Hepatitis E, a liver disease caused by infection with the hepatitis E virus (HEV), is a worldwide emerging disease. The diagnosis is based on the detection of viral RNA and of HEV-specific immunoglobulins (Ig). For the latter, various assays are commercially available but still lack harmonization. In this study, a Luminex-based multiplex serological assay was established that measures the presence of total IgG, IgA, and IgM antibodies, targeting a short peptide derived from the viral E2 protein.
KEYWORD: S hepatitis E virus, E2 antigen, multiplex assay, anti-HEV immunoglobulins, antibody prevalence
ABSTRACT
Hepatitis E, a liver disease caused by infection with the hepatitis E virus (HEV), is a worldwide emerging disease. The diagnosis is based on the detection of viral RNA and of HEV-specific immunoglobulins (Ig). For the latter, various assays are commercially available but still lack harmonization. In this study, a Luminex-based multiplex serological assay was established that measures the presence of total IgG, IgA, and IgM antibodies, targeting a short peptide derived from the viral E2 protein. For the validation, 160 serum samples with a known HEV serostatus were used to determine the assay cutoff and accuracy. Thereby, HEV IgG- and RNA-positive sera were identified with a sensitivity of 100% and a specificity of 98% (95% confidence interval [CI], 94% to 100%). Application of the assay by retesting 514 serum samples previously characterized with different HEV-IgG or total antibody tests revealed a high level of agreement between the assays (Cohen’s kappa, 0.58 to 0.99). The established method is highly sensitive and specific and can be easily implemented in a multiplex format to facilitate rapid differential diagnostics with a few microliters of sample input.
INTRODUCTION
The hepatitis E virus (HEV) represents a nonenveloped positive-stranded RNA virus that belongs to the genus Orthohepevirus within the family Hepeviridae (1). The viral genome is organized in three open reading frames (ORFs) flanked by nontranslated regions. ORF1 codes for nonstructural proteins necessary for viral replication. ORF2 bears the information for a major capsid protein and overlaps with ORF3, which encodes a phosphoprotein of various functions (2–5).
In 1978, hepatitis E was recognized for the first time as a distinct disease during the course of an outbreak of a non-A non-B hepatitis in Kashmir, India (6). A few years later, HEV was identified as the causative agent of this infection in a self-experimentation (7).
Nearly all human HEV infections are caused by one of four HEV genotypes (gt), named HEV-1, -2, -3, and -4, which all belong to the Orthohepevirus A species (5). gt HEV-1 and HEV-2 are restricted to humans. Both are highly endemic in developing countries, and infections mainly occur through consumption of contaminated freshwater (8, 9). In contrast, gt HEV-3 and HEV-4 exhibit a wide host range, including other mammalian species such as pigs, wild boar, and deer (1, 10). gt HEV-3 is of major importance in industrialized countries. Here, it accounts for autochthonous infections, mainly through ingestion of raw or undercooked pork but also via contaminated blood products and direct contact with infected animals (11–15). Most HEV infections are asymptomatic and self-limiting (16).
However, estimates from a global burden of disease study for 2005 show that HEV-1/-2 infection can lead to up to 3.4 million symptomatic cases and 70,000 deaths per year in selected regions (17–19). Acute infections by HEV-3/-4 predominantly occur in middle-aged or elderly patients with preexisting liver conditions. In immunocompromised patients, HEV-3/-4 infection may even take a chronic course, which requires reduction of immunosuppression and treatment with ribavirin or interferon α (20, 21). Furthermore, extrahepatic manifestations with neurological or renal symptoms have been reported (21).
For prevention of HEV infection, vaccine development was initiated. Virus-like particles (VLP) overexpressed from baculovirus vectors carrying a partial HEV genome are highly immunogenic. Capsid proteins were also generated by expression of ORF2 in Escherichia coli. The VLP p239, or ORF2-E2, was successfully tested in clinical trials and has been licensed in China as a commercial vaccine, named Hecolin. This vaccine shows high efficacy in areas with cocirculation of HEV-1 and -4 (22, 23). There is also indirect evidence that the vaccine will be protective against HEV-2/-3 infection (24).
Serological diagnosis of acute HEV infection is based on the detection of HEV-specific IgM antibodies or the recent appearance or significant increase in titers of virus-specific IgG antibodies (25). Commercially available HEV antibody assays, however, exhibit marked variations in sensitivity and specificity (26). These tests mainly use purified proteins of ORF2 and -3 as antigens (27–30).
The World Health Organization now recognizes hepatitis E infection as a public health issue in many developing countries, particularly among at risk populations such as pregnant women, individuals living in camps for displaced persons, and in outbreak situations (31). This emphasizes the need for reliable assays for the detection of acute or past HEV infection. The similar clinical pictures of acute hepatitis caused by diverse hepatotropic viruses and microorganisms can be a diagnostic challenge. Some of these antigens could be detected simultaneously in a multiplex assay.
Here, we developed a method for the detection of HEV antibodies based on Luminex technology (32). This test utilizes the ORF2-E2 peptide conjugated with fluorophore-labeled magnetic beads to identify total anti-HEV immunoglobulins. Assay validation was performed with sera of known HEV IgG status. Our assay needs only a few microliters of serum input. In addition, antigens of other hepatotropic viruses, bacteria, and parasites can be included to facilitate rapid differential diagnosis of liver disease.
MATERIALS AND METHODS
A multiplex serological assay for HEV was established according to the previously published method for the detection of hepatitis A virus (HAV)-specific antibodies (33).
Spectrally distinct fluorescent beads were coupled to recombinant HEV proteins, and bound serum antibodies were subsequently quantified. Using two sets of validation sera with predetermined HEV serostatus, a threshold was calculated that allows the classification of samples as either HEV seronegative or positive, and the accuracy of the assay was determined.
Generation of recombinant HEV proteins.
The complete HEV sequence of the gt1 Burma strain is available in the GenBank database (L08816.1) (34). Nucleotide sequences of ORF2 (nucleotides [nt] 5123 to 7105) and ORF3 (nt 5082 to 5453) were codon optimized for expression in E. coli and commercially synthesized (Eurofins Genomics, Ebersberg, Germany). In addition, a short version of ORF2, named ORF2-E2 (nt 6303 to 6941) was PCR amplified from the ORF2 sequence. All clones were verified by sequence analysis. These antigens were expressed as N-terminal glutathione transferase (GST) fusion proteins using a modified pGEX4T3 vector, as described previously (35). For this, transformed E. coli BL21 cells (Amersham Pharmacia, Amersham, UK) were grown at 20°C overnight in Terrific broth, followed by lysis using a high-pressure homogenizer (HTU-DIGI-Press; G. Heinemann Ultraschall und Labortechnik, Schwäbisch Gmünd, Germany). Full-length antigen expression was verified by Western blotting and GST capture enzyme-linked immunosorbent assay (ELISA) to estimate concentrations of the specific antigens, as previously described by Sehr et al. (35). A concentration of ≤20 μg/ml total protein lysate was found to be sufficient to reach antigen saturation, and beads were coupled with antigen-containing lysates diluted to 1 mg/ml.
Multiplex serology.
Carboxylated, fluorescence-labeled magnetic beads (MagPlex; Luminex Corporation, Austin, TX, USA) were coupled with glutathione-casein (GC) according to the descriptions by Waterboer et al. (32) and Bohm et al. (33) by using a magnetic separator (Dyna Mag-2; Life Technologies, Carlsbad, CA, USA) for the washing steps. Recombinant HEV antigens were bound to and affinity purified on the GC-coupled beads. The complete bead set was used to analyze serum (2 μl) in a final dilution of 1:100 in PVX buffer (phosphate-buffered saline [PBS], 0.8% polyvinylpyrrolidone, 0.5% polyvinyl alcohol, 1 mg/ml casein, 2.5% CBS-K100 [Millipore]). A biotinylated polyvalent secondary antibody [goat anti-human IgA, IgM, IgG(H+L); Dianova, Hamburg, Germany) diluted 1:1,000 in PBS (pH 7.4) containing 1 g/liter casein, showed the presence of total human antibody bound to the HEV antigens coupled to the beads. The Luminex xPONENT software package for Luminex LX100 was used for analysis (version 3.1 build 971).
Assay validation and statistical analysis.
Two serum panels were used for the assay validation, which was conducted in accordance with previous reports (33, 36). A third serum panel was used to compare our HEV multiplex assay to commercially available assays (37). Panel 1, independently tested by four technicians, was used to calculate the individual positive/negative mean fluorescence intensity (MFI) cutoff values for antigens ORF2, ORF2-E2, and ORF3, as well as the assay cutoff values needed to correctly identify the predefined sera as negative or positive for HEV immunoglobulins. Normal distribution for all antigens was assessed after transformation into natural logarithm (ln) scale and exclusion of outliers (±1.5 interquartile range [IQR]). The assay run mean and variance values for each antigen were compared using analysis of variance (ANOVA). When the mean and variance were the same for all runs, a fixed cutoff (mean + 1.645 standard deviation) could be determined; otherwise, a floating cutoff value was assigned. The floating cutoff was calculated anew for each run based on the use of normalization sera in each run (see Table 3 for the run of panel 2 as an example). Although the mean run values for ORF2-E2 were the same in all runs, for continuity and to facilitate analysis of multiplexed assays, a floating cutoff was used.
In the first validation step, the normalization factor (fixed cutoff − mean of anti-HEV IgG negative normalization sera) was determined and was used in every subsequent assay run. Each new run contained the anti-HEV IgG-negative normalization sera, and the floating cutoff was calculated as the mean of the normalization sera plus the normalization factor determined in panel 1, thus adjusting the cutoff for lower or higher run-specific mean fluorescence intensities (MFIs). Panel 2 was used to assess the sensitivity and specificity of the assay based on the normalization factor, the prerequisite for the floating cutoff, generated with panel 1. Table 1 shows a detailed description of the serum panels, while Table 2 depicts the results of the validation, including the normalization factors and floating cutoffs calculated for panel 2.
TABLE 1.
Panels 1 and 2 of pretested sera were used for the validation (n = 160) and panel 3 was used to compare HEV multiplex serology to commercial assays (n = 514)
| Serum sample | No. of samples |
|||
|---|---|---|---|---|
| Antigen cutoff, panel 1 | Assay cutoff and accuracy, panel 2 | Application, panel 3 | Total | |
| HEV IgG negative | 50 | 50 | 100 | |
| HEV IgG positive | 25 | 25 | 50 | |
| HEV RNA positive | 5 | 5 | 10 | |
| Serosurvey | 514 | 514 | ||
TABLE 2.
Results from the assay validation and statistical analysis
| Antigen | Normal distribution | Same mean | Same variance | Normalization factora | Mean of normalization sera | Floating cutoffb |
|---|---|---|---|---|---|---|
| ORF2 | YES | NO | YES | 1,88 | 4.04 | 5.92 |
| ORF2-E2 | YES | YES | YES | 2.15 | 5.76 | 7.91 |
| ORF3 | YES | NO | YES | 1.78 | 3.28 | 5.06 |
Normalization factors for the HEV antigens were calculated from four individual measurements of 50 IgG-negative serum samples (panel 1).
As only the variance but not the mean was the same between the four runs for two of the three antigens, a floating cutoff for all antigens was calculated anew, adding the mean of normalization sera and normalization factor for each subsequent assay (here illustrated for the run of panel 2).
All statistical tests were performed two sided, and P values of <0.05 were considered significant. Box plots depict ln-transformed MFIs. The lines inside the boxes represent the medians, and the diamonds indicate the mean values. The boxes are delimited by the first and third quartiles, and whiskers extend to the 1.5× IQR, respectively. Outliers are presented as circles. Statistical analysis to calculate the antigen and assay cutoffs was performed with SAS 9.4 software, while assay analysis was performed with Microsoft Excel 2007.
Samples.
Serum samples that were initially tested in order to differentially diagnose viral hepatitis or suspected to be HEV infected (and thus with a predetermined HEV antibody or RNA status) were obtained from the National Reference Laboratory for hepatitis A and E virus infections (University Hospital Regensburg, Germany) and used for assay validation. Original results were obtained by two commercially available assays, the Axiom HEV IgG enzyme immunoassay (EIA) (Axiom Diagnostics, Worms, Germany, developed by Wantai, Beijing, China) and the Mikrogen recomLine HEV IgG immunoblot (Mikrogen, Neuried, Germany). The presence of viral RNA was demonstrated using the RealStar HEV reverse transcriptase PCR (RT-PCR) kit 2.0 (Altona Diagnostics, Hamburg, Germany).
The 160 reference serum samples were split into two serum panels, each consisting of 50 HEV antibody-negative samples, 25 HEV IgG-positive samples, and 5 HEV RNA-positive samples to be used for assay validation.
Panel 3, consisting of 514 serum samples initially collected and tested with six commercial HEV tests as part of an HEV serosurvey (37), was retested with the developed multiplex assay. Human standard IgG (Privigen, 100 μg/ml infusion solution; CSL Behring) and three different pools of up to five individual HEV IgG-positive sera were included as controls in this setting.
Ethical approval for inclusion of stored samples from the serosurvey was obtained from the ethics commission of the University Hospital Jena (4537-08/15).
Additionally, 23 serum samples of acute HEV-infected patients were tested, provided by the Medizinische Hochschule Hannover (MHH), where the serostatus had been initially determined using the Wantai assay.
Multiplex approach.
To evaluate how the antigens react in a multiplexed assay, serum with defined serostatus for HEV and HAV was measured in a HAV-HEV multiplex serology, adding two antigens for the differentiation of HAV. The HEV reference sera were cross-validated with the Abnova HAV ELISA kit (KA0284), while HAV reference sera used in the validation for HAV-multiplex serology (33) were cross-validated with the Axiom HEV IgG EIA and the Mikrogen recomLine HEV IgG immunoblot.
RESULTS
To establish and validate the HEV multiplex serological assay, we used 160 predefined samples known to be HEV IgG negative, IgG positive, or containing HEV RNA (unknown serostatus). These were equally divided into two groups, named panel 1 and panel 2 (Table 1). Panel 1 was used to calculate the positive/negative cutoff MFI values for each antigen, resulting in the determination of a normalization factor for each of the antigens ORF2, ORF-E2, and ORF3. This normalization factor will be used to calculate a floating cutoff MFI value in all future assays and is exemplarily shown for panel 2 (Table 2).
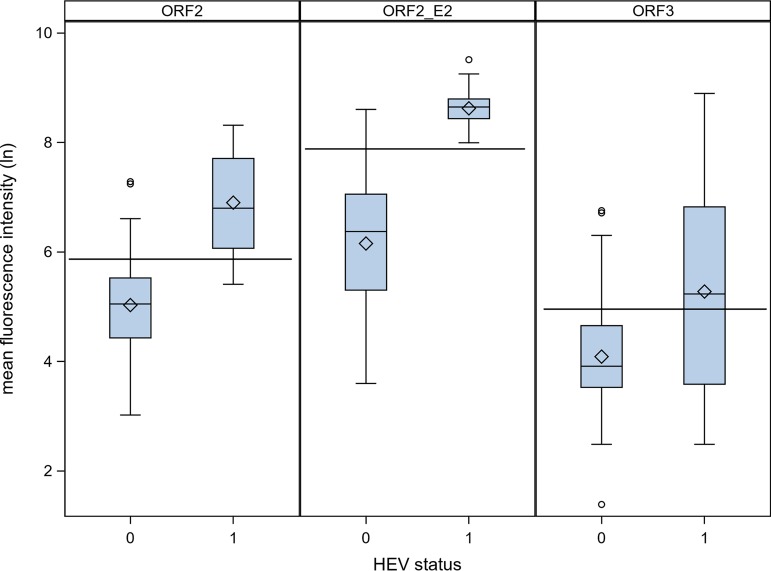
Use of the antigen ORF-E2 was shown to be sufficient to determine the HEV serostatus of a sample (Fig. 1). With a sensitivity of 100% and a specificity of 98% (95% confidence interval [CI], 94% to 100%), reactivity against ORF-E2 was used to establish a positive/negative cutoff MFI value for the assay (Table 3). In contrast, antigens ORF2 (sensitivity of 83% [95% CI, 70% to 97%], specificity of 84% [95% CI, 74% to 94%]) and ORF3 (sensitivity of 53% [95% CI, 36% to 71%], specificity of 84% [95% CI, 74% to 94%]) did not contribute to the accuracy of the assay.
FIG 1.
Validation of multiplex serology for the different HEV antigens with negative (0; n = 50) and IgG/RNA+ (1; n = 30) serum samples in serum panel 2. Horizontal lines indicate the floating cutoffs for each antigen.
TABLE 3.
Sensitivity and specificity of individual recombinant HEV antigensa
| Recombinant HEV antigens | Sensitivity (% [95% CI]) | Specificity (% [95% CI]) |
|---|---|---|
| ORF2 | 83 (70–97) | 84 (74–94) |
| ORF2-E2 | 100 | 98 (94–100) |
| ORF3 | 53 (36–71) | 84 (74–94) |
Calculated using the normalization factor determined from panel 1 and mean of the normalization sera, which were run together with the serum samples to be analyzed in panel 2, to calculate the floating cutoff.
Next, we used the established and validated assay to screen a large set of 514 serum samples collected during an HEV serosurvey conducted by Krumbholz et al. (37). These 514 serum samples were designated panel 3. All samples were initially tested with the Mikrogen recomWell HEV IgG ELISA (i) and the Axiom HEV-Ab ELISA (ii), among others, covering all classes of HEV antibodies. In addition, IgG antibodies in the sera were also measured using the recomLine HEV IgG assay (iii) (Sebastian Joel, personal communication). Analysis of results obtained from testing panel 3 with our HEV multiplex serological assay revealed that, overall, 423 serum samples tested HEV antibody negative and 91 serum samples tested HEV antibody (IgG, IgA, IgM) positive. Thus, the HEV seroprevalence rate was determined to be 17.7% seropositive (95% CI, 14.6% to 21.2%) using our HEV multiplex serological assay.
As our assay was validated with serum samples measured both with recomLine HEV IgG and Axiom HEV IgG/-Ab assays, the serosurvey results for both these assays for the 514 samples are compared in Table 4.
TABLE 4.
Concordance of results from 514 serum samples tested with Axiom HEV ELISA and recomLine HEV IgGa
| Axiom HEV-Ab result | No. of samples tested with recomLine HEV IgG |
||
|---|---|---|---|
| Neg result | Pos result | Total | |
| Neg | 374 | 2 | 376 |
| Pos | 95 | 41 | 136 |
| Borderline | 2 | 0 | 2 |
| Total | 471 | 43 | 514 |
Reported by Krumbholz et al. (37); 415 samples were tested concordantly with the two assays, while 97 samples show different results depending on which test was used. Pos, positive; Neg, negative.
While 41 serum samples were positive in both assays, 374 were concordantly negative. Of the remaining 99 samples, 95 were positive in the Axiom assay only, two were positive in the recomLine test only, and two samples were borderline positive in the Axiom assay. When tested using our HEV multiplex serological assay, 414 of the 415 concordant samples gave the same overall result. This meant that, overall, there was a 99.8% (95% CI, 98.8% to 99.9%) agreement rate between the HEV multiplex serology and results generated with both Axiom HEV-Ab and recomLine HEV IgG tests. Of the remaining 99 samples which measured inconsistently with the two tests, 49 tested positive in the HEV multiplex serological assay, including one of the two sera that was positive for the recomLine test only, while 50 were negative for HEV antibodies, including the two sera which were borderline positive in the Axiom HEV-Ab assay.
Compared to the single tests only (Table 5), our assay corresponds most closely to recomWell HEV IgG with a 90.3% (95% CI, 87.5% to 92.6%) agreement rate; 422/471 Ig-negative and 42/43 Ig-positive serum samples were confirmed in the multiplex HEV serological assay. The five serum samples that measured borderline in the recomWell assay tested positive in the HEV multiplex serological assay. The Axiom HEV-Ab ELISA resulted in a 90.4% (95% CI, 87.7% to 92.8%) agreement rate, with 374/376 Ig-negative and 89/136 Ig-positive serum samples showing the same results. Agreement with the recomLine HEV IgG test was 90.3% (95% CI, 87.5% to 92.6%), with 422/471 Ig-negative and 42/43 Ig-positive serum samples being confirmed in the multiplex HEV serological assay.
TABLE 5.
Agreement of novel multiplex HEV seroassay with selected tests applied in a previous serosurveya
| HEV-IgG or total antibody assay | No. of samples (n borderline)b | HEV multiplex serology |
||
|---|---|---|---|---|
| Agreement |
Cohens kappa | |||
| n | % | |||
| (i) recomWell HEV IgG | 509 (5) | 478 | 93.9 | 0.75 |
| (ii) Axiom HEV-Ab ELISA | 512 (2) | 463 | 90.4 | 0.73 |
| (iii) recomLine HEV IgG | 514 | 464 | 90.3 | 0.58 |
| Axiom ELISA & recomLine IgGc | 415 | 414 | 99.8 | 0.99 |
Reported by Krumbholz et al. (37).
The numbers of serum samples exclude specimens with borderline values in the former tests, which were excluded from the analysis of agreement with the HEV multiplex serology.
Specimens that were concordant by Axiom ELISA and recomLine IgG assays in the previous study.
The HEV multiplex assay was validated with serum samples from HEV suspected cases and applied to serum samples collected during an HEV seroprevalence study. In addition, 23 serum samples from acute HEV patients were also measured with the HEV multiplex serological assay. With the Wantai assay, 20/23 samples were initially determined as IgG-positive sera. With HEV multiplex serology, the IgG status was confirmed for 18 of these 20 samples, including one genotype 1 case, which was confirmed seropositive.
To show that our HEV multiplex serological assay is also functional in a multiplex approach, 18 of the HEV reference samples and 29 HAV reference samples were tested against HEV and previously published HAV antigens (33) in a HAV-HEV Luminex-based multiplex approach. The reference status of 8 HEV-negative, 10 HEV-positive, 9 HAV-negative, 10 HAV-positive, and 10 HAV-vaccinated serum samples were confirmed (Table 6). Additionally, seven (24%) of the HAV reference samples tested positive against HEV and 14 (78%) HEV reference samples tested positive against HAV, of which 2 (11%) were determined as vaccinated with HAV multiplex serology. The nonreference samples were cross-validated using the same assays that were used to determine the initial serostatus. For the tested samples, the agreement rate between HAV-HEV multiplex and AXIOM HEV IgG EIA was 76% (95% CI, 55% to 91%), with RecomLine HEV IgG it was 95% (95% CI, 78% to 100%), with both tests it was 100%, and with Abnova HAV ELISA it was 100%.
TABLE 6.
Reference serum samples with predefined serostatus for HAV or HEV were used in a HAV-HEV multiplex approach with antigens targeting HEV and previously published HAV (33)
| Reference status (n) | No. of samplesa
|
||||
|---|---|---|---|---|---|
| HAV neg | HAV inf | HAV vacc | HEV neg | HEV pos | |
| HEV neg (8) | 2 | 5 | 1 | 8 | |
| HEV pos (10) | 2 | 7 | 1 | 10 | |
| HAV neg (9) | 9 | 9 | 0 | ||
| HAV inf (10) | 10 | 8 | 2 | ||
| HAV vacc (10) | 10 | 7 | 3 | ||
| Total (47) | 13 | 22 | 12 | 32 | 15 |
pos, positive; neg, negative; inf, infected; vacc, vaccinated.
DISCUSSION
In this study, an innovative assay for HEV antibody detection was established, which can be upgraded to also detect antibodies against other hepatotropic pathogens, as shown here in combination with HAV antigen-coupled beads, requiring a total of less than 5 μl of serum (33, 38, 39). We validated our assay with precharacterized serum samples that were previously tested both with the Wantai HEV IgG ELISA, distributed as Axiom Diagnostics HEV IgG EIA, and the recomLine HEV IgG test. The assay developed by Wantai was one of the first HEV assays and shows a high sensitivity in HEV IgG detection (40–43). It uses recombinant HEV antigens, but their genomic origins and their genotypes are unclear (44). Existing patents indicate the inclusion of a peptide corresponding to amino acid (aa) residues 394 to 630 of ORF2 of HEV-1 or to aa residues 394 to 606 (45). The latter conforms to the ORF2-E2 peptide included in the present study. The recomLine HEV IgG is an immunoblot with seven recombinant antigens expressed from ORF2 (N-terminal, middle, or C-terminal parts of the capsid protein) and ORF3 of HEV-1 and -3 (41, 46). Our assay uses a short recombinant peptide of gt HEV-1, similar to that for the Wantai assay and the vaccine Hecolin (22). It protects against HEV-1 and HEV-4 infection and shows evidence for being protective against HEV-2/-3 (24). Thus, the specificity of our assay is expected to be against all four genotypes, of which we observed reactivity to gt HEV-3 (endemic in Europe) and one tested case of gt HEV-1.
The determination of assay sensitivity and specificity relies on commonly used tests. Depending on the type of samples used for assay validation (acute HEV infection, follow-up on HEV RNA-positive blood donors, suspected HEV cases, and the presence of liver disease), the performance of commercial kits may vary markedly (41, 43, 47) and bias the results of seroprevalence studies (26).
Axiom HEV-Ab and IgG ELISA (Wantai) have been shown to be very reactive toward the detection of anti-HEV IgG antibodies. Among the available HEV ELISAs, they were shown to have the highest sensitivity and specificity (26, 43, 47).
A previous German survey included 537 serums samples which were tested with five HEV antibody assays, resulting in a seroprevalence range from 4.1% to 27.9% (37). The recomLine HEV IgG test places more weight on high specificity, while Axiom HEV-Ab EIA shows higher sensitivity, resulting in a seroprevalence of 9.9% compared to 27.9%, respectively. Assuming that recomLine HEV IgG is underestimating the HEV seroprevalence in a population-based sample set, while Axiom HEV-Ab/IgG EIA is overestimating it, our assay, validated against both tests (recomLine HEV IgG/Axiom HEV IgG EIA), shows high sensitivity and specificity, resulting in a seroprevalence of 17.7%.
Using 160 diagnostic samples, our qualitative assay archived a sensitivity of 100% and specificity of 98%. These numbers are reflected in the application of our assay to a population-based serum set (>400 samples), which was tested with a range of different assays. Of concordant samples measured both with Axiom HEV-Ab and recomLine HEV IgG, similarly to the procedures at the National Reference Laboratory for hepatitis A and E virus infections, 99% were confirmed with our assay. One-half of the remaining 99 serosurvey samples which were initially measured positive with only Axiom or recomLine (and vice versa negative) were determined by our HEV multiplex serological assay to be HEV antibody positive (49%, 49 serum samples), the remaining were negative (51%, 50 serum samples).
Our assay, therefore, offers several advantages over existing tests, especially when it comes to testing large sets of serum samples to estimate HEV prevalence. It is applicable in the diagnostic setting, as we observed a similar sensitivity to the Wantai/Axiom assay for acute-phase serum samples. For only one of the acute samples, information on HEV RNA positivity was available, which was the gt HEV-1 sample, confirmed antibody positive. The samples tested negative with Wantai and/or HEV multiplex serology had very low MFIs in the multiplex assay. Thus, there is the possibility that the clinical diagnosis for HEV was incorrect, since RNA positivity was not tested or that for early acute cases, antibody titers were too low to be detected in any serological assay.
Most important, the multiplex assay is an especially useful tool for the analysis of samples collected during seroprevalence surveys, as it combines the easy handling of ELISA plates with the specificity of line immunoassays.
Large sets of serum samples can therefore be analyzed without losing specificity, as the multiplex approach combines the advantages of two or more assays in one. The combination of several assays in one in a multiplexed approach enables the development of, for example, a full hepatitis panel containing antigens against all pathogens known to cause viral hepatitis. Such a panel would greatly facilitate differential diagnosis of viral hepatitis as well as seroprevalence studies, which could both be carried out much more efficiently.
Additionally, only a small volume of serum (2 μl) is needed, and the multiplex approach allows the analysis of serum for antibodies against different pathogens simultaneously and reliably, without cross-reactivity between antigens and antibodies.
Conclusions.
The reduction to only one differentiation factor, such as the ORF2-E2 antigen, opens the door to the integration of HEV antibody detection into multidetection methods, such as a Luminex-based hepatitis panel. The immune responses against hepatitis A, B, C, and E can be validated with very small volumes of serum that generate a characteristic antibody pattern, which could be used for an in-depth characterization of individuals in large prevalence or cohort studies. In conclusion, our assay is particularly helpful for systematic screening purposes and large epidemiological studies, as the test system uses a small volume of serum and one measurement is sufficient to obtain results for different pathogens and for differential diagnosis.
ACKNOWLEDGMENTS
We thank Mathias Schemmerer, Jürgen Wenzel, and Wolfgang Jilg from the National Reference Laboratory for HAV and HEV, Regensburg, Germany, for providing the reference sera, Sebastian Joel from the Division of Experimental Virology, Institute of Medical Microbiology, Jena University, Germany, for sharing unpublished data on the testing of all serosurvey samples with the recomLine assay, and Rebecca Surtees from the Robert Koch Institute for language editing and proofreading.
We declare no potential conflicts of interest.
REFERENCES
- 1.Smith DB, Simmonds P, Jameel S, Emerson SU, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WHM, Purdy MA. 2014. Consensus proposals for classification of the family Hepeviridae. J Gen Virol 95:2223–2232. doi: 10.1099/vir.0.068429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ropp SL, Tam AW, Beames B, Purdy M, Frey TK. 2000. Expression of the hepatitis E virus ORF1. Arch Virol 145:1321–1337. doi: 10.1007/s007050070093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou YH, Purcell RH, Emerson SU. 2005. A truncated ORF2 protein contains the most immunogenic site on ORF2: antibody responses to non-vaccine sequences following challenge of vaccinated and non-vaccinated macaques with hepatitis E virus. Vaccine 23:3157–3165. doi: 10.1016/j.vaccine.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 4.Zafrullah M, Ozdener MH, Panda SK, Jameel S. 1997. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol 71:9045–9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ICTV. 2018. Virus taxonomy. Genus: Orthohepevirus. https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/w/hepeviridae/728/genus-orthohepevirus. Accessed 20 May 2019.
- 6.Khuroo MS. 1980. Study of an epidemic of non-A, non-B hepatitis. Possibility of another human hepatitis virus distinct from post-transfusion non-A, non-B type. Am J Med 68:818–824. doi: 10.1016/0002-9343(80)90200-4. [DOI] [PubMed] [Google Scholar]
- 7.Nalin DR, Kuter BJ, Brown L, Patterson C, Calandra GB, Werzberger A, Shouval D, Ellerbeck E, Block SL, Bishop R. 1993. Worldwide experience with the CR326F-derived inactivated hepatitis A virus vaccine in pediatric and adult populations: an overview. J Hepatol 18 Suppl 2:S51–S55. doi: 10.1016/S0168-8278(05)80379-4. [DOI] [PubMed] [Google Scholar]
- 8.Wedemeyer H, Pischke S, Manns MP. 2012. Pathogenesis and treatment of hepatitis E virus infection. Gastroenterology 142:1388.e1–1397.e1. doi: 10.1053/j.gastro.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Elduma A, Zein M, Karlsson M, Elkhidir I, Norder H. 2016. A single lineage of hepatitis E virus causes both outbreaks and sporadic hepatitis in Sudan. Viruses 8:273. doi: 10.3390/v8100273. [DOI] [Google Scholar]
- 10.Batts W, Yun S, Hedrick R, Winton J. 2011. A novel member of the family Hepeviridae from cutthroat trout (Oncorhynchus clarkii). Virus Res 158:116–123. doi: 10.1016/j.virusres.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 11.Khuroo MS, Khuroo MS, Khuroo NS. 2016. Hepatitis E: discovery, global impact, control and cure. World J Gastroenterol 22:7030–7045. doi: 10.3748/wjg.v22.i31.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreier J, Juhl D. 2014. Autochthonous hepatitis e virus infections: a new transfusion-associated risk? Transfus Med Hemother 41:29–39. doi: 10.1159/000357098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izopet J, Lhomme S, Chapuy-Regaud S, Mansuy JM, Kamar N, Abravanel F. 2017. HEV and transfusion-recipient risk. Transfus Clin Biol 24:176–181. doi: 10.1016/j.tracli.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 14.King NJ, Hewitt J, Perchec-Merien AM. 2018. Hiding in plain sight? It’s time to investigate other possible transmission routes for hepatitis E virus (HEV) in developed countries. Food Environ Virol 10:225–252. doi: 10.1007/s12560-018-9342-8. [DOI] [PubMed] [Google Scholar]
- 15.Yue N, Wang Q, Zheng M, Wang D, Duan C, Yu X, Zhang X, Bao C, Jin H. 2019. Prevalence of hepatitis E virus infection among people and swine in mainland China: a systematic review and meta-analysis. Zoonoses Public Health 66:265–275. doi: 10.1111/zph.12555. [DOI] [PubMed] [Google Scholar]
- 16.Khuroo MS, Khuroo MS. 2016. Hepatitis E: an emerging global disease - from discovery towards control and cure. J Viral Hepat 23:68–79. doi: 10.1111/jvh.12445. [DOI] [PubMed] [Google Scholar]
- 17.Rein DB, Stevens GA, Theaker J, Wittenborn JS, Wiersma ST. 2012. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology 55:988–997. doi: 10.1002/hep.25505. [DOI] [PubMed] [Google Scholar]
- 18.Shinde N, Patil T, Deshpande A, Gulhane R, Patil M, Bansod Y. 2014. Clinical profile, maternal and fetal outcomes of acute hepatitis e in pregnancy. Ann Med Health Sci Res 4:133. doi: 10.4103/2141-9248.138033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patra S, Kumar A, Trivedi SS, Puri M, Sarin SK. 2007. Maternal and fetal outcomes in pregnant women with acute hepatitis E virus infection. Ann Intern Med 147:28–33. doi: 10.7326/0003-4819-147-1-200707030-00005. [DOI] [PubMed] [Google Scholar]
- 20.Kamar N, Selves J, Mansuy J-M, Ouezzani L, Péron J-M, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, Durand D, Vinel J-P, Izopet J, Rostaing L. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med 358:811–817. doi: 10.1056/NEJMoa0706992. [DOI] [PubMed] [Google Scholar]
- 21.Kamar N, Pischke S. 2018. Acute and persistent hepatitis E virus genotype 3 and 4 infection: clinical features, pathogenesis, and treatment. Cold Spring Harb Perspect Med 9:a031872. doi: 10.1101/cshperspect.a031872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu F-C, Zhang J, Zhang X-F, Zhou C, Wang Z-Z, Huang S-J, Wang H, Yang C-L, Jiang H-M, Cai J-P, Wang Y-J, Ai X, Hu Y-M, Tang Q, Yao X, Yan Q, Xian Y-L, Wu T, Li Y-M, Miao J, Ng M-H, Shih J-K, Xia N-S. 2010. Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: a large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet 376:895–902. doi: 10.1016/S0140-6736(10)61030-6. [DOI] [PubMed] [Google Scholar]
- 23.WHO. 2015. Efficacy of hepatitis E vaccination in immunocompetent individuals against hepatitis E disease. https://www.who.int/immunization/policy/position_papers/hepe_grad_efficacy_disease.pdf.Accessed 20 May 2019.
- 24.Gu Y, Tang X, Zhang X, Song C, Zheng M, Wang K, Zhang J, Ng MH, Hew CL, Li S, Xia N, Sivaraman J. 2015. Structural basis for the neutralization of hepatitis E virus by a cross-genotype antibody. Cell Res 25:604–620. doi: 10.1038/cr.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khudyakov Y, Kamili S. 2011. Serological diagnostics of hepatitis E virus infection. Virus Res 161:84–92. doi: 10.1016/j.virusres.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Wenzel JJ, Preiss J, Schemmerer M, Huber B, Jilg W. 2013. Test performance characteristics of anti-HEV IgG assays strongly influence hepatitis E seroprevalence estimates. J Infect Dis 207:497–500. doi: 10.1093/infdis/jis688. [DOI] [PubMed] [Google Scholar]
- 27.Meng J, Dai X, Chang JC, Lopareva E, Pillot J, Fields HA, Khudyakov YE. 2001. Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology 288:203–211. doi: 10.1006/viro.2001.1093. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Zhang W, Gu H, Chen W, Zeng M, Ji C, Song R, Zhang G. 2017. Identification and characterization of two linear epitope motifs in hepatitis E virus ORF2 protein. PLoS One 12:e0184947. doi: 10.1371/journal.pone.0184947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Zhao C, Tian Y, Xu N, Wang Y. 2016. Characteristics and functions of HEV proteins, p 17–38. In Cohen I, Lajtha A, Paoletti R, Lambris JD. (ed), Advances in experimental medicine and biology. Springer Nature, London, United Kingdom. [DOI] [PubMed] [Google Scholar]
- 30.Khudyakov YE, Khudyakova NS, Fields HA, Jue D, Starling C, Favorov MO, Krawczynski K, Polish L, Mast E, Margolis H. 1993. Epitope mapping in proteins of hepatitis E virus. Virology 194:89–96. doi: 10.1006/viro.1993.1238. [DOI] [PubMed] [Google Scholar]
- 31.WHO. 2015. Hepatitis E vaccine: WHO position paper, May 2015. https://www.who.int/wer/2015/wer9018.pdf.
- 32.Waterboer T, Sehr P, Michael KM, Franceschi S, Nieland JD, Joos TO, Templin MF, Pawlita M. 2005. Multiplex human papillomavirus serology based on in situ-purified glutathione S-transferase fusion proteins. Clin Chem 51:1845–1853. doi: 10.1373/clinchem.2005.052381. [DOI] [PubMed] [Google Scholar]
- 33.Bohm K, Filomena A, Schneiderhan-Marra N, Krause G, Sievers C. 2017. Validation of HAV biomarker 2A for differential diagnostic of hepatitis A infected and vaccinated individuals using multiplex serology. Vaccine 35:5883–5889. doi: 10.1016/j.vaccine.2017.08.089. [DOI] [PubMed] [Google Scholar]
- 34.Tam AW, Smith MM, Guerra ME, Huang C-C, Bradley DW, Fry KE, Reyes GR. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sehr P, Zumbach K, Pawlita M. 2001. A generic capture ELISA for recombinant proteins fused to glutathione S-transferase: validation for HPV serology. J Immunol Methods 253:153–162. doi: 10.1016/s0022-1759(01)00376-3. [DOI] [PubMed] [Google Scholar]
- 36.Shankar G, Devanarayan V, Amaravadi L, Barrett YC, Bowsher R, Finco-Kent D, Fiscella M, Gorovits B, Kirschner S, Moxness M, Parish T, Quarmby V, Smith H, Smith W, Zuckerman LA, Koren E. 2008. Recommendations for the validation of immunoassays used for detection of host antibodies against biotechnology products. J Pharm Biomed Anal 48:1267–1281. doi: 10.1016/j.jpba.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 37.Krumbholz A, Joel S, Dremsek P, Neubert A, Johne R, Durrwald R, Walther M, Muller TH, Kuhnel D, Lange J, Wutzler P, Sauerbrei A, Ulrich RG, Zell R. 2014. Seroprevalence of hepatitis E virus (HEV) in humans living in high pig density areas of Germany. Med Microbiol Immunol 203:273–282. doi: 10.1007/s00430-014-0336-3. [DOI] [PubMed] [Google Scholar]
- 38.Dondog B, Schnitzler P, Michael KM, Clifford G, Franceschi S, Pawlita M, Waterboer T. 2015. Hepatitis C virus seroprevalence in Mongolian women assessed by a novel multiplex antibody detection assay. Cancer Epidemiol Biomarkers Prev 24:1360–1365. doi: 10.1158/1055-9965.EPI-15-0351. [DOI] [PubMed] [Google Scholar]
- 39.Michel A, Waterboer T, Kist M, Pawlita M. 2009. Helicobacter pylori multiplex serology. Helicobacter 14:525–535. doi: 10.1111/j.1523-5378.2009.00723.x. [DOI] [PubMed] [Google Scholar]
- 40.Abravanel F, Chapuy-Regaud S, Lhomme S, Miedougé M, Peron J-M, Alric L, Rostaing L, Kamar N, Izopet J. 2013. Performance of anti-HEV assays for diagnosing acute hepatitis E in immunocompromised patients. J Clin Virol 58:624–628. doi: 10.1016/j.jcv.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Vollmer T, Diekmann J, Eberhardt M, Knabbe C, Dreier J. 2016. Monitoring of anti-hepatitis E virus antibody seroconversion in asymptomatically infected blood donors: systematic comparison of nine commercial anti-HEV IgM and IgG assays. Viruses 8:232. doi: 10.3390/v8080232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kmush BL, Labrique AB, Dalton HR, Ahmed ZB, Ticehurst JR, Heaney CD, Nelson KE, Zaman K. 2015. Two generations of “gold standards”: the impact of a decade in hepatitis E virus testing innovation on population seroprevalence. Am J Trop Med Hyg 93:714–717. doi: 10.4269/ajtmh.15-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norder H, Karlsson M, Mellgren Å, Konar J, Sandberg E, Lasson A, Castedal M, Magnius L, Lagging M. 2016. Diagnostic performance of five assays for anti-hepatitis E virus IgG and IgM in a large cohort study. J Clin Microbiol 54:549–555. doi: 10.1128/JCM.02343-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Sadeq DW, Majdalawieh AF, Mesleh AG, Abdalla OM, Nasrallah GK. 2018. Laboratory challenges in the diagnosis of hepatitis E virus. J Med Microbiol 67:466–480. doi: 10.1099/jmm.0.000706. [DOI] [PubMed] [Google Scholar]
- 45.Ningshao X, Jun Z, Shaowei L, Shengxiang G, Ying Gu ZH. 2014. Polypeptide fragments of the hepatitis E virus, the vaccine composition comprising said fragments and the diagnostic kits. Publication number 20100143410; patent grant number 8715695.
- 46.Wu W-C, Su C-W, Yang J-Y, Lin S-F, Chen J-Y, Wu J-C. 2014. Application of serologic assays for diagnosing acute hepatitis E in national surveillance of a nonendemic area. J Med Virol 86:720–728. doi: 10.1002/jmv.23785. [DOI] [PubMed] [Google Scholar]
- 47.Pas SD, Streefkerk RH, Pronk M, de Man RA, Beersma MF, Osterhaus AD, van der Eijk AA. 2013. Diagnostic performance of selected commercial HEV IgM and IgG ELISAs for immunocompromised and immunocompetent patients. J Clin Virol 58:629–634. doi: 10.1016/j.jcv.2013.10.010. [DOI] [PubMed] [Google Scholar]