Abstract
Over the course of roughly a decade, the lasso peptide field has been transformed. Whereas new compounds were discovered infrequently via activity-driven approaches, now the vast majority of lasso peptide discovery is driven by genome mining approaches. This paper starts with a historical overview of the first genome mining approaches for lasso peptide discovery, and then covers new tools that have emerged. Several examples of novel lasso peptides that have been discovered via genome mining are presented as are examples of new enzymes found associated with lasso peptide gene clusters. Finally, the paper concludes with future directions and unsolved challenges in lasso peptide genome mining.
Introduction
Lasso peptides are a class of ribosomally-derived natural products categorized by their 1-rotaxane structure [40,20]. This slipknotted structure is close in size to many chemically-synthesized rotaxanes [15,18] and often endows these peptides with proteolytic resistance and thermal resistance [50,3]. The lasso structure can be thought of as two macrocycles. The first is formed via a covalent isopeptide bond between the N-terminus of the peptide and either a glutamate or aspartate sidechain. The second macrocycle is generated via a mechanical bond established by threading the C-terminal tail of the peptide through the isopeptide-bonded macrocycle (Figure 1). The first report of a lasso peptide was in 1991 with the discovery of anantin via an activity-based approach [66]. Shortly thereafter, microcin J25 (MccJ25), the most well-studied lasso peptide, was discovered in a search for new antimicrobial compounds in infant fecal bacteria [51]. Another compelling example of an antimicrobial lasso peptide discovered by activity-guided approaches is lariatin, which has activity against mycobacteria including M. tuberculosis [25,24,23]. Lasso peptides that have been discovered by activity-guided approaches have been reviewed elsewhere extensively [40,49,20]. Here instead we focus on genomics-guided approaches for lasso peptide discovery. We last reviewed this topic in 2014 [39], so this review will focus primarily on new lasso peptide genome mining tools and experimental validation from 2015 onward.
Figure 1:
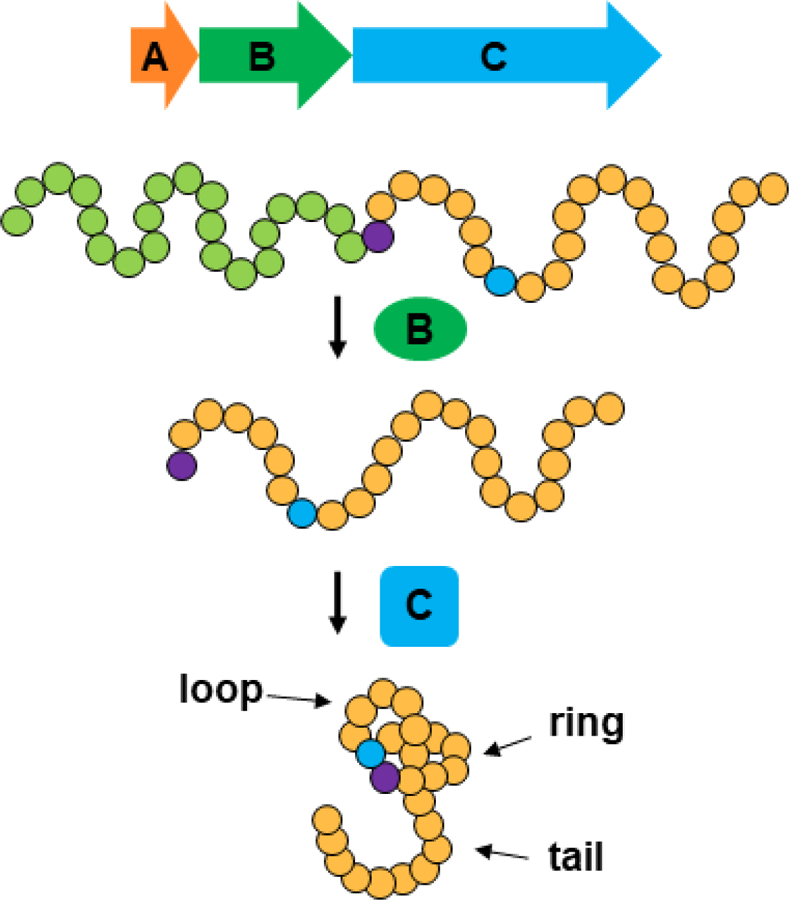
Schematic of lasso peptide biosynthesis. Top, a minimal gene cluster for lasso peptide biosynthesis. The A gene encodes the lasso peptide precursor, the B gene encodes a cysteine peptidase, and the C gene encodes an asparagine synthetase homolog that forms the isopeptide bond. Middle: schematic of lasso peptide biosynthesis; each circle represents a single amino acids. Bottom: the topology of the lasso peptide includes an isopeptide-bonded ring, a loop held in place via mechanical bonding, and a C-terminal tail.
Early efforts at lasso peptide genome mining
Like genome mining for other microbial natural products, lasso peptide genome mining depends on the existence of gene clusters encoding the biosynthesis of lasso peptides. The first lasso peptide gene cluster sequenced was for MccJ25, and appeared in 1999 [55]. This relatively early example of gene cluster sequencing was facilitated by the fact that the gene cluster for MccJ25 is found on a plasmid [51]. This early work laid out the blueprints for lasso peptide biosynthesis. Like other RiPPs (ribosomally synthesized and post-translationally modified peptides) [4], lasso peptides are born from a precursor peptide, or A protein. This precursor is processed by a peptidase, the B protein, with a cysteine catalytic triad into leader and core peptides. Finally, the lasso is formed via the action of an asparagine synthetase homolog, the C protein, that presumably both activates the Glu or Asp residue with ATP and templates the lasso structure (Figure 1). The colocalization of these genes within a cluster on prokaryotic chromosomes or plasmids is the foundation for lasso peptide genome mining.
The first indications of genome mining for lasso peptides appear in the literature in 2007. The group of Konstantin Severinov, who carried out seminal work in characterizing the antimicrobial function of MccJ25 [1,48,69], suggested that gene clusters resembling that of MccJ25 were present in the genomes of several different bacteria, primarily clustered in proteobacteria and actinobacteria [52]. The first isolation of a new lasso peptide from genome mining approaches followed shortly from the group of Mohamed Marahiel, who published the gene cluster and structure of the antimicrobial lasso peptide capistruin in 2008 [30]. Both of these approaches relied on BLAST searches for colocalized B and C enzymes, followed by a manual search for short open reading frames (ORFs) adjacent to the enzymes (Figure 2).
Figure 2:
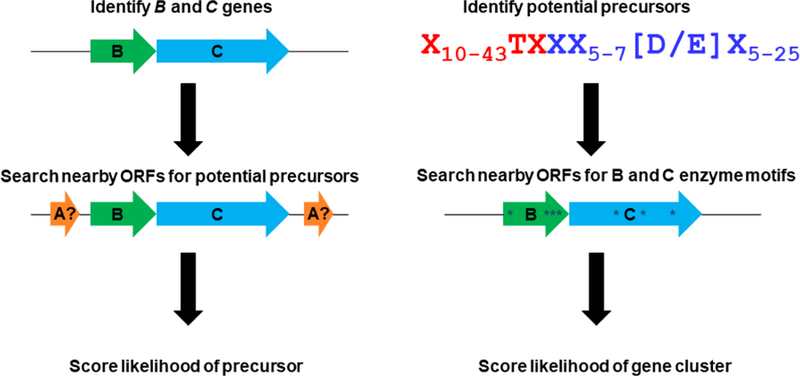
Approaches to lasso peptide genome mining. Left: the enzyme-centric approach pioneered by Marahiel and co-workers. Putative B and C enzymes are identified first followed by searches for short ORFs nearby these enzymes. Subsequently developed tools such as PRISM and RODEO also use this approach. Right: the precursor-centric approach developed by Link and co-workers. Putative precursors are identified first via pattern matching followed by scoring of clusters based on the presence of conserved motifs within the B and C enzymes (asterisks).
Our group took a different approach, and searched for lasso peptides based on a pattern of conserved amino acids within the precursor protein. This pattern built on our previous observations that the conserved Thr residue in the penultimate positon of the MccJ25 and capistruin leader peptides is important for correct processing of these peptides [11,47]. Beyond this, we specified that the N-terminal amino acid of the lasso peptide should be Gly, since all known examples of lasso peptides other than the siamycin-like class I lasso peptides [12,13,61] had Gly at the first position. Finally, we specified that an Asp or Glu residue be present 7–11 amino acids away from the Gly to form the isopeptide bond (Figure 2). Software that de novo translates genomes in all six frames and matches putative ORFs to this pattern was written. On the set of 3200 genomes available in public databases in 2011, the pattern matching software found on the order of 3 × 108 matches. This list was rapidly winnowed down to a more manageable size by imposing the restriction that conserved motifs from the B and C enzymes must be found in nearby ORFs. This first attempt at understanding the global prevalence of lasso peptide in prokaryotic genomes led to the prediction of 98 new lasso peptides in 76 prokaryotic genomes. As predicted by Severinov et al., the largest clades of lasso peptides were found in proteobacteria and actinobacteria, though examples were found across 9 bacterial phyla and even one archaeal phylum. We experimentally validated our precursor-centric approach to lasso peptide discovery with the isolation of astexin-1 from Asticcacaulis excentricus [41], a peptide also subsequently studied by Marahiel and colleagues [71].
This approach to lasso peptide genome mining is not without its limitations. For example, the Marahiel group demonstrated that lasso peptides can accommodate residues other than Gly at the N-terminus [72] and that lasso peptide with ring sizes of 7 aa can be produced [22]. With these discoveries, we have altered the precursor pattern in our genome mining approach (Figure 2). Likewise, our group recognized that one of the four conserved motifs found near the N-terminus of B proteins occasionally appeared in its own ORF, resulting in a B protein split between two ORFs [38]. Lasso peptide gene cluster with this so-called “split-B” protein are especially prevalent in actinobacteria and firmicutes. Marahiel and colleagues suggested naming these ORFs B1 and B2 [20]. The precursor-centric genome mining approach has been updated to account for this possibility.
Big data leads to new genome mining approaches
The pace of prokaryotic genome sequencing has quickened dramatically with the introduction of next generation sequencing technology. As of this writing, GenBank now contains more than 200,000 distinct genome sequences. Concurrent with this explosion in genome sequence data is increased interest in lasso peptides and other RiPPs. Platforms such as BAGEL [62,63], antiSMASH [6,7,43,65], RiPPER [8], and RiPPMiner [2] now can efficiently search for lasso peptides in genomic sequence data with high accuracy. Two additional genome mining tools, PRISM and RODEO, have carried out a global survey of lasso peptides and will be discussed in more detail below.
Magarvey and co-workers originally developed PRISM for genome mining of polyketides and non-ribosomal peptides [53], and expanded it to search for diverse families of RiPPs in 2016 [54]. At the heart of the PRISM approach is a set of hidden Markov models coupled with structure prediction to search for RiPP gene clusters. In this 2016 study, ~65,000 genome sequences were analyzed for the presence of lasso peptides and other RiPPs. For the identification of lasso peptide gene clusters, both B and C enzymes must be present. A hidden Markov model was used to predict precursor sequences with pattern matching similar to our approach as a backup. PRISM identified 1466 lasso peptides from ~65,000 genomes. The uniqueness of this set of lasso peptides was assessed using the Tanimoto coefficient, resulting in the assignment of roughly 500 unique lasso peptides. Applying this analysis across all RiPP families analyzed, lasso peptides had the largest number of unique members.
The RODEO approach, published by Mitchell and coworkers in 2017 [60], uses hidden Markov models to identify the B and C proteins within a genome sequence. This is followed by six-frame translation of intergenic regions and scoring of potential precursors via a set of heuristics. A support vector machine was trained in order to more accurately identify lasso peptide precursors. Final scoring of putative precursor included a combination of heuristics, the support vector machine, and motif matching via MEME [5]. The RODEO approach identified ~1300 different lasso peptides, a number comparable to the PRISM approach described above. Interestingly, since RODEO identifies B and C proteins first, ~100 putative gene clusters were identified that did not have a discernible precursor. It remains to be seen if these clusters function on precursors that are located elsewhere in the genome or if the precursor genes have simply been lost to genetic drift.
Experimental validation of lasso peptides from genome mining
Beyond knowing the extent of lasso peptides across genomes, the major motivation of genome mining efforts is to discover new examples of lasso peptides along with testing for bioactivity. Once predicted within a genome, lasso peptides can either be isolated from the native producer strain or produced via heterologous expression. While robust tools have been developed for heterologous expression of lasso peptides from proteobacteria [46,21,10], heterologous expression of lasso peptides from actinobacteria remains challenging, with some notable exceptions [36,33,45,42]. In this section we will highlight new experimentally validated lasso peptides that have been isolated by these approaches from 2015 onward. A selection of the NMR structures of the peptides discussed in this section are presented in Figure 3. For a comprehensive list of lasso peptides isolated up to 2015, please see the review article by Hegemann et al. [20].
Figure 3:
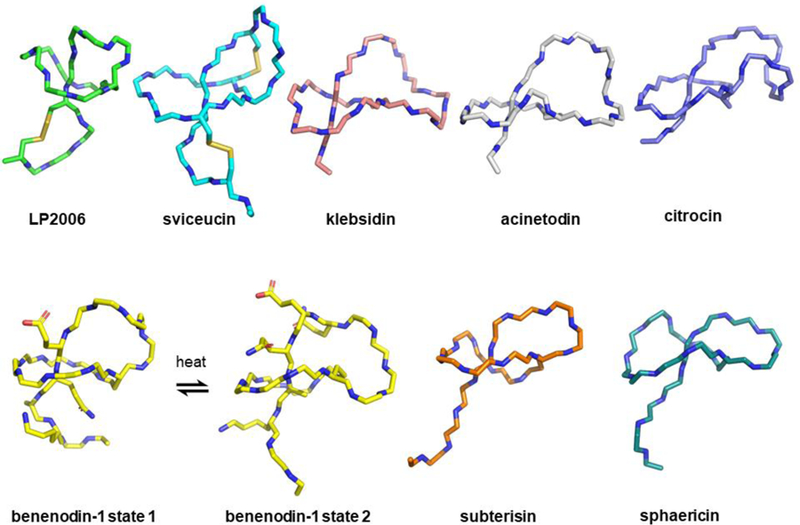
NMR structures of lasso peptides identified by genome mining. LP 2006 (PDB 5JPL) and sviceucin (PDB 2LS1) are buttressed by disulfide bonds (yellow). Klebsidin (PDB 5UI7), acinetodin (5UI6) and citrocin (6MW6) are all RNA polymerase inhibitors. Benenodin-1 establishes a thermodynamic equilibrium between 2 distinct threaded conformers (PDB 5TJ1 and 6B5W). Subterisin was isolated from Sphingomonas (PDB 5XM4) and sphaericin was isolated from an actinobacterium and is antimicrobial (PDB 5GVO).
In validation studies for the RODEO approach, Mitchell and colleagues selected five gene clusters from actinobacteria to be produced either from their native hosts or via heterologous expression. This effort led to the discovery of citrulassin, a lasso peptide in which arginine is deamidated to citrulline. Another novel lasso peptide architecture experimentally validated from the RODEO approach is LP2006, which includes a disulfide bonded macrocycle in the C-terminal tail of the peptide. A new producing strain for anantin [66,67] was identified and validated. Of this set of peptides, two (anantin and LP2006) exhibited antimicrobial activity.
Several other new examples of lasso peptides from both actinobacteria and proteobacteria have been published recently by Kodani and co-workers. The lasso peptides specialicin [28], achromosin [29], sphaericin [32], and actinokineosin [59] have been isolated from various actinobacteria. Each of these peptides exhibits antimicrobial activity. Another example, cattlecin, was isolated from Streptomyces cattleya [58], but does not exhibit antimicrobial activity. This same peptide was isolated by Mitchell and colleagues as part of the RODEO validation, and they named it “moomysin.” The bovine connotation in both of these names is a bit of a misnomer, since “cattleya” in the name of the producing organism refers to a genus of orchid. The producing organism S. cattleya produces orchid-colored mycelia [26]. From proteobacteria Kodani and colleagues have produced two novel lasso peptides: subterisin [34], from a Sphingomonas strain and brevunsin [31], from a Brevundimonas. Of particular note is that brevunsin was heterologously expressed in the subterisin producing strain, S. subterranea. No antimicrobial activity was observed for these two peptides.
While antimicrobial activity is believed to be the natural function of many lasso peptides, other bioactivities are often observed as well. Two new lasso peptides, chaxapeptin [14] and ulleungdin [56], inhibit the migration of mammalian cell lines. These peptides were isolated from actinobacteria that live in vastly different environments. The producing strain of chaxapeptin, Streptomyces leeuwenhoekii strain C58, was isolated from the arid Atacama desert, while the ulleungdin producer, another Streptomyces strain, was isolated from soil on Ulleung Island, a volcanic island in Korea that receives more than a meter of rainfall each year on average.
Severinov and coworkers used genome mining to discover two new lasso peptides from human bacterial isolates, a strain of Klebsiella pneumonia and a strain of Acinetobacter gyllenbergii. The resulting peptides were referred to as klebsidin and acinetodin, respectively [44]. The structures of both of these peptides were determined by NMR and both peptides inhibit RNA polymerase (RNAP) in vitro, showing that these peptides have a similar mode of action to MccJ25 and capistruin. Despite this, acinetodin exhibited no antimicrobial activity against a panel of bacterial strains while klebsidin was only able to kill other K. pneumoniae strains at relatively high concentrations. Our group has also recently published a report of a new RNAP-inhibiting lasso peptide, citrocin [10]. In contrast to MccJ25, which is unable to kill cells knocked out in the TonB-dependent transporter FhuA, citrocin retains antimicrobial activity against all TonB-dependent transporter knockouts in E. coli. Citrocin is a very potent inhibitor of RNAP, exhibiting activity roughly 100-fold more potent than MccJ25 in abortive transcription assays. Despite this potent activity in vitro, citrocin has only modest activity against a panel of enterobacteria.
Our laboratory became interested in several lasso peptides encoded in the genomes of thermophilic actinobacteria in the Thermobifida genus. We have characterized one such peptide in depth, fuscanodin, from T. fusca [33]. Our initial interest in this peptide stemmed from its unprecedented Trp residue at position 1 of the core peptide. Heterologous expression of fuscanodin was successful in E. coli, but the organic solvents used to isolate and purify fuscanodin led to its unthreading into a branched cyclic form, preventing structural and functional characterization. Perhaps the most noteworthy aspect of this work is that the fuscanodin biosynthetic enzymes can be expressed solubly with good yields, allowing for the first ever studies into the kinetics of lasso peptide biosynthesis. These studies showed that, at least in vitro, the rate of lasso peptide biosynthesis is insensitive to the concentration of the precursor protein A [33].
Following up on our laboratory’s work on lasso peptides from Asticcacaulis excentricus [38,41], we also became interested in a lasso peptide encoded in the genome of Asticcacaulis benevestitus, an alpha-proteobacterium isolated from peat in Vorkuta, Russia [64]. The NMR structure of this peptide, named benenodin-1, revealed steric lock residues of Glu and Gln (Figure 3), which differ from the more common aromatic residues that maintain the lasso in its threaded conformation. Upon heating, we ultimately determined that this peptide establishes an equilibrium between two distinct threaded conformers [73], generating a switch analogous to those observed in the synthetic rotaxane field [68]. In addition to providing interesting new topologies and bioactivites, this work shows that genome mining for new lasso peptides will also uncover molecules with unprecedented biophysical behaviors.
Discovery of new enzymology associated with lasso peptide gene clusters
Another benefit of genome mining for lasso peptides is the discovery of additional enzymes adjacent or nearby the core biosynthetic genes that further interact with the lasso peptide (Figure 4). Our group published a seminal example of this type of analysis in 2013 with the discovery of lasso peptide isopeptidase, an enzyme that recognizes the threaded lasso shape and cleaves it back into a linear peptide [38,37,9,73]. Lasso peptide isopeptidase represents the first example of lasso peptide catabolic enzyme. We have hypothesized that this isopeptidase is part of a metal trafficking system in which lasso peptides act like siderophores, and the isopeptidase releases the metal from the lasso scaffold. This assertion is based on colocalization of a TonB-dependent transporter and FecI/RecR proteins with the isopeptidase. Another possibility is that these lassoes function as signaling molecules, and the isopeptidase attenuates these signals.
Figure 4:
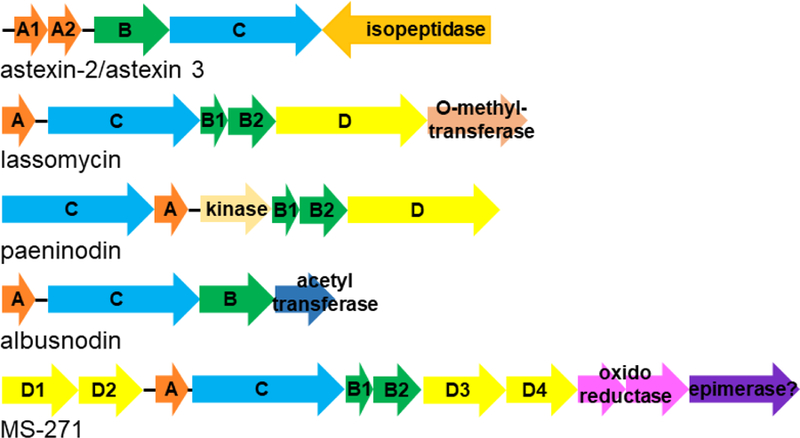
Lasso peptide gene clusters with additional tailoring or catabolic enzymes. The A, B, and C genes are coded consistently throughout. The D genes encode ABC transporters. The function or proposed function of all other genes is presented on the figure.
The “guilt by association” with lasso peptide gene clusters approach has also uncovered a series of enzymes that add posttranslational modifications to lasso peptides beyond the isopeptide bond that defines the family. The lasso peptide paeninodin includes a kinase embedded in its gene cluster which was shown to phosphorylate the C-terminus of the peptide [70]. Our group studied a lasso peptide encoded in the genome of Streptomyces albus DSM 41398 with an acetyltransferase adjacent to the gene cluster. The peptide encoded by this cluster, albusnodin, includes a single lysine acetylation in the loop of the peptide [74]. Via heterologous expression in Streptomyces, we showed that albusnodin could only be expressed when the acetyltransferase was coexpressed. This suggests that acetylation is an obligate PTM for this peptide. Unfortunately, the expression level of albusnodin is quite low via heterologous expression, thus far precluding further analysis of the role of acetylation on the structure and/or function of the peptide.
Genome mining has also uncovered secrets about PTMs that occur on lasso peptides initially discovered by activity guided approaches. For example, the lasso peptide lassomycin was reported in 2014 and includes methylation of its C-terminus catalyzed by an O-methyltransferase [17,19,35]. Recently Zhu and colleagues searched genomes for additional examples of “lassomycin-like” lasso peptides in order to study the function of the methyltransferase, identifying two new strains that produce homologs of lassomycin [57]. Though the lasso peptides encoded by these clusters were recalcitrant to heterologous expression, the methyltransferase enzyme could be purified and studied. This work showed that the methyltransferase functions on linear precursor peptides, suggesting a potential order of posttranslational modifications. Another peptide that carries a modification at its C-terminus is MS-271, also known as siamycin I and NP-06, originally reported in 1996 [27]. MS-271 includes a D-Trp residue at the end of its core peptide. More recently, the genome of the producing organism, Streptomyces sp. M-271, was sequenced, revealing the lasso peptide gene cluster [16]. This cluster included all requisite genes for lasso peptide biosynthesis, plus oxidoreductases presumably involved in accelerating the formation of the two disulfide bonds found in the structure. In addition to these genes, a gene of unknown function, mslH, was present, and the authors suggest that this gene product may be responsible for Trp epimerization. When the mslH gene was deleted, MS-271 production ceased, suggested a possible obligate role for Trp epimerization in the biosynthesis of this lasso peptide.
Future outlook and unsolved challenges for lasso peptide genome mining
It is not an exaggeration to say that the lasso peptide field has been completely transformed by genome mining approaches. With continued advances in genome sequencing, these approaches will continue to uncover new examples of lasso peptides for years to come. As we look to future applications of genome mining for lasso peptides, studying the regulation of lasso peptides should be a top priority. Lasso peptide gene clusters are often flanked by regulatory proteins, but these have been rarely studied given the robustness of heterologous expression approaches for lasso peptides. Understanding the cues that trigger lasso peptide biosynthesis in their native contexts will provide new insights into the true functions of these peptides. Even more important is the challenge of leveraging genome mining approaches to help predict which lasso peptides will have compelling bioactivities, antimicrobial or otherwise. This challenge is likely only to be supplanted with careful study into the mechanism of action of lasso peptides coupled with activity studies on variants of the peptide. Such sequence-structure-activity relationships will allow not just the rapid identification of lasso peptides using genome mining approaches, but also prioritization of those peptide most likely to serve as drug-like molecules.
Acknowledgements
Lasso peptide research in the Link Lab is supported by the NIH (GM107036) and a grant from the Princeton University School of Engineering and Applied Sciences (SEAS) for the Focused Research Team on Precision Antibiotics.
References
- 1.Adelman K, Yuzenkova J, La Porta A, Zenkin N, Lee J, Lis JT, Borukhov S, Wang MD, Severinov K (2004) Molecular mechanism of transcription inhibition by peptide antibiotic microcin J25. Mol Cell 14:753–762. 10.1016/j.molcel.2004.05.017 [DOI] [PubMed] [Google Scholar]
- 2.Agrawal P, Khater S, Gupta M, Sain N, Mohanty D (2017) RiPPMiner: a bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res 45:W80–W88. 10.1093/nar/gkx408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen CD, Chen MY, Trick AY, Le DT, Ferguson AL, Link AJ (2016) Thermal Unthreading of the Lasso Peptides Astexin-2 and Astexin-3. ACS Chemical Biology 11:3043–3051. 10.1021/acschembio.6b00588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey TL, Elkan C (1995) Unsupervised Learning of Multiple Motifs in Biopolymers Using Expectation Maximization. Machine Learning 21:51–80 [Google Scholar]
- 6.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T (2013) antiSMASH 2.0-a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 41:W204–W212. 10.1093/nar/gkt449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blin K, Wolf T, Chevrette MG, Lu XW, Schwalen CJ, Kautsar SA, Duran HGS, Santos E, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema MH (2017) antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45:W36–W41. 10.1093/nar/gkx319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandra G, Santos-Aberturas J, Frattaruolo L, Vior NM, Lacret R, Pham TH, Eyles TH, Truman AW (2019) Uncovering the unexplored diversity of thioamidated ribosomal peptides in Actinobacteria using the RiPPER genome mining tool 10.1093/nar/gkz192 [DOI] [PMC free article] [PubMed]
- 9.Chekan JR, Koos JD, Zong C, Maksimov MO, Link AJ, Nair SK (2016) Structure of the Lasso Peptide Isopeptidase Identifies a Topology for Processing Threaded Substrates. J Am Chem Soc 138:16452–16458. 10.1021/jacs.6b10389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung-Lee WL, Parry ME, Cartagena AJ, Darst SA, Link AJ (2019) Discovery and structure of the antimicrobial lasso peptide citrocin. J Biol Chem 294:6822–6830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung WL, Pan SJ, Link AJ (2010) Much of the Microcin J25 Leader Peptide is Dispensable. J Am Chem Soc 132:2514–2515 [DOI] [PubMed] [Google Scholar]
- 12.Constantine KL, Friedrichs MS, Detlefsen D, Nishio M, Tsunakawa M, Furumai T, Ohkuma H, Oki T, Hill S, Bruccoleri RE, Lin PF, Mueller L (1995) High-resolution solution structure of siamycin-II - Novel amphipathic character of a 21-residue peptide that inhibits HIV fusion. J Biomol NMR 5:271–286 [DOI] [PubMed] [Google Scholar]
- 13.Detlefsen DJ, Hill SE, Volk KJ, Klohr SE, Tsunakawa M, Furumai T, Lin PF, Nishio M, Kawano K, Oki T, Lee MS (1995) Siamycins I and II, new Anti-HIV-1 peptides .2. Sequence analysis and structure determination of siamycin I. J Antibiot 48:1515–1517 [DOI] [PubMed] [Google Scholar]
- 14.Elsard SS, Trusch F, Deng H, Raab A, Prokes I, Busarakam K, Asenjo JA, Andrews BA, van West P, Bull AT, Goodfellow M, Yi Y, Ebel R, Jaspars M, Rateb ME (2015) Chaxapeptin, a Lasso Peptide from Extremotolerant Streptomyces leeuwenhoekii Strain C58 from the Hyperarid Atacama Desert. J Org Chem 80:10252–10260. 10.1021/acs.joc.5b01878 [DOI] [PubMed] [Google Scholar]
- 15.Erbas-Cakmak S, Leigh DA, McTernan CT, Nussbaumer AL (2015) Artificial Molecular Machines. Chem Rev 115:10081–10206. 10.1021/acs.chemrev.5b00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Z, Ogasawara Y, Nomura S, Dairi T (2018) Biosynthetic Gene Cluster of a D-Tryptophan-Containing Lasso Peptide, MS-271. Chembiochem 19:2045–2048. 10.1002/cbic.201800315 [DOI] [PubMed] [Google Scholar]
- 17.Gavrish E, Sit CS, Cao SG, Kandror O, Spoering A, Peoples A, Ling L, Fetterman A, Hughes D, Bissell A, Torrey H, Akopian T, Mueller A, Epstein S, Goldberg A, Clardy J, Lewis K (2014) Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2. Chem Biol 21:509–518. 10.1016/j.chembiol.2014.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gil-Ramirez G, Leigh DA, Stephens AJ (2015) Catenanes: Fifty Years of Molecular Links. Angew Chem-Int Edit 54:6110–6150. 10.1002/anie.201411619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris PWR, Cook GM, Leung IKH, Brimble MA (2017) An Efficient Chemical Synthesis of Lassomycin Enabled by an On-Resin Lactamisation-Off-Resin Methanolysis Strategy and Preparation of Chemical Variants. Aust J Chem 70:172–183. 10.1071/ch16499 [DOI] [Google Scholar]
- 20.Hegemann JD, Zimmermann M, Xie X, Marahiel MA (2015) Lasso Peptides: An Intriguing Class of Bacterial Natural Products. Acc Chem Res 48:1909–1919. 10.1021/acs.accounts.5b00156 [DOI] [PubMed] [Google Scholar]
- 21.Hegemann JD, Zimmermann M, Zhu SZ, Klug D, Marahiel MA (2013) Lasso Peptides From Proteobacteria: Genome Mining Employing Heterologous Expression and Mass Spectrometry. Biopolymers 100:527–542. 10.1002/bip.22326 [DOI] [PubMed] [Google Scholar]
- 22.Hegemann JD, Zimmermann M, Zhu SZ, Steuber H, Harms K, Xie XL, Marahiel MA (2014) Xanthomonins I-III: A New Class of Lasso Peptides with a Seven-Residue Macrolactam Ring. Angew Chem-Int Edit 53:2230–2234. 10.1002/anie.201309267 [DOI] [PubMed] [Google Scholar]
- 23.Inokoshi J, Koyama N, Miyake M, Shimizu Y, Tomoda H (2016) Structure-Activity Analysis of Gram-positive Bacterium-producing Lasso Peptides with Anti-mycobacterial Activity. Scientific Reports 6:9 10.1038/srep30375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inokoshi J, Matsuhama M, Miyake M, Ikeda H, Tomoda H (2012) Molecular cloning of the gene cluster for lariatin biosynthesis of Rhodococcus jostii K01-B0171. Appl Microbiol Biotechnol 95:451–460. 10.1007/s00253-012-3973-8 [DOI] [PubMed] [Google Scholar]
- 25.Iwatsuki M, Tomoda H, Uchida R, Gouda H, Hirono S, Omura S (2006) Lariatins, antimycobacterial peptides produced by Rhodococcus sp K01-B0171, have a lasso structure. J Am Chem Soc 128:7486–7491 [DOI] [PubMed] [Google Scholar]
- 26.Kahan JS, Kahan FM, Goegelman R, Currie SA, Jackson M, Stapley EO, Miller TW, Miller AK, Hendlin D, Mochales S, Hernandez S, Woodruff HB, Birnbaum J (1979) THIENAMYCIN, A NEW BETA-LACTAM ANTIBIOTIC .1. DISCOVERY, TAXONOMY, ISOLATION AND PHYSICAL-PROPERTIES. J Antibiot 32:1–12 [DOI] [PubMed] [Google Scholar]
- 27.Katahira R, Yamasaki M, Matsuda Y, Yoshida M (1996) MS-271, a novel inhibitor of calmodulin-activated myosin light chain kinase from Streptomyces sp .2. Solution structure of MS-271: Characteristic features of the ‘lasso’ structure. Bioorg Med Chem 4:121–129. 10.1016/0968-0896(95)00176-x [DOI] [PubMed] [Google Scholar]
- 28.Kaweewan I, Hemmi H, Komaki H, Harada S, Kodani S (2018) Isolation and structure determination of a new lasso peptide specialicin based on genome mining. Bioorg Med Chem 26:6050–6055. 10.1016/j.bmc.2018.11.007 [DOI] [PubMed] [Google Scholar]
- 29.Kaweewan I, Ohnishi-Kameyama M, Kodani S (2017) Isolation of a new antibacterial peptide achromosin from Streptomyces achromogenes subsp achromogenes based on genome mining. J Antibiot 70:208–211. 10.1038/ja.2016.108 [DOI] [PubMed] [Google Scholar]
- 30.Knappe TA, Linne U, Zirah S, Rebuffat S, Xie XL, Marahiel MA (2008) Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J Am Chem Soc 130:11446–11454. 10.1021/ja802966g [DOI] [PubMed] [Google Scholar]
- 31.Kodani S, Hemmi H, Miyake Y, Kaweewan I, Nakagawa H (2018) Heterologous production of a new lasso peptide brevunsin in Sphingomonas subterranea. J Ind Microbiol Biotechnol 45:983–992. 10.1007/s10295-018-2077-6 [DOI] [PubMed] [Google Scholar]
- 32.Kodani S, Inoue Y, Suzuki M, Dohra H, Suzuki T, Hemmi H, Ohnishi-Kameyama M (2017) Sphaericin, a Lasso Peptide from the Rare Actinomycete Planomonospora sphaerica. European Journal of Organic Chemistry:1177–1183. 10.1002/ejoc.201601334 [DOI]
- 33.Koos JD, Link AJ (2019) Heterologous and in vitro reconstitution of fuscanodin, a lasso peptide from Thermobifida fusca. J Am Chem Soc 141:928–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuroha M, Hemmi H, Ohnishi-Kameyama M, Kodani S (2017) Isolation and structure determination of a new lasso peptide subterisin from Sphingomonas subterranea. Tetrahedron Lett 58:3429–3432. 10.1016/j.tetlet.2017.07.064 [DOI] [Google Scholar]
- 35.Lear S, Munshi T, Hudson AS, Hatton C, Clardy J, Mosely JA, Bull TJ, Sit CS, Cobb SL (2016) Total chemical synthesis of lassomycin and lassomycin-amide. Organic & Biomolecular Chemistry 14:4534–4541. 10.1039/c6ob00631k [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Ducasse R, Zirah S, Blond A, Goulard C, Lescop E, Giraud C, Hartke A, Guittet E, Pernodet J-L, Rebuffat S (2015) Characterization of Sviceucin from Streptomyces Provides Insight into Enzyme Exchangeability and Disulfide Bond Formation in Lasso Peptides. ACS Chemical Biology 10:2641–2649. 10.1021/acschembio.5b00584 [DOI] [PubMed] [Google Scholar]
- 37.Maksimov MO, Koos JD, Zong C, Lisko B, Link AJ (2015) Elucidating the Specificity Determinants of the AtxE2 Lasso Peptide Isopeptidase. J Biol Chem 290:30806–30812. 10.1074/jbc.M115.694083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maksimov MO, Link AJ (2013) Discovery and Characterization of an Isopeptidase That Linearizes Lasso Peptides. J Am Chem Soc 135:12038–12047. 10.1021/ja4054256 [DOI] [PubMed] [Google Scholar]
- 39.Maksimov MO, Link AJ (2014) Prospecting genomes for lasso peptides. J Ind Microbiol Biotechnol 41:333–344. 10.1007/s10295-013-1357-4 [DOI] [PubMed] [Google Scholar]
- 40.Maksimov MO, Pan SJ, Link AJ (2012) Lasso peptides: structure, function, biosynthesis, and engineering. Nat Prod Rep 29:996–1006 [DOI] [PubMed] [Google Scholar]
- 41.Maksimov MO, Pelczer I, Link AJ (2012) Precursor-centric genome-mining approach for lasso peptide discovery. Proc Natl Acad Sci U S A 109:15223–15228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin-Gomez H, Linne U, Albericio F, Tulla-Puche J, Hegemann JD (2018) Investigation of the Biosynthesis of the Lasso Peptide Chaxapeptin Using an E. coli-Based Production System. J Nat Prod 81:2050–2056. 10.1021/acs.jnatprod.8b00392 [DOI] [PubMed] [Google Scholar]
- 43.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R (2011) antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39:W339–W346. 10.1093/nar/gkr466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metelev M, Arseniev A, Bushin LB, Kuznedelov K, Artamonova TO, Kondratenko R, Khodorkovskii M, Seyedsayamdost MR, Severinov K (2017) Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae. Acs Chemical Biology 12:814–824. 10.1021/acschembio.6b01154 [DOI] [PubMed] [Google Scholar]
- 45.Mevaere J, Goulard C, Schneider O, Sekurova ON, Ma HY, Zirah S, Afonso C, Rebuffat S, Zotchev SB, Li YY (2018) An orthogonal system for heterologous expression of actinobacterial lasso peptides in Streptomyces hosts. Scientific Reports 8 10.1038/s41598-018-26620-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan SJ, Cheung WL, Link AJ (2010) Engineered gene clusters for the production of the antimicrobial peptide microcin J25. Protein Expr Purif 71:200–206. 10.1016/j.pep.2009.12.010 [DOI] [PubMed] [Google Scholar]
- 47.Pan SJ, Rajniak J, Maksimov MO, Link AJ (2012) The Role of a Conserved Threonine Residue in the Leader Peptide of Lasso Peptide Precursors. Chem Commun 48:1880–1882 [DOI] [PubMed] [Google Scholar]
- 48.Pavlova O, Mukhopadhyay J, Sineva E, Ebright RH, Severinov K (2008) Systematic structure-activity analysis of microcin J25. J Biol Chem 283:25589–25595. 10.1074/jbc.M803995200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rebuffat S, Blond A, Destoumieux-Garzon D, Goulard C, Peduzzi J (2004) Microcin J25, from the macrocyclic to the lasso structure: Implications for biosynthetic, evolutionary and biotechnological perspectives. Curr Protein Pept Sci 5:383–391 [DOI] [PubMed] [Google Scholar]
- 50.Rosengren KJ, Blond A, Afonso C, Tabet JC, Rebuffat S, Craik DJ (2004) Structure of thermolysin cleaved microcin J25: Extreme stability of a two-chain antimicrobial peptide devoid of covalent links. Biochemistry 43:4696–4702. 10.1021/bi0361261 [DOI] [PubMed] [Google Scholar]
- 51.Salomon RA, Farias RN (1992) Microcin-25, a Novel Antimicrobial Peptide Produced by Escherichia coli. J Bacteriol 174:7428–7435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Severinov K, Semenova E, Kazakov A, Kazakov T, Gelfand MS (2007) Low-molecular-weight post-translationally modified microcins. Mol Microbiol 65:1380–1394. 10.1111/j.1365-2958.2007.05874.x [DOI] [PubMed] [Google Scholar]
- 53.Skinnider MA, Dejong CA, Rees PN, Johnston CW, Li HX, Webster ALH, Wyatt MA, Magarvey NA (2015) Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res 43:9645–9662. 10.1093/nar/gkv1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skinnider MA, Johnston CW, Edgar RE, Dejong CA, Merwin NJ, Rees PN, Magarvey NA (2016) Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proc Natl Acad Sci U S A 113:E6343–E6351. 10.1073/pnas.1609014113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Solbiati JO, Ciaccio M, Farias RN, Gonzalez-Pastor JE, Moreno F, Salomon RA (1999) Sequence analysis of the four plasmid genes required to produce the circular peptide antibiotic microcin J25. J Bacteriol 181:2659–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Son S, Jang M, Lee B, Hong YS, Ko SK, Jang JH, Ahn JS (2018) Ulleungdin, a Lasso Peptide with Cancer Cell Migration Inhibitory Activity Discovered by the Genome Mining Approach. J Nat Prod 81:2205–2211. 10.1021/acs.jnatprod.8b00449 [DOI] [PubMed] [Google Scholar]
- 57.Su Y, Han M, Meng XB, Feng Y, Luo SZ, Yu CY, Zheng GJ, Zhu SZ (2019) Discovery and characterization of a novel C-terminal peptide carboxyl methyltransferase in a lassomycin-like lasso peptide biosynthetic pathway. Appl Microbiol Biotechnol 103:2649–2664. 10.1007/s00253-019-09645-x [DOI] [PubMed] [Google Scholar]
- 58.Sugai S, Ohnishi-Kameyama M, Kodani S (2017) Isolation and identification of a new lasso peptide cattlecin from Streptomyces cattleya based on genome mining. Applied Biological Chemistry 60:163–167. 10.1007/s13765-017-0268-x [DOI] [Google Scholar]
- 59.Takasaka N, Kaweewan I, Ohnishi-Kameyama M, Kodani S (2017) Isolation of a new antibacterial peptide actinokineosin from Actinokineospora spheciospongiae based on genome mining. Lett Appl Microbiol 64:150–157. 10.1111/lam.12693 [DOI] [PubMed] [Google Scholar]
- 60.Tietz JI, Schwalen CJ, Patel PS, Maxson T, Blair PM, Tai HC, Zakai UI, Mitchell DA (2017) A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nature Chemical Biology 13:470–478. 10.1038/nchembio.2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsunakawa M, Hu SL, Hoshino Y, Detlefson DJ, Hill SE, Furumai T, White RJ, Nishio M, Kawano K, Yamamoto S, Fukagawa Y, Oki T (1995) Siamycin-I and Siamycin-II, new anti-HIV peptides .1. Fermentation, isolation, biological-activity and initial characterization. J Antibiot 48:433–434 [DOI] [PubMed] [Google Scholar]
- 62.van Heel AJ, de Jong A, Montalban-Lopez M, Kok J, Kuipers OP (2013) BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res 41:W448–W453. 10.1093/nar/gkt391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Heel AJ, de Jong A, Song CX, Viel JH, Kok J, Kuipers OP (2018) BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res 46:W278–W281. 10.1093/nar/gky383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vasilyeva LV, Omelchenko MV, Berestovskaya YY, Lysenko AM, Abraham WR, Dedysh SN, Zavarzin GA (2006) Asticcacaulis benevestitus sp nov., a psychrotolerant, dimorphic, prosthecate bacterium from tundra wetland soil. International Journal of Systematic and Evolutionary Microbiology 56:2083–2088. 10.1099/ijs.0.64122-0 [DOI] [PubMed] [Google Scholar]
- 65.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Muller R, Wohlleben W, Breitling R, Takano E, Medema MH (2015) antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243. 10.1093/nar/gkv437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weber W, Fischli W, Hochuli E, Kupfer E, Weibel EK (1991) Anantin - A Peptide Antagonst of the Atrial-Natriuretic-Factor (ANF) 1. Producing Organism, Fermentation, Isolation and Biological Activity. J Antibiot 44:164–171 [DOI] [PubMed] [Google Scholar]
- 67.Wyss DF, Lahm HW, Manneberg M, Labhardt AM (1991) Anantin - A Peptide Antagonist of the Atrial-Natriuretic Factor (ANF) 2. Determination of the Primary Sequence by NMR on the Basis of Proton Assignments. J Antibiot 44:172–180 [DOI] [PubMed] [Google Scholar]
- 68.Yang WL, Li YJ, Liu HBA, Chi LF, Li YL (2012) Design and Assembly of Rotaxane-Based Molecular Switches and Machines. Small 8:504–516. 10.1002/smll.201101738 [DOI] [PubMed] [Google Scholar]
- 69.Yuzenkova J, Delgado M, Nechaev S, Savalia D, Epshtein V, Artsimovitch I, Mooney RA, Landick R, Farias RN, Salomon R, Severinov K (2002) Mutations of bacterial RNA polymerase leading to resistance to microcin J25. J Biol Chem 277:50867–50875 [DOI] [PubMed] [Google Scholar]
- 70.Zhu SZ, Hegemann JD, Fage CD, Zimmermann M, Xie XL, Linne U, Marahiel MA (2016) Insights into the Unique Phosphorylation of the Lasso Peptide Paeninodin. J Biol Chem 291:13662–13678. 10.1074/jbc.M116.722108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zimmermann M, Hegemann Julian D, Xie X, Marahiel Mohamed A (2013) The Astexin-1 Lasso Peptides: Biosynthesis, Stability, and Structural Studies. Chem Biol 20:558–569. 10.1016/j.chembiol.2013.03.013 [DOI] [PubMed] [Google Scholar]
- 72.Zimmermann M, Hegemann JD, Xie X, Marahiel MA (2014) Characterization of caulonodin lasso peptides revealed unprecedented N-terminal residues and a precursor motif essential for peptide maturation. Chemical Science 5:4032–4043. 10.1039/c4sc01428f [DOI] [Google Scholar]
- 73.Zong C, Wu MJ, Qin JZ, Link AJ (2017) Lasso Peptide Benenodin-1 is a Thermally Actuated [1]Rotaxane Switch. J Am Chem Soc 139:10403–10409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zong CH, Cheung-Lee WL, Elashal HE, Raj M, Link AJ (2018) Albusnodin: an acetylated lasso peptide from Streptomyces albus. Chem Commun 54:1339–1342. 10.1039/c7cc08620b [DOI] [PMC free article] [PubMed] [Google Scholar]


