found that the accumulation of a lipophilic antibiotic (bedaquiline) in lipid droplets enhances its antibacterial action against Mycobacterium tuberculosis 22. In both our studies on LasA and the published work on bedaquiline, lipid droplets contribute to drug efficacy but are not absolutely required: the IC50 of LasA is reduced by 2–3-fold by treatments that deplete lipid droplets (Fig. 4f) but by ~20 fold when LDAH is ablated (Fig.1d). Thus, LDAH is likely capable of converting LasA to LasF even in the absence of lipid droplets. Since membrane localization of LDAH is required for cell sensitivity to LasA (Fig. 4e), it may also be able to activate LasA at the surface of the ER. More generally, our work shows that the sub-cellular localization or accumulation of drugs can influence their efficacies. A challenge that must be overcome is to develop better analytic methods, such as ion microscopy or imaging mass spectrometry, to assess the distribution of small molecules in cells at sub-cellular or even sub-organelle resolution22–23.
Lipid droplets are dynamic organelles that accumulate in diverse pathological states, especially those linked to inflammatory stress24. Activated myeloid cells, such as the “foamy” macrophages seen in atherosclerotic lesions, are characterized by large numbers of lipid droplets25. Indeed, this may be the reason bedaquiline is particularly effective against mycobacteria that have infected macrophages22. Pathological lesions of Alzheimer’s disease have also been noted to contain high levels of lipid droplets for over a century26. Lipid droplets are increased in several types of cancer cells27–28, including clear cell renal cancer29, prostate cancer30 and breast cancer31, and this increase has been correlated with tumor aggressiveness32. Nutrient deprivation, hypoxia and chemotherapy exposure, in addition to inflammatory stress, have been postulated to promote lipid droplet biogenesis in cancer cells. Cancer types with increased lipid droplet number may be particularly vulnerable to drugs like LasA that exploit a lipid-droplet based mechanism of toxicity. Medicinal chemistry strategies to modify small molecules to drive their accumulation in lipid droplets or to take advantage of the enzymes found in lipid droplets could allow this approach to be applied to other small molecule therapeutics for cancer, inflammation and neurodegeneration.
The use of alkyl esters to make prodrugs with improved cell permeability is a well-established strategy: nearly one-third of the approved prodrugs in the last decade utilize cleavage of an ester bond for drug activation33. In most cases, these esterified pro-drugs are activated promiscuously by the many esterases present in cells. In contrast, the cleavage of LasA is not a generic ester hydrolysis reaction but instead is highly selective in its requirement for LDAH. This selectivity suggests that it may be possible to achieve significant tissue or tumor selectivity in prodrug design by matching the prodrug and structure of the ester side-chain to the substrate specificity of a hydrolase expressed specifically in the target tissue. In summary, our work highlights the power of genetic screens to provide actionable insights, not just into drug targets, but also into the mechanisms of drug uptake and activation.
ONLINE METHODS
Cell lines
The Hap1 cell line10 was kindly provided by Dr.Thijn Brummelkamp, Netherlands Cancer Institute, and validated by confirming its haploid genomic DNA content at regular intervals during the screens. The 293FT cell line used to generate high-titer lentiviruses was obtained from ThermoFisher Scientific (Grand Island, NY); the MCF7, H1650, A549 and RKO cell lines were purchased from ATCC (Manassas, VA). Purchased cells lines came with a certificate of authenticity. We did not further validate cell lines but used them at passages <10. All cell lines were confirmed to be negative for Mycoplasma contamination. Hap1 cells were grown in IMDM with 10% FBS and 6mM L-Glutamine; 293FT in DMEM with 10% FBS, 1 mM Sodium Pyruvate, 1x MEM-NEAA and 2 mM L-Glutamine; MCF7 cells in RPMI with 10% FBS and 2mM L-Glutamine.
Constructs and alleles
Null alleles for LDAH were constructed using the CRISPR/Cas9 system. For Hap1 cells the oligos encoding the guide RNAs were cloned into pSpCas9(BB)-2A-GFP (PX458, Addgene Plasmid #48138) kindly provided by Dr. Feng Zhang34. The sequences for target sites for the two sgRNAs are Guide1: GACCTACTTACCGGGAGCTC and Guide2: GTAGCTAATGCTGCCTACCT. Single cells were sorted using flow cytometry, expanded and clones bearing null alleles were identified by Sanger sequencing and immunoblotting. For gene disruption MCF7 cells, the following four gRNAs targeting LDAH were introduced into LentiCRISPR v2 (Addgene Plasmid #52961), kindly provided by Dr. Feng Zhang35 for lentiviral-mediated delivery. Lenti-CR1:CAAGTCCTTGCTAATCAGAA, Lenti-CR2:CCTGAGAACTCATGTGCCAA, Lenti-CR3:CGAATGTCTGAGTCACCCAA, Lenti-CR4:GAAGATTCTTACAACATCAG. For gene disruption in H1650, A549 and RKO cell lines, Lenti-CR2 was used.
LDAH constructs were based on the human LDAH gene purchased from GeneCopoeia14. LDAH-GFP fusion constructs were created using Gibson assembly and S139C mutants were made using site-directed mutagenesis (Quickchange). LDAH mutants lacking the lipid droplet localizing hairpin motifs (Fig.4B) were made using Gibson assembly where the residues from 176–219 or 188–203 were replaced with a linker sequence “SSSGGGGSGGGGS”. Mutants were subcloned into pLenti-CMV-Puro-DEST36 (Plasmid #17452) from Addgene kindly provided by Dr. Eric Campeau for stable expression. Lentivirus (generated in 293FT) infected cells were selected with 2 μg/mL of puromycin for 2 weeks.
Chemical Compounds
Synthesis and characterization of LasA, Ces-73, Ces-24a and Ces-24b have been described previously6, 37. LasA, Ces-73, Ces-24a and Ces-24b are annotated as (-)-lasonolide-A, compounds 106, 105 and 104 (respectively) in these prior manuscripts. ChemDraw Professional v.16 was used to calculate LogP values of LasA and LasF. T-863 (≥98%), PF-06424439 (≥98%) and Triacsin-C (≥98%) were purchased from Milipore Sigma (St. Louis, MO).
To generate LasF from LasA at microscale, Tetrakis(triphenylphosphine)palladium(0) (1.0 equiv.) was added to a solution of lasonolide-A (LasA) (0.1 mg, 0.14 μM) and morpholine (10.0 equiv.) in THF (0.5 mL) at room temperature. After 30 minutes, the reaction was concentrated under a stream of nitrogen. The LC-MS of the residue confirmed full conversion of LasA into LasF (LasF ~ 98.3% pure) and this material was used in subsequent experiments without further purification. HRMS: (ESI) calcd for C33H47O9 [M+H]+ 587.3220 found 587.3332.
Haploid Genetic Screens
Methods for the execution of haploid genetic screens using insertional retroviral mutagenesis and the bioinformatic pipeline to map the distribution of these insertions in the genome have been described previously8–9. One hundred million mutagenized Hap1 cells were treated with LasA (80 nM) or Ces-73 (400 nM) for a total of 8 days, replenishing the media after the first two days only with fresh LasA or Ces-73. Surviving cells were allowed to recover and expand for 1 week in the absence of LasA or Ces-73 prior to harvest. Genome-wide mapping of insertions was performed in a pool of 30 million LasA/Ces-73-resistant cells and compared to insertions mapped in an equal number of cells from the mutant library prior to drug selection. Results of the screen were analyzed as detailed previously9–10 and the computational pipeline is available on github: https://github.com/RohatgiLab/BAIMS-Pipeline.
Cytotoxicity (MTT) assays
20,000 cells were plated in each well of a 96 well plate, allowed to grow for 24 hours and then treated with various concentrations of LasA, Ces-24a, Ces-24b, Ces-73 or LasF in replicates; the concentration of the solvent (DMSO) in which these compounds were dissolved was normalized across all wells to 0.1% (v/v). Assays were performed after either short-term drug exposure (30 min), to measure the known immediate effects on LasA on cell attachment, or after a long-term drug exposure (45 hours) to measure viability. After washing plates to remove unattached or dead cells, the assay was performed by adding 10 μL of 5 mg/mL MTT reagent to each well. Cells were incubated for 3 hrs with the MTT reagent, media was removed and 50 μL of DMSO was added to each well to solubilize the purple colored formazan. Absorbance was read at 570 nm on Biotek Synergy HT microplate reader.
For inhibitor studies, cells were treated with a combination of T-863 (20 μM) and PF06424439 (10 μM), Triacsin-C alone (1 μM) or Oleic Acid (200 μM ) for 24 hrs prior to LasA exposure. For oleic acid treatment, 20 μL of a 200 mM stock of Oleic Acid in ethanol was first added to 2 mL 10% BSA (w/v) and incubated at 37°C for 30 min until the solution cleared. This mixture was added to 18 mL normal growth medium to achieve Oleic Acid concentration of 200 μM and BSA fraction of 1% (w/v).
Immunoblotting
Cells were scraped into ice-cold phosphate buffered saline (PBS) containing 1x Sigma Fast Protease Inhibitor Cocktail (Roche) and collected as a pellet by centrifugation (500xg, 5 min, 4°C). Cells were lysed (45 min., 4°C) by agitation in modified RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2% NP-40, 0.25% Deoxycholate, 0.1% SDS, 1mM NaF, 1mM DTT, 1X SigmaFast, 10% glycerol), followed by centrifugation at 20,000xg for 30 min. at 4°C and determination of the total protein concentration in each lysate using the Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). Equal amounts of each lysate (50 μg) were fractionated by 8% PAGE, transferred to nitrocellulose membrane and blocked with 0.1% Casein in 0.2x PBS for 1 hour at room temperature. Membranes were incubated with protein-specific antibodies overnight at 4°C followed by 1 hour treatment with secondary antibodies prior to quantification using chemiluminescence or LiCOR Odyssey. Antibodies used are as follows: anti-LDAH (described previously14); anti-GFP (NB600–308) from Novus Biologicals, Centennials, CO; anti-Calnexin (ADI-SPA-865-D) from Enzo Life Sciences, Farmingdale, NY; anti-ATGL (2138S) from Cell Signaling Technology, Danvers, MA; anti-αTubulin (T6199) from Sigma-Aldrich, St. Louis, MO.
Measurement of LasA and LasF in cell extracts by quantitative mass spectrometry
Sample Preparation
Cells expressing LDAH variants grown in 15 cm culture plates to ~80% confluency were treated with 200 nM LasA for 1 hour and washed with cold PBS. Cells from each 15cm plate were scraped into 5 mL of a Methanol/Chloroform (9:1) mixture (5 mL) in the cold room, transferred to a 15 mL tube, mixed by thorough pipetting, and centrifuged at 2000xg to remove insoluble debris. The supernatant was transferred to a 20 mL scintillation vial and concentrated under vacuum to remove the methanol/chloroform mixture. The samples were stored at -80°C prior to analysis by liquid chromatography coupled to mass spectrometry (LC-MS).
Sub-cellular fractionation studies (Fig.5) were performed with cells grown in three 15 cm plates and treated with 200 μM Oleic acid for 24 hrs followed by LasA (200 nM) for 1 hour prior to harvest. Cells were washed with ice-cold PBS and harvested by scraping in 1 mL of Phosphate Buffered Saline (PBS) per 15 cm plate into a 15 mL tube. The cells were pelleted by centrifugation and resuspended at 4°C in HLM (Hypotonic Lysis Medium) buffer (20mM Tris, pH 7.4, 1 mM EDTA, 10 mM NaF) for 15 min. The cells were lysed by subjecting them to 20 strokes in a Dounce homogenizer using tight pestle, followed by two cycles of centrifugation (1000xg for 10 min) to pellet nuclei and large debris. The resultant post-nuclear supernatant (PNS) was brought to 20% sucrose (w/w) by supplementing from 60% (w/w) sucrose stock and then layered at the bottom of an ultracentrifuge tube (Beckman, Ultra-clear 16×102 mm). The PNS (2.5 ml in 20% sucrose) was overlaid sequentially with 3 ml of 5% sucrose in HLM and finally with HLM solution lacking any sucrose (Fig.5A). After centrifugation in (SW28 rotor) at 120,000xg for 1 hour at 4°C, the lipid-droplet layer on the top of the gradient was removed with a tube slicer (Beckman). The remaining solution in the tube was taken as the cytoplasm and the membrane-containing pellet was collected separately in 2 mL HLM (Fig.5A). The entirety of the three cell fractions (lipid droplet, membrane and cytoplasm) were extracted using a chloroform/ethyl acetate (1:1) mixture and the organic phase was transferred to a 20 mL scintillation vial for solvent removal under vacuum. Samples were stored at -80°C prior to analysis by LC-MS.
To fully recover the samples prior to mass spectrometry, each scintillation vial was washed down with 1mL of methanol, sonicated for 1 minute, transferred to an eppendorf vial and evaporated to dryness. All standards and samples were reconstituted in 150 uL methanol and transferred to autosampler vials.
Liquid chromatography and mass spectrometry
Samples were analyzed using a 1290 Infinity II UPLC integrated with a 6560 IM-QTOF (Agilent Technologies). Liquid chromatography was performed on a 2.1 × 50 mm Acquity UPLC BEH C18 column (1.7 μm particle size) with gradient elution at a flow rate of 0.50 ml/min and temperature of 35°C. Mobile phases were 50 mM NH4Ac in water (A) and 0.1% formic acid in acetonitrile (B). Gradient elution profile: initial hold at 35% B for 2 minutes, followed by a linear gradient of 35%-95% B in 3 minutes, then hold at 95% B for 1 minutes before equilibrating back to 35% B; total run time was 7 minutes. The mass spectrometer acquired full scan (m/z 100 – 1700) using dual AJS ESI source in negative mode. While LasA and LasF ionize in both positive and negative modes, higher ionization efficiency was achieved in negative mode. Measurements were done at MS1 level (full mass scan). To maximize LC/MS method sensitivity, LasA (HAc adduct, M+HAc-H; m/z=755.01) and LasF (M-H; m/z=585.3) were monitored using targeted EIC channels collecting full scan on the parent and metabolite ions, with 1 m/z isolation widths. To quantify the amount of LasA/LasF in cell extracts, we generated a standard curve using external standards. Varying (known) amounts of purified LasA and LasF were quantified using the same LC-MS conditions as used for the cell extracts and linear regression was used to find the best-fit line, which was then used to estimate the amount of LasA/LasF in cell extracts. LOQ for LasA = 34 ng/mL, LOD for LasA ~ 17 ng/mL; LOQ for LasF = 26 ng/mL and LOD for LasF ~ 10 ng/mL.
Amount of genomic DNA present in the cell extracts was taken as a measure of the number of cells to normalize the measurements of LasA and LasF across samples. (Note that the LasA/LasF ratio is an internally controlled value that should not depend on the number of cells harvested for each sample and we present data in both ways in Fig.3B and 3C). For normalization, genomic DNA was extracted and quantitated from 1/10’th of each cell sample; the remaining 9/10’th of each sample was used for LasA and LasF extraction.
Analysis of NCI-60 panel cell lines
The LasA toxicity data in NCI-60 cell lines was obtained from a previously published3 study that reported data from the NCI Therapeutics Program where LasA toxicity was measured in two similar experiments NSC: D-674673-X/0–1/17, Experiment ID: 9502RM14 and NSC: D-674673-X/0–1/35, Experiment ID: 9412MD01. The GI50 values from both the experiments were averaged for each cell line. NCI defines the GI50 as the drug concentration resulting in a 50% reduction in the net protein increase (as measured by Sulforhodamine B staining) in control cells during the drug incubation. LDAH expression levels from RNAseq were obtained from Broad Institute’s Cancer Cell Line Encyclopedia (CCLE). We were able obtained expression data for 45 cell lines out of the NCI-60 panel for which LasA toxicity data was also available. The LasA GI50 was plotted against the LDAH RPKM value and Pearson Correlation Coefficient was calculated using Graphpad Prism 8.
RNAseq analysis of serine hydrolases in Hap1 cells
Hap1 RNAseq data has been reported in our previous publications9 (NCBI-GEO-GSE75515). The RPKM values representing mRNA expression levels were obtained using Partek Flow software. The RPKM values of 117 metabolic serine hydrolases reported earlier11–12 were plotted using Graphpad Prism8.
Fluorescence Microscopy
The cells expressing eGFP-tagged LDAH were used to capture fluorescent images to study LDAH localization. The cells (40–80×103 cells per well) were plated on ibidi μslide-8well plate. After 24 hours the live cells were imaged using Leica TCS SP8 confocal imaging system equipped with a 63× oil immersion objective and captured using the Leica Application Suite X software. For LipidTOX (Deep Red) staining, 40,000 Hap1 cells were plated in each well of an ibidi μslide-8well plate. Cells were incubated with various small molecules for 24 hours prior to fixation in 4% PFA for 10 min at room temperature. 200 μL LipidTOX (1:1000 dilution in PBS) was added to the cells. The images were captured 45 min. after addition of LipidTOX and processed using Fiji (ImageJ) software.
Statistics
Data analysis and data visualization were performed in GraphPad Prism 8. The only exception is analysis of RNAseq data, which was performed in Partek Flow, and analysis of screen data, which was performed using a published pipeline (code available at https://github.com/RohatgiLab/BAIMS-Pipeline). For LasA dose-response experiments, data were fit using the “[inhibitor] vs normalized response – variable slope” option in Graphpad Prism8 software, which uses the equation Y=100/(1+(IC50/X)ĤillSlope. All IC50’s shown in the paper are derived from these curve fits and include the standard error reported for these curve fits by GraphPad Prism 8. Information about error bars, statistical tests and n values are reported in each figure legend and were calculated using GraphPad Prism 8. All experiments were repeated at least three times with concordant results, with the exception of the experiment depicted in Figure 5, which was repeated twice due to the scarcity of LasA.
Data Availability
The complete lists of the hits from the genetic screens are given in Supplementary Data 1. RNAseq data from Hap1 cells is freely available at NCBI-GEO-GSE75515. The GI50 data for LasA and the RNAseq data for cancer cell lines is publicly available (accession numbers given in the appropriate Methods section). Software for analysis of screen results has been described previously 9–10 and is freely available on github: https://github.com/RohatgiLab/BAIMS-Pipeline.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Herschlag for bringing the LasA project to our attention, C. Pataki and R. Kopito for comments and advice on lipid droplet fractionation experiments, and A. Lebensohn for advice on the project. The work was funded by DP2 GM105448 (R. Rohatgi), R35 GM118082 (R. Rohatgi), DP2 AI104557 (J.E. Carette), American Heart Association (A. Paul), Transformational Research Project # 18TPA34230103 (A. Paul), Dominic Ferraioli Foundation (A. Paul), Transformational Research Project # 18TPA34230086 (Y. Goo). R. Rohatgi is a Josephine Q. Berry Faculty Scholar in Cancer Research at Stanford, J.E. Carette is a David and Lucile Packard Foundation fellow, and R. Dubey was supported by fellowships from the Stanford Dean's Fund and Alex's Lemonade Stand Foundation.
Footnotes
COMPETING FINANCIAL INTERESTS
None of the authors have any competing financial interests to report.
REFERENCES
- 1.Horton PA; Koehn FE; Longley RE; McConnell OJ, Lasonolide-A A new cytotoxic macrolide from the marine sponge forcepia sp. J. Am. Chem. Soc. 1994, 116 (13), 6015–6016. [Google Scholar]
- 2.Wright AE; Chen Y; Winder PL; Pitts TP; Pomponi SA; Longley RE, Lasonolides C-G, five new lasonolide compounds from the sponge Forcepia sp. J. Nat. Prod. 2004, 67 (8), 1351–1355. [DOI] [PubMed] [Google Scholar]
- 3.Isbrucker RA; Guzman EA; Pitts TP; Wright AE, Early Effects of Lasonolide A on Pancreatic Cancer Cells. J. Pharmacol. Exp. Ther. 2009, 331 (2), 733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang YW; Ghosh AK; Pommier Y, Lasonolide A, a potent and reversible inducer of chromosome condensation. Cell Cycle 2012, 11 (23), 4424–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Josse R; Zhang YW; Giroux V; Ghosh AK; Luo J; Pommier Y, Activation of RAF1 (c-RAF) by the Marine Alkaloid Lasonolide A Induces Rapid Premature Chromosome Condensation. Mar. Drugs 2015, 13 (6), 3625–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trost BM; Stivala CE; Fandrick DR; Hull KL; Huang A; Poock C; Kalkofen R, Total Synthesis of (-)-Lasonolide A. J. Am. Chem. Soc. 2016, 138 (36), 11690–11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carette JE; Guimaraes CP; Varadarajan M; Park AS; Wuethrich I; Godarova A; Kotecki M; Cochran BH; Spooner E; Ploegh HL; Brummelkamp TR, Haploid Genetic Screens in Human Cells Identify Host Factors Used by Pathogens. Science 2009, 326 (5957), 1231–1235. [DOI] [PubMed] [Google Scholar]
- 8.Carette JE; Guimaraes CP; Wuethrich I; Blomen VA; Varadarajan M; Sun C; Bell G; Yuan BB; Muellner MK; Nijman SM; Ploegh HL; Brummelkamp TR, Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat. Biotechnol. 2011, 29 (6), 542–U108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubey R; Lebensohn AM; Bahrami-Nejad Z; Marceau C; Champion M; Gevaert O; Sikic BI; Carette JE; Rohatgi R, Chromatin-Remodeling Complex SWI/SNF Controls Multidrug Resistance by Transcriptionally Regulating the Drug Efflux Pump ABCB1. Cancer Res. 2016, 76 (19), 5810–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carette JE; Raaben M; Wong AC; Herbert AS; Obernosterer G; Mulherkar N; Kuehne AI; Kranzusch PJ; Griffin AM; Ruthel G; Cin PD; Dye JM; Whelan SP; Chandran K; Brummelkamp TR, Ebola virus entry requires the cholesterol transporter Niemann–Pick C1. Nature 2011, 477, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon GM; Cravatt BF, Activity-based Proteomics of Enzyme Superfamilies: Serine Hydrolases as a Case Study. Journal of Biological Chemistry 2010, 285 (15), 11051–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachovchin DA; Cravatt BF, The pharmacological landscape and therapeutic potential of serine hydrolases. Nat. Rev. Drug Discov. 2012, 11 (1), 52–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebenstreit D; Fang M; Gu M; Charoensawan V; van Oudenaarden A; Teichmann SA, RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol Syst Biol 2011, 7, 497–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goo YH; Son SH; Kreienberg PB; Paul A, Novel Lipid Droplet-Associated Serine Hydrolase Regulates Macrophage Cholesterol Mobilization. Arterioscler. Thromb. Vasc. Biol. 2014, 34 (2), 386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thiel K; Heier C; Haberl V; Thul PJ; Oberer M; Lass A; Jäckle H; Beller M, The evolutionarily conserved protein CG9186 is associated with lipid droplets, required for their positioning and for fat storage. Journal of Cell Science 2013, 126 (10), 2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchler-Bauer A; Derbyshire MK; Gonzales NR; Lu S; Chitsaz F; Geer LY; Geer RC; He J; Gwadz M; Hurwitz DI; Lanczycki CJ; Lu F; Marchler GH; Song JS; Thanki N; Wang Z; Yamashita RA; Zhang D; Zheng C; Bryant SH, CDD: NCBI's conserved domain database. Nucleic acids research 2015, 43 (Database issue), D222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Currall BB; Chen M; Sallari RC; Cotter M; Wong KE; Robertson NG; Penney KL; Lunardi A; Reschke M; Hickox AE; Yin Y; Wong GT; Fung J; Brown KK; Williamson RE; Sinnott-Armstrong NA; Kammin T; Ivanov A; Zepeda-Mendoza CJ; Shen J; Quade BJ; Signoretti S; Arnos KS; Banks AS; Patsopoulos N; Liberman MC; Kellis M; Pandolfi PP; Morton CC, Loss of LDAH associated with prostate cancer and hearing loss. Human Molecular Genetics 2018, ddy310–ddy310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kory N; Grond S; Kamat SS; Li Z; Krahmer N; Chitraju C; Zhou P; Fröhlich F; Semova I; Ejsing C; Zechner R; Cravatt BF; Farese RV; Walther TC, Mice lacking lipid droplet-associated hydrolase, a gene linked to human prostate cancer, have normal cholesterol ester metabolism. Journal of Lipid Research 2017, 58 (1), 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olzmann JA; Carvalho P, Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20 (3), 137–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brasaemle DL; Wolins NE, Isolation of Lipid Droplets from Cells by Density Gradient Centrifugation. Current Protocols in Cell Biology 2016, 72 (1), 3.15.1–3.15.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banani SF; Lee HO; Hyman AA; Rosen MK, Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenwood DJ; Dos Santos MS; Huang S; Russell MRG; Collinson LM; MacRae JI; West A; Jiang H; Gutierrez MG, Subcellular antibiotic visualization reveals a dynamic drug reservoir in infected macrophages. Science 2019, 364 (6447), 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng N; Tsai HN; Zhang X; Rosania GR, The Subcellular Distribution of Small Molecules: From Pharmacokinetics to Synthetic Biology. Molecular Pharmaceutics 2011, 8 (5), 1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.den Brok MH; Raaijmakers TK; Collado-Camps E; Adema GJ, Lipid Droplets as Immune Modulators in Myeloid Cells. Trends in immunology 2018, 39 (5), 380–392. [DOI] [PubMed] [Google Scholar]
- 25.Fowler S; Shio H; Haley NJ, Characterization of lipid-laden aortic cells from cholesterol-fed rabbits. IV. Investigation of macrophage-like properties of aortic cell populations. Laboratory investigation; a journal of technical methods and pathology 1979, 41 (4), 372–8. [PubMed] [Google Scholar]
- 26.Foley P, Lipids in Alzheimer's disease: A century-old story. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2010, 1801 (8), 750–753. [DOI] [PubMed] [Google Scholar]
- 27.Delikatny EJ; Chawla S; Leung D-J; Poptani H, MR-visible lipids and the tumor microenvironment. NMR in Biomedicine 2011, 24 (6), 592–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petan T; Jarc E; Jusović M, Lipid Droplets in Cancer: Guardians of Fat in a Stressful World. Molecules 2018, 23 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sundelin JP; Ståhlman M; Lundqvist A; Levin M; Parini P; Johansson ME; Borén J, Increased Expression of the Very Low-Density Lipoprotein Receptor Mediates Lipid Accumulation in Clear-Cell Renal Cell Carcinoma. PLOS ONE 2012, 7 (11), e48694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hager MH; Solomon KR; Freeman MR, The role of cholesterol in prostate cancer. Current opinion in clinical nutrition and metabolic care 2006, 9 (4), 379–85. [DOI] [PubMed] [Google Scholar]
- 31.Aboumrad MH; Horn RC Jr.; Fine G, Lipid-secreting mammary carcinoma. Report of a case associated with Pageťs disease of the nipple. Cancer 1963, 16, 521–5. [DOI] [PubMed] [Google Scholar]
- 32.Ramos CV; Taylor HB, Lipid-rich carcinoma of the breast. A clinicopathologic analysis of 13 examples. Cancer 1974, 33 (3), 812–9. [DOI] [PubMed] [Google Scholar]
- 33.Rautio J; Meanwell NA; Di L; Hageman MJ, The expanding role of prodrugs in contemporary drug design and development. Nat. Rev. Drug Discov. 2018, 17, 559. [DOI] [PubMed] [Google Scholar]
Online methods references
- 34.Ran FA; Hsu PD; Wright J; Agarwala V; Scott DA; Zhang F, Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8 (11), 2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanjana NE; Shalem O; Zhang F, Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11 (8), 783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campeau E; Ruhl VE; Rodier F; Smith CL; Rahmberg BL; Fuss JO; Campisi J; Yaswen P; Cooper PK; Kaufman PD, A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 2009, 4 (8), e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trost BM; Stivala CE; Hull KL; Huang A; Fandrick DR, A Concise Synthesis of (-)-Lasonolide A. J. Am. Chem. Soc. 2014, 136 (1), 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete lists of the hits from the genetic screens are given in Supplementary Data 1. RNAseq data from Hap1 cells is freely available at NCBI-GEO-GSE75515. The GI50 data for LasA and the RNAseq data for cancer cell lines is publicly available (accession numbers given in the appropriate Methods section). Software for analysis of screen results has been described previously 9–10 and is freely available on github: https://github.com/RohatgiLab/BAIMS-Pipeline.