Pregenomic RNA (pgRNA) is a direct transcription product of hepatitis B virus (HBV) covalently closed circular DNA (cccDNA), and it plays important roles in viral genome amplification and replication. This study was designed to investigate whether serum pgRNA is a strong alternative marker for reflecting HBV cccDNA levels and to analyze the correlation between serum pgRNA, serum HBV DNA, and hepatitis B surface antigen (HBsAg).
KEYWORDS: real-time fluorescence quantitative PCR, hepatitis B virus, pregenomic RNA, covalently closed circular DNA, clinical outcomes
ABSTRACT
Pregenomic RNA (pgRNA) is a direct transcription product of hepatitis B virus (HBV) covalently closed circular DNA (cccDNA), and it plays important roles in viral genome amplification and replication. This study was designed to investigate whether serum pgRNA is a strong alternative marker for reflecting HBV cccDNA levels and to analyze the correlation between serum pgRNA, serum HBV DNA, and hepatitis B surface antigen (HBsAg). A total of 400 HBV-infected patients who received nucleos(t)ide analog (NA) therapy with different clinical outcomes were involved in this research. Case groups included asymptomatic hepatitis B virus carrier (ASC), chronic hepatitis B (CHB), liver cirrhosis (LC), and hepatocellular carcinoma (HCC) patients, with 100 patients in each group. The results showed that the levels of HBV pgRNA had significant differences between these 4 groups. Serum pgRNA levels correlated well with serum HBV DNA and HBsAg levels (HBV pgRNA levels versus HBV DNA levels, r = 0.58, P < 0.001; HBV pgRNA levels versus HBsAg levels, r = 0.47, P < 0.001). In addition, we focused on the 108 HBV-infected patients with HBV DNA levels of <500 IU/ml; it was surprising to find that in 17.57% (13/74) of cases, HBV pgRNA could be detected even when the HBV DNA level was below 20 IU/ml. In conclusion, HBV pgRNA levels in serum can be a surrogate marker for intrahepatic HBV cccDNA compared with serum HBV DNA and HBsAg. The detection of serum HBV pgRNA levels may provide a reference for clinical monitoring of cccDNA levels and the selection of appropriate timing for discontinuing antiviral therapy, especially when HBV DNA levels are below the detection limit.
INTRODUCTION
Hepatitis B is a potentially life-threatening liver infection caused by hepatitis B virus (HBV) (1). Hepatitis B creates a serious global public health problem. About 2 billion people worldwide are infected with HBV, including about 350 million patients with chronic hepatitis B (CHB) (2). CHB is closely related to the occurrence of liver cirrhosis and liver cancer; therefore, the control and treatment of HBV infection has important medical and social significance. While the covalently closed circular DNA (cccDNA) in liver tissue is a key factor reflecting the replication of HBV and the formation of an infectious state, the complete elimination of HBV cccDNA has become the gold standard for evaluating the cure of CHB patients (3–6). In theory, as long as there is one replication-capable HBV cccDNA remaining in the infected liver cells, there may be a risk of recurrence once antiviral therapy is stopped (7). Previous studies (8–10) have proposed a “parafunctional cure” as the new endpoint of HBV treatment. It is based on the disappearance or silencing of HBV cccDNA and uses a sustained level of HBV RNA below the limit of detection and a low level of hepatitis B surface antigen (HBsAg) as the criteria. This is of great significance in guiding new antiviral drug research and safe drug withdrawal. Therefore, it is important to comprehensively monitor the transcriptional activity of HBV cccDNA in infected cells. However, the detection of intrahepatic HBV cccDNA relies on liver biopsy. Unfortunately, many factors, including damage caused by invasive examination, low specimen yield, the subjectivity of different observers, and potential complications of hepatic puncture, hinder the widespread application of liver biopsy (11–13).
HBV pregenomic RNA (pgRNA) is an intermediate of HBV replication (14). In recent years, many researchers have confirmed that the HBV pgRNA in serum is derived from the active transcription of HBV cccDNA in the infected hepatocytes (15–17). In particular, the serum HBV RNA level in patients receiving nucleoside(t)ide analog (NA) treatment can reflect the presence of the cccDNA and its transcriptional activity in hepatocytes, when the reverse transcription and DNA synthesis of the virus are inhibited (8, 18). These pgRNAs are present in the nucleocapsid of mature viral particles in the form of HBV RNA virus-like particles and are associated with persistent viral infection and virological rebound risk (19). To further verify whether HBV pgRNA in serum is a good alternative marker for reflecting HBV cccDNA levels in hepatocytes, we collected serum samples from 400 patients with different clinical outcomes after HBV infection and who received NA treatment. We examined the serum levels of HBV pgRNA, HBV DNA, HBsAg, and hepatitis B e antigen (HBeAg), and performed statistical analysis of these serological indicators. The results will facilitate better understanding of the infection status of HBV and better evaluation of the antiviral treatment efficacy. The promising findings reported here will provide a new potential predictive index for the disease progression of HBV-infected patients with different clinical outcomes and for judgement of the treatment endpoint.
MATERIALS AND METHODS
Study subjects.
The subjects of this study were 400 HBV-infected patients with different clinical outcomes in the department of the Liver Disease Center of the First Affiliated Hospital of Fujian Medical University who received entecavir (ETV) or tenofovir (TDF) for more than 6 months in 2015 to 2018. Case group included ASC, CHB, LC, and HCC patients, with 100 cases in each of the four groups. The clinical specimens were collected according to diagnostic criteria of the Chinese Medical Association’s 2013 Expert Consensus on Standardized Diagnosis and Treatment of Primary Liver Cancer and the 2015 Guidelines for Prevention and Treatment of Chronic Hepatitis B. Serum samples were rapidly stored at −80°C until further use. It should be noted that for all patients in the ASC group, HBV DNA results were positive, and the histological activity index (HAI) or fibrosis index (F) were greater than or equal to 2 according to the Metavir scoring system. The control group included healthy people and people with other viral infections, such as patients infected with hepatitis C virus (HCV), herpes simplex virus (HSV), and Epstein-Barr virus (EBV). The studies described above are in line with the ethical principles of the Helsinki Declaration and were approved by the Ethics Committee of Fujian Medical University. Each patient enrolled signed an informed consent form.
Cell lines.
The stable HBV-expressing human liver cancer cell line HepG2.2.15 was maintained in Dulbecco’s modified Eagle medium (Thermo Fisher Scientific, Shanghai, China) supplemented with 10% fetal bovine serum (Gibco, Mexico) at 37°C in 5% CO2.
Real-time quantitative PCR to detect serum pgRNA level.
The EasyPure viral RNA kit (TransGen Biotech, Beijing, China) was used for extraction of total HBV RNA from HepG 2.2.15 cell supernatant. After treated with DNase I (Thermo Fisher Scientific, Waltham, MA, USA), the sample was then reverse transcribed into cDNA using a RevertAid First Strand DNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA) with a HBV RT primer. The specific upstream and downstream primers of HBV pgRNA were used in the PCR amplification. After regular PCR amplification, the PCR product was purified with a Universal purification and recovery kit (Tiangen, Beijing, China), then ligated with pEASY-Blunt plasmid (TransGen Biotech, Beijing, China). The construct was transformed into Trans1-T1 competent cells. Positive colonies were picked and further screened by PCR. The positive plasmids were isolated using a TIANprep mini plasmid kit (Tiangen, Beijing, China) and were sequenced. A standard was prepared in 10-fold dilutions by serial dilution of the plasmid with Easy Dilution Buffer (Tiangen, Beijing, China) at 1 × 101 to 1 × 1010 copies/ml. A StepOne Plus fluorescence quantitative PCR (qPCR) machine (Life Technologies, USA) was used to detect the serum pgRNA level of the HBV-infected patients. The specific primers and TaqMan probe used are the same as those mentioned in a previous article (15) (Table 1).
TABLE 1.
Primers and probe for real-time quantitative PCRa
| Primer name | Sequence (5′–3′) |
|---|---|
| Forward primer | AYAGACCATCAAATGCCC |
| Reverse primer | ATTCTCAGACCGTAGCACACGACAC |
| Probe | CTTATCAACACTTCCGGARACTACTGTTGTTAGAC |
PCR product length, 167 bp.
Detection of serum HBV DNA, HBsAg, and HBeAg levels.
HBV DNA was extracted using a viral genomic DNA extraction kit (Beijing Xinnuo Company, China) according to the manufacturer’s instructions and then stored at −80°C until use. The levels of HBV DNA and high-sensitivity HBV DNA were quantified with a commercially available real-time fluorescence quantitative kit (Shengxiang Biotech Company, Hunan Province, China), then measured on a LightCycler 480 fluorescence quantitative PCR machine (F. Hoffmann-La Roche AG, Basel, Switzerland) and an ABI 7500 real-time PCR machine (Life Technologies, USA). The levels of HBV serum markers HBsAg and HBeAg were quantitatively examined with a commercially available kit (Abbott Laboratories, USA) using the Architect i4000 microparticle chemiluminescence immunoassay analyzer (Abbott Laboratories, USA).
Statistical analysis.
Serum HBV DNA levels and HBsAg and HBeAg concentrations were log transformed; data statistics and analysis were performed using SPSS 22.0 software (SPSS Inc., Chicago, IL, USA). Figures was prepared using Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). The data were subjected to the one-sample Kolmogorov-Smirnov normality test. Data with a normal distribution were presented as mean plus or minus standard deviation, and the mean between the two groups was compared using a t test. The one-way analysis of variance (ANOVA) test was used to compare normal distribution variables. Data with a nonnormal distribution were presented as the median (25th percentile [P25], 75th percentile [P75]), and the Kruskal-Wallis test was used to compare abnormal variables. Degree of correlation was analyzed using Pearson’s correlation analysis. The difference was statistically significant at a P value of <0.05.
RESULTS
Successfully established real-time qPCR method to detect serum pgRNA levels.
The established real-time qPCR method was used to evaluate linear range, lower detection limit, repeatability, specificity, and sensitivity. The linear range of the plasmid standard detected by real-time qPCR was ∼1 × 1010 to 1 × 103 copies/ml; the detection limit was 1 × 103 copies/ml; the intra-assay coefficient of variation was between 0.13% and 0.73%; the interassay coefficient of variation was between 0.16% and 0.63%. The specificity was tested with 15 different serum specimens from the control group, including healthy people and people with other viral infections, such as HCV, HSV and EBV. The results showed that the serum levels of HBV pgRNA of those samples were below the limit of detection and were very specific. In consideration of the complex background of serum samples, we also did recovery experiments to verify the accuracy of the method. According to the calculation formula of recovery rate = recovery concentration/addition concentration × 100%, the average recovery rate was 93.785%, in line with the requirements of the PRC Pharmaceutical Industry Standards (20).
Analysis and comparison of serum HBV pgRNA, HBV DNA, and HBsAg levels in patients with different clinical outcomes of HBV infection.
This study involved 400 HBV-infected patients with different clinical outcomes, comprising 100 cases each in ASC, CHB, LC, and HCC groups. Essential information, including gender, age, HBV DNA level, HBV pgRNA level, HBsAg level, and HBeAg level in each group, is presented in Table 2.
TABLE 2.
Clinical characteristics of patients with different clinical outcomes of HBV infection
| Variableb | Patient groupa
|
|||
|---|---|---|---|---|
| ASC | CHB | LC | HCC | |
| Male/female | 67/33 | 56/44 | 72/28 | 87/13 |
| Age (yrs) | 34.87 ± 12.5 | 36.75 ± 12.02 | 54.9 ± 12.69 | 55.79 ± 12.39 |
| HBV viral load (log10 copies/ml) | 4.42 ± 1.72 | 4.7 ± 1.77 | 3.68 ± 0.89 | 3.64 ± 0.65 |
| HBV pgRNA (log10 copies/ml) | 4.32 ± 2.36 | 4.41 ± 2.51 | 3.22 ± 2.13 | 3.14 ± 2.12 |
| HBsAg (IU/ml) | 7,393.32 ± 16,302.77 | 6,157.9 ± 11,583.95 | 1,831.56 ± 6,005.84 | 1,276.72 ± 1,697.07 |
| HBeAg (S/CO) | 251.15 ± 537.47 | 207.48 ± 485.05 | 52.39 ± 215.95 | 9.15 ± 77.28 |
ASC, asymptomatic hepatitis B virus carrier; CHB, chronic hepatitis B; LC, liver cirrhosis; HCC, hepatocellular carcinoma.
pgRNA, pregenomic RNA; HBsAg, hepatitis B virus surface antigen; HBeAg, hepatitis B virus e antigen; S/CO, specimen/cutoff ratio.
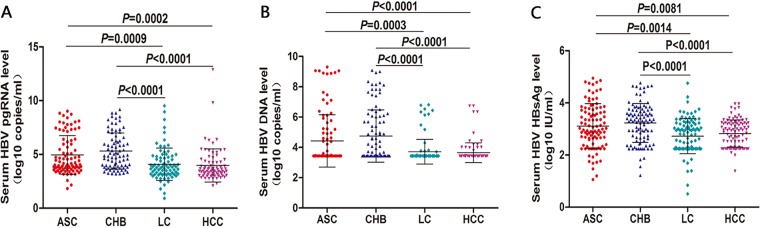
In the analysis of the changes in different serological markers of HBV-infected patients with different clinical outcomes, we found that there was a significant difference in HBV pgRNA levels between ASC and LC groups (4.32 ± 2.36 log10 copies/ml versus 3.22 ± 2.13 log10 copies/ml; t = 3.389, P = 0.0009), between ASC and HCC groups (4.32 ± 2.36 log10 copies/ml versus 3.14 ± 2.12 log10 copies/ml; t = 3.766, P = 0.0002), between CHB and LC groups (4.41 ± 2.51 log10 copies/ml versus 3.22 ± 2.13 log10 copies/ml; t = 4.911, P < 0.001), and between CHB and HCC groups (4.41 ± 2.51 log10 copies/ml versus 3.22 ± 2.13 log10 copies/ml; t = 5.275, P < 0.001) (Fig. 1A).
FIG 1.
Analysis and comparison of serum HBV pgRNA, HBV DNA, and HBsAg levels in patients with different clinical outcomes of HBV infection. The baseline serum levels of HBV pgRNA (A), HBV DNA (B), and HBV HBsAg (C) were compared between the ASC group (n = 100), CHB group (n = 100), LC group (n = 100), and HCC group (n = 100).
In the analysis of the HBV DNA levels in HBV-infected patients with different clinical outcomes, a significant difference in HBV DNA levels was found between ASC and LC groups (4.42 ± 1.72 log10 copies/ml versus 3.68 ± 0.89 log10 copies/ml; t = 3.700, P = 0.0003), between ASC and the HCC groups (4.42 ± 1.72 log10 copies/ml versus 3.64 ± 0.65 log10 copies/ml; t = 4.238, P < 0.001), between CHB and LC groups (4.7 ± 1.77 log10 copies/ml versus 3.68 ± 0.89 log10 copies/ml; t = 5.375, P < 0.001), and between CHB and HCC groups (4.7 ± 1.77 log10 copies/ml versus 3.64 ± 0.65 log10 copies/ml; t = 5.980, P < 0.001) (Fig. 1B).
In the analysis of the HBsAg levels of HBV-infected patients with different clinical outcomes, the results showed that there was a significant difference in HBsAg levels between ASC and LC groups (7,393.32 ± 16,302.77 IU/ml versus 1,831.56 ± 6,005.84 IU/ml; t = 3.552, P = 0.0014), between ASC and the HCC groups (7,393.32 ± 16,302.77 IU/ml versus 1,276.72 ± 1,697.07 IU/ml; t = 2.784, P = 0.0081), between CHB and LC groups (6,157.9 ± 11,583.95 IU/ml versus 1,831.56 ± 6,005.84 IU/ml; t = 5.076, P < 0.001), and between HCC and CHB groups (6,157.9 ± 11,583.95 IU/ml versus 1,276.72 ± 1,697.07 IU/ml; t = 4.405, P < 0.001) (Fig. 1C).
Correlation analysis of different serological indicators in HBV-infected patients.
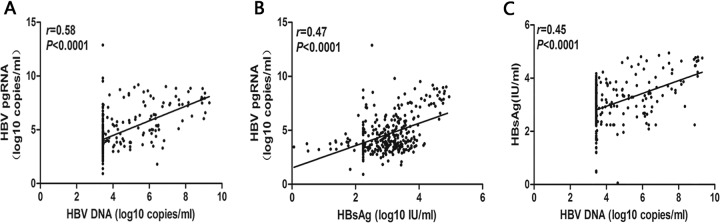
By analyzing the correlation between different serological indicators of HBV-infected patients, positive correlations was found between HBV DNA, HBV pgRNA, and HBsAg levels (HBV pgRNA group versus HBV DNA group: r = 0.58, P < 0.001, Fig. 2A; HBV pgRNA group versus HBsAg group: r = 0.47, P < 0.001, Fig. 2B; and HBsAg group versus HBV DNA group: r = 0.45, P < 0.001, Fig. 2C).
FIG 2.
Correlation analysis of different serological indicators in HBV-infected patients. The correlation between HBV pgRNA and HBV DNA was analyzed in patients with HBV infection (A). The correlation between HBV pgRNA and HBsAg was analyzed in patients with HBV infection (B). The correlation between the HBsAg group and HBV DNA was analyzed in patients with HBV infection (C).
Correlation analysis of different serological indicators of HBV-infected patients with different clinical outcomes.
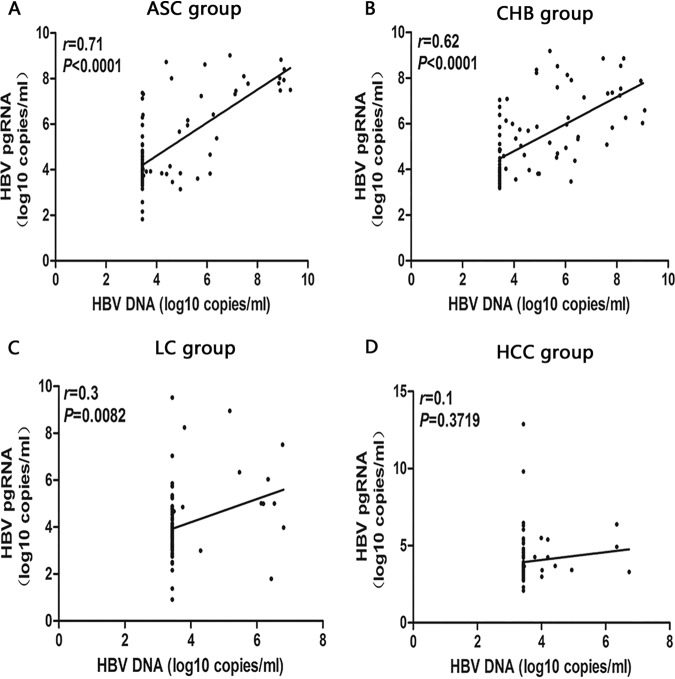
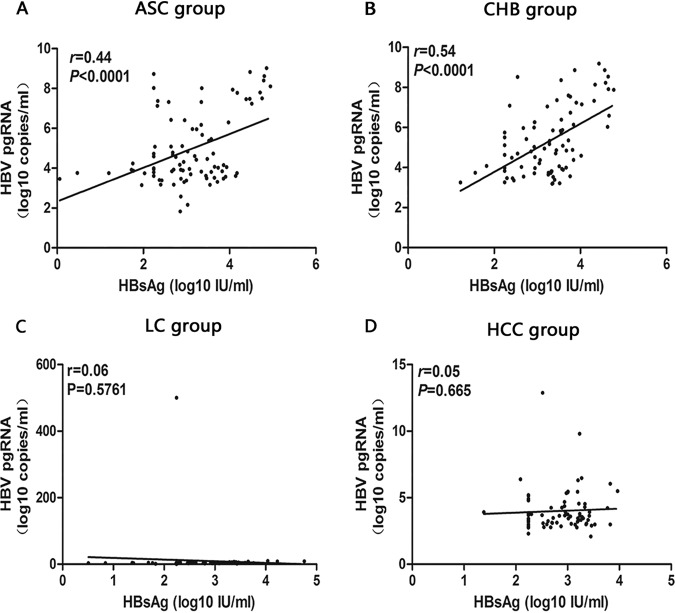
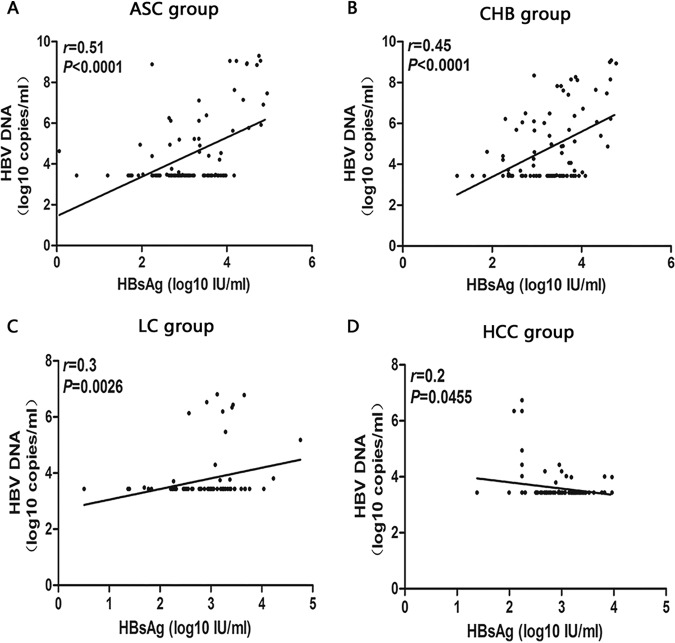
The correlation analysis of different serological markers of HBV-infected patients with different clinical outcomes showed that there was a positive correlation between HBV pgRNA and HBV DNA levels in ASC and CHB groups (ASC group: r = 0.71, P < 0.001, Fig. 3A; CHB group: r = 0.62, P < 0.001, Fig. 3B), between HBV pgRNA and HBsAg levels in ASC and CHB groups (ASC group: r = 0.44, P < 0.001, Fig. 4A) CHB group: r = 0.54, P < 0.001, Fig. 4B), and between HBV DNA and HBsAg levels in ASC and CHB groups (ASC group: r = 0.51, P < 0.001, Fig. 5A; CHB group: r = 0.45, P < 0.001, Fig. 5B).
FIG 3.
Analysis of the correlation between serum HBV pgRNA and HBV DNA in patients with ASC (A), CHB (B), LC (C), and HCC (D).
FIG 4.
Analysis of the correlation between serum HBV pgRNA and HBsAg in patients with ASC (A), CHB (B), LC (C), and HCC (4D).
FIG 5.
Analysis of the correlation between serum HBV DNA and HBsAg in patients with ASC (5A), CHB (B), LC (C), and HCC (5D).
Comparing the levels of HBV pgRNA and HBV DNA and their correlation in HBeAg-positive and -negative groups.
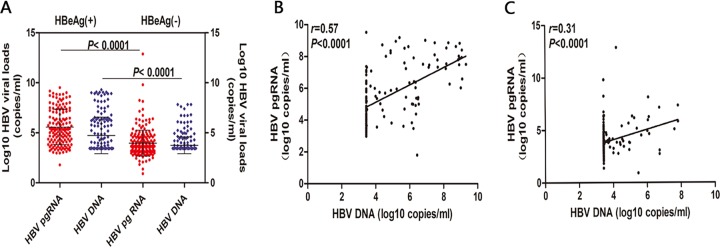
The specimens of 400 HBV patients were divided into an HBeAg-positive group and an HBeAg-negative group. Analysis showed that HBV pgRNA and HBV DNA levels were significantly different in the HBeAg-positive group (5.04 ± 2.49 log10 copies/ml and 4.85 ± 1.97 log10 copies/ml) from those in the HBeAg-negative group (3.11 ± 1.99 log10 copies/ml and 3.72 ± 0.91 log10 copies/ml) (HBV pgRNA group: t = 9.987, P < 0.001; HBV DNA group: t = 7.545, P < 0.001; Fig. 6A). HBV pgRNA and HBV DNA levels were positively correlated in both HBeAg-positive and HBeAg-negative groups, but the correlation was stronger in the former than in the latter (HBeAg-positive group: r = 0.57, P < 0.001, Fig. 6B; HBeAg-negative group: r = 0.31, P < 0.001, Fig. 6 C).
FIG 6.
Comparison of serum levels of HBV pgRNA and HBV DNA according to HBeAg classification and correlation of serum levels of HBV pgRNA and HBV DNA classified by HBeAg. The baseline serum levels of HBV pgRNA and HBV DNA were compared between the HBeAg-positive [HBeAg(+)] and HBeAg-negative [HBeAg(−)] patients (A). The correlation of serum levels of HBV pgRNA and HBV DNA was analyzed in HBeAg(+) patients (B). The correlation of serum levels of HBV pgRNA and HBV DNA was analyzed in HBeAg(−) patients (C).
Analysis of detection rate of HBV pgRNA in specimens with HBV DNA levels of <500 IU/ml.
Detection and statistical analysis of serological markers in 400 HBV-infected patients indicated that the detection rate of HBV pgRNA in serum samples with HBV DNA levels of <500 IU/ml was 78.9%, and the undetected rate was 21.1%. Based on this result, we selected specimens from 108 HBV-infected and HBeAg-negative patients for further analysis, including 74 specimens with HBV DNA levels of <20 IU/ml and 34 specimens with HBV DNA levels of >20 IU/ml but <500 IU/ml. Statistical analysis indicated detection rates of HBV pgRNA in both groups of 17.57% (13/74) and 41.18% (14/34), respectively. The undetected rates of HBV pgRNA in both groups were 82.43% (61/74) and 58.82% (20/34), respectively. Essential information about these 108 patients is presented at Table 3.
TABLE 3.
Clinical characteristics of patients with an HBV DNA level of <500 IU/ml
| Clinical characteristic | HBV DNA level |
|
|---|---|---|
| 20–500 IU/ml | <20 IU/ml | |
| No. male (no. female) | 25 (9) | 54 (20) |
| Age (yrs) | 44.5 ± 12.20 | 45 ± 14.58 |
| HBV viral load (log10 copies/ml) | 2.70 ± 0.37 | 2.04 ± 0.39 |
| HBV pgRNA (log10 copies/ml) | 2.04 (1.68, 2.04)a | 0 (0, 0.63)a |
| HBsAg (IU/ml) | 2,241.47 ± 717.09 | 1,394.34 ± 370.55 |
| HBeAg (S/CO) | 0.47 ± 0.25 | 0.39 ± 0.19 |
aNonnormal distribution data, results expressed as the median (25th percentile [P25], 75th percentile [P75]).
DISCUSSION
Chronic hepatitis B affects approximately 2 billion people worldwide, with an estimated 15 to 40% progressing to cirrhosis or hepatocellular carcinoma (21). Early diagnosis and treatment of CHB can reduce the incidence of cirrhosis and HCC. It always has been a challenge to find biological markers in peripheral blood that can effectively reflect the transcriptional activity of hepatitis virus and allow observation of the therapeutic effect of drugs. Currently, we rely mainly on the levels of HBsAg, HBeAg, HBV DNA, and alanine aminotransferase (ALT) in serum, combined with liver fibrosis examination, in order to distinguish different periods of HBV infection, determine the timing of treatment, and judge the timing of drug withdrawal. In this study, we successfully established a real-time qPCR method to quantitatively detect HBV pgRNA levels in sera of HBV-infected patients. Analysis of the correlation between pgRNA and traditional serological and molecular markers helps to determine whether it can be used as an alternative marker.
Studies have reported that the level of serum HBsAg in CHB patients is closely related to the transcriptional status of HBV cccDNA in the nucleus of hepatocytes (22). The decrease of serum HBsAg level was correlated with the decrease of serum HBV DNA level or HBV cccDNA level in hepatocyte nucleus, suggesting that, to a certain extent, the level of serum HBsAg could reflect the clearance of HBV cccDNA in hepatocyte nucleus. Our results showed that HBV DNA and HBsAg levels were significantly different among the four groups. It is worth noting that both HBV DNA and HBsAg levels were higher in the CHB group than those in the LC group (t = 5.375, P < 0.001; t = 5.076, P < 0.001), which is consistent with the literature mentioned above. In addition, the expression of serum HBV pgRNA in the CHB and LC groups (t = 4.911, P < 0.001) was also consistent with that of HBV DNA and HBsAg. The disappearance of serum HBsAg is considered to be the standard for CHB “functional cure” (13, 23). However, due to the frequent integration of HBV DNA into HBV-infected hepatocytes (18, 24), it may be the reason for the sustained low expression of serum HBsAg. Under these circumstances, serum HBsAg may not reflect the activity of cccDNA in the liver. Unlike serum HBsAg, 3.5-kb serum pgRNA is produced only from cccDNA. Therefore, serum HBV RNA can accurately reflect the status of cccDNA in the liver.
Studies have shown that there is a positive correlation between full-length HBV RNA and HBV DNA levels in serum of CHB patients (25). Our results also confirmed that serum HBV pgRNA level and HBV DNA expression level were well correlated in HBV-infected patients (r = 0.58, P < 0.001). The serum HBV pgRNA level was also positively correlated with HBsAg level (r = 0.47, P < 0.001). There were some correlations between serum HBV pgRNA, HBV DNA, and HBsAg levels in HBV-infected patients with different clinical outcomes. In particular, in the ASC group and CHB group, the levels of HBV pgRNA and HBV DNA (ASC group: r = 0.71, P < 0.001; CHB group: r = 0.62, P < 0.001), HBV pgRNA and HBsAg (ASC group: r = 0.44, P < 0.001; CHB group: r = 0.54, P < 0.001), and HBV DNA and HBsAg (ASC group: r = 0.51, P < 0.001; CHB group: r = 0.45, P < 0.001) were positively correlated. These results suggest that the detection of HBV pgRNA levels in sera of HBV-infected patients is comparable with the detection of other traditional diagnostic indicators. It is worth noting that, despite positive correlations among the various HBV serologic and molecular markers, the degree of correlation was not good. The weaker correlation coefficient value might be attributed to the accumulation of viral variation, as well as to the diversity of host backgrounds in clinical research.
At present, the evaluation of the efficacy of antiviral therapy relies on the detection of HBV DNA levels. However, HBV DNA in HBV-infected patients below the detection limit only indicates that reverse transcription of the virus is inhibited, and cannot be used as a clinical index for drug withdrawal (7, 26). To validate this theory, we analyzed the serological markers of 400 HBV-infected patients with HBV DNA levels of <500 IU/ml, from which the detection rate of HBV pgRNA was 78.9%. Intrigued by this finding, we selected 108 HBeAg-negative patients with HBV DNA levels of <500 IU/ml and further divided these clinical specimens into two groups according to HBV DNA levels. In one group, 74 patients had HBV DNA levels below 20 IU/ml. In another group, 34 patients had HBV DNA levels above 20 IU/ml but below 500 IU/ml. We were surprised to find that in 17.57% (13/74) of cases, HBV pgRNA could be detected even when the HBV DNA level was below 20 IU/ml. On the other hand, this suggested that an HBV DNA level below the detection limit cannot be used as a clinical index for drug withdrawal and that it provides a favorable proof for the previously proposed concept of parafunctional cure (27, 28). Therefore, the detection of HBV DNA copy number alone to assess the efficacy of antiviral therapy has certain limitations. Compared with HBV DNA, serum HBV pgRNA has an advantage in monitoring changes in sustained viral response and cccDNA levels during treatment.
In summary, the detection of serum HBV pgRNA levels in HBV-infected patients can be a potential alternative marker and provides a reference for clinical monitoring of cccDNA levels and the selection of appropriate timing for discontinuing antiviral therapy. Especially when HBV DNA levels are below the detection limit, the detection of HBV pgRNA levels can help to determine whether the criteria for parafunctional cure are met and to evaluate the efficacy of antiviral therapy. It can also provide meaningful guidance for rational clinical medication.
ACKNOWLEDGMENTS
The study was supported by grants from the National Natural Science Foundation of China (grant 81971996 and 81572067) and by Joint Funds for the Innovation of Science and Technology, Fujian Province (grant 2016Y9020).
REFERENCES
- 1.Krause A, Haberkorn U, Mier W. 2018. Strategies for the treatment of HBV/HDV. Eur J Pharmacol 833:379–391. doi: 10.1016/j.ejphar.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 2.Jung KS, Park JY, Chon YE, Kim HS, Kang W, Kim BK, Kim SU, Kim do Y, Han KH, Ahn SH. 2016. Clinical outcomes and predictors for relapse after cessation of oral antiviral treatment in chronic hepatitis B patients. J Gastroenterol 51:830–839. doi: 10.1007/s00535-015-1153-1. [DOI] [PubMed] [Google Scholar]
- 3.Lai CL, Wong D, Ip P, Kopaniszen M, Seto WK, Fung J, Huang FY, Lee B, Cullaro G, Chong CK, Wu R, Cheng C, Yuen J, Ngai V, Yuen MF. 2017. Reduction of covalently closed circular DNA with long-term nucleos(t)ide analogue treatment in chronic hepatitis B. J Hepatol 66:275–281. doi: 10.1016/j.jhep.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 4.Liang LB, Zhu X, Yan LB, Du LY, Liu C, Liao J, Tang H. 2016. Quantitative intrahepatic HBV cccDNA correlates with histological liver inflammation in chronic hepatitis B virus infection. Int J Infect Dis 52:77–82. doi: 10.1016/j.ijid.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 5.Gill US, Kennedy P. 2017. Current therapeutic approaches for HBV infected patients. J Hepatol 67:412–414. doi: 10.1016/j.jhep.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Nassal M. 2015. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 7.Levrero M, Testoni B, Zoulim F. 2016. HBV cure: why, how, when? Curr Opin Virol 18:135–143. doi: 10.1016/j.coviro.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Jiang M, Xue J, Yan H, Liang X. 2019. Serum HBV RNA quantification: useful for monitoring natural history of chronic hepatitis B infection. BMC Gastroenterol 19:53. doi: 10.1186/s12876-019-0966-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lok AS, Zoulim F, Dusheiko G, Ghany MG. 2017. Hepatitis B cure: from discovery to regulatory approval. J Hepatol 67:847–861. doi: 10.1016/j.jhep.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Lucifora J, Protzer U. 2016. Attacking hepatitis B virus cccDNA—the holy grail to hepatitis B cure. J Hepatol 64:S41–S48. doi: 10.1016/j.jhep.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Allweiss L, Dandri M. 2017. The role of cccDNA in HBV maintenance. Viruses 9:156. doi: 10.3390/v9060156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong J, Ying J, Qiu X, Lu Y, Zhang M. 2018. Advanced strategies for eliminating the cccDNA of HBV. Dig Dis Sci 63:7–15. doi: 10.1007/s10620-017-4842-1. [DOI] [PubMed] [Google Scholar]
- 13.Chang J, Guo F, Zhao X, Guo JT. 2014. Therapeutic strategies for a functional cure of chronic hepatitis B virus infection. Acta Pharm Sin B 4:248–257. doi: 10.1016/j.apsb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan Z, Pionek K, Unchwaniwala N, Maguire ML, Loeb DD, Zlotnick A. 2015. The interface between hepatitis B virus capsid proteins affects self-assembly, pregenomic RNA packaging, and reverse transcription. J Virol 89:3275–3284. doi: 10.1128/JVI.03545-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Shen T, Huang X, Kumar GR, Chen X, Zeng Z, Zhang R, Chen R, Li T, Zhang T, Yuan Q, Li PC, Huang Q, Colonno R, Jia J, Hou J, McCrae MA, Gao Z, Ren H, Xia N, Zhuang H, Lu F. 2016. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol 65:700–710. doi: 10.1016/j.jhep.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 16.Bai F, Yano Y, Fukumoto T, Takebe A, Tanaka M, Kuramitsu K, Anggorowati N, Rinonce HT, Widasari DI, Saito M, Hirano H, Hayakumo T, Seo Y, Azuma T, Ku Y, Hayashi Y. 2013. Quantification of pregenomic RNA and covalently closed circular DNA in hepatitis B virus-related hepatocellular carcinoma. Int J Hepatol 2013:849290. doi: 10.1155/2013/849290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Wen J, Xiao W, Zhang B. 2019. Pregenomic RNA: how to assist the management of chronic hepatitis B? Rev Med Virol 29:e2051. doi: 10.1002/rmv.2051. [DOI] [PubMed] [Google Scholar]
- 18.Wei L, Kao JH. 2017. Benefits of long-term therapy with nucleos(t)ide analogues in treatment-naive patients with chronic hepatitis B. Curr Med Res Opin 33:495–504. doi: 10.1080/03007995.2016.1264932. [DOI] [PubMed] [Google Scholar]
- 19.Huang H, Wang J, Li W, Chen R, Chen X, Zhang F, Xu D, Lu F. 2018. Serum HBV DNA plus RNA shows superiority in reflecting the activity of intrahepatic cccDNA in treatment-naive HBV-infected individuals. J Clin Virol 99–100:71–78. doi: 10.1016/j.jcv.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 20.China Food and Drug Administration. 2010. Reagents for nucleic acid amplification. PRC Pharmaceutical Industry Standard YY_T1182-2010 China Food and Drug Administration, Beijing, China. [Google Scholar]
- 21.Rapti I, Hadziyannis S. 2015. Risk for hepatocellular carcinoma in the course of chronic hepatitis B virus infection and the protective effect of therapy with nucleos(t)ide analogues. World J Hepatol 7:1064–1073. doi: 10.4254/wjh.v7.i8.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson AJ, Nguyen T, Iser D, Ayres A, Jackson K, Littlejohn M, Slavin J, Bowden S, Gane EJ, Abbott W, Lau GK, Lewin SR, Visvanathan K, Desmond PV, Locarnini SA. 2010. Serum hepatitis B surface antigen and hepatitis B e antigen titers: disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology 51:1933–1944. doi: 10.1002/hep.23571. [DOI] [PubMed] [Google Scholar]
- 23.Petersen J, Thompson AJ, Levrero M. 2016. Aiming for cure in HBV and HDV infection. J Hepatol 65:835–848. doi: 10.1016/j.jhep.2016.05.043. [DOI] [PubMed] [Google Scholar]
- 24.Tu T, Budzinska MA, Shackel NA, Urban S. 2017. HBV DNA integration: molecular mechanisms and clinical implications. Viruses 9:E75. doi: 10.3390/v9040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatakeyama T, Noguchi C, Hiraga N, Mori N, Tsuge M, Imamura M, Takahashi S, Kawakami Y, Fujimoto Y, Ochi H, Abe H, Maekawa T, Kawakami H, Yatsuji H, Aisaka Y, Kohno H, Aimitsu S, Chayama K. 2007. Serum HBV RNA is a predictor of early emergence of the YMDD mutant in patients treated with lamivudine. Hepatology 45:1179–1186. doi: 10.1002/hep.21581. [DOI] [PubMed] [Google Scholar]
- 26.Sarin SK, Kumar M, Lau GK, Abbas Z, Chan HL, Chen CJ, Chen DS, Chen HL, Chen PJ, Chien RN, Dokmeci AK, Gane E, Hou JL, Jafri W, Jia J, Kim JH, Lai CL, Lee HC, Lim SG, Liu CJ, Locarnini S, Al Mahtab M, Mohamed R, Omata M, Park J, Piratvisuth T, Sharma BC, Sollano J, Wang FS, Wei L, Yuen MF, Zheng SS, Kao JH. 2016. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int 10:1–98. doi: 10.1007/s12072-015-9675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giersch K, Allweiss L, Volz T, Dandri M, Lutgehetmann M. 2017. Serum HBV pgRNA as a clinical marker for cccDNA activity. J Hepatol 66:460–462. doi: 10.1016/j.jhep.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Du M, Huang H, Chen R, Niu J, Jiang J, Zhuang H, Lu F. 2017. Reply to: “Serum HBV pgRNA as a clinical marker for cccDNA activity”: consistent loss of serum HBV RNA might predict the “para-functional cure” of chronic hepatitis B. J Hepatol 66:462–463. doi: 10.1016/j.jhep.2016.10.034. [DOI] [PubMed] [Google Scholar]