Abstract
Background:
Bicuspid aortic valve (BAV) disease is a congenital defect that affects 0.5–1.2% of the population and is associated with co-morbidities including ascending aortic dilation and calcific aortic valve stenosis (CAVS). To date, while a few causal genes have been identified, the genetic basis for the vast majority of BAV cases remains unknown, likely pointing to complex genetic heterogeneity underlying this phenotype. Identifying genetic pathways versus individual gene variants may provide an avenue for uncovering additional BAV causes and consequent co-morbidities.
Methods:
We performed a genome wide association (GWAS) and replication study using cohorts of 2,131 BAV and 2,728 control patients, respectively, which identified primary cilia genes as associated with the BAV phenotype. GWAS hits were prioritized based on p-value and validated through in vivo loss of function and rescue experiments, 3D-immunohistochemistry (IHC), histology and morphometric analyses during aortic valve morphogenesis and in aged animals in multiple species. Consequences of these genetic perturbations on cilia dependent pathways were analyzed by Western and IHC analyses, and assessment of aortic valve and cardiac function were determined by echocardiography.
Results:
GWAS revealed an association between BAV and genetic variation in human primary cilia. The most associated SNPs were identified in or near genes that are important in regulating ciliogenesis through the exocyst, a shuttling complex that chaperones cilia cargo to the membrane. Genetic dismantling of the exocyst resulted in impaired ciliogenesis, disrupted ciliogenic signaling and a spectrum of cardiac defects in zebrafish, and aortic valve defects including BAV, valvular stenosis, and valvular calcification in murine models.
Conclusions:
These data support the exocyst as required for normal ciliogenesis during aortic valve morphogenesis and implicate disruption of ciliogenesis and its downstream pathways as contributory to BAV and associated co-morbidities in humans.
Keywords: exocyst, primary cilia, bicuspid aortic valve, calcific aortic valve, aortic stenosis
Introduction
Congenital heart disease (CHD) is one of the most common birth defects1, 2. Bicuspid aortic valve (BAV) is the most frequently observed congenital valvular heart defect, having an overall frequency of 0.5%–1.2%3. BAVs result from abnormal aortic cusp formation during valvulogenesis, whereby cusps fail to form or adjacent cusps fuse into a single large cusp. The fusion events are phenotypically heterogeneous with significant variability in fusion patterns. Regardless of these fusion patterns, patients with BAV frequently have profound deleterious long-term consequences on heart function and can develop progressive aortic valve calcification and stenosis as well as thoracic aortic aneurysm and dissection4. As such, patients with BAV frequently require valvular surgeries to repair or replace damaged tissue in the cardiac outlet segment.
Due to its clinical prevalence and severity, concerted efforts by many groups have led to new genetic discoveries underlying disease etiology in humans5, 6. BAV is a known phenotypic component in patients with rare syndromic disease such as Marfan, Loeys-Dietz and Turner Syndromes for which causal genes are known7. Genetic studies on families displaying inherited non-syndromic BAV have proven fruitful and have revealed causative mutations in specific transcription factors5, signaling molecules6, and structural genes8. However, no single-gene model clearly explains BAV etiology in patients, thus supporting a complex network of interacting genes and pathways. We, therefore, hypothesized that population-based genetic studies would identify new pathways involved in BAV etiology. Thus, genome wide association studies were undertaken and revealed modest association with genes involved in the generation of primary cilia. Cilia are small microtubule-based organelles that are found on most mammalian cell types and can be classified as either motile or non-motile (primary). Primary cilia are solitary and function as mechano- and chemosensory organelles9. Although previously viewed as vestigial, recent studies have clearly demonstrated the functional importance of primary cilia in human disease. These diseases, termed ciliopathies, are a collection of congenital syndromic diseases (e.g. Joubert Syndrome, Bardet-Biedl Syndrome, Meckel-Gruber Syndrome and Autosomal Dominant Polycystic Kidney Disease) that affect many tissues, including the heart. Cardiac phenotypes are observed in most types of ciliopathy and are most commonly associated with BAV and aortic stenosis10. Additionally, our previous work has shown that knockout of intraflagellar transport protein 88 (IFT88) in mice leads to loss of cilia and a 70% penetrant BAV phenotype11. These clinical data support our hypothesis that cilia gene variants are associated with BAV in the population and are critical for aortic valve development. Genetic studies in zebrafish, mice, and humans identified a molecular trafficking complex (known as the exocyst) that is critical for cilia biogenesis in cardiac valves, and when dysregulated, can cause outflow tract defects, BAV and calcific aortic stenosis.
Methods
The data that support the findings of this study are available from the corresponding author upon request.
Materials.
All chemicals, unless stated otherwise, were purchased from Sigma-Aldrich.
Animal approval.
All experiments on zebrafish and mice were approved by the Institutional Animal Care and Use Committee (IACUC) of the Medical University of South Carolina and/or the Ralph H. Johnson VAMC.
Zebrafish husbandry and genotyping.
Adult zebrafish were maintained and raised in an Aquatic Habitats recirculating water system (Tecniplast) in a 14:10-hour light-dark cycle. The exoc5 mutant line was purchased from the Zebrafish International Resource Center (ZIRC, exoc5-sa23168). The exoc5 C377T point-nonsense mutation was verified by PCR and direct sequencing of both strands in heterozygote adults and mutant larvae progeny. Genomic DNA from clipped fins, or whole 3.5 dpf zebrafish, was extracted in 50 μL 1x lysis buffer (10 mM Tris-HCl pH 8.0, 50 mM KCl, 0.3% Tween 20, 0.3% NP40), denatured at 98 °C for 10 minutes, digested at 55 °C for 6 hours with 10 μg/mL proteinase K, and the reaction was stopped at 98 °C for 10 minutes. The PCR primers were: forward primer, 5’-CTATATAGACATGGAGCGGCAAT-3’; reverse primer: 5’- CCAACAATTCCTCACCTTCC-3’. Sequencing was performed by Genewiz with the forward PCR primer. The CRISPR mutant exoc5 zebrafish line was generated with the assistance of Dr. John Parant in Core B of the UAB P30 Hepatorenal Fibrocystic Disease Core Center. The CRISPR mutation was identified as a 13 bp insertion in exon 4, leading to a frameshift and premature stop codon. To confirm the mutation, genomic DNA from clipped fins or whole 3 dpf zebrafish with phenotypes was extracted and processed as described above. The PCR primers were: forward primer, 5’-CAAAGGTAGCCTTCCAGCAC-3’; reverse primer: 5’-CCTCTGTCTCGGGGTATTGA −3’. Sequencing was performed by Eurofins with the forward primer, 5’-TCCAGCATGTTTTTCTGGTG −3’(Eurofins.com).
Western blot analysis.
Dechorionated zebrafish embryos at 3.5 or 4 dpf were homogenized in SDS buffer containing protease inhibitor cocktail (Sigma) and phosphatase inhibitor (ThermoScientific) to perform Western blot analysis. The homogenized lysates were boiled for 5 minutes at 95 °C followed by centrifugation at 13,500 rpm for 20 minutes at 4 °C, and the supernatants were collected and mixed with 3x Laemmli sample buffer for the protein electrophoresis. The protein samples were separated on NuPage 4–12% Bis-Tris gels (Novex) and then transferred to a nitrocellulose membrane (Novex). The antibodies included: rabbit polyclonal anti-EXOC5 that we generated12, mouse anti-EXOC4 (Enzo), and mouse anti-GAPDH monoclonal antibody (Sigma), all at 1:1000 dilution. Secondary antibodies were from Jackson Immunoresearch Laboratories and Thermo Fisher Scientific.
Exoc5 mutant mRNA rescue experiments.
For rescue experiments of zebrafish exoc5 mutants, capped and polyadenylated mRNA of wild-type (WT) and ciliary-targeting sequence mutated human EXOC5 was synthesized in vitro using the mMESSAGE mMACHINE kit (Ambion). Two doses of EXOC5 mRNA (low: 100 pg or high: 250 pg) were injected, using a Sutter Instruments microinjector, into 100 embryos at the one-cell stage. At 3.5 dpf, twelve randomly selected larvae were imaged and individually genotyped by direct sequencing as outlined above.
Mouse husbandry and genotyping.
Animals were kept in a 12-hour light-dark cycle with food and water ad libitum. The generation and genotyping of our Exoc5f/f (aka Sec10) mice have been described previously13. The conditional Exoc5 knockout mice were generated by crossing Exoc5f/f with Nfatc1Cre(+) (a kind gift from Dr. Bin Zhou) and Tie2Cre(+) mice (Jackson Laboratories), and are designated as NfatC1Cre(+);Exoc5f/+, NfatC1Cre(+);Exoc5f/f, Tie2Cre(+);Exoc5f/+, and Tie2Cre(+);Exoc5f/f in the manuscript. Genotyping was performed by Transnetyx or inhouse with the KAPA Mouse Genotyping Kit (Kapabiosystems, #KK7301).
Mouse immunohistochemistry and fluorescence imaging.
Immunohistochemical and fluorescence stains were performed on 5 um, paraffin-embedded sections from E13.5, E15.5, P0, and adult wildtype and conditional Exoc5 mice as previously described11. Antibodies and their dilutions include: acetylated tubulin (Sigma, #T6793, 1:500), gamma tubulin (Abcam, #T6793, 1:1000), RUNX2 (Abcam, #ab192256, 1:250), ERK1/2 (Cell Signaling, #4695, 1:50), phospho-ERK1/2 (Cell Signaling, #4370, 1:40), cleaved NOTCH1 (Cell Signaling, #4147, 1:200), Hoescht (Life Technologies, #H3569, 1:10,000), and Life Technologies Alexa Fluors: 488 (#A-11029, #A-11034) 568 (#A-11004, #A-11036) and Cy5 (#A-10524) at 1:100. Slides were coverslipped using Invitrogen SlowFade Gold Antifade Reagent (#S36936). Images were captured using: Leica TCS SP5 AOBS Confocal Microscope System and LAS AF v2.6.3 Build 8173 Acquisition and Analysis Software, Zeiss Axioscope M2, or Olympus BH-2 brightfield microscope.
Mouse immunohistochemical stain quantification.
Cilia length: Measurements were taken from z-stack confocal images using Imaris 9.0 software as previously described11. Measurements were taken from all visible aortic cusps in the tissue section. N = 3 animals/genotype (>1500 cells total) with slides selected from the front, middle, and back of each aortic valve.
Alizarin red:
Guide slides from adult mice were stained using a standard alizarin red protocol. A total of 13 conditional heterozygous mice (6 with TAV and 7 with BAV) and 4 control mice were stained.
pERK/ERK:
Cells were counted with CellProfiler software. A pipeline was created to separate the channels of florescent images and then count the total number of pERK/ERK positive cells (red channel) and the total number cells (blue channel). Histological sections from similar regions of aortic valve were selected (n=3 animals/genotype), and images were edited to include only cells from the aortic cusps before counting.
Human BAV Genetic Studies.
Discovery Cohort.
480 Caucasian BAV cases collected with IRB approval and written individual patient consent were genotyped with the Omni2.5 chip, yielding 2,379,855 genetic markers. From the Framingham Heart Study cohort, available at dbGap, 2,477 Caucasians without BAV were genotyped, using the HumanOmni5.0 bead chip with 4,271,233 genetic markers from dbGaP, and used as controls. Quality control (QC) of the genotype data from both cohorts was performed by jointly and separately re-calling all genotypes using GenomeStudio and PLINK. We considered markers with a MAF>1% and performed extensive principal components-based filtering for population stratification. After merging cases and controls and further QC, we used 452 BAV cases and 1,834 Caucasian controls for a common set of 1,355,128 single nucleotide polymorphisms (SNPs). Genotyping of the 15 most significant SNPs identified in the Discovery cohort, including those containing EXOC4,6,8, was performed upon a Replication cohort of 1,679 BAV cases and 894 control patients undergoing coronary artery bypass grafting. These studies were approved by the IRB at Harvard University.
Pathway Analysis.
Pathway analysis was performed using GeNets for the 33 genes +/− 150 kb from the 15 SNPs that were used for the Replication study. Genets statistically tested the connectivity of our gene set and then visualized how the significantly enriched pathways from the Molecular Significant Data Base, a collection of annotated gene sets for use with GSEA software (MSigDB database v6.2 updated July 2018), intersected with our gene set. In order to find any significant pathway between the genes, we ran the 33 genes of interest in the web-based program and identified the PID_ARF6_TRAFFICKINGPATHWAY. The number of candidate genes in this pathway was 49. The p-value of 1.48E-05 indicated that the network was significantly more connected than one would expect for a gene set of this size and the global connectivity of its genes, i.e. the degree distribution of the genes in the entire network.
Site-directed mutagenesis.
To inhibit the ciliary targeting sequence VxPx in human EXOC5, the QuikChange Site-Directed Mutagenesis Kit (Stratagene, #200518) was used for in vitro site-directed mutagenesis of proline 668 to alanine. The primers were:
5’ CTTCTGGTAGTTGCCGCAGATAATTTAAAGCAAGTCTGC 3’
5’ GCAGACTTGCTTTAAATTATCTGCGGCAACTACCAGAAG 3’
Plasmid pcDNA3-hEXOC5 was used as template. Sequencing confirmed the specific amino acid change.
Echocardiography.
Echocardiographic images were acquired in the parasternal long axis, short axis, and apical views (40 MHz probe, Visualsonics 2100, Fujifilm, Ontario, Canada). Two-dimensional images were used for quantification of the diameter of the aortic root. Pulse wave and color Doppler modes in conjunction with timing of the cardiac cycle (electrocardiogram) were used to visualize flow patterns at the aortic valves. A total of 5 conditional heterozygous mice and 11 controls were imaged.
Statistics
GWAS:
A logistic regression analysis based on an additive genetic model was performed for association analysis adjusted for gender and 8 PCAs using PLINK. Additional statistical analyses are described above, and in the text of the manuscript.
Cilia length:
A generalized linear mixed model was used to compare the likelihood of cilia presence across genotypes employing a logit link function and litter, mouse, and genotype by litter interaction as random effects in the model. For mice with cilia present, mixed model analysis of variance was used to compare mouse average cilia length across genotypes, with litter serving as a random effect.
pERK/ERK:
Student t-test was used to compare the fold change in pERK positive cells between genotypes.
Results
We performed a GWAS study using a Discovery cohort of 452 BAV patients and 1,834 Caucasian population-controls (Figure S1A), examining a common set of 1,355,128 SNPs (demographic data, and clinical variables are shown in Figure S1B). 734,104 SNPs were assigned to 26,290 coding genes. We identified 1,889 SNPs associated with BAV at p<1×10−4. The Manhattan and QQ plots are shown in Figures S2A & B. Working on the hypothesis that SNPs in/near cilia genes may be over-represented in the BAV population, we analyzed the ciliome in the context of our dataset. A large number of cilia genes (28/303, or 9.2%), from the SysCilia set, were found +/−150 kb from the 1,889 most significant SNPs, including members of the exocyst complex, required for promoting cilia biogenesis (EXOC4, EXOC6, and EXOC8 (Figure S3A)). Another well-characterized cilia list14, yielded 156 cilia genes (156/1,925, or 8.1%) within 150 kb of the 1,889 SNPs (Figure S3B), also including exocyst members: EXOC4, 6, and 8. From the Discovery GWAS, three of the eight exocyst members (EXOC4, p=7.78 X 10−7; EXOC6, p=5.63 X 10−5; and EXOC8, p=6.64 X 10−5) and an exocyst regulator, CDC4215–17 (p=5.88 X 10−5) were associated with BAV at p<1×10−4. EXOC4, EXOC6, EXOC8 were 17.11 kb, 148.4 kb, and 79.41 kb from their respective SNPs. Regional plots +/− 500 kb from the associated SNPs are shown in Figure S4. Hypergeometric probability analysis showed that the overlap between the exocyst genes and the genes located within 150 kb of the 1,889 SNPs most associated with BAV (using the cutoff p<1×10−4) was significantly higher than expected by chance alone (p=0.033).
Genotyping of the 15 most significant SNPs identified in the Discovery cohort, including those containing EXOC4,6,8, was performed upon a replication cohort of 1,679 BAV cases and 894 controls (Table 1). The combined p-value for the exocyst components was 3.63 X 10−4. As these data suggested a modest association between BAV, the exocyst and ciliogenesis, we performed additional analyses in animal models to bolster support for the cilia biogenesis pathway as a molecular link to BAV etiology.
Table 1.
GWAS Studies.
| CHR | SNP | Ref. Allele | Minor Allele | MAF BAV Cases Discovery |
MAF Controls Discovery |
MAF BAV Cases Replication |
MAF Controls Replication |
Combined P–Values | CHR:Position | Near Gene |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs2807347 | A | C | 0.1051 | 0.1392 | 0.1355 | 0.1439 | 0.07796 | 1:22533608 | CDC42 |
| 1 | rs7543130 | C | A | 0.4354 | 0.4927 | 0.4419 | 0.5101 | 2.37E–08 | 1:100049785 | PALMD |
| 1 | rs17645143 | C | T | 0.3639 | 0.2965 | 0.321 | 0.2943 | 2.62E–05 | 1:109835757 | CELSR2 |
| 1 | rs3738777 | G | A | 0.3650 | 0.2977 | 0.3142 | 0.2951 | 0.000208 | 1:109836835 | CELSR2 |
| 1 | rs2491417 | G | A | 0.1648 | 0.1365 | 0.1538 | 0.1549 | 0.2332 | 1:231547888 | EXOC8 |
| 2 | rs1830321 | T | C | 0.4181 | 0.3957 | 0.4163 | 0.3736 | 0.001566 | 2:145825555 | TEX41 |
| 2 | rs62169409 | G | A | 0.2882 | 0.2849 | 0.3168 | 0.2876 | 0.0264 | 2:145904907 | TEX41 |
| 4 | rs73248479 | A | G | 0.0951 | 0.0462 | 0.04172 | 0.04083 | 0.000394 | 4:24754220 | LGI2 |
| 6 | rs10455872 | G | A | 0.0814 | 0.0600 | 0.07539 | 0.08669 | 0.7497 | 6:161010118 | LPA |
| 7 | rs10279531 | C | T | 0.2777 | 0.2166 | 0.2266 | 0.2122 | 0.000872 | 7:132920710 | EXOC4 |
| 10 | rs2488087 | A | G | 0.4303 | 0.4749 | 0.4633 | 0.4821 | 0.01966 | 10:94446041 | EXOC6 |
| 10 | rs12355604 | G | A | 0.1228 | 0.1585 | 0.1521 | 0.1396 | 0.326 | 10:94987241 | EXOC6 |
| 11 | rs41310330 | A | G | 0.0332 | 0.0153 | 0.02443 | 0.01566 | 0.000193 | 11:6631300 | DCHS1 |
| 13 | rs61965893 | A | G | 0.3385 | 0.3944 | 0.3536 | 0.3841 | 0.001709 | 13:95824227 | ABCC4 |
| 13 | rs9590221 | C | T | 0.4779 | 0.4198 | 0.4571 | 0.4418 | 0.006648 | 13:95907223 | ABCC4 |
Ref.=Reference, MAF=minor allele frequency
Fifteen top BAV GWAS hits from the Discovery Study of 452 BAV patients and 1,834 Caucasian population-controls were used in the Replication Study, with a separate cohort of 1,679 BAV patients and 894 control patients undergoing coronary artery bypass grafting.
The exocyst complex is a holocomplex of eight unique subunits18. Disruption of a key linker protein, EXOC5, is known to effectively dismantle this complex and impair ciliogenesis12. Using zebrafish, we generated homozygous exoc5 mutants (Figure S5) to test whether disruption of the exocyst complex results in impaired cardiac outlet development (Figures 1, S6). By Western blot analysis, we confirmed that exoc5 protein was virtually undetectable in exoc5 mutant larvae, and that exoc4 protein was also significantly decreased, when compared to WT siblings (Figure 1A). Mutant exoc5 zebrafish displayed gross phenotypes consistent with defects in other ciliary genes19. At 3.5 days post fertilization (dpf), all exoc5 mutants had a similar phenotype to our previous studies in exoc5 morphants15, 20, including pericardial edema, when compared to wild-type (WT) siblings (Figure 1B). The morphologic phenotype was consistently observed in ~25% of the progeny from heterozygous crosses, as would be expected by Mendelian inheritance. The exoc5 homozygous mutants only survived to 4 dpf and exhibited severe cardiac outflow obstruction (Figure 1C, D), as well as disruption of cardiac function (Supplementary Movies 1–3). The outflow tract of the exoc5 mutants appeared stenotic with dramatic narrowing of the arterial lumen, as is observed in patients with aortic stenosis. To ensure exoc5 loss caused the phenotype, an additional exoc5 mutant was generated using CRISPR gene editing21, and the resulting phenotype was identical to the exoc5 mutant described above (Figure S6).
Figure 1. Exoc5 mutant zebrafish display ciliopathic phenotypes and cardiac outflow tract stenosis.
(A) By Western blot analysis, exoc5 protein was virtually undetectable in exoc5 mutant larvae, and exoc4 protein was also significantly decreased, when compared to WT siblings. Quantification is shown below. (B) Lateral view of representative wild-type (exoc5+/+) and exoc5 homozygous mutant (exoc5−/−) zebrafish at 3.5 dpf. Exoc5 mutants showed cilia defects, including pericardial edema (arrowheads indicate pericardium). Scale bars 0.171 um. (C, D) Low magnification (left) and high magnification (right) of hematoxylin and eosin (H&E)-stained hearts of 3.5 dpf wild-type (C) and mutant (D) zebrafish. Orange arrowheads indicate the outflow tract between the ventricle and bulbus arteriosus, highlighting the severe stenosis in the exoc5 mutant. BA=bulbus arteriosus, V=ventricle, A=atrium. (E) Injection of wild-type human EXOC5 mRNA rescued the exoc5 mutant phenotype in zebrafish in a dose dependant manner. (F) Human EXOC5 mRNA with a mutated ciliary targeting sequence (VxPx to VxAx), in contrast to wild-type EXOC5 mRNA, was unable to efficiently rescue the cardiac phenotype. *= p < 0.05.
To confirm that the ciliary phenotypes observed in exoc5 mutants were caused specifically by loss of exoc5, we performed rescue experiments by injecting WT human EXOC5 mRNA into control and exoc5 mutant embryos at the 1–2 cell stage. At 3.5 dpf, we found that low dose (150 pg) reconstitution of EXOC5 mRNA, in exoc5 mutant embryos, partially rescued the mutant phenotypes, including cardiac edema; while high dose (250 pg) EXOC5 mRNA showed near complete phenotypic rescue (Figure 1E). EXOC5 contains a highly-conserved VxPx ciliary targeting sequence22 (Figure S7A). To further strengthen the link between primary cilia and the cardiac phenotype, we used site-directed mutagenesis to disrupt the ciliary targeting sequence of human EXOC5 (Figure S7B). Rescue of the exoc5 mutant zebrafish was significantly decreased using the mutated ciliary targeting sequence EXOC5 mRNA compared to WT EXOC5 mRNA (Figure 1F). Mutagenesis of the ciliary targeting sequence did not affect the stability of the EXOC5 protein, or its ability to bind other exocyst components23. These data demonstrate that dismantling the exocyst ciliary-trafficking complex during zebrafish development results in cardiac phenotypes that are reminiscent of patients with BAV and aortic stenosis24.
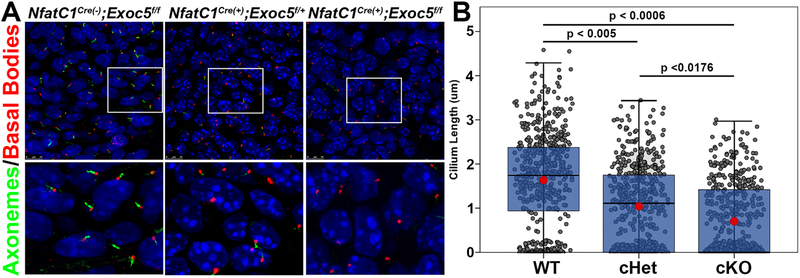
Based on our zebrafish findings, and our previous report that loss of cilia in outlet valves can cause a BAV phenotype in mice11, we decided to conditionally ablate the Exoc5 gene, using Exoc5f/f mice that we previously generated13. Aortic valves are built through integration of various cell types (endothelial, anterior heart field, and neural crest), with endothelial-derivatives providing the largest contribution to the aortic cusps3. Thus, knocking out Exoc5 in endocardial and endocardial-derived cells, using an NfatC1-Cre driver25, permits analyses of the function of the exocyst in a majority of cells that make up the aortic valves. Conditional homozygous knockout in EXOC5 mice (NfatC1Cre(+);Exoc5f/f) resulted in zero out of fifty-eight pups surviving to birth, and genotyping data revealed non-Mendelian birth ratios consistent with embryonic lethality (Figure 2A). Lethality of the NfatC1Cre(+);Exoc5f/f embryos occurred between E13.5 and E15.5. At these timepoints, NfatC1Cre(+);Exoc5f/f conditional knockout mice exhibited ventricular septal defects with large cellular voids at the junction between the interventricular membranous septum and atrioventricular septal complex, as well as evidence of apical coronary vascular hemorrhaging (Figure 2B). Due to the lack of P0 NfatC1Cre(+);Exoc5f/f mice, subsequent analyses were performed on conditional heterozygous animals (NfatC1Cre(+);Exoc5f/+). Three-dimensional reconstruction of P0 aortic valves revealed increased incidence of bicuspid aortic valves resulting from the fusion of the right and non-coronary cusps in conditional heterozygous (NfatC1Cre(+);Exoc5f/+) mice (Figure 2C). Whereas five out of eleven (45%) NfatC1Cre(+);Exoc5f/+ mice displayed BAV, with 4/5 being right, non-coronary (RNC) fusions and 1/5 with left, non-coronary (LNC) fusions, 100% of the conditional heterozygotes displayed dysmorphic aortic valves in addition to other cardiac phenotypes including ventricular septal defects and vascular hemorrhages. High-resolution confocal microscopy of E13.5 aortic cushions, confirmed a significant effect of exocyst loss on ciliogenesis, which preceded the BAV phenotype observed at P0 (Figure 3).
Figure 2. Exoc5 endocardial-specific knockout mice display homozygous embryonic lethality and BAV.
(A) Mice that are homozygous for Exoc5 deletion in endocardial and endocardial-derived cells (NfatC1Cre(+);Exoc5f/f) display embryonic lethality. WT refers to NfatC1Cre(+);Exoc5+/+, NfatC1Cre(−);Exoc5f/+, and NfatC1Cre(−);Exoc5f/f mice, while Het refers to NfatC1Cre(+);Exoc5f/+ mice, and KO refers to NfatC1Cre(+);Exoc5f/f mice. (B) Homozygous KO NfatC1Cre(+);Exoc5f/f embryos died between E13 and E15, most likely due to ventricular septal defects (first row, indicated by black arrows and dotted line) and progressive vascular haemorrhage (third row, indicated by black arrows). Embryos also showed anomalous attachment of the heart to the body cavity (second row, indicated by black arrows). (C) Representative 3D reconstructions of H&E-stained control (NfatC1Cre(−);Exoc5f/f) and conditional heterozygous KO (NfatC1Cre(+);Exoc5f/+) P0 aortic valves. Conditional heterozygous mice had an increased prevalence of bicuspid aortic valves (45%, n=11) and partially fused valves (data not shown). NC=non-coronary, RC=right coronary, LC=left coronary, RNC=right non-coronary fusion.
Figure 3. Exoc5 endocardial-specific knockout mice show disrupted ciliogenesis.
(A) Representative high-resolution confocal microscopy images of E13.5 aortic cushion cells. Axonemes (acetylated alpha tubulin) are shown in green and basal bodies (gamma tubulin) are shown in red. The bottom row is a higher magnification of the boxed area from the top row. (B) Graphic representation of all cilia measurements from E13.5 aortic cushion cells. Each gray dot represents the cilia length from a single cell (including dots at zero, which are indicative of no cilia). The blue boxes show the cilia length lower, median, and upper quartiles for each genotype and the red dot is the mean cilia length. WT control (NfatC1Cre(−);Exoc5f/f) mice had longer cilia, as well as a lower percentage of cells without cilia, than did the conditional heterozygous and homozygous Exoc5 knockout mice, indicating that ablation of Exoc5 disrupts ciliogenesis. P-values indicate the mean cilia length was statistically significant between all genotypes. The number of cells without cilia was also significant between the controls and knockouts (p < 0.007) and between controls and heterozygotes (p < 0.05). n=3 animals per genotype and >1500 cells.
Cilia and the exocyst are known to regulate MAPK signaling and disruption of exocyst-mediated ciliogenesis leads to MAPK pathway activation13, 20, 26. Additionally, recent work has shown involvement of the MAPK pathway in murine27, 28 and human BAV and calcific aortic stenosis29. As a readout for altered ciliogenic signaling, we analyzed whether the final downstream component of MAPK signaling, ERK (pERK1/2) was altered in the EXOC5 BAV model. In E13.5 control aortic valves, ERK phosphorylation was prominent in valve endocardium and interstitial cells, while in NfatC1Cre(+);Exoc5f/+ and NfatC1Cre(+);Exoc5f/f valves, ERK1/2 activation was significantly downregulated (Figure S8). Human BAV and CAVS samples have shown elevation of the MAPK pathway as a potential marker for disease progression29. Thus, we examined time points after which BAV is evident (P0) and CAVS is observed in the adult. As shown in Figure S9, pERK1/2 is elevated at neonatal and adult time points, indicating aberrant activation of the MAPK pathway, similar to results observed in human disease.
As our model of EXOC5 deficiency demonstrated fusion of the aortic cusps and molecular signatures consistent with the disease process observed in humans with calcific aortic stenosis, we evaluated long-term effects of the aortic valve phenotype on cardiac function. Echocardiography was performed on aged (18-month) NfatC1Cre(+);Exoc5f/+ mice and demonstrated evidence of stenosis and calcification (Figure 4A), biphasic depolarization in the QRS complex (Figure 4B), and significant aortic root dilation (Figure 4C). As the histological, molecular and echocardiographic data in both the mouse and zebrafish models suggested aortic stenosis, we further analyzed aged mice for evidence of calcification. Adult NfatC1Cre(+);Exoc5f/+ (six with TAV, five with RNC and two with LNC) and four control aortic valves were isolated from our aged mice and processed for histology and immunohistochemistry. In contrast to control valves (Figure 4D), 4/7 (57%) BAV and 3/6 (50%) TAV showed evidence of calcification, being positive for alizarin red (Figure 4E). Aortic valves were further analyzed for evidence of calcification by performing IHC staining for RUNX2 on control (NfatC1Cre(−);Exoc5f/f) and alizarin red-positive conditional heterozygote (NfatC1Cre(+);Exoc5f/+) aortic valves (N=3). RUNX2 signal by IHC was below the level of detection in control aortic valves (N=3). However, in each conditional heterozygote analyzed, positive RUNX2 expression was detected in areas of alizarin-red, indicating aberrant osteoblastic differentiation had occurred (Figure 4E).
Figure 4. Conditionally heterozygous Exoc5 adult mice have calcific valves.
Echocardiographic images were acquired in five conditional heterozygous mice and eleven controls, revealing several functional abnormalities in aortic blood flow in theNfatC1Cre(+);Exoc5f/+mice. (A) Color Doppler images of the aortic valve in an aged (18 month)NfatC1Cre(+);Exoc5f/+mouse and a control sibling from a 2D short axis view (within the dashed circle). Blood flow through the aortic valve in the control mouse was uniform (laminar; shown in blue) and was initiated soon after the QRS complex of the electrocardiogram (ECG; green trace (B)). In contrast, in theconditional heterozygous mice, aortic blood flow during the same phase of the cardiac cycle showed jets of high velocity flow around the aortic cusps (red and blue mixinghighlighted by white arrowheads) providing evidence of stenosis and calcification. (B) The biphasic depolarization pattern seen in the QRS complex of the conditional heterozygous mouse (right) could be suggestive of slower electrical conduction through one of the bundle branches. (C) Aortic rootdiameters of conditional heterozygous mice were around 24% larger than control mice. (D) Control valves did not stain positive for alizarin red or RUNX2. (E) In contrast,NfatC1Cre(+);Exoc5f/+adult (6 weeks old) aortic valves stained positive for alizarin red along the cusps and showed positive RUNX2 staining on sequential histological sections indicating areas of valve calcification. Brightfield images were taken to ensure melanocyte autofluorescence was not responsible for the positive staining.
Discussion
Bicuspid aortic valve disease is a frequent valvular abnormality that can lead to calcific aortic valve stenosis (CAVS). CAVS is the most prevalent acquired valvular heart disorder in the Western world and accounts for greater than $10 billion in health care costs in the U.S. each year30. Aortic stenosis displays increased severity and prevalence with age, with >12% of individuals over the age of 75 having CAVS24. 30–50% of CAVS patients have a BAV31, consistent with our GWAS patient population, in which 42% were diagnosed with aortic stenosis (data not shown). The genetic, molecular and cellular mechanisms that contribute to BAV and CAVS are poorly understood and few applicable models have been developed to study this disease5, 6. This paucity of data is likely reflected in the fact that there are currently no preventative medical therapies for CAVS and, in North America alone, 80,000 patients progress annually to severe symptomatic CAVS requiring aortic valve replacement (AVR).
We initiated our studies with patients that have BAV as a unifying disease phenotype. We hypothesized that BAV in the population may not commonly be caused by single damaging variants, but rather be caused by an association of various SNPs in converged genetic pathways. This appears to be the case from our current study, whereby the most significant SNPs generated from GWAS analyses fit within a cilia biogenesis regulatory network. Although these data do not provide definitive genetic evidence, they do support cilia and the exocyst as a target worthy of further validation. Additionally, these data suggest that this population of BAV patients may not harbor single damaging SNPs as is evident in inherited BAV families, but rather it is the combination of genetic variants in this pathway that lead to disease. Thus, it is possible that the combination of multiple SNPs in/near cilia genes is more relevant to BAV pathology than any one individual SNP.
Prior reports support cilia and the exocyst in the etiology of BAV/CAVS based on the following observations. First, patients with ciliopathies, which are syndromic diseases caused by mutations in functional components of primary cilia (e.g., Meckel-Gruber, Joubert and Bardet-Biedl Syndromes, autosomal dominant polycystic kidney disease), frequently present with BAV and calcific aortic valve stenosis10. In fact, a family with Joubert Syndrome, a well-described ciliopathy, has recently been identified with a damaging mutation in EXOC832, one of the exocyst genes identified in our GWAS analysis. Second, NOTCH1, the first gene identified in patients with inherited forms of aortic valve disease and CAVS, depends on primary cilia for cleavage and proper function6, 33–35. However, our analysis of NOTCH1 cleavage within the EXOC5 deficient valves failed to observe any changes in NOTCH1 activation (Figure S10). This result is consistent with our previous studies, which demonstrated that primary cilia are rarely observed on the aortic valve endocardium, where NOTCH1 is expressed11. Third, primary cilia are expressed during aortic valve development, and genetic ablation of cilia, by removal of IFT88, results in a highly penetrant (>70%) BAV phenotype11, the single most important predictor of aortic valve disease and aortic stenosis. Combined with the supportive GWAS-driven finding of the exocyst in BAV etiology, we sought to confirm these genetic findings and bolster support for this protein complex and pathway in human BAV by performing genetic ablation and rescue experiments in zebrafish and mice. Here we show that disrupting the exocyst through deletion of the key linker protein, EXOC5, results in a severe cardiac phenotype in zebrafish and mice, highlighting the importance of this trafficking pathway in normal heart development across species. Deletion of Exoc5 in mice results in shorter and less abundant cilia within the aortic valve, increased incidence of BAV, and calcification of aortic valves concomitant with elevation of pERK1/2, a marker of disease phenotype.
Further genomic analysis of the exocyst GWAS data also supports a role for the exocyst in BAV. GenoCanyon36 was used to measure the functional importance of genomic loci. GenoCanyon scores for BAV-associated SNPs near EXOC4, EXOC6, and EXOC8 were 0.94, 0.92, and 1.00, respectively, implying potential functional impact. We examined the functional role of all associated SNPs using the Human Enhancer Disease Database (HEDD)37, identifying the EXOC4 SNP (rs10279531) as a putative enhancer38. Pathway analysis using GeNets for the 33 genes +/−150 kb from the 15 SNPs that were used for the Replication study, showed that the ARF6 trafficking pathway was significantly associated at p=1.48 X 10−5. We have previously shown that ARF proteins interact with the exocyst to control ciliogenesis and MAPK signaling26, and others have shown that EXOC5 interacts directly with ARF639.
The embryonic lethality in the context of deletion of the exocyst from endocardium and endocardial-derived cells is intriguing and implicates a role for primary cilia not only in valvulogenesis, but also in endothelial cells within the interventricular septum and ventricular myocardium. Unlike the valve endocardium, primary cilia are present on ventricular endocardial cells (Figure S11), which may help to explain the vascular hemorrhaging that is the likely cause of death at E15.5.
The developmental origins of BAV and CAVS are still poorly understood. Motivated by the severity and high prevalence of BAV/CAVS, numerous groups have endeavoured to understand the embryological development of the aortic valve in an attempt to relate these findings back to patients with BAV. Included in the findings are the temporal-spatial mapping of various cell lineages that make up the aortic valve, as well as the identification of molecular and biomechanical pathways that regulate cell phenotype and function during the organization and maturation of the aortic cusps. It is now clear that formation of the cusps requires an interplay between mechanical forces and the cell types that contribute to its development, including: neural crest, endocardial, myocardial, and circulating cells. The majority of cells within the aortic valves originate from endothelial cells that undergo endothelial to mesenchymal transformation (EndoMT). Consistent with this finding, endocardial-specific removal of Exoc5 in mice is sufficient to generate a highly penetrant BAV phenotype that proceeds to CAVS in adulthood. A similar phenotype was observed when we genetically removed Exoc5 with the pan-endothelial Cre, Tie2Cre, demonstrating that the BAV phenotype and lethality are due to defects in endothelial cell function within the heart (Figure S12).
Currently, only a few, human genetic-based models of BAV/CAVS have been reported. Here, we report the EXOC5 model as an applicable model to study mechanisms underlying BAV formation and its progression to a stenotic valve. Through the use of zebrafish, mouse, and human genetics, we have demonstrated that BAV, and its related phenotypes, which affect between 38 to 91 million people worldwide3, can occur in association with defects in exocyst-mediated ciliogenic programs. As included in this study, GWAS analyses on BAV populations may need to invoke combinatorial SNP analyses to define convergent, parallel, or overlapping pathways that contribute to disease etiology. Future studies will take advantage of these new data and animal models to further tease apart the underlying mechanisms that could inform new, non-surgical therapies to benefit BAV/CAVS patients.
Supplementary Material
Clinical Perspective.
What is new?
Combinatorial genetic, molecular and biochemical data reveal the ciliogenic pathway as a common cause for bicuspid aortic valve.
Ablation of the ciliogenic shuttling complex (“exocyst”) results in primary cilia defects and penetrant bicuspid aortic valve disease and aortic stenosis in zebrafish, mice and humans.
What are the clinical implications?
Here we report novel, human genetic-based models, which develop BAV and aortic stenosis with high penetrance, and can now be used to understand the progression of the disease as well as serve as models for identifying therapies for patient benefit.
Acknowledgments
The UAB P30 Hepatorenal Fibrocystic Disease Core Center (NIH P30DK074038) is gratefully acknowledged for generating the second exoc5 zebrafish mutant line using CRISPR gene editing, as well as for previously generating the Exoc5f/f mouse line. Dr. Martina Brueckner is gratefully acknowledged for critical reading of the manuscript. R.A.N., J.H.L., S.B., and D.B.F. designed the research studies and wrote the manuscript. D.B.F., L.G., G.P.L., Y.S., Y.D., X.Z., R.M., R.M., R.S., K.A.T., K.M., J.G., and R.S. conducted experiments and acquired data. R.A.N., D.B.F., B.F., J.H.L., G.P.L., R.A.S., S.B., M.H., P.G., and D.C. analyzed and interpreted the data, and reviewed the manuscript.
Sources of Funding
This work was supported in part by grants from the Veterans Affairs (Merit Award I01 BX000820 to J.H.L), NIH (GM103444 to R.A.N, P30DK074038 to J.H.L., R21EY025034 to G.P.L., R01HL131546 to R.A.N., P20GM103444 to R.A.N., R01HL127692 to R.A.N., K01DK087852 to B.F., P20GM103457–8293 to B.F., T32HL007260 to D.B.F and K.M, and F31HL142159 to D.B.F.,R01GM122078 to D.C., and R21CA209848 to D.C.), and American Heart Association (17CSA33590067 to R.A.N, S.B., J.L. and 18PRE34080172 to L.G.).
Non-standard Abbreviations and Acronyms:
- BAV
bicuspid aortic valve
- CAVS
calcific aortic valve stenosis
- IHC
immunohistochemistry
- CHD
congenital heart disease
- IFT88
intraflagellar transport protein 88
- TAV
tricuspid aortic valve
- WT
wildtype
- Dpf
days post fertilization
- RNC
right non-coronary
- LNC
left non-coronary
- E#
embryonic day #
- P0
postnatal day 0
- AVR
aortic valve replacement
Footnotes
Disclosures
None. The authors declare that there are no conflicts of interest.
References
- 1.Michelena HI, Prakash SK, Della Corte A, Bissell MM, Anavekar N, Mathieu P, Bosse Y, Limongelli G, Bossone E, Benson DW, Lancellotti P, Isselbacher EM, Enriquez-Sarano M, Sundt TM, 3rd, Pibarot P, Evangelista A, Milewicz DM, Body SC and Investigators BA. Bicuspid aortic valve: identifying knowledge gaps and rising to the challenge from the International Bicuspid Aortic Valve Consortium (BAVCon). Circulation. 2014;129:2691–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ and Roos-Hesselink JW. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58:2241–2247. [DOI] [PubMed] [Google Scholar]
- 3.Martin PS, Kloesel B, Norris RA, Lindsay M, Milan D and Body SC. Embryonic Development of the Bicuspid Aortic Valve. J Cardiovasc Dev Dis. 2015;2:248–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mordi I and Tzemos N. Bicuspid aortic valve disease: a comprehensive review. Cardiol Res Pract. 2012;2012:196037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonachea EM, Chang SW, Zender G, LaHaye S, Fitzgerald-Butt S, McBride KL and Garg V. Rare GATA5 sequence variants identified in individuals with bicuspid aortic valve. Pediatr Res. 2014;76:211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD and Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. [DOI] [PubMed] [Google Scholar]
- 7.Giusti B, Sticchi E, De Cario R, Magi A, Nistri S and Pepe G. Genetic Bases of Bicuspid Aortic Valve: The Contribution of Traditional and High-Throughput Sequencing Approaches on Research and Diagnosis. Front Physiol. 2017;8:612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pepe G, Nistri S, Giusti B, Sticchi E, Attanasio M, Porciani C, Abbate R, Bonow RO, Yacoub M and Gensini GF. Identification of fibrillin 1 gene mutations in patients with bicuspid aortic valve (BAV) without Marfan syndrome. BMC Med Genet. 2014;15:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badano JL, Mitsuma N, Beales PL and Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. [DOI] [PubMed] [Google Scholar]
- 10.Karp N, Grosse-Wortmann L and Bowdin S. Severe aortic stenosis, bicuspid aortic valve and atrial septal defect in a child with Joubert Syndrome and Related Disorders (JSRD) - a case report and review of congenital heart defects reported in the human ciliopathies. Eur J Med Genet. 2012;55:605–610. [DOI] [PubMed] [Google Scholar]
- 11.Toomer KA, Fulmer D, Guo L, Drohan A, Peterson N, Swanson P, Brooks B, Mukherjee R, Body S, Lipschutz JH, Wessels A and Norris RA. A role for primary cilia in aortic valve development and disease. Dev Dyn. 2017;246:625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuo X, Guo W and Lipschutz JH. The exocyst protein Sec10 is necessary for primary ciliogenesis and cystogenesis in vitro. Mol Biol Cell. 2009;20:2522–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fogelgren B, Polgar N, Lui VH, Lee AJ, Tamashiro KK, Napoli JA, Walton CB, Zuo X and Lipschutz JH. Urothelial Defects from Targeted Inactivation of Exocyst Sec10 in Mice Cause Ureteropelvic Junction Obstructions. PLoS One. 2015;10:e0129346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Jr., Rux JJ, Speicher DW and Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi SY, Baek JI, Zuo X, Kim SH, Dunaief JL and Lipschutz JH. Cdc42 and sec10 Are Required for Normal Retinal Development in Zebrafish. Invest Ophthalmol Vis Sci. 2015;56:3361–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi SY, Chacon-Heszele MF, Huang L, McKenna S, Wilson FP, Zuo X and Lipschutz JH. Cdc42 deficiency causes ciliary abnormalities and cystic kidneys. J Am Soc Nephrol. 2013;24:1435–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuo X, Fogelgren B and Lipschutz JH. The small GTPase Cdc42 is necessary for primary ciliogenesis in renal tubular epithelial cells. J Biol Chem. 2011;286:22469–22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipschutz JH and Mostov KE. Exocytosis: the many masters of the exocyst. Curr Biol. 2002;12:R212–R214. [DOI] [PubMed] [Google Scholar]
- 19.Stoetzel C, Bar S, De Craene JO, Scheidecker S, Etard C, Chicher J, Reck JR, Perrault I, Geoffroy V, Chennen K, Strahle U, Hammann P, Friant S and Dollfus H. A mutation in VPS15 (PIK3R4) causes a ciliopathy and affects IFT20 release from the cis–Golgi. Nat Commun. 2016;7:13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fogelgren B, Lin SY, Zuo X, Jaffe KM, Park KM, Reichert RJ, Bell PD, Burdine RD and Lipschutz JH. The exocyst protein Sec10 interacts with Polycystin-2 and knockdown causes PKD-phenotypes. PLoS Genet. 2011;7:e1001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobo GP, Fulmer D, Guo L, Zuo X, Dang Y, Kim SH, Su Y, George K, Obert E, Fogelgren B, Nihalani D, Norris RA, Rohrer B and Lipschutz JH. The exocyst is required for photoreceptor ciliogenesis and retinal development. J Biol Chem. 2017;292:14814–14826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazelova J, Astuto-Gribble L, Inoue H, Tam BM, Schonteich E, Prekeris R, Moritz OL, Randazzo PA and Deretic D. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 2009;28:183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuo X, Lobo G, Fulmer D, Guo L, Dang Y, Su Y, Ilatovskaya DV, Nihalani D, Rohrer B, Body SC, Norris RA and Lipschutz JH. The exocyst acting through the primary cilium is necessary for renal ciliogenesis, cystogenesis, and tubulogenesis. J Biol Chem. 2019. 294(17):6710–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michelena HI, Khanna AD, Mahoney D, Margaryan E, Topilsky Y, Suri RM, Eidem B, Edwards WD, Sundt TM, 3rd and Enriquez-Sarano M. Incidence of aortic complications in patients with bicuspid aortic valves. JAMA. 2011;306:1104–1112. [DOI] [PubMed] [Google Scholar]
- 25.Wu B, Zhang Z, Lui W, Chen X, Wang Y, Chamberlain AA, Moreno-Rodriguez RA, Markwald RR, O’Rourke BP, Sharp DJ, Zheng D, Lenz J, Baldwin HS, Chang CP and Zhou B. Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell. 2012;151:1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seixas C, Choi SY, Polgar N, Umberger NL, East MP, Zuo X, Moreiras H, Ghossoub R, Benmerah A, Kahn RA, Fogelgren B, Caspary T, Lipschutz JH and Barral DC. Arl13b and the exocyst interact synergistically in ciliogenesis. Mol Biol Cell. 2016;27:308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu X and Masters KS. Role of the MAPK/ERK pathway in valvular interstitial cell calcification. Am J Physiol Heart Circ Physiol. 2009;296:H1748–H1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munjal C, Jegga AG, Opoka AM, Stoilov I, Norris RA, Thomas CJ, Smith JM, Mecham RP, Bressan GM and Hinton RB. Inhibition of MAPK-Erk pathway in vivo attenuates aortic valve disease processes in Emilin1-deficient mouse model. Physiol Rep. 2017;5:e13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadaba JR, Martinez-Martinez E, Arrieta V, Alvarez V, Fernandez-Celis A, Ibarrola J, Melero A, Rossignol P, Cachofeiro V and Lopez-Andres N. Role for Galectin-3 in Calcific Aortic Valve Stenosis. J Am Heart Assoc. 2016;5:e004360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajamannan NM, Gersh B and Bonow RO. Calcific aortic stenosis: from bench to the bedside--emerging clinical and cellular concepts. Heart. 2003;89:801–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts WC and Ko JM. Frequency by decades of unicuspid, bicuspid, and tricuspid aortic valves in adults having isolated aortic valve replacement for aortic stenosis, with or without associated aortic regurgitation. Circulation. 2005;111:920–925. [DOI] [PubMed] [Google Scholar]
- 32.Dixon-Salazar TJ, Silhavy JL, Udpa N, Schroth J, Bielas S, Schaffer AE, Olvera J, Bafna V, Zaki MS, Abdel-Salam GH, Mansour LA, Selim L, Abdel-Hadi S, Marzouki N, Ben-Omran T, Al-Saana NA, Sonmez FM, Celep F, Azam M, Hill KJ, Collazo A, Fenstermaker AG, Novarino G, Akizu N, Garimella KV, Sougnez C, Russ C, Gabriel SB and Gleeson JG. Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med. 2012;4:138ra78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ezratty EJ, Stokes N, Chai S, Shah AS, Williams SE and Fuchs E. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell. 2011;145:1129–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grisanti L, Revenkova E, Gordon RE and Iomini C. Primary cilia maintain corneal epithelial homeostasis by regulation of the Notch signaling pathway. Development. 2016;143:2160–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsao PN, Vasconcelos M, Izvolsky KI, Qian J, Lu J and Cardoso WV. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development. 2009;136:2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.GenoCanyon, http://genocanyon.med.yale.edu/.
- 37.Human Enhancer Disease Database, http://zdzlab.einstein.yu.edu/1/hedd.php.
- 38.Identification of the EXOC4 SNP (rs10279531) as a putative enhancer from the Human Enhancer Disease Database, http://zdzlab.einstein.yu.edu/1/hedd/detail.action.php?idno=1269828.
- 39.Prigent M, Dubois T, Raposo G, Derrien V, Tenza D, Rosse C, Camonis J and Chavrier P. ARF6 controls post-endocytic recycling through its downstream exocyst complex effector. J Cell Biol. 2003;163:1111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.