Mycobacterium tuberculosis infection and nontuberculous mycobacteria (NTM) infections exhibit similar clinical symptoms; however, the therapies for these two types of infections are different. Therefore, the rapid and accurate identification of M. tuberculosis and NTM species is very important for the control of tuberculosis and NTM infections. In the present study, a Cas12a/guide RNA (gRNA)-based platform was developed to identify M. tuberculosis and most NTM species.
KEYWORDS: Mycobacterium tuberculosis, nontuberculous mycobacteria, CRISPR/Cas12a, gRNA probe, fluorescence, identification, biosensing
ABSTRACT
Mycobacterium tuberculosis infection and nontuberculous mycobacteria (NTM) infections exhibit similar clinical symptoms; however, the therapies for these two types of infections are different. Therefore, the rapid and accurate identification of M. tuberculosis and NTM species is very important for the control of tuberculosis and NTM infections. In the present study, a Cas12a/guide RNA (gRNA)-based platform was developed to identify M. tuberculosis and most NTM species. By designing species-specific gRNA probes targeting the rpoB sequence, a Cas12a/gRNA-based platform successfully identified M. tuberculosis and six major NTM species (Mycobacterium abscessus, Mycobacterium intracellulare, Mycobacterium avium, Mycobacterium kansasii, Mycobacterium gordonae, and Mycobacterium fortuitum) without cross-reactivity. In a blind assessment, a total of 72 out of 73 clinical Mycobacterium isolates were correctly identified, which is consistent with previous rpoB sequencing results. These results suggest that the Cas12a/gRNA-based platform is a promising tool for the rapid, accurate, and cost-effective identification of both M. tuberculosis and NTM species.
INTRODUCTION
Tuberculosis is a serious infectious disease that endangers human health and is mainly caused by Mycobacterium tuberculosis infection. In recent years, some investigations have shown an increased incidence of nontuberculosis Mycobacterium infections. NTMs have become recognized as important human pathogens causing public health problems (1–3). NTM infections are insensitive to most antituberculosis drugs, and the treatment regimen for patients infected with M. tuberculosis versus an NTM is completely different (4). However, it is difficult to distinguish M. tuberculosis infection from NTM infections, since they exhibit similar clinical symptoms. Therefore, the rapid and accurate identification of M. tuberculosis and NTM is of great significance for the early diagnosis, treatment, and control of tuberculosis and NTM infections.
The identification of M. tuberculosis and NTM species mainly depends on laboratory diagnosis. Conventional etiological methods used in microbiology laboratories are complex, time-consuming and labor-intensive, and have poor specificity and sensitivity, which is not conducive to the timely guidance of clinical treatment (5). In recent decades, the introduction of molecular diagnostic methods has overcome some of these disadvantages, resulting in a remarkable improvement in the direct detection of mycobacteria. A variety of molecular techniques have been developed for the identification of M. tuberculosis and NTM from clinical specimens and culture. These mainly include multiplex PCR (6–8), real-time PCR (9–11), DNA probe assays (12–15), PCR restriction fragment length polymorphism (RFLP) analysis (16–19), target gene sequencing (20–24), and microarray technology (25–27). Among them, microarray technology is currently popular because it can analyze thousands of genes at the same time and in a short time, which is very helpful for phylogenetic analysis and species identification. However, most of these techniques are not cost-effective, and some of them require expensive equipment. Moreover, most of these assays have been used for the detection or quantification of a certain Mycobacterium species only, such as M. tuberculosis, or the detection of drug resistance.
Cas12a (Cpf1) was first characterized by the Zhang Feng group as an RNA-guided endonuclease that can directly bind and cut target DNA (28). Recently, two independent groups found Cas12a possessed collateral DNA cleavage activity (29, 30). This process was immediately developed into two Cas12a-based nucleic acid detection methods called HOLMES (one-hour low-cost multipurpose highly efficient system) and DETECTR (DNA endonuclease-targeted CRISPR trans reporter) (30, 31). Both platforms could achieve attomolar sensitivity for detecting DNA targets with single-base resolution. Here, by designing a variety of species-specific gRNA probes, we attempted to rapidly and accurately identify M. tuberculosis and the most common NTM species (M. abscessus, M. intracellulare, M. avium, M. kansasii, M. gordonae and M. fortuitum) using a Cas12a/gRNA-based platform.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Clinical isolates, including 10 M. tuberculosis, 15 M. abscessus, 15 M. intracellulare, 10 M. avium, 10 M. fortuitum, 7 M. kansasii and 6 M. gordonae were obtained from Shenzhen Third People’s Hospital, Shenzhen, China, and were confirmed by rpoB sequencing. Seven reference strains, including M. tuberculosis H37Rv, M. abscessus (ATCC 19977), M. intracellulare (ATCC 13950), M. avium (ATCC 25291), M. kansasii (ATCC 12478), M. gordonae (ATCC 14470), and M. fortuitum (ATCC 6841) were a kind gift from the Chinese Center for Disease Control and Prevention. Strains were cultured in Middlebrook 7H9 liquid medium and incubated at 37°C. Escherichia coli, Pseudomonas aeruginosa, Actinobacter baumannii, Salmonella enteritidis and Klebsiella pneumoniae were obtained from Shenzhen Third People’s Hospital, Shenzhen, China, and cultured on blood agar plates.
Design and preparation of gRNA probes.
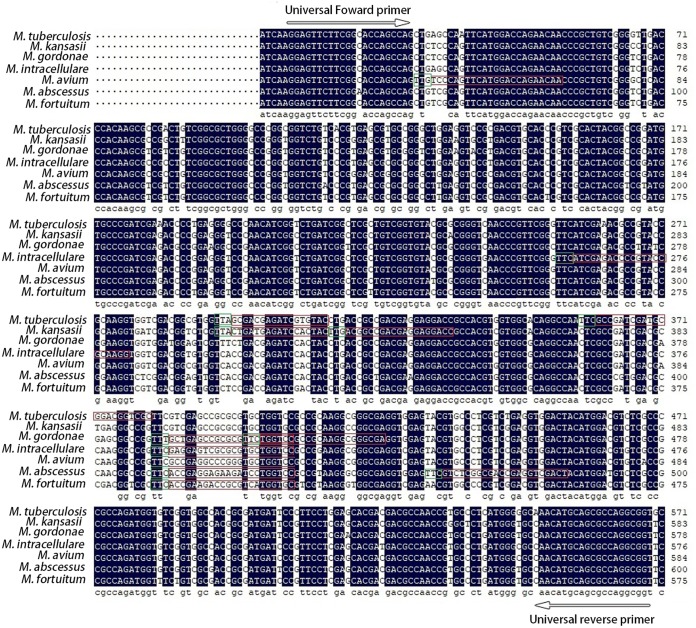
Since the rpoB gene has been reported to be a suitable locus for the identification of Mycobacterium species (24, 32–34), we chose it as the target sequence for the design of gRNA probes. The rpoB gene sequences of M. tuberculosis H37Rv and the most common NTM (M. abscessus ATCC 19977, M. intracellulare ATCC 13950, M. avium ATCC 25291, M. kansasii ATCC 12478, M. gordonae ATCC 14470, and M. fortuitum ATCC 6841) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). Multiple alignments of rpoB gene sequences were performed using DNAMAN 8 software (Fig. 1). Species-specific gRNA probes were designed based on the divergence of rpoB genes (Fig. 1). We designed two specific gRNA probes for each Mycobacterium species except M. fortuitum (Fig. 1). A universal primer pair (RpoB-F and RpoB-R) for amplification of target sequences was designed based on the conserved sequences flanking the gRNA probe targeting regions (Fig. 1). All gRNA probes were prepared by in vitro transcription using a T7 High Yield RNA transcription kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. The DNA templates for gRNA transcription were synthesized by Sangon Biotech (Shanghai, China). The transcribed RNA was purified using VAHTS RNA Clean Beads (Vazyme, Nanjing, China) and quantified with NanoDrop 2000. All the oligonucleotides are listed in Table S1 in the supplemental material.
FIG 1.
Design of species-specific gRNA probes based on rpoB sequences. rpoB sequences were downloaded from M. tuberculosis H37Rv, M. abscessus (ATCC 19977), M. intracellulare (ATCC 13950), M. avium (ATCC 25291), M. kansasii (ATCC 12478), M. gordonae (ATCC 14470), and M. fortuitum (ATCC 6841). Multiple sequence alignment was performed using DNAMAN 8 software. Conserved sequences are blank. Red boxes represent gRNA probe targeted regions. Green boxes represent the PAM motif. Universal primers used for amplification of rpoB fragments are indicated by arrows.
DNA preparation and PCR amplification.
DNA was extracted from cultured strains using a TIANamp Bacteria DNA kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s protocol or a simple boiling method (35). DNA concentration was determined using a Qubit dsDNA HS assay kit (Thermo Fisher Scientific, MA, USA). Target sequences were amplified by PCR using the universal primer pairs described above. PCR was performed as follows: one cycle at 94°C for 3 min, then 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by one cycle at 72°C for 5 min, and the reactions were cooled at 4°C. PCR products were examined by 1% agarose gel electrophoresis. The PCR product concentration was determined using a Qubit dsDNA HS assay kit (Thermo Fisher Scientific, MA, USA).
Cas12a/gRNA transcleavage assay.
Recombinant Francisella novicida Cas12a (FnCas12a) protein purchased from Tolo Biotech (Shanghai, China) was used for the Cas12a transcleavage assay. The recognition site of FnCas12a is “TTN,” not “TTTN” (29), which provided more candidate loci for design of gRNA probes. Cas12a transcleavage assays were performed mainly according to Li’s description (31). In brief, the FnCas12a transcleavage assay was performed in reaction buffer consisting of 0.5 pmol FnCas12a, 100 nM purified gRNA, target DNA (1 μl unpurified PCR products), 500 nM collateral single-stranded DNA (ssDNA) (quenched fluorescent single-strand DNA reporter), and 10 U RNase inhibitor (TaKaRa, Dalian, China) in a 20 μl volume at 37°C for 1 h. The reaction was stopped by adding 2 μl of 0.25 M EDTA. Fluorescence emission was excited at 535 nm and detected at 560 nm using a Varioskan Flash (Thermo Fisher Scientific, MA, USA), and reactions with no target DNA were used as the background.
Evaluation of the specificity of gRNA probes and determination of the limit of detection with reference strains.
The seven mycobacterial reference strains and five other bacterial species, including E. coli, P. aeruginosa, A. baumannii, S. enteritidis, and K. pneumoniae, were used to evaluate the specificity of gRNA probes.
The limit of detection (LOD) was determined in both DNA and quantified CFU. DNA was extracted from the reference strains using a TIANamp Bacteria DNA kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s protocol and then serially diluted ranging from 10 ng to 0.01 pg. Serial dilutions with the indicated mycobacterial numbers were spiked into the artificial sputum samples. Sputum samples were treated with a 3% NaOH solution and then neutralized with a phosphate buffer followed by centrifugation. DNA was extracted from the pellets by a rapid boiling method (35). After PCR amplification, 1 μl unpurified PCR products were used for DNA detection via an FnCas12a/gRNA-based platform.
RESULTS
Schematic overview of the FnCas12a/gRNA-based platform.
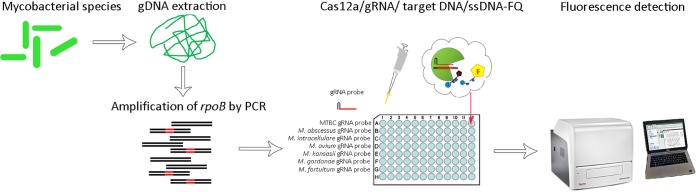
Recently, two revolutionary DNA detection methods named DNA DETECTR and HOLMES, respectively, were developed by two independent groups based on the Cas12a transcleavage activity (30, 31). Both platforms could specifically and accurately detect target DNA by designing a specific gRNA, thereby achieving attomolar sensitivity for DNA detection with single-base resolution. Thus, we reasoned that a Cas12a-based nucleic acid detection assay could be developed to identify Mycobacterium species. Fig. 2 shows a schematic overview of the FnCas12a/gRNA-based platform for the identification of Mycobacterium species. Using this platform, target DNA was amplified by PCR and then incubated with FnCas12a, a gRNA probe, and quenched fluorescence reporter ssDNA. If target DNA is detected by the species-specific gRNA probe in the reaction mixture, the formation of the FnCas12a/gRNA/target DNA ternary complex transcleaves the reporter ssDNA, resulting in fluorescence.
FIG 2.
Illustration of FnCas12a/gRNA-based platform for identification of Mycobacterium species. rpoB fragments were specifically amplified from extracted DNA of bacteria by PCR. Species-specific gRNA probes were designed to target rpoB fragments. Unpurified PCR product was directly mixed with FnCas12a/gRNA/ssDNA-FQ (quenched fluorescent single-strand DNA reporter). Once gRNA probes match the target DNA and form a complex, FnCas12a will transcleave the quenched fluorescent ssDNA reporter, illuminating the fluorescence.
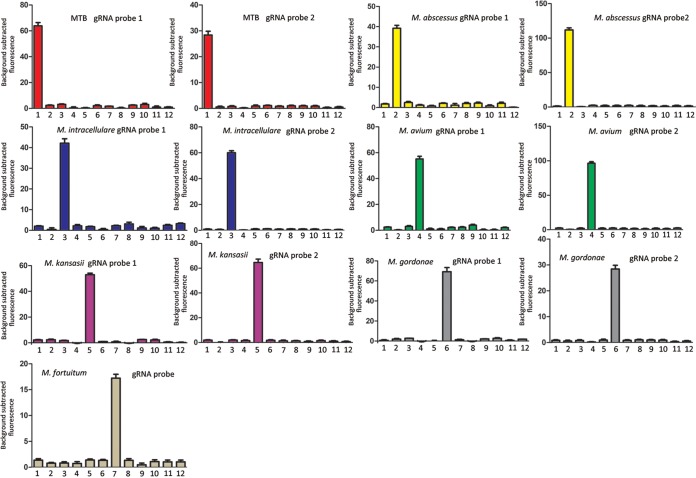
Evaluation of the FnCas12a/gRNA-based platform with Mycobacterium reference strains.
We selected two gRNA probes for each mycobacterial species to be analyzed, except M. fortuitum. The specificity of each gRNA probe was evaluated preliminarily using reference strains. From the fluorescence detection results shown in Fig. 3, each designed gRNA probe was able to identify the specific mycobacterial species without cross-reaction. In addition, we observed that fluorescence intensities varied with gRNA probe sequences (Fig. 3).
FIG 3.
Determination of the specificity of the designed gRNA probes using reference Mycobacterium species. Bars 1 to 7 represent M. tuberculosis H37Rv, M. abscessus (ATCC 19977), M. intracellulare (ATCC 13950), M. avium (ATCC 25291), M. kansasii (ATCC 12478), M. gordonae (ATCC 14470), and M. fortuitum (ATCC 6841), respectively. Bars 8 to 12 represent E. coli, P. aeruginosa, A. baumannii, S. enteritidis, and K. pneumoniae, respectively. Error bars represent mean ± standard error of the mean (SEM), with n = 3 technical replicates.
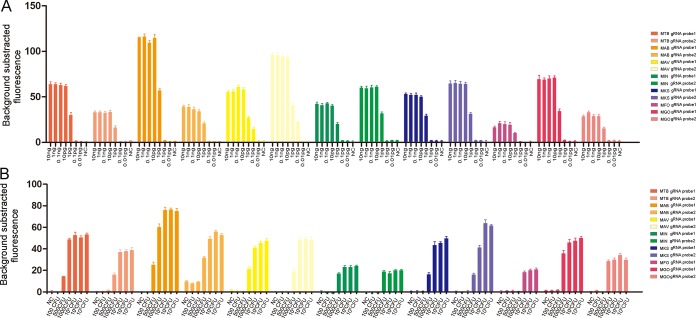
To determine the sensitivity of the FnCas12a/gRNA-based platform, DNA extracted from reference strains was serially diluted ranging from 10 ng to 10 fg. Each gRNA probe showed a similar LOD, ranging from 1 pg to 100 fg (Fig. 4A). The ratios of fluorescence intensity from LODs to the negative controls were set as the cutoffs used for subsequent clinical strain detection. To further evaluate the LOD in CFU, serial dilutions with the indicated mycobacterial numbers were spiked into the artificial sputum samples. We found the FnCas12a/gRNA-based platform had an LOD of 500 to 10,000 CFU/ml (Fig. 4B).
FIG 4.
Evaluation of LOD with purified DNA and quantified CFU. (A) DNA extracted from pure- cultured reference strains were serially diluted in the range of 10 ng to 10 fg. (B) Serial quantified bacilli were spiked into artificial sputum. E. coli was used as negative control (NC). Error bars represent mean ± SEM, with n = 3 technical replicates.
Application of FnCas12a/gRNA-based platform in clinical Mycobacterium isolates.
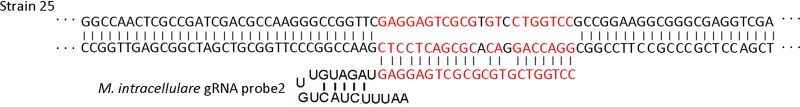
A total of 73 clinical isolates, including 10 M. tuberculosis, 15 M. abscessus, 15 M. intracellulare, 10 M. avium, 7 M. kansasii, 10 M. gordonae, and 6 M. fortuitum, were examined in a blind assessment to determine the feasibility of the FnCas12a/gRNA-based platform in the identification of the major Mycobacterium species. For each test, E. coli was used as a negative control (NC). Fluorescence signals from samples were normalized against the negative controls. The fold changes in samples above or below the cutoff in FnCas12a/gRNA-based assays were considered positive or negative, respectively. Using these screening criteria, a total of 72 out of 73 clinical strains were correctly identified, showing almost 100% agreement with previous rpoB sequencing results (Table 1). Only one strain thought to be M. intracellulare was misidentified by the M. intracellulare gRNA probe2 (MIN gRNA probe2). We further investigated the sequence of this misidentified strain and found that it has two base mutations in the M. intracellulare gRNA probe2 targeting region (Fig. 5), which resulted in false negatives.
TABLE 1.
Identification of clinical isolates using FnCas12a/gRNA-based platform
| Clinical organism | No. of strains | No. correctly identified (%) | No. misidentified | gRNA probe name |
|---|---|---|---|---|
| M. tuberculosis | 10 | 10 (100) | 0 | MTB gRNA probe1 |
| M. tuberculosis | 10 | 10 (100) | 0 | MTB gRNA probe2 |
| M. abscessus | 15 | 15 (100) | 0 | MAB gRNA probe1 |
| M. abscessus | 15 | 15 (100) | 0 | MAB gRNA probe2 |
| M. intracellulare | 15 | 15 (100) | 0 | MIN gRNA probe1 |
| M. intracellulare | 15 | 14 (93.33) | 1 | MIN gRNA probe2 |
| M. avium | 10 | 10 (100) | 0 | MAV gRNA probe1 |
| M. avium | 10 | 10 (100) | 0 | MAV gRNA probe2 |
| M. gordonae | 10 | 10 (100) | 0 | MGO gRNA probe1 |
| M. gordonae | 10 | 10 (100) | 0 | MGO gRNA probe2 |
| M. kansasii | 7 | 7 (100) | 0 | MKA gRNA probe1 |
| M. kansasii | 7 | 7 (100) | 0 | MKA gRNA probe2 |
| M. fortuitum | 6 | 6 (100) | 0 | MFO gRNA probe |
FIG 5.
Partial sequences of rpoB gene from the two misidentified strains. Strain 25 was previously identified as M. intracellulare by rpoB sequencing. Red nucleotides indicate the M. intracellulare gRNA probe 2 targeting region. Black nucleotides within the red region indicate the mutated bases.
DISCUSSION
Tuberculosis (TB) is one of the top 10 causes of death and the leading cause by a single infectious agent (above HIV/AIDS). Millions of people continue to fall sick with TB each year.
A report from the World Health Organization (WHO) indicates that TB caused an estimated 1.3 million deaths, and 10.0 million people developed TB disease in 2017. Nontuberculous mycobacteria (NTM) species are ubiquitous organisms distributed in ambient environments which opportunistically cause human diseases (36). In recent years, the incidence and prevalence of NTM diseases have continued to increase worldwide (37). Although the causative agents of NTM diseases vary with geographical location, the most common pulmonary NTM pathogens are M. abscessus, M. intracellulare, M. avium, M. fortuitum, M. kansasii, and M. gordonae. The clinical symptoms of NTM diseases and M. tuberculosis infection are often very similar. However, the mycobacterial species differ dramatically from one another in terms of treatment outcomes and antibiotic susceptibility (37–39), which seriously hinders the diagnosis and treatment of M. tuberculosis- and NTM-caused diseases. The most effective strategy to control mycobacterial infection is early diagnosis and treatment of the disease. Therefore, it is important to develop methods to rapidly and accurately detect and identify the infecting mycobacterial species for specific chemotherapy and better patient management.
Traditional detection methods based on growth characteristics, colony morphology, pigment production, and biochemical reactions are laborious, tedious, and unable to categorize NTM species. Nucleic acid amplification-based methods have made great improvements in the rapid and accurate diagnosis of mycobacterial infections. Several commercially available assays based on molecular detection, such as GeneXpert M. tuberculosis /RIF Ultra test, Roche M. tuberculosis -RIF/INH test, and IS6110 PCR, are widely used for the direct detection of M. tuberculosis in clinical samples. Although these methods are rapid, highly sensitive, and specific, they exclusively focus on the identification of M. tuberculosis. The development of multiplex PCR and multiplex real-time PCR assays, which are able to detect a number of organisms in a single reaction, has dramatically accelerated the detection of mycobacterial species (8, 9, 11). However, it is difficult to identify closely related species, and the interpretations of the results are complicated. To date, Sanger sequencing is still regarded as the gold standard for identifying M. tuberculosis and NTM infections, but it is not cost-effective. Recently, the development of Cas13- and Cas12a-based CRISPR/Cas biosensing systems has enabled sensitive, robust, and low-cost tools for detecting nucleic acids, displaying great potential for the diagnosis of pathogens (29–31, 40). Since Cas13- and Cas12a-based CRISPR/Cas biosensing systems can discriminate single-base differences, we reasoned that these methods could be promising tools to identify Mycobacterium species.
Here, we attempted to identify M. tuberculosis and six major NTM species using an FnCas12a/gRNA-based platform. Through designing species-specific gRNA probes targeting the rpoB gene, 72 of the 73 clinical isolates were correctly identified, and there were no false-positive results, indicating the accuracy of this platform. However, there was a strain that was misidentified by the M. intracellulare gRNA probe 2 due to two base mutations in the targeting sequence, indicating that two base mismatches could lead to inactivation of the FnCas12a/gRNA complex. This result suggested that variation in the target sequences potentially caused detection failure or misidentification. In addition, we observed that different gRNA probes caused different fluorescence intensities. However, we did not find any nucleotide preferences for highly active gRNAs through analysis of gRNA probe sequences. Moreover, the Mycobacterium complex could also be identified with a package of gRNA probes (not shown). In addition, the FnCas12a-based platform exhibited high sensitivity in the detection of Mycobacterium species, ranging from 500 to 10,000 CFU/ml, which is comparable to real-time PCR. This method is easy to operate without any expensive equipment, which contributes to its application in poor countries and regions. The entire process of this method could be finished within 3 h, and the experimental results can be easily and intuitively read out according to the fluorescence intensity.
This study also has several limitations. For example, the clinical isolates of some NTM species may not be sufficient to test the effectiveness of gRNA probes due to the limited incidences of NTM species infection. Another limitation of this study is that we did not use direct clinical specimens to evaluate the FnCas12a/gRNA-based platform. Further study is needed on clinical samples, such as sputum, blood, tissues, and bronchoalveolar lavage fluid. In conclusion, this method allowed for the rapid, highly sensitive, and specific identification of M. tuberculosis and most NTM species.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Zhao Yanlin at the Chinese Center for Disease Control and Prevention for providing the type strains of Mycobacterium species.
This work was supported by the China Postdoctoral Science Foundation (no. 2019M653108), the National Natural Science Foundation of China (no. 81873958), the National Science and Technology Major Project for Control and Prevention of Major Infectious Diseases of China (no. 2017ZX10103004), the Shenzhen Scientific and Technological Foundation (no. JCYJ20170412151620658, JCYJ20170307095003051, JCYJ20180228162453330, and JCYJ20180228162511084), the State Key Laboratory of Respiratory Diseases Open Project (no. SKLRD-OP-201919), and the Shenzhen Special Funds for introduced high level medical team on constructing a systematic network against tuberculosis.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Donohue MJ, Wymer L. 2016. Increasing prevalence rate of nontuberculous mycobacteria infections in five states, 2008–2013. Annals Ats 13:2143 AnnalsATS.201605-353OC. doi: 10.1513/AnnalsATS.201605-353OC. [DOI] [PubMed] [Google Scholar]
- 2.Donohue MJ. 2018. Increasing nontuberculous mycobacteria reporting rates and species diversity identified in clinical laboratory reports. Bmc Infect Dis 18:163. doi: 10.1186/s12879-018-3043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu X, Liu P, Liu G, Zhao L, Hu Y, Wei G, Luo J, Huang H. 2016. The prevalence of non-tuberculous mycobacterial infections in mainland China: systematic review and meta-analysis. J Infect 73:558. doi: 10.1016/j.jinf.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 4.Frieden TR, Sterling TR, Munsiff SS, Watt CJ, Dye C. 2003. Tuberculosis. Lancet 362:887–899. doi: 10.1016/S0140-6736(03)14333-4. [DOI] [PubMed] [Google Scholar]
- 5.Bae E, Im JH, Kim SW, Yoon N-S, Sung H, Kim M-N, Shim TS. 2008. Evaluation of combination of BACTEC mycobacteria growth indicator tube 960 system and Ogawa media for mycobacterial culture. Korean J Lab Med 28:299. doi: 10.3343/kjlm.2008.28.4.299. [DOI] [PubMed] [Google Scholar]
- 6.Lima AS, Duarte RS, Montenegro LML, Schindler HC, Lima AS, Duarte RS, Montenegro LML, Schindler HC. 2013. Rapid detection and differentiation of mycobacterial species using a multiplex PCR system. Rev Soc Bras Med Trop 46:447–452. doi: 10.1590/0037-8682-0097-2013. [DOI] [PubMed] [Google Scholar]
- 7.Gupta S, Bandyopadhyay D, Paine SK, Gupta S, Banerjee S, Bhattacharya S, Gachhui R, Bhattacharya B. 2010. Rapid identification of mycobacterium species with the aid of multiplex polymerase chain reaction (PCR) from clinical isolates. Open Microbiol J 4:93–97. doi: 10.2174/1874285801004010093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chae H, Han SJ, Kim SY, Ki CS, Huh HJ, Yong D, Koh WJ, Shin SJ. 2017. Development of a one-step multiplex PCR assay for differential detection of major Mycobacterium species. J Clin Microbiol 55:2736. doi: 10.1128/JCM.00549-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang HY, Kim H, Kim S, Kim DK, Cho SN, Lee H. 2015. Performance of a real-time PCR assay for the rapid identification of Mycobacterium species. J Microbiol 53:38. doi: 10.1007/s12275-015-4495-8. [DOI] [PubMed] [Google Scholar]
- 10.Lim JH, Kim CK, Bae MH. 2019. Evaluation of the performance of two real‐time PCR assays for detecting Mycobacterium species. J Clin Lab Anal 33:e22645. doi: 10.1002/jcla.22645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim JU, Ryu DS, Cha CH, Park SH. 2018. Paradigm for diagnosing mycobacterial disease: direct detection and differentiation of Mycobacterium tuberculosis complex and non-tuberculous mycobacteria in clinical specimens using multiplex real-time PCR. J Clin Pathol 71:774. doi: 10.1136/jclinpath-2017-204945. [DOI] [PubMed] [Google Scholar]
- 12.Troesch A, Nguyen H, Miyada CG, Desvarenne S, Gingeras TR, Kaplan PM, Cros P, Mabilat C. 1999. Mycobacterium species identification and rifampin resistance testing with high-density DNA probe arrays. J Clin Microbiol 37:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali RM, Alsudani AA. 2016. Discordance between GeneXpert assay and conventional drug-susceptibility testing in detecting rifampicin-resistant tuberculosis: a perspective of the line probe assay. Int J Mycobacteriol 5 Suppl 1:S193. doi: 10.1016/j.ijmyco.2016.09.039. [DOI] [PubMed] [Google Scholar]
- 14.Singh AK, Maurya AK, Umrao J, Kant S, Kushwaha RAS, Nag VL, Dhole TN. 2013. Role of GenoType Mycobacterium common mycobacteria/additional species assay for rapid differentiation between Mycobacterium tuberculosis complex and different species of non-tuberculous mycobacteria. J Lab Physicians 5:83–89. doi: 10.4103/0974-2727.119847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cristina R, Enrico T, Donato M. 2006. Evaluation of the new GenoType Mycobacterium assay for identification of mycobacterial species. J Clin Microbiol 44:334. doi: 10.1128/JCM.44.2.334-339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nour-Neamatollahie A, Ebrahimzadeh N, Siadat SD, Vaziri F, Eslami M, Akhavan Sepahi A, Khanipour S, Masoumi M, Sakhaee F, Ghazanfari Jajin M, Bahrmand A, Fateh A. 2017. Distribution of non-tuberculosis mycobacteria strains from suspected tuberculosis patients by heat shock protein 65 PCR–RFLP. Saudi J Biol Sci 24:1380–1386. doi: 10.1016/j.sjbs.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saifi M, Jabbarzadeh E, Bahrmand AR, Karimi A, Pourazar S, Fateh A, Masoumi M, Vahidi E. 2013. HSP65-PRA identification of non-tuberculosis mycobacteria from 4892 samples suspicious for mycobacterial infections. Clin Microbiol Infect 19:723–728. doi: 10.1111/j.1469-0691.2012.04005.x. [DOI] [PubMed] [Google Scholar]
- 18.Tsu-Lan W, Ju-Hsin C, An-Jing K, Lin-Hui S, Ting-Shu W, Hsin-Chih L. 2008. Rapid identification of mycobacteria from smear-positive sputum samples by nested PCR-restriction fragment length polymorphism analysis. J Clin Microbiol 46:3591. doi: 10.1128/JCM.00856-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong K, Kim SH, Shim TS, Kim MN, Bai GH, Park YG, Lee SH, Cha CY, Kook YH, Kim BJ. 2005. PCR restriction fragment length polymorphism analysis (PRA)-algorithm targeting 644 bp Heat Shock Protein 65 (hsp65) gene for differentiation of Mycobacterium spp. J Microbiol Methods 62:199–209. doi: 10.1016/j.mimet.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Kim BR, Kim JM, Kim BJ, Jang Y, Ryoo S, Kook YH, Kim BJ. 2014. Identification of nontuberculous mycobacteria isolated from Hanwoo (Bos taurus coreanae) in South Korea by sequencing analysis targeting hsp65, rpoB and 16S rRNA genes. Vet Microbiol 173:385–389. doi: 10.1016/j.vetmic.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 21.Xiao-Li Y, Lian L, Gao-Zhan C, Zhi-Guo L, Hang L, Yan-Zheng S, Shu-Lin Z. 2014. Identification and characterization of non-tuberculous mycobacteria isolated from tuberculosis suspects in Southern-central China. PLoS One 9:e114353. doi: 10.1371/journal.pone.0114353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jagielski T, Minias A, van Ingen J, Rastogi N, Brzostek A, Żaczek A, Dziadek J. 2016. Methodological and clinical aspects of the molecular epidemiology of Mycobacterium tuberculosis and other mycobacteria. Clin Microbiol Rev 29:239–290. doi: 10.1128/CMR.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Si HK, Shin JH. 2018. Identification of nontuberculous mycobacteria using multilocous sequence analysis of 16S rRNA, hsp65, and rpoB. J Clin Lab Anal 32:e22184. doi: 10.1002/jcla.22184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Zwaan R, van Ingen J, van Soolingen D. 2014. Utility of rpoB gene sequencing for identification of nontuberculous mycobacteria in the Netherlands. J Clin Microbiol 52:2544–2551. doi: 10.1128/JCM.00233-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimenkov DV, Kulagina EV, Antonova OV, Krasnova MA, Chernyaeva EN, Zhuravlev VY, Kuz'min AV, Popov SA, Zasedatelev AS, Gryadunov DA. 2015. Evaluation of a low-density hydrogel microarray technique for mycobacterial species identification. J Clin Microbiol 53:1103. doi: 10.1128/JCM.02579-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prapaporn S, Angkana C, Katsushi T, Nishida N, Therdsak P. 2014. Novel DNA chip based on a modified DigiTag2 assay for high-throughput species identification and genotyping of Mycobacterium tuberculosis complex isolates. J Clin Microbiol 52:1962–1968. doi: 10.1128/JCM.00153-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linger Y, Knickerbocker C, Sipes D, Golova J, Franke M, Calderon R, Lecca L, Thakore N, Holmberg R, Qu P, Kukhtin A, Murray MB, Cooney CG, Chandler DP. 2018. Genotyping multidrug-resistant Mycobacterium tuberculosis from primary sputum and decontaminated sediment with an integrated microfluidic amplification microarray test. J Clin Microbiol 56:e01652-17. doi: 10.1128/JCM.01652-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li SY, Cheng QX, Liu JK, Nie XQ, Zhao GP, Wang J. 2018. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res 28:491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. 2018. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li SY, Cheng QX, Wang JM, Li XY, Zhang ZL, Gao S, Cao RB, Zhao GP, Wang J. 2018. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov 4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nie W, Duan H, Huang H, Lu Y, Bi D, Chu N. Species identification of Mycobacterium abscessus subsp. abscessus and Mycobacterium abscessus subsp. bolletii using rpoB and hsp65, and susceptibility testing to eight antibiotics. Int J Infectious Diseases doi: 10.1016/j.ijid.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 33.Adekambi T, Colson P, Drancourt M. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing Mycobacteria. J Clin Microbiol 41:5699–5708. doi: 10.1128/jcm.41.12.5699-5708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben Salah I, Adekambi T, Raoult D, Drancourt M. 2008. rpoB sequence-based identification of Mycobacterium avium complex species. Microbiology 154:3715–3723. doi: 10.1099/mic.0.2008/020164-0. [DOI] [PubMed] [Google Scholar]
- 35.Mikaeili F, Kia EB, Sharbatkhori M, Sharifdini M, Jalalizand N, Heidari Z, Zarei Z, Stensvold CR, Mirhendi H. 2013. Comparison of six simple methods for extracting ribosomal and mitochondrial DNA from Toxocara and Toxascaris nematodes. Exp Parasitol 134:155–159. doi: 10.1016/j.exppara.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Brown-Elliott BA, Griffith DE, Wallace RJ. 2002. Diagnosis of nontuberculous mycobacterial infections. Clinics in Laboratory Medicine 22:911–925. doi: 10.1016/S0272-2712(02)00018-5. [DOI] [PubMed] [Google Scholar]
- 37.Stout JE, Koh WJ, Yew WW. 2016. Update on pulmonary disease due to non-tuberculous mycobacteria. Int J Infect Dis 45:123–134. doi: 10.1016/j.ijid.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 38.Nie W, Duan H, Huang H, Lu Y, Chu N. 2015. Species identification and clarithromycin susceptibility testing of 278 clinical nontuberculosis mycobacteria isolates. Biomed Res International 2015:506598. doi: 10.1155/2015/506598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryu YJ, Koh WJ, Daley CL. 2016. Diagnosis and treatment of nontuberculous mycobacterial lung disease: clinicians’ perspectives. Tuberc Respir Dis (Seoul) 79:74–84. doi: 10.4046/trd.2016.79.2.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F. 2017. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.