Summary
Recent progress in the genomics of non-syndromic autism spectrum disorder (nsASD) highlights rare large-effect heterozygous de novo coding mutations. This distinguishes nsASD from later-onset psychiatric disorders where gene discovery efforts have predominantly yielded common alleles of small effect. These differences point to distinctive opportunities for clarifying the neurobiology of nsASD and developing novel treatments. We argue that the path ahead also presents key challenges, including distinguishing human pathophysiology from the potentially pleiotropic neurobiology mediated by established risk genes. We present our view of some of the conceptual limitations of traditional studies of model organisms, suggest a strategy focused on investigating the convergence of multiple nsASD genes, and propose that the detailed characterization of the molecular and cellular landscapes of developing human brain is essential to illuminate underlying mechanisms. Finally, we address how recent advances are leading to novel strategies for therapeutics that target various points along the path from genes to behavior.
Keywords: Autism Spectrum Disorder, de novo mutation, genomics, neurodevelopmental disorders, convergence, non-syndromic autism spectrum disorder, human brain development, transcriptomics, gene therapy, convergence neuroscience
In Brief:
Sestan and State propose that the distinctive genomic findings in Autism Spectrum Disorder, the deepening understanding of developing human brain, and the identification of spatiotemporal convergence of large-effect risk genes will help clarify pathophysiological mechanisms and identify novel treatment approaches.
Over the past two decades, the genetics of autism spectrum disorder (ASD) has advanced spectacularly. From the early hard-won successes cloning genes for monogenic forms of intellectual and social disability (European Chromosome 16 Tuberous Sclerosis, 1993; van Slegtenhorst et al., 1997; Verkerk et al., 1991) to current large-scale studies utilizing whole-genome sequencing (Brandler et al., 2018; Sanders et al., 2017; Turner et al., 2017; Werling et al., 2018; Yuen et al., 2015) the field has evolved to a point where discovery in so-called idiopathic or “non-syndromic” forms of the syndrome (nsASD) is now highly reliable and reproducible, yielding dozens of well-established risk genes (De Rubeis et al., 2014; Dong et al., 2014; Iossifov et al., 2014; Iossifov et al., 2012; Neale et al., 2012; O’Roak et al., 2011; O’Roak et al., 2012b; Sanders et al., 2015; Sanders et al., 2012). Importantly, in contrast to later-onset psychiatric disorders, such as schizophrenia, bipolar disorder and major depression, where genomic studies aimed at identifying individual genes have mainly highlighted a highly polygenic risk architecture involving the simultaneous contribution of multiple alleles of very small effect (Bipolar et al., 2018; Cross-Disorder Group of the Psychiatric Genomics et al., 2013; Schizophrenia Working Group of the Psychiatric Genomics, 2014; Wray et al., 2018), progress in nsASD has been notable for the discovery of rare, de novo, coding, heterozygous mutations carrying large effects in the individual. And while these variants contribute to about 20–30% of clinical cases of nsASD (De Rubeis et al., 2014; Iossifov et al., 2014; Sanders et al., 2015), we propose that they offer distinctive opportunities to illuminate ASD-related biology and critically important avenues for the development of novel and precision therapies.
Despite the obvious advantages at the bench of modeling large-effect likely gene disrupting (LGD) coding mutations (that is stop codons, canonical splice site mutations and frameshifts) versus the typical results of a genome-wide association studies (GWAS) (see for review (Visscher et al., 2017), navigating the path from genomics to therapeutics in nsASD nonetheless poses considerable obstacles. For instance, the phenotypes of greatest interest, including emotion, social communication and complex behavior, are highly-derived in humans, presenting challenges for faithful recapitulation in other species, including those model systems most tractable for genetic studies. Moreover, the tissue of greatest interest, living human brain, remains rarely accessible for direct investigation or manipulation. Further complicating the issue, the genes so-far identified tend to be biologically pleiotropic and drive multiple diverse functions across developmental time and anatomical distribution, suggesting that only a subset of all the potentially observable neurobiological changes present in any mutant model system may be relevant for the human disorder. Similarly, any attempt to disentangle nsASD pathology must contend with the observation that many risk genes are ubiquitously expressed, as well as biologically pleiotropic, and yet all have been identified because they lead to shared core deficits that point to dysfunction of specific human brain regions and circuits. Finally, risk variants in humans show variable penetrance, a great deal of phenotypic heterogeneity -- with a single variant potentially leading to multiple distinct neurodevelopmental outcomes in different individuals -- and may be sexually dimorphic, given the well-established male predominance in nsASD (Halladay et al., 2015; Volkmar et al., 2014).
We argue that these considerations suggest that the widely-used approach of manipulating individual risk genes and characterizing resultant biology in rodents or other evolutionarily-distant organisms, gene-by-gene, is unlikely to be sufficient, in isolation, to clarify the mechanisms underlying nsASD -- despite the large effect size of nsASD associated mutations and the evidence that often a single mutation in the individual confers the lion’s share of risk. This is not to say that such mechanisms would necessarily not be observable or manipulatable in distant species; only that differentiating the basis of pathophysiology in human from the broader set of neurobiological processes related to these genes and mutations in any non-human model poses key conceptual and experimental challenges.
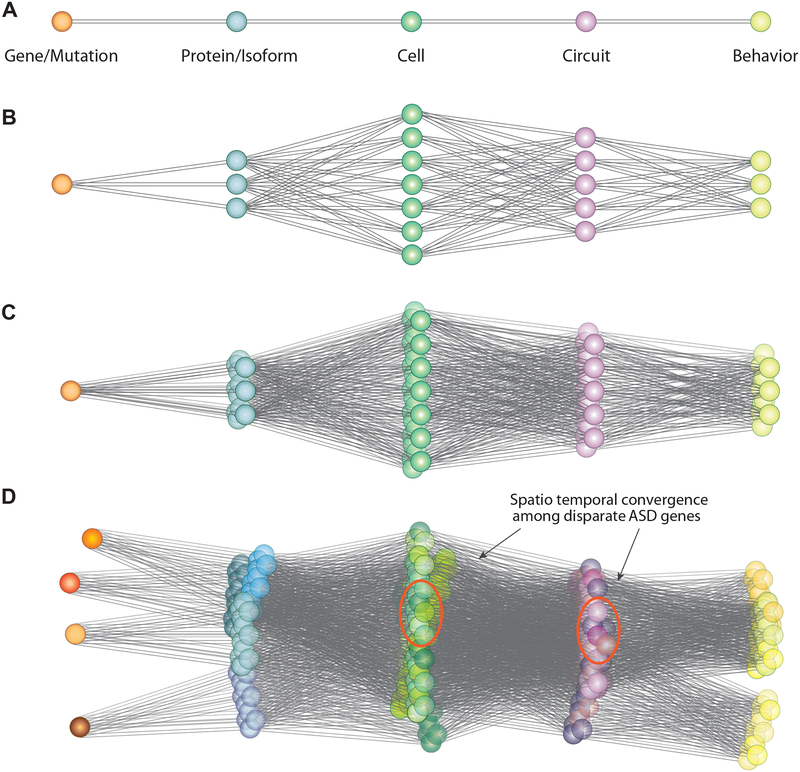
We have proposed a strategy that augments more traditional model systems approaches by the search for “convergence” among disparate risk genes (State and Sestan, 2012; Willsey et al., 2018; Willsey et al., 2013). We use this term to convey the idea that studying multiple nsASD genes in parallel can potentially contribute to a clearer understanding of human pathology, based on the hypothesis that at some point on the path from genes to behavior, subsets of distinct ASD large-effect risk genes intersect in a meaningful way to lead to a shared, reproducible set of phenotypic features we define as nsASD. We propose that this convergence may exist at multiple levels of analysis, from proteins, to cells, to circuits (Figure 1), and that subsets of risk genes will intersect at different points along this trajectory (Figure 1D). Moreover, we argue that derangements in a variety of overtly-distinct biological processes can lead to identical consequences by impacting a particular point of developmental or functional vulnerability. In short, we propose that strategies aimed at clarifying pathophysiological mechanisms must explore overlap not only “what” multiple, diverse nsASD genes do but “when and where” they do them (i.e., “spatiotemporal” convergence) (State and Sestan, 2012; Willsey et al., 2018; Willsey et al., 2013; Willsey and State, 2015).
Figure 1: Spatiotemporal convergence among high confidence ASD risk genes.
The figure provides a conceptual illustration of the path from risk genes to behavior in nsASD. Panel A illustrates an idealized path from genes to proteins to cells, circuits and behavior. This is shown as a line connecting color spheres representing the noted levels of analysis (orange = cells, blue = proteins; green =cell etc.); Panel B shows the complexity added by consideration of isoform diversity and biological pleiotropy, with a single gene/mutation leading to multiple transcripts and proteins and corresponding to multiple cell types and circuits. Panel C represents the further complexity added by the consideration of spatial and temporal influence on expression and function during brain development. This is shown as a three-dimensional space connecting a single gene to multiple isoforms, protein, cell types and circuits. Sexual dimorphism potentially adds another dimension of complexity. Panel D represents the strategy of using multiple independent risk genes in an effort to triangulate on specific cell types and circuits that show overlap among functionally diverse risk genes.
What follows is our perspective on recent progress in the genomics of nsASD, the evolving understanding of the molecular landscape of developing human brain, and a convergence strategy for moving beyond gene discovery -- including the critical issue of how this approach can help define and constrain future model systems experiments, offer an avenue to begin to distinguish between mutation-related biology and pathophysiology, and suggest hypotheses regarding the selective vulnerability of specific cells and circuits underlying the manifestation of nsASD. It is not intended as a comprehensive review of the genetics or biology of the autism spectrum, what has been gleaned from model system studies to date, or a compendium of current advances in the neurobiology of brain development. There are many outstanding reviews of these topics (see for example: (Bae et al., 2015; Bourgeron, 2015, 2016; Cameron et al., 2017; de la Torre-Ubieta et al., 2016; De Rubeis and Buxbaum, 2015; Di Lullo and Kriegstein, 2017; Krumm et al., 2014; Lein et al., 2017; Lodato and Arlotta, 2015; Nord et al., 2015; Silbereis et al., 2016; Vorstman et al., 2017). Instead we address selected recent discoveries regarding allelic architecture– that is the types and distribution of genetic variants conferring risk -- and argue that this is particularly well-suited in nsASD for convergence approaches. We present our view of some of the conceptual limitations of studying model organisms in series, describe how advances in the basic understanding of normal developing human brain may lend a deeper understanding of pathophysiological mechanisms, and argue for the importance of complementing more traditional approaches with those that leverage high-throughput, parallel studies and hypothesis-free analyses of multi-scale omics data derived from typically-developing human post-mortem brain. Finally, we consider how progress in genomics and in approaches assessing the convergence of nsASD risk genes across multiple levels of analysis have led to promising strategies for the development of novel therapeutics in nsASD, including near-term opportunities for gene-targeting based on the genomic characteristics of a substantial subset of nsASD-related mutations.
Gene Discovery in Syndromic versus Non-syndromic (ns)ASD
Before delving into a description of the distinctive character of recent genomic findings, a clarification of the notion of “idiopathic,” or “non-syndromic,” versus “syndromic” ASD is in order. In general, the term syndromic is used to denote individuals who meet diagnostic criteria – that is show fundamental deficits in reciprocal social communication and highly restrictive interests and/or repetitive behaviors (Volkmar et al., 2014; Volkmar et al., 2009)– and who also have evidence of a genetic syndrome, typically involving distinctive dysmorphology and other associated findings, and/or a known monogenic form of social, and very often concomitant intellectual disability (ID). Non-syndromic ASD refers to diagnosed individuals without these associated features.
While such a distinction is commonly employed by both clinicians and researchers, there are obvious definitional challenges. For example, successes in gene discovery among cohorts of individuals with putative nsASD have shown that the lines between syndromic and non-syndromic forms are decidedly blurry. Some of the first risk genes identified in well-characterized nsASD cases (Autism Genome Project et al., 2007; Jamain et al., 2003; Marshall et al., 2008; Neale et al., 2012; O’Roak et al., 2012b; Sanders et al., 2012) show very-high penetrance, approaching that seen in monogenic forms of ASD and ID. These nsASD genes point to pathways and processes that show obvious areas of overlap with what is known about syndromic biology (De Rubeis et al., 2014; Iossifov et al., 2014; Iossifov et al., 2012; Zoghbi, 2003); and, when phenotypically characterized in depth, some nsASD individuals with deleterious mutations in a confirmed risk gene have shown clusters of associated physical features suggesting a previously uncharacterized ASD syndrome (Bernier et al., 2014; Earl et al., 2017; O’Roak et al., 2012a).
In short, evidence for a reliable distinction between affected individuals with and without dysmorphology, with and without highly penetrant mutations, or based on specific pathways to disease has not been forthcoming. Alternatively, in retrospect, it’s clear that genes such as FMR1 (Verkerk et al., 1991) and TSC2 (European Chromosome 16 Tuberous Sclerosis, 1993) were cloned decades before the recent flurry of gene discovery in nsASD largely due to a combination of the nature of the underlying genetic lesion and its “visibility” to the available technology of the time; because affected individuals represented the extreme end of the distribution with regard to the reliability of the relationship of genotype to phenotype; and due to the prominence and predictability of associated physical signs and symptoms.
Nonetheless, despite these definitional challenges and the leading role that syndromic ASD has played in elaborating both the genetics and biology of neurodevelopmental disorders, we have elected to focus this perspective on nsASD and associated issues for translational science. This is in part because, as a group, these individuals make up the vast majority of those affected and, until quite recently – long after the initial successes in cloning monogenic causes of intellectual and social disability-- continued to present daunting obstacles to systematic gene discovery. Consequently, these more recent findings offer the first definitive molecular clues to underlying mechanisms in a large group of patients who fall more into the center of the distribution: where the presence or absence of ID is more variable, the associated physical features, if present, are subtler, the penetrance of the mutations may be lower, and the range of diagnostic outcomes, broader. The important question of whether studying this subgroup of individuals with nsASD will ultimately lead to distinct conclusions regarding underlying biology and/or generalizable therapeutic targets remains to be seen. However, these are important and now at least theoretically tractable, questions that underscore the value of continuing work on the genetics and neurobiology of both syndromic and non-syndromic autism, and of studying both rare as well as common risk variants.
The Advantageous Allelic Architecture of nsASD
The genomics of ASD has only recently matured to a point where gene discovery is now routine and systematic. The number of risk genes has increased from less than a handful at the beginning of this decade (Autism Genome Project et al., 2007; Durand et al., 2007; Jamain et al., 2003; Marshall et al., 2008) to dozens at present. In 2015, a comprehensive analysis combining data from multiple research groups that all studied de novo mutations in simplex families (those with only a single affected individual) identified 71 independent risk genes and loci (Sanders et al., 2015). And in short order, additional publications from large sequencing consortia will dramatically increase this number. The consensus, based on existing data, is that in toto between several hundred to approximately a thousand genes will be found to confer liability due to rare heterozygous de novo protein damaging mutations (Iossifov et al., 2012; Sanders et al., 2015; Sanders et al., 2012). As described in more detail below, these genes and mutations only explain a small fraction of the population risk for ASD (Gaugler et al., 2014; Klei et al., 2012; Weiner et al., 2017) but account for a considerable proportion of patients seen in clinic (Iossifov et al., 2014; Sanders et al., 2015). While not representing the majority of genetic risk for nsASD in the population, we propose that focusing on this particular segment of the allelic distribution provides key advantages for convergence strategies, for translation to model organisms, and potentially for the development of novel therapies:
De novo LGD mutations confer quite large effects
While there has been evidence for the contribution of rare transmitted alleles from the earliest successful studies of nsASD, (Jamain et al., 2003; Laumonnier et al., 2004), the relatively much larger effect sizes of de novo LGD mutations have now been repeatedly confirmed (De Rubeis et al., 2014; Iossifov et al., 2014). And the associated statistical power has been critical for the evolution of reliable and reproducible discovery (He et al., 2013; Liu et al., 2014; Sanders et al., 2011).
From a theoretical standpoint, this observation is not terribly surprising. Germ line de novo mutations arise prior to fertilization or immediately thereafter, and consequently, the window for purifying natural selection to impact these changes before they can be observed in the incident generation is quite limited. For this reason, mutations carrying large effects on early brain development that otherwise would impair reproductive fitness and be removed from the genome over time can nonetheless be detected in an affected proband. Conversely, transmitted heterozygous mutations that carry risk across multiple generations would on average be expected to remain in the genome only if they carried much smaller effects. And, indeed, a recent meta-analysis of GWAS data, involving a discovery sample of nearly 8000 ASD cases failed to identify a single locus meeting the threshold for genome wide significance (Autism Spectrum Disorders Working Group of The Psychiatric Genomics, 2017) despite incontrovertible evidence for the major contribution of polygenic inheritance (Brainstorm et al., 2018; Gaugler et al., 2014; Klei et al., 2012; Robinson et al., 2016; St Pourcain et al., 2018). These results strongly suggest that even larger samples will be required to provide the power necessary to detect individual risk alleles due to their extremely modest effects.
In contrast, the marked effect size of heterozygous de novo mutations has been a defining feature of nsASD gene discovery. The earliest analyses of rates of de novo copy number variations (CNVs)—submicroscopic changes in chromosomal structure -- detected statistically significant differences in simplex versus multiplex cases versus controls in cohorts consisting of only several hundred families (Sebat et al., 2007). Similarly, the first finding of an increased rate of LGD mutations using whole exome sequencing, as well as the first identification of significantly-associated risk genes, was accomplished in cohorts of similar size (Iossifov et al., 2012; Neale et al., 2012; O’Roak et al., 2012b; Sanders et al., 2012). As these findings have now been widely replicated, they point to quite large overall effects for this type of mutation. More directly, estimates of the increased risk contributed by individual common risk single nucleotide polymorphisms (SNPs) to psychiatric disorders generally don’t exceed 20–30 percent; the highest confidence nsASD genes show increases in risk that are many-fold greater (De Rubeis et al., 2014). An illustrative comparison is that one would have to be in the top decile of cumulative polygenic risk for schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics, 2014) to approach the effect size of single de novo LGD mutation in one of the highest-confidence nsASD risk genes (De Rubeis et al., 2014; Sanders et al., 2015).
Typically, only a single variant is responsible for the majority of risk in de novo LGD mutation carriers
The cumulative evidence to date points to the conclusion that de novo LGD risk events generally involve a single locus at a time in the affected individual and are acting in cis. With regard to this first observation, LGD, and to a lesser extent missense heterozygous de novo mutations, carry nsASD risk, but silent point mutations that have no predicted impact on the encoded proteins do not (Iossifov et al., 2012; Neale et al., 2012; O’Roak et al., 2012b; Sanders et al., 2012). This suggests that the increase in frequency in de novo mutation seen in cases is the result of ascertaining for causal or contributory events, not a result of DNA hyper-mutability. In addition, overall the frequency and distribution of individuals with multiple non-synonymous de novo sequence variants does not differ between cases and controls (Sanders et al., 2012). And while some studies have identified examples of putative “two-hit” mechanisms (Girirajan et al., 2010), large-scale sequencing studies have consistently supported the conclusion that, as a general proposition, a single de novo mutation in a confirmed ASD-gene, in an individual, carries the majority of risk (Sanders et al., 2015). Finally, the functional impact of these single de novo heterozygous mutations appears in most cases to be acting in cis. Sequencing studies have identified a small percentage of cases resulting from compound heterozygous mutations, but generally have not suggested that cryptic recessive inheritance is a widely applicable mechanism (Lim et al., 2013).
The value of pursuing the biology of large-effect heterozygous de novo coding mutations in nsASD is not without debate. For example, there is compelling evidence that these types of variants are acting in the context of a highly polygenic risk background (Weiner et al., 2017) leading to the assertion that nsASD cannot be understood without a full elaboration of this polygenic risk. There are tremendously important questions regarding how rare and common variants interact and how this might influence disease course, treatment response, and phenotypic outcomes or variability. Indeed, one of the most intriguing questions in this regard is whether, and if so, how common polygenic risk might act as a disease-modifier and potentially offer novel paths to therapeutics development. However, the data noted above supporting the impact of a single de novo mutation, coupled with very high penetrance for the highest confidence nsASD risk genes, strongly suggests that these individual mutations and genes confer substantial main effects and, consequently, we argue that modeling them holds the potential to provide important insights into central pathophysiological mechanisms and potentially novel avenues to therapeutics.
De novo mutations account for a small percentage of population risk but a substantial subset of patients seen in clinic
Current estimates of the proportion of explained clinical cases related to de novo mutations range from about 10–40% of individuals diagnosed with ASD (Gaugler et al., 2014; Iossifov et al., 2014; Sanders et al., 2015). Because this calculation uses a clinically-defined population as the denominator, it is dependent on ascertainment. For example, identifying affected females, individuals with a history of seizure, families with only a single affected, and multiple unaffected, siblings, and the presence of lower IQ (Sanders et al., 2015) all significantly increase rates of detection of a de novo mutation.
These observations are fully consistent with studies that demonstrate the vast majority of population risk for autism is highly polygenic involving the cumulative impact of many small effect SNPs (Gaugler et al., 2014; Klei et al., 2012). The explanation for what might seem a contradiction is that in the general population, most individuals carrying genetic liability for nsASD will have no overt evidence of a clinical syndrome. Consequently, while only a small proportion of all individuals will carry a de novo risk mutation, this equates to a relatively large percentage of those who cross the diagnostic threshold for nsASD.
In sum, the recent advances in nsASD genomics have resulted in the identification of an ample subset of clinical cases in which a single, heterozygous, functional, coding de novo mutation accounts for the majority of the observed risk. For those not following these developments closely, this statement might appear to run counter to the prevailing view of psychiatric genetics – which are generally thought of as synonymous with highly polygenic inheritance and the absence of genes of main effect. As a consequence, with later onset disorders, the most immediate challenges involve confirming the relationship between hundreds of associated non-coding SNPs and the specific genes they may be influencing; the complexity of modeling many alleles simultaneously and the resulting very large number of potential combinations; and the gap between the types of mutations that are typically modeled in evolutionarily distant organisms, that is constitutive or conditional knockouts, versus the much more subtle small-effect, often non-coding, SNPs that carry common-variant risk in the human context.
We propose that nsASD provides an important counterpoint, with distinctive opportunities for translation: first, the ability to interpret the result on protein structure or function of a frameshift or canonical stop codon is far easier than for most common non-coding alleles. Second, modeling heterozygous loss-of-function mutations in animal models provides an opportunity to faithfully recapitulate the human genomic “lesion.” For strategies that rely on investigating multiple models in parallel, the ability to manipulate one main variable in an organism or culture at a time is far more experimentally tractable versus assessing for convergence while modeling hundreds of small- effect alleles simultaneously.
Of course, many challenges remain: the genomic architecture of nsASD has already been shown to be remarkably heterogeneous, the population frequency of mutations in any given risk gene is very small - raising issues of statistical power for genotype-phenotype analyses and introducing questions about the generalizability of any related findings-- and there is ample reason to suspect that modeling heterozygous as opposed to homozygous mutations will be relatively more difficult, in part because the phenotypes will be expected to be subtler on average. Finally, as we address below, there are inherent challenges in studying and modelling human neurodevelopmental phenotypes that impact complex behavior, emotion, and communication, regardless of the types of underlying genetic variations.
The Challenges of Clarifying nsASD Pathophysiology
The foregoing has highlighted a genetic substrate that would seem particularly well-suited for traditional model systems analyses. The immediate path from the discovery of a coding, large-effect mutation to the bench is comparatively straight forward. The armamentarium to illuminate the biochemistry, molecular and cellular biology, and physiology of these types of mutations --including new nucleic acid editing techniques and improved tools and methods to study and manipulate neural circuit-level functioning-- is both mature and still rapidly advancing. Moreover, it is difficult to overstate the contribution of a wide variety of studies of model organisms to the understanding of neurobiology broadly, including human brain. At this point it should not be controversial, but it is always worth pointing out, that these contributions have not been restricted to models with extensive repertoires of higher order behaviors. From fly, worm and fish, to rodent and non-human primate, induced pluripotent stem cell (iPSC) models and neural organoids, a diverse range of models have all made important contributions to our evolving understanding of the human nervous system, its function, and its dysfunction.
With regard to ASD, neurobiologists have undertaken extensive modeling of monogenic forms of ASD and many of the emerging nsASD genes both in vitro and in vivo (see for review (Varghese et al., 2017). Yet, despite the inarguable contribution of such investigations for highlighting biological sequelae, important issues remain regarding the limitations of these methods, particularly the typical practice of modeling one gene/mutation at a time in an evolutionarily distant species, such as the mouse.
While there is little question that these approaches can both identify and illuminate key aspects of biology that are conserved across species, there are some conceptual gaps as well. For example, the nsASD risk genes so far identified are typically biologically pleiotropic and the molecular and cellular underpinnings of human brain development are both dynamic and exquisitely regulated spatially and temporally. Not only the overt function of the gene, but the consequences of a given mutation may vary considerably depending on when in development and where in the nervous system it is operating (and being examined). Consequently, while studies of individual genes may identify dysfunction in a given process, cell-type or brain region in a mouse, the question of how one can determine if this relates to human social disability is not self-evident. Similarly, the observation of a consistent but unexplained male predominance in ASD prevalence suggests that sexual dimorphism adds another dimension of complexity. At present, regardless of how extensive the examination of a single rodent model, it is simply not feasible to examine all brain regions, all cell types, and all time points in development in both male and female in an unbiased fashion. Moreover, it is not clear even if this could be accomplished how one would know in isolation which of the many observations to pursue to illuminate causal mechanisms in the human.
Some investigations have relied on model system behaviors that appear to recapitulate features of ASD, i.e. face-validity (resemblance to the human behavioral phenotypes), to try to answer this question. For example, if deleting a bona-fide ASD risk gene in mouse leads to observable molecular, cellular or circuit phenomenon as well as to a social phenotype, this might be offered as evidence that the intermediate phenotype is part of a causal chain related to the human disorder.
However, face validity has serious limitations with regard to the study of psychiatric phenotypes, both empirically and theoretically (Nestler and Hyman, 2010). Indeed the departure of large pharmaceutical companies from investment in psychiatric drug discovery can be traced in part to the challenges of relying on face-valid behavioral models to establish treatment targets showing efficacy in human (Hyman, 2013). Moreover, it’s clear that mutations in well-established nsASD risk genes can lead to a wide range of non-overlapping behavioral manifestations, some of which have no overt social content (Rothwell et al., 2014) --pointing to the absence of a reliable 1:1 relationship from genotype to social phenotype. Consequently, it is problematic to rely on the presence or absence of any specific behavioral manifestation in mouse as definitive evidence of a link (or lack thereof) to core human pathology.
Clearly, face validity may be confounded by differences between humans and specific model organisms. While the use of these models has revolutionized neurodevelopment and biomedical sciences, they are not sufficient to fully reproduce all of the features of human, particularly those affecting the most distinctly human aspects of cognition and behavior. This fact is evidenced by the numerous cases of mouse models of human disease mutations that lack a comparable or even observable brain phenotype (Barak et al., 2011; Liao and Zhang, 2008). These discrepancies can be explained in part by evolutionary distance and, consequently, differences in specific gene sequences and the spatiotemporal regulation of gene expression. Alternatively, species differences in the underlying developmental, molecular, cellular, and physiological context may be also responsible, with contextual differences present even when the normal sequence and expression profile of a particular gene appears to be conserved. Indeed, in the limited data available for direct comparison of genome-wide expression profiles in human and non-human brains, major differences are evident even between human and chimpanzee, our closest living relative (Geschwind and Konopka, 2012; Sousa et al., 2017b).
Another critical limitation of current model organisms in drawing direct conclusions for human brain is the compressed time window of development, for example in the case of rodents. Comparative studies have shown that mammals with large brains require more time to build and mature circuits and thus have far more pronounced regional and, in the case of the cerebral cortex, areal, differences in the timing of major neurodevelopmental processes (Figure 3). In addition, many of the neurodevelopmental processes that are temporally dispersed in humans occur concurrently in rodents (Sousa et al., 2017a). These differences are especially pronounced during the prenatal and early postnatal periods, crucial developmental windows for the formation of neural circuits as well as for the understanding of many neuropsychiatric disorders (Figure 3). Species differences seem to be particularly pronounced during prenatal development, as several studies have indicated that perhaps an order of magnitude more genes exhibit differential spatial or regional expression within the prenatal human neocortex, for example, than in the mouse neocortex at a comparable period of development--comparing data from (Johnson et al., 2009) for human and (Kudo et al., 2007) for mouse. Together, species differences underscore the need for a deeper understanding of molecular and cellular process in the developing human nervous system and how this specially relates to the various model organisms used to examine risk genes. Illuminating both the unique character of the human nervous system as well as developing a far deeper understanding of when and where molecules, cells and circuits overlap across species will be a critical foundational element in the search for pathophysiological mechanisms underlying disorders of behavior, communication and emotion.
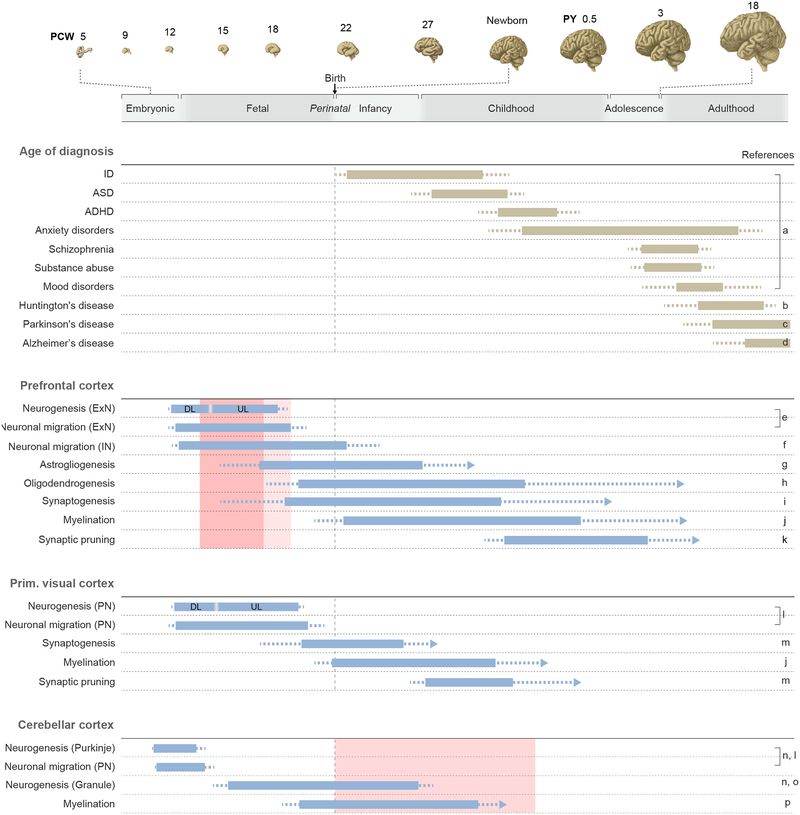
Figure 3. A timeline of major human neurodevelopmental processes and neuropsychiatric disorders.
The illustrations in the top panel demonstrate the dramatic changes in the anatomical features of human brain over the course of prenatal and postnatal development. The gray bar provides a timeline of major phases of human development (Kang et al., 2011), which are plotted on a logarithmic scale. Age is shown in postconceptional weeks (PCW) and postnatal years (PY). The second panel shows the age of onset for selected psychiatric and neurological disorders. The beige bars indicate the age range at which each disorder is most commonly diagnosed, with less frequent ages of diagnosis denoted as dotted lines. Note that the age of diagnosis is highly variable between disorders. The schematic in the third, fourth and fifth panel details the approximate timing and sequence of key cellular processes in the prefrontal cortex, the primary visual cortex and cerebellum, respectively. Bars indicate the peak developmental window in which each feature is acquired. Dotted lines indicate that feature acquisition occurs at these ages, though to a relatively minor degree. Arrows indicate that the feature is present throughout postnatal life. DL, deep layer excitatory projection (pyramidal) neurons; ExN, excitatory projection neurons; Granule, granule cells; IN, Interneurons; PN, Purkinje neurons; and UL, upper layer excitatory projection neurons. Based on figures from Silbereis et al., 2016. Light red squares denote two spatiotemporal windows (i.e., the prefrontal and motor-somatosensory cortex during mainly the midfetal development, and thalamus/cerebellar cortex during infancy and early childhood) that were identified in Willsey et al., 2013 to be significantly enriched for risk among high-confidence ASD genes. The shade of red denotes the significance level of the enrichment after correction for multiple comparisons (darker shade denotes greater enrichment). Relevant references pertaining to information detailed in the panels are provided in the rightmost column: a (Kessler et al., 2007) and (Lee et al., 2014), b (Myers, 2004), c (Pagano et al., 2016), d (van der Flier et al., 2011), e (Clancy et al., 2001) (Bystron et al., 2006), f (Paredes et al., 2016), g (Kang et al., 2011); (deAzevedo et al., 2003);(Choi and Lapham, 1978), h (Yeung et al., 2014); (Kang et al., 2011)), i (Petanjek et al., 2011); (Huttenlocher and Dabholkar, 1997);(Kostovic and Rakic, 1990); (Kwan et al., 2012), j (Yakovlev and Lecours 1967); (Gilles, 1976); (Miller et al., 2012), k (Petanjek et al., 2011); (Huttenlocher and Dabholkar, 1997)), l (Clancy et al., 2001)), m (Huttenlocher and Dabholkar, 1997), n (Sidman and Rakic, 1973), o (Kiessling et al., 2014; Riedel et al., 1989), p (Yakovlev and Lecours 1967); (Gilles, 1976)).
The Evolving Molecular and Cellular Landscape of Developing Human Brain
Advances in high-throughput genomic technologies and analytic approaches have enabled genome-wide analyses of gene expression and other genomic features across a wide range of tissues, cells, conditions and species. One field that has greatly benefited from adopting these technologies, and in turn has provided fresh insights into neurodevelopmental disorders, is research using postmortem human brain tissue. Utilizing oligonucleotide microarrays, RNA-sequencing (RNA-Seq), and other genomic technologies, a number of studies have been conducted using bulk tissue samples to contrast gene expression patterns, epigenetic modifications and regulatory elements in a variety of ways, such as comparing expression and activity between multiple brain regions within a given time period; comparing expression in a single brain region between time periods; and comparing those features between sexes, species or disease states. This work has revealed that, compared to other human organs, the brain has a distinct transcriptomic profile including the second highest number of tissue-enriched genes (after testis) and a greater diversity in expressed transcripts (Uhlen et al., 2016). Comparisons of human and closely-related primate brain transcriptomes have also yielded insights into the uniqueness of the human brain (Sousa et al., 2017b), while comparisons of healthy adult profiles to those of individuals affected with neurodevelopmental and neuropsychiatric disorders have identified key differences which may contribute to disease symptoms or whose mature properties may be altered by the condition (Bryois et al., 2018; Fromer et al., 2016; Hauberg et al., 2018; Psych et al., 2015; Voineagu et al., 2011) – though extending such studies to distinguishing whether such changes reflect cause or effect in the etiology of childhood-onset neuropsychiatric disorders may be problematic.
Aiming to capture spatiotemporal molecular signatures underlying the development of the human nervous system, our and other groups have initially focused on specific prenatal and early postnatal time windows. Extensive spatial coverage of the mid-fetal human brain was first provided by a microarray analysis of thirteen brain regions: the hippocampus, striatum, mediodorsal nucleus of the thalamus, cerebellar cortex, and nine neocortical areas (Johnson et al., 2009). Even greater spatial and temporal resolution of gene expression in the developing human brain, has been generated by subsequent studies (Colantuoni et al., 2011; Kang et al., 2011; Miller et al., 2014), yielding valuable public resources. A common finding of these and other relevant investigations is that human brain transcriptomes differ more prominently across time and space than they do between sexes, ethnicities or individuals, despite the underlying genetic variations between people and populations and epigenetic differences between the sexes.
The spatiotemporal dynamism of gene expression mainly reflects regional differences in cellular composition, biological processes and developmental timing. Given this, it’s not surprising that transcriptomic differences across brain regions and neocortical areas were particularly prominent during prenatal development and included specific transcriptomic signatures associated with prospective prefrontal and perisylvian areas (Kang et al., 2011), thought to be involved in complex cognitive behavior, personality expression, decision making, moderating social behavior, and language comprehension and expression (Gazzaniga and Mangun, 2014). These robust regional transcriptomic differences of the prenatal period are also transient and diminish during late fetal development and infancy (Pletikos et al., 2014), likely reflecting the reorganization of the cellular architecture and changes in the maturational states of cells of the human brain during that time.
Gene co-expression analyses have also revealed that the developing human brain transcriptome can be organized into distinct co-expression networks with often prominent spatial or temporal patterns and enriched for specific biological functions (Johnson et al., 2009; Kang et al., 2011; Miller et al., 2014; Pletikos et al., 2014). Interestingly, genetic variations in some of the most well-connected genes in the networks with dynamic patterns have previously been linked to psychiatric or neurological disorders (http://www.szgene.org; https://gene.sfari.org; http://alsgene.org; http://pdgene.org) suggesting that they may have converging functions in specific brain regions and developmental periods (Gulsuner et al., 2013; Parikshak et al., 2013; Willsey et al., 2013).
Analyses of both male and female tissue samples have revealed that more genes exhibit sex-biased expression during prenatal development than during postnatal life, with the adult brain having the greatest similarity between males and females (Kang et al., 2011). However, although hundreds of genes show sex-biased expression and splicing, included those previously implicated in brain development and function as well as some disease-related genes, no evidence for systematic sex-differential expression of ASD risk genes has been found (Kang et al., 2011; Werling et al., 2016). Consequently, there is at present no simple explanation for the male bias in ASD prevalence based merely on sex-biased gene expression.
Until a few years ago, the majority of functional genomics studies of the human brain examined whole-tissue samples. As a result, observed differences in gene expression or regulation represent some combination of differences in the underlying transcription within cells as well as differences in the cell composition between brain regions. The cellular composition of the human brain and the molecular profiles, anatomic locations, and physiological properties of the cells that make up the human brain change throughout development, in response to neural stimuli, environmental and external stimuli, and cell-intrinsic variation in gene expression and activity.
Historically, challenges in deciphering cellular heterogeneity have hindered our understanding of human brain development and pathology. For example, some common brain disorders, such as Alzheimer, Huntington and Parkinson’s diseases, are associated with dysfunction of particular cell types (Saxena and Caroni, 2011), which may be traditionally defined based on anatomical location, morphology, connectivity, neurotransmitter utilization, electrophysiological properties, or the expression of one to a few marker genes (Fishell and Heintz, 2013; Jones, 1986). Consequently, incomplete knowledge regarding the cell types present in any given region will limit the conclusions that can be drawn from the analysis of that region. Unfortunately, studies conducted at the level of the tissue do not readily offer the resolution necessary for identifying and studying cell types and their corresponding functions.
Recent advances in genomic technologies offer the ability to examine gene expression and other genomic features of individual cells or pooled cell populations at unprecedented resolution and in a largely unbiased fashion. These cell population-specific and single cell approaches have not only characterized rare and not well understood cells types in the developing human, but also complemented the existing tissue level data through global or comprehensive deconvolution of individual cell types and cellular heterogeneity. This approach takes us one step closer to creating a complete molecular inventory of human neural cell types and, consequently, identifying differences in those cell types across time and region. Systems biology approaches and ASD research will likely benefit immeasurably from these single cell-level resources (for example see: (Camp et al., 2015; Darmanis et al., 2015; Fan et al., 2018; Nowakowski et al., 2017; Onorati et al., 2016; Pollen et al., 2015; Pollen et al., 2014; Zhong et al., 2018).
Combining Genomics and Transcriptomics to Identify Convergence in nsASD
While there has been some consternation regarding the sheer number of distinct nsASD risk genes, for example whether “1000 different ASD genes will require 1000 different treatments,” we argue that it may well be this extensive heterogeneity, combined with the deepening view of the molecular and cellular properties of developing human nervous system, that will prove the key to illuminating pathophysiology. The presence of so many large-effect mutations in genes provides the opportunity to interrogate for convergence among disparate risk genes and potentially triangulate pathological mechanisms. Similarly, given that various types of mutations or proteins may be differentially targetable from a therapeutics development standpoint, the heterogeneity of individually important genes may allow multiple diverse paths forward simultaneously.
The search for convergence can take place across multiple levels of analysis. Some of the earliest efforts to investigate overlap of emerging risk genes focused on their overt functions. These have consistently pointed to two categories encompassing the majority of those associated with nsASD risk: genes encoding synaptic proteins and those involved in transcriptional regulation (Figure 2) (De Rubeis et al., 2014; Iossifov et al., 2014). Indeed, some of the earliest discoveries in both syndromic and nsASD were noted at the time to implicate convergence at the synapse (Zoghbi, 2003). And the very first significantly-associated genes identified in exome sequencing prima face suggested important roles for synaptic function and chromatin modification (Iossifov et al., 2014; Iossifov et al., 2012; Neale et al., 2012; O’Roak et al., 2012b; Sanders et al., 2012). Additional findings with regard to functional intersection of nsASD risk genes have highlighted overlap with the earliest discoveries in syndromic ASD. For example, there has been a consistent observation of enrichment of confirmed nsASD risk genes among targets of the Fragile X Mental Retardation Protein (De Rubeis et al., 2014; Iossifov et al., 2014; Iossifov et al., 2012). Of course, these categorizations do not capture all nsASD risk genes, and it may be that outliers, for example SCN2A – one of the most frequently mutated genes in nsASD – will turn out to provide critically important biological and therapeutic insights (Sanders et al., 2018).
Figure 2: High confidence ASD risk genes encode synaptic proteins and chromatin and transcriptional regulators.
Genetic studies have identified a large number of risk genes for ASD, many of which have pleiotropic functional properties. Synapse function, chromatin modification and transcriptional regulation top the list of statistically enriched functional categories. On the left, a simplified schematic of the major cellular components of neural circuits in the cerebral cortex: Pyramid-shaped glutamatergic excitatory projections neurons, GABAergic inhibitory interneurons, and glial cells. On the right, diverse intracellular distribution and pleiotropic roles of ASD risk genes (Sanders et al., 2015). Red circles depict a view of the synapse with its many protein products of ASD risk genes (top) and the ASD proteins in the nucleus (bottom). Proteins in synaptic signaling pathways encompass cell adhesion, scaffolding and signaling molecules. Nuclear protein products of ASD risk genes are mainly associated with chromatin modification and transcriptional control, suggesting that alterations in chromatin structure and gene expression may contribute to ASD.
However, these results reflect something of a watershed for a field that historically has been devoid of experimentally-validated molecular genetic findings. At the same time, the insights provided by grouping genes based on existing ontologies and functional categorizations have some inherent limitations. For example, finding that risk genes contributing to nsASD, a paradigmatic neurodevelopment disorder, parse into functions involving neurons and development may be viewed as something of a glimpse into the obvious. In addition, the data underlying these categorizations is often not based on direct experimentation in central nervous system but rather on pre-existing databases and a variety of assays with variable relevance for human brain (Willsey et al., 2018). Finally, simply because a protein has been assigned a function based on prior hypothesis driven studies does not confirm that this is its only role or one that is necessarily relevant to the emergence of nsASD.
We have proposed that an interrogation of spatiotemporal convergence of nsASD genes may be as important as assessing overlap in their overt functions. We hypothesize that at some point in the trajectory from the molecular scale to complex behavior (Figure 1) subsets of multiple, functionally diverse molecular effectors encoded by disease-risk genes are bound to converge at the cellular or circuit level (or both) (Figure 1). Identifying the points of convergence of risk genes in typically developing human brain may therefore begin to offer a means to constrain both in vitro and in vivo experiments aimed at understanding disease mechanisms. That is, data-driven hypotheses can be developed regarding which specific sets of mutations might be studied in specific anatomical and developmental contexts to look for overlapping properties/phenotypes that reflect shared nsASD pathology.
Early efforts to pursue this strategy, by our group and others, were constrained by multiple difficulties including tissue acquisition, limited pre-existing reference datasets, technical limitations, and the general infancy of bioinformatics approaches. In retrospect, given the tremendous cellular diversity, developmental dynamism, and functional complexity of the human central nervous system, the questions of time and space we sought to investigate might easily have been fatally confounded. Similarly, other limitations, including the reliance exclusively on bulk tissue gene expression; the absence of cell-type specific data across development; and the paucity of information on male versus female brain development could well have thwarted our and other contemporaneous analyses facing similar challenges.
Nevertheless, efforts by multiple laboratories to capture anatomical and developmental variables in systems analyses, have yielded strong evidence for convergence among nsASD risk genes in glutamatergic excitatory projection (pyramidal) neurons during mid fetal development (Ben-David and Shifman, 2013; Chang et al., 2015; Lin et al., 2015; Parikshak et al., 2013; Pinto et al., 2014; Uddin et al., 2014; Willsey et al., 2013; Xu et al., 2014). Our initial efforts in this regard (Willsey et al., 2013) demonstrated convergence of the highest confidence nsASD risk genes in frontal lobe (i.e., prefrontal and primary motor cortex) in deep layer projection neurons from 10–24 postconceptional weeks (PCW) or approximately spanning the end of the early fetal period and the entire mid-fetal period. Among the 16 regions of the developing human brain we initially analyzed, we also observed a weaker convergence signal in early post-natal cerebellum and the medial dorsal nucleus of the thalamus. Similar systems biological analysis that included transcriptomic data from mice and humans has found evidence for enrichment in the neocortex and striatum, as well as specific cell types within those two regions (i.e., neocortical deep layer projection neurons and striatal medium spiny neurons) (Xu et al., 2014).
Even though limited by the breadth and depth of available datasets, these findings associating the mid-fetal timeframe with specific ASD genes and LGD mutations offer concrete avenues for further studies aimed at identifying pathophysiological mechanisms. They suggest that, for example, characterizing specific heterozygous LGD mutations in projection neurons in the mid-fetal prefrontal and motor cortex promises may offer insights into the neurodevelopmental underpinnings of ASD and that future mechanistic studies should involve these cell types and developmentally relevant time points. Importantly, the foregoing points to a general experimental strategy, not a unifying theory of nsASD causality. For instance, as expected, there is already evidence for convergence at other time points and brain regions (Parikshak et al., 2013; Willsey et al., 2013; Willsey and State, 2015; Xu et al., 2014) suggesting that some genes and mutations should be interrogated in specific contexts other than cortical projection neurons. Crucially, these studies also suggest that the neurodevelopmental processes leading to ASD start in utero, well before ASD is diagnosed (Figure 3).
These initial insights into the spatiotemporal dimensions of genetic risk may also turn out to contribute significantly to an understanding of when and under what circumstances other factors, including gene-environment interactions and the impacts of maternal and placental health (Bauman et al., 2013; Cattane et al., 2018; Meltzer and Van de Water, 2017; Thompson and Levitt, 2010), may be operative. However, importantly, this timeframe does not necessarily identify a unique window of vulnerability that opens and closes in mid-fetal development. For example, other regions, such as thalamus and cerebellum, exhibit a later convergence with the same gene set. Moreover, it is not clear from the available evidence whether the convergence identified relates to what would classically be considered developmental abnormalities or some combination of fixed and plastic functional deficits. A wide range of investigations of monogenic neurodevelopmental disorders that show phenotypic overlap with ASD have successfully rescued the sequelae of human causative mutation(s) even into adulthood (Sztainberg and Zoghbi, 2016). These surprisingly consistent results across disorders are one of several lines of evidence suggesting that some optimism is warranted with regard to post-natal intervention.
Examining spatiotemporal convergence may also help clarify some of the more confounding aspects of nsASD, including how seemingly common and essential molecular and cellular processes can be implicated so specifically in a well-recognized and clinically defined disorder. NsASD risk genes generally fall into two large categories (Figure 2). One involves synaptic proteins and includes cell adhesion, scaffolding and signaling molecules that can affect synapses at various times in synapse formation and elimination, synaptic transmission and plasticity, as well as proteins mediating the levels of synapse-related proteins. The expression of these synapse-related genes increases during late mid-fetal and late fetal development and peaks just after birth in the cerebral cortex (State and Sestan, 2012), as would be predicted given their function in synapse development (Huttenlocher and Dabholkar, 1997). The second group of high-confidence ASD risk gene products include various chromatin remodeling proteins and transcriptional regulators with a broad expression pattern in the developing human nervous system. Of course, both synaptic function and chromatin/transcriptional regulation are necessary for all brain functions, raising the critical question of how mutations in these genes might lead in some individuals to a specific disorder such as ASD, to a specific spatiotemporal location within the brain or even to a specific species such as ours? Why would neural circuits and networks involved in social and linguistic functions, sub-served largely by prefrontal and temporal association areas and interconnected subcortical regions, be more selectively affected than those involved in, for example, visual processing?
The integrated analysis of molecular and cellular atlases/maps suggest an answer: that the spatiotemporal specificity we observe in ASD is a consequence of the spatiotemporal specificity of the gene regulatory networks and neurobiological processes in which those genes are involved. Many of the synapse and chromatin/transcription-related genes are present in co-expression networks converging in mid-fetal human frontal cortex and deep layer projection neurons (Willsey et al., 2013).
Foremost, the human nervous system develops heterochronologically and hierarchically (reviewed in (Silbereis et al., 2016). The fact that distinct areas mature at different rates (heterochrony) helps explain many of the intellectual and emotional changes seen in children, teens and young adults. Furthermore, the development of certain structures of the human nervous system unfolds over a long time, with some processes such as synaptic pruning and myelination lasting to the middle of the third postnatal decade in the prefrontal cortex and higher-order cortical regions (Figure 3) (Petanjek et al., 2011; Rapoport and Gogtay, 2008). This extended period of synaptic pruning and axon myelination allows these regions to retain juvenile characteristics (i.e., plasticity) well into postnatal life, while also rendering these regions potentially vulnerable to a broad range of developmental disorders.
Furthermore, this prolonged development amplifies regional differences in maturation, providing another level of potential specificity as to why certain neural circuits are affected in nsASD. In general, the assembly of cortical circuits progresses in an orderly spatiotemporal pattern such that the neurons in the anterior regions (e.g., the prefrontal cortex) are chronologically older, and start to form synapses earlier, than the neurons of equivalent subtypes in the posterior regions (e.g., the primary visual cortex) (Kostovic and Rakic, 1990; Rakic, 2002). Indeed, some of the earliest signs of neuronal differentiation and synaptic circuits are present in the regions of the early and mid-fetal prefrontal and limbic cortices (Kostovic and Rakic, 1990; Kwan et al., 2012) that ultimately give rise to neural circuits underlying executive control, social affective processing, and language - all functions affected in ASD. Interestingly, the long-range axonal projections to and from the primary motor and sensory cortices are the first to myelinate, followed by adjacent cortical areas, and with delayed axonal myelination in higher-order cortical regions (Yakovlev and Lecours 1967), indicating that the period of circuit development and maturation starts early and ends late in prefrontal and limbic areas of the cortex, making them vulnerable to developmental disorders where intrinsic genetic or extrinsic environmental stimuli provide insults altering the native context.
Regional and laminar differences in the timing of processes such as neurogenesis, synaptogenesis and myelination may also help explain why neural circuits that begin to develop earlier and end later, and the genes that are involved in these processes, are particularly vulnerable in nsASD. For example, the subplate and prospective deep layers (layers 5 and 6) of the cortical plate contain neurons born early in development, during the embryonic, early fetal, and mid-fetal period, and that belong to diverse projectional and physiological phenotypes. For example, many of these neurons send long-range axonal projections that target sub-cerebral regions such as thalamus, brainstem and spinal cord (Han and Sestan, 2013; Lodato and Arlotta, 2015) and another subset of deep layer neurons project exclusively to other cortical areas within the same hemisphere (i.e., associational axons) or opposite hemisphere (i.e., commissural axons). Conversely, the later born projection neurons of the upper layers (L2–4) that project exclusively to form synaptic connections within the cerebrum (e.g., cerebral cortex and basal ganglia) are still being generated or migrating toward the outer parts of the cortical plate (the future layers 2 to 6) during mid-fetal development. As such, their projections are much less developed compared to those in the deep layers and largely lack synaptic connections at this developmental period (reviewed in (Kostovic and Judas, 2010; Silbereis et al., 2016)
The rarity and functional immaturity of nascent synapses, such as those in the mid-fetal frontal cortex, may make them especially susceptible to perturbed function of nsASD-related genes, which appear to be dynamically regulated during the same developmental window. Crucially, deep layer projection neurons, especially those in prospective layer 5, form synapses, differentiate (Kostovic and Judas, 2010; Kwan et al., 2012), and establish the first descending projections from the fetal neocortex (Clasca et al., 1995) during this time, perhaps making them susceptible in ways not possible for upper layer neurons that are still being born or migrating. The precise spatiotemporal timing of convergence may also explain why some areas of the neocortex are particularly affected in ASD while other areas, such as those involved in visual perception, are less affected.
Finally, building a neural circuit requires coordinated gene expression and electrochemical activity among developing cells. Indeed, studies in model organisms have shown that early in development, neurons form spontaneously electrochemically-active networks whose activity patterns are important for properly wiring-up different synaptic circuits (Katz and Shatz, 1996; Sur and Rubenstein, 2005). Perturbing the precise spatiotemporal sequence of gene expression in cells within these circuits as well as various non-genetic factors can thus lead to disruptions in synaptic function and connectivity. We conjecture that due to the high level of interconnectedness between gene regulatory networks and between molecular pathways active within a developing neuron, alterations in many of the genes expressed in a given cell type during the critical period in circuit formation (i.e., developmental vulnerability), may affect the neuron’s connectivity and function. Genes highly specified for neuronal functions such as synaptic transmission or those having a broad effect on gene expression (e.g., chromatin remodelers and transcription factors), may be more likely to alter the development and consequently the function of those circuits.
The Path Ahead to Therapeutics Development
The foregoing has highlighted some of the recent progress in the genomics of nsASD and the neuroscience of human brain development as well as the challenges in distinguishing between the biology of risk genes and relevant pathophysiological mechanisms underlying the syndrome. And clearly, despite the advances noted, the most pressing and still-elusive goal for the next epoch of ASD research is the development of novel, more effective therapies. Current pharmaco-therapeutics do not target the defining social deficits of ASD, structured educational and behavioral interventions provide important but generally limited relief of core symptoms, and the prospect for effective prevention or cure remains a distant aspiration. However, as the genetics of nsASD begins to come into focus, avenues are emerging to conceptualize new approaches to somatic treatment development that, until very recently, would likely have been considered out of reach.
The notion that there might be a straight-line path between genes on the one hand and a complex behavioral disorder on the other (Figure 1A) is clearly implausible (Geschwind and Levitt, 2007; State and Levitt, 2011). Instead, the search space for therapeutic targets is dictated by a host of complicating factors, including isoform variation and biological pleiotropy (Figure 1B), sexual dimorphisms and the spatial and temporal dynamics of human brain development and ASD risk (Figure 1C). However, despite the expansive territory existing between gene and behavior, it is becoming increasingly possible to conceptualize strategies to attack points along this path. Three examples are introduced below: one leveraging functional genomics and proteomics to target “close to the gene”; a second using systems biological/convergence approaches to identify intermediate phenotypes, and, third, electrocorticography (ECoG) and deep brain stimulation to move “distally,” directly targeting circuit mechanisms.
Haploinsufficiency and Protein-Focused Therapeutics
The foregoing has highlighted that a significant percentage of the identified nsASD risk genes share a common genomic mechanism, namely haploinsufficiency. Given the evidence in nsASD that a single LGD mutation in the individual is conferring the lion’s share of risk and in cis, a broad therapeutic strategy could focus directly on enhancing expression from the remaining normal allele(s). With the development of advanced nucleic acid editing, antisense oligonucleotide (ASO) technologies, viral targeting, traditional small molecule screening and repurposing of existing pharmacotherapeutics, along with a widening variety of higher throughput model systems, including iPSCs, multiple approaches to address this goal are now increasingly plausible (Dugger et al., 2018).
The identification of interventions that return the level of the nsASD- related protein in haploinsufficient cells to wild-type levels offers a tractable screening strategy. And, to date, the absence of clarity on what a reliable high-throughput screening phenotype would consist of has been a key rate limiting step in nsASD therapeutics development. An additional advantage of such an approach is that strategies might be pursued that restore expression to wild-type in the mutant while leaving intact many native regulatory mechanisms, preserving developmental expression patterns and isoform diversity.
Some of the most exciting work in this area at present involves efforts either to reactivate silent alleles or to repress over-expression contributing to syndromic ASD (Sztainberg and Zoghbi, 2016) for example in Angelman and MECP2 duplication syndrome (Sztainberg et al., 2015). With regard to the former, loss of function mutations in UBE3A have been found to be causal for Angelman syndrome due in part to silencing of the remaining normal allele via imprinting. Several studies have shown that topoisomerase inhibitors (Huang et al., 2011) and anti-sense oligonucleotides (ASOs) (Meng et al., 2015) can de-repress the silent allele. Recent studies have moved to the screening of compounds with sustained penetration into the CNS (Huang et al., 2011; Lee et al., 2018). While in this case, the pathology is related to the functional equivalent of homozygous loss of the UBE3A gene, the concept of leveraging the biology of the normal intact allele is highly analogous to the approach we are proposing in nsASD.
Similarly, dramatic progress in the treatment of spinal muscular atrophy (SMA) (Finkel et al., 2016; Mendell et al., 2017; Mercuri et al., 2018) mirrors this approach and has been transformative for severely ill children and their families. Recently both ASO and viral vector approaches have shown substantial evidence for efficacy. The ASO strategy is particularly interesting vis-a-vis the notion of leveraging the remaining normal allele in nsASD. ASO approaches in SMA rely on existing genetic substrate, in this case the gene Survival Motor Neuron 2 (SMN2) which differs at only a single base pair from SMN1, homozygous loss of which results in the disease phenotype. The single base change differentiating SMN1 from SMN2 results in the exclusion of a critical functional exon. ASOs targeting the splice site variation retain the key exon, allowing SMN2 to substitute functionally for the loss of SMN1. This strategy has leveraged intrathecal injection and targeted anterior horn cells. Clearly this initial success is far from a guarantee that, for instance, deep layer cortical neurons could be effectively targeted in nsASD. However, progress in this devastating early onset pediatric neurological disorder is likely to pave the way forward for experimentation in a range of other conditions, including neurodegeneration and epilepsy, that may offer additional insights into the challenges and opportunities for broader use in nsASD.
Of course, there are significant challenges that would confront any effort to deploy a similar approach in nsASD even if the appropriate cell type(s) could be identified and targeted: First, the ability to predict the specific phenotypic consequences of highly-penetrant nsASD mutations is still limited. Second, the nature of the behavioral or cognitive pathology, as well as the severity, can vary widely. For this reason, early studies would likely have to focus on a very small number of risk genes with the greatest penetrance for serious neurodevelopmental outcomes, for example CHD8, SCN2A or ARID1B have all been shown to confer very high degrees of risk due to putative loss-of-function de novo heterozygous mutations.
An additional complexity arises due to the extreme rarity of de novo LGD mutations in any given gene even in the affected population, raising questions regarding how potential research subjects might be identified. The increasing frequency of pre-natal or early post-natal exome or genome sequencing in large health systems appear poised to help address such challenges. The extreme rarity of mutations in any given gene could also conceivably be a practical/financial obstacle to therapeutics development. However, the recent examples of SMA (Finkel et al., 2017) and of Chimeric Antigen Receptor (CAR) T-Cell therapy (June et al., 2018) suggests this might not be insurmountable – though the ultimate costs of treatment and related ethical concerns would not be trivial.
The timing of intervention would also pose difficult questions. While there is now good evidence for convergent risk for a subset of high-effect LGD mutations in mid-fetal neocortical development, one would imagine that for a variety of reasons -- including the variable outcomes from identical risk mutations -- that participation in a trial would most likely be considered only postnatally and after symptoms emerged. With continually advancing early detection approaches, this would still conceivably be within the first several postnatal months, well before clinical diagnosis is typically made but substantially after the mid-fetal period (Hazlett et al., 2017; Jones and Klin, 2013; June et al., 2018; Shultz et al., 2018; Zwaigenbaum and Penner, 2018).
Of course, waiting on intervention would risk increasing the liability for type II error. It could well be that early post-natal intervention would still be too late to be effective. Conversely, it’s important to recall that it’s far from certain how the emergence of vulnerability in mid-fetal brain development might relate to a therapeutic window. Several lines of evidence suggest that such a window might be open for longer than expected: as noted, studies in a variety of models of Mendelian neurodevelopmental syndromes indicate that ongoing functional abnormalities are rescuable into adulthood (Sztainberg and Zoghbi, 2016). In addition, despite the current limited armamentarium of therapies in humans, and even more limited understanding of the specific circuits that are being targeted, affected children can improve with structured educational and behavioral intervention (Volkmar et al., 2014), suggesting that ASD pathology is not set in stone in utero.
Finally, even if one could identify strategies to increase expression of the normal allele and address all the challenges noted above, measuring the trajectory of social functioning in the developing child would remain an obstacle. Similarly, developing a large enough case and control group to confidently assess outcomes in the face of interindividual variability, and identifying what time period would be required to assess change, would all constitute substantial logistical challenges.
Nonetheless it is worth reflecting on the foregoing discussion: the breakthroughs in reliably identifying large- effect heterozygous LGD mutations in nsASD, the use of systems approaches to begin to identify spatiotemporal vulnerability, and advances in functional genomics and gene therapies now allow for a detailed consideration of a plausible, if challenging, path forward toward rational therapies.
Systems Biology and Convergence:
As discussed above, systems biological approaches focusing on the intersection of nsASD risk genes, with regard to function and interaction, can refine hypotheses regarding pathophysiological mechanisms. In this regard, higher throughput model systems promise to be quite useful in interrogating many established nsASD genes and mutations, in parallel, in search of experimentally-validated, overlapping pathways (Willsey et al., 2018).
Moreover, we argue that there is likely to be particular traction in investigating spatiotemporal convergence. In this regard, the data from studies of human transcriptome have already begun to provide initial clues with regard to when and where nsASD risk genes may be having an effect. Genes identified based on de novo LGD mutations have been combined with transcriptome data from overtly normal developing human brain leading to the identification of glutamatergic projections neurons during mid-fetal cortical development as one key point of intersection for a subset of high-confidence nsASD risk genes. Moreover, while there has been some variability in the precise localization of this signal, deeper layers (V and VI) have been a common denominator (Parikshak et al., 2013; Willsey et al., 2013). It is not difficult to imagine that with the rapidly advancing characterization of the molecular landscape of human brain, including at single-cell resolution, studies such as these may well be able to refine further and to resolve the particular cell-types and/or circuits implicated by distinct sets of functionally-divergent nsASD risk genes.
This initial step in identifying spatiotemporal convergence can help constrain subsequent experiments in a variety of model systems by restricting analyses to specific genes carrying specific mutations and having a putative impact in a specific anatomical region, cell-type or circuit. Of course, while is seems reasonable to suppose that having a data-driven hypothesis about when and where to model a set of mutations is better than the alternative, this strategy is clearly not devoid of serious challenges: For example, important questions have yet to be answered with regard to how closely one would need to match the human context in a given model system in order to be able to generate useful data from these types of experiments. This issue of recapitulating relevant contextual properties across model systems speaks to the critical importance of foundational data regarding the molecular, cellular and circuit-level developmental properties of a wide range of organisms. Similarly, it points to the importance of a broad experimental armamentarium -- as some questions may be tractable in for example, frog or fly while others might require iPSCs, marmoset or other non-human primates.
Moreover, if a cell type and developmental stage could be adequately – if not precisely – recapitulated in a particular model, one would still need to establish a robust statistical framework to evaluate whether any observed overlap in phenotype from divergent risk genes was shared more often than would be expected by chance. In addition, even if one observed a convergent cellular or circuit level phenotype, ultimate confirmation of the relationship of this to nsASD pathology in human would still potentially remain a question. One could imagine gaining some confidence regarding the import of a statistically significant overlapping phenotype based on the observation across either multiple sets of nsASD risk genes and/or across multiple models. In this regard both the expansion of the current armamentarium to include iPSCs, neural organoids and non-human primates, in addition to improved methods for the parallel investigation of multiple risk genes are welcome developments. While it seems likely that no single model, apart from living human brain, can provide a complete answer, leveraging a wide variety of systems, while recognizing the strengths and limitations of each, may be the most productive approach to identifying convergent pathology among nsASD risk genes.
Electrocorticography (ECoG) and Closed Loop Stimulation
The examples above target different points of entry in the complex path from genes to behaviors. A third approach could conceivably leverage ECoG arrays in human patients and deep brain stimulation to directly modify brain activity in order to target human behavior and emotion.
Over the last several years the development of thin film electrode arrays and sophisticated analytic approaches have demonstrated the tremendous potential to dissect complex neural processes. The spatial and temporal resolution of such approaches compared to non-invasive imaging and EEG is remarkable – leading to transformative findings. For example, recent work has dissected the microanatomy of language perception and the regulation of prosody (the musical quality of vocal pitch)– abnormalities of which are a central feature of higher-functioning ASD (Chang et al., 2011; Chartier et al., 2018; Dichter et al., 2018; Mesgarani and Chang, 2012).
Given the high rate of seizures in individuals with ASD it is certainly plausible that subjects could be ascertained for studies, in cases where telemetry to localize epileptic foci and/or surgical resection is clinically indicated. Very recently, emerging data regarding speech and language as well as mood regulation (Kirkby et al., 2018) suggests that quite complex higher-order phenomena can be effectively studied and manipulated in the human. Still the additional question of whether persistent deficits in social communication could be either detected, targeted or mediated remains to be seen. And, of course, all of the aforementioned challenges regarding phenotyping, assessing change and deriving sufficient statistical power would remain. However, the notion that there may already be emerging machine brain interface technology that could allow efforts to treat ASD to circumvent the complexities of elaborating molecular and cellular mechanisms in favor of manipulating circuits directly is quite intriguing.
Conclusion:
Recent progress in the genomics of complex human disorders and in the characterization of the molecular, cellular and circuit landscapes of developing human brain have opened new avenues to an actionable understanding of nsASD. Recent genomic findings have been remarkable for the identification of rare, de novo coding heterozygous mutations carrying large effects. This contrasts with the state-of-the-science with regard to typical later-onset psychiatric disorders where GWAS has been tremendously successful. In nsASD, discovery cohorts, reaching more than 7000 cases, still have not had adequate power to identify individual common alleles, given their extremely small effects. Consequently, these studies provide no immediate avenue to illuminate molecular mechanisms.
In contrast, the earliest studies of syndromic ASD and, more recently, the systematic identification of de novo LGD mutations in nsASD has revealed an allelic architecture that is particularly tractable for model systems approaches. Traditional strategies to pursue these large-effect, heterozygote LGD variants have provided critical insights into the underlying biology of risk genes. However, differences between the developing human brain and other more evolutionarily distant model systems have led to a still incomplete picture, particularly with regard to the distinction between nsASD gene related biology and pathophysiological processes in the human. Likewise, assessing gene expression or epigenetic regulation in adult postmortem ASD tissue, relative to neurotypical controls, has identified intriguing differences, but whether these are primary drivers of ASD, moderators of clinical course, or reflections of compensatory mechanisms is difficult to determine.
Though not without challenges, integrating data from genetics studies with that from developmentally timed and spatially (i.e., brain regions and cell types) distinct postmortem human tissue provides an avenue to begin to constrain experiments aimed at elaborating causal mechanisms. Already, these approaches have yielded something of a consensus about the importance of mid-fetal cortical glutamatergic projection neurons for a subset of the highest confidence nsASD risk genes. This example suggests that with the rapid elaboration of the developing human brain at the single cell and circuit level it is conceivable that specific mutations may, in the near future, be assignable to increasingly more precise biological contexts relevant to disease pathology – and in turn set the stage for a more informed assessment of convergence among functionally distinct nsASD risk genes across a range of model systems.
Importantly, these recent advances are beginning to reveal novel avenues to advance treatment. Whether these strategies leverage the common observation of haploinsufficiency as a genomic mechanism in nsASD, the identification of therapeutic targets based on a deeper understanding of cellular and circuit-level contexts and mechanisms, or the development of advanced machine brain interfaces, the field is poised for a profound transformation.
Acknowledgments
The authors wish to thank Forrest Gulden and the anonymous reviewers for their extremely helpful comments. We also wish to thank Sarah Pyle for her illustrations. Dr. State wishes to thank A. Jeremy Willsey and Stephan J. Sanders for their many contributions to developing the conceptual framework, data and analyses supporting this perspective as well as their invaluable ongoing collaborations on related projects. This work was support in part by NIH grants MH106934 (to NS and MWS), MH106874 (to NS), MH109904 (to NS), MH109901 (to MWS), MH110926 (to NS), MH110928 (to MWS), MH116487 (to MWS), MH116488 (to NS), MH102342, MH111662, MH105575, MH115747 (to MWS), the Overlook International Foundation (to MWS) and the Simons Foundation (to NS and MWS). Finally, we wish apologize to our many colleagues whose important works we could not cite due to space limitations. Matthew W. State serves on the scientific advisory boards and has stock or stock options for BlackThorn therapeutics and ARett pharmaceuticals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
Matthew W. State serves on the scientific advisory boards and has stock or stock options for BlackThorn therapeutics and ARett pharmaceuticals.
References
- Autism Genome Project C, Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, et al. (2007). Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 39, 319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autism Spectrum Disorders Working Group of The Psychiatric Genomics, C. (2017). Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol Autism 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae BI, Jayaraman D, and Walsh CA (2015). Genetic changes shaping the human brain. Dev Cell 32, 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak T, Kwan KY, Louvi A, Demirbilek V, Saygi S, Tuysuz B, Choi M, Boyaci H, Doerschner K, Zhu Y, et al. (2011). Recessive LAMC3 mutations cause malformations of occipital cortical development. Nat Genet 43, 590–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman MD, Iosif AM, Ashwood P, Braunschweig D, Lee A, Schumann CM, Van de Water J, and Amaral DG (2013). Maternal antibodies from mothers of children with autism alter brain growth and social behavior development in the rhesus monkey. Transl Psychiatry 3, e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David E, and Shifman S (2013). Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism. Mol Psychiatry 18, 1054–1056. [DOI] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout AT, et al. (2014). Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bipolar, D., Schizophrenia Working Group of the Psychiatric Genomics Consortium. Electronic address, d.r.v.e., Bipolar, D., and Schizophrenia Working Group of the Psychiatric Genomics, C. (2018). Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell 173, 1705–1715 e1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron T (2015). From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci 16, 551–563. [DOI] [PubMed] [Google Scholar]
- Bourgeron T (2016). Current knowledge on the genetics of autism and propositions for future research. C R Biol 339, 300–307. [DOI] [PubMed] [Google Scholar]
- Brainstorm C, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L, Escott-Price V, Falcone GJ, Gormley P, et al. (2018). Analysis of shared heritability in common disorders of the brain. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandler WM, Antaki D, Gujral M, Kleiber ML, Whitney J, Maile MS, Hong O, Chapman TR, Tan S, Tandon P, et al. (2018). Paternally inherited cis-regulatory structural variants are associated with autism. Science 360, 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryois J, Garrett ME, Song L, Safi A, Giusti-Rodriguez P, Johnson GD, Shieh AW, Buil A, Fullard JF, Roussos P, et al. (2018). Evaluation of chromatin accessibility in prefrontal cortex of individuals with schizophrenia. Nat Commun 9, 3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I, Rakic P, Molnar Z, and Blakemore C (2006). The first neurons of the human cerebral cortex. Nat Neurosci 9, 880–886. [DOI] [PubMed] [Google Scholar]
- Cameron JL, Eagleson KL, Fox NA, Hensch TK, and Levitt P (2017). Social Origins of Developmental Risk for Mental and Physical Illness. J Neurosci 37, 10783–10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, et al. (2015). Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A 112, 15672–15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattane N, Richetto J, and Cattaneo A (2018). Prenatal exposure to environmental insults and enhanced risk of developing Schizophrenia and Autism Spectrum Disorder: focus on biological pathways and epigenetic mechanisms. Neurosci Biobehav Rev. [DOI] [PubMed] [Google Scholar]
- Chang EF, Edwards E, Nagarajan SS, Fogelson N, Dalal SS, Canolty RT, Kirsch HE, Barbaro NM, and Knight RT (2011). Cortical spatio-temporal dynamics underlying phonological target detection in humans. J Cogn Neurosci 23, 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Gilman SR, Chiang AH, Sanders SJ, and Vitkup D (2015). Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci 18, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier J, Anumanchipalli GK, Johnson K, and Chang EF (2018). Encoding of Articulatory Kinematic Trajectories in Human Speech Sensorimotor Cortex. Neuron 98, 1042–1054 e1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BH, and Lapham LW (1978). Radial glia in the human fetal cerebrum: a combined Golgi, immunofluorescent and electron microscopic study. Brain Res 148, 295–311. [DOI] [PubMed] [Google Scholar]
- Clancy B, Darlington RB, and Finlay BL (2001). Translating developmental time across mammalian species. Neuroscience 105, 7–17. [DOI] [PubMed] [Google Scholar]
- Clasca F, Angelucci A, and Sur M (1995). Layer-specific programs of development in neocortical projection neurons. Proc Natl Acad Sci U S A 92, 11145–11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, et al. (2011). Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics, C., Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, Mowry BJ, Thapar A, Goddard ME, et al. (2013). Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 45, 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, and Quake SR (2015). A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A 112, 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Won H, Stein JL, and Geschwind DH (2016). Advancing the understanding of autism disease mechanisms through genetics. Nat Med 22, 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, and Buxbaum JD (2015). Recent advances in the genetics of autism spectrum disorder. Curr Neurol Neurosci Rep 15, 36. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deAzevedo LC, Fallet C, Moura-Neto V, Daumas-Duport C, Hedin-Pereira C, and Lent R (2003). Cortical radial glial cells in human fetuses: depth-correlated transformation into astrocytes. J Neurobiol 55, 288–298. [DOI] [PubMed] [Google Scholar]
- Di Lullo E, and Kriegstein AR (2017). The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci 18, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichter BK, Breshears JD, Leonard MK, and Chang EF (2018). The Control of Vocal Pitch in Human Laryngeal Motor Cortex. Cell 174, 21–31 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Walker MF, Carriero NJ, DiCola M, Willsey AJ, Ye AY, Waqar Z, Gonzalez LE, Overton JD, Frahm S, et al. (2014). De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep 9, 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugger SA, Platt A, and Goldstein DB (2018). Drug development in the era of precision medicine. Nat Rev Drug Discov 17, 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, et al. (2007). Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet 39, 25–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl RK, Turner TN, Mefford HC, Hudac CM, Gerdts J, Eichler EE, and Bernier RA (2017). Clinical phenotype of ASD-associated DYRK1A haploinsufficiency. Mol Autism 8, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Chromosome16 Tuberous Sclerosis, C. (1993). Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell 75, 1305–1315. [DOI] [PubMed] [Google Scholar]
- Fan X, Dong J, Zhong S, Wei Y, Wu Q, Yan L, Yong J, Sun L, Wang X, Zhao Y, et al. (2018). Spatial transcriptomic survey of human embryonic cerebral cortex by single-cell RNA-seq analysis. Cell Res 28, 730–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, Yamashita M, Rigo F, Hung G, Schneider E, et al. (2016). Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet 388, 3017–3026. [DOI] [PubMed] [Google Scholar]
- Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, Chiriboga CA, Saito K, Servais L, Tizzano E, et al. (2017). Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N Engl J Med 377, 1723–1732. [DOI] [PubMed] [Google Scholar]
- Fishell G, and Heintz N (2013). The neuron identity problem: form meets function. Neuron 80, 602–612. [DOI] [PubMed] [Google Scholar]
- Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR, et al. (2016). Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 19, 1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Mahajan M, Manaa D, Pawitan Y, Reichert J, et al. (2014). Most genetic risk for autism resides with common variation. Nat Genet 46, 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzaniga MS, and Mangun GR (2014). The cognitive neurosciences (5th ed.) (Cambridge, MA, US: MIT Press.). [Google Scholar]
- Geschwind DH, and Konopka G (2012). Neuroscience: Genes and human brain evolution. Nature 486, 481–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, and Levitt P (2007). Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol 17, 103–111. [DOI] [PubMed] [Google Scholar]
- Gilles FH (1976). Myelination in the neonatal brain. Hum Pathol 7, 244–248. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, et al. (2010). A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet 42, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H, Consortium on the Genetics of, S., Group, P.S., et al. (2013). Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halladay AK, Bishop S, Constantino JN, Daniels AM, Koenig K, Palmer K, Messinger D, Pelphrey K, Sanders SJ, Singer AT, et al. (2015). Sex and gender differences in autism spectrum disorder: summarizing evidence gaps and identifying emerging areas of priority. Mol Autism 6, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, and Sestan N (2013). Cortical projection neurons: sprung from the same root. Neuron 80, 1103–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauberg ME, Fullard JF, Zhu L, Cohain AT, Giambartolomei C, Misir R, Reach S, Johnson JS, Wang M, Mattheisen M, et al. (2018). Differential activity of transcribed enhancers in the prefrontal cortex of 537 cases with schizophrenia and controls. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett HC, Gu H, Munsell BC, Kim SH, Styner M, Wolff JJ, Elison JT, Swanson MR, Zhu H, Botteron KN, et al. (2017). Early brain development in infants at high risk for autism spectrum disorder. Nature 542, 348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Sanders SJ, Liu L, De Rubeis S, Lim ET, Sutcliffe JS, Schellenberg GD, Gibbs RA, Daly MJ, Buxbaum JD, et al. (2013). Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet 9, e1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, Sciaky N, Dutton JW Jr., Lee HM, Chen X, et al. (2011). Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature 481, 185–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher PR, and Dabholkar AS (1997). Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol 387, 167–178. [DOI] [PubMed] [Google Scholar]
- Hyman SE (2013). Psychiatric drug development: diagnosing a crisis. Cerebrum 2013, 5. [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. (2014). The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. (2012). De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, et al. (2003). Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet 34, 27–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, Geschwind DH, Mane SM, State MW, and Sestan N (2009). Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62, 494–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EG (1986). Neurotransmitters in the cerebral cortex. J Neurosurg 65, 135–153. [DOI] [PubMed] [Google Scholar]
- Jones W, and Klin A (2013). Attention to eyes is present but in decline in 2–6-month-old infants later diagnosed with autism. Nature 504, 427–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June CH, O’Connor RS, Kawalekar OU, Ghassemi S, and Milone MC (2018). CAR T cell immunotherapy for human cancer. Science 359, 1361–1365. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. (2011). Spatio-temporal transcriptome of the human brain. Nature 478, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LC, and Shatz CJ (1996). Synaptic activity and the construction of cortical circuits. Science 274, 1133–1138. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Amminger GP, Aguilar-Gaxiola S, Alonso J, Lee S, and Ustun TB (2007). Age of onset of mental disorders: a review of recent literature. Curr Opin Psychiatry 20, 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling MC, Buttner A, Butti C, Muller-Starck J, Milz S, Hof PR, Frank HG, and Schmitz C (2014). Cerebellar granule cells are generated postnatally in humans. Brain Struct Funct 219, 1271–1286. [DOI] [PubMed] [Google Scholar]
- Kirkby LA, Luongo FJ, Lee MB, Nahum M, Van Vleet TM, Rao VR, Dawes H, Chang E, and Sohal VS (2018). An amygdala-1 hippocampus subnetwork that encodes variation in human mood. Cell In Press. [DOI] [PubMed] [Google Scholar]
- Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey AJ, Moreno-De-Luca D, Yu TW, Fombonne E, Geschwind D, et al. (2012). Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostovic I, and Judas M (2010). The development of the subplate and thalamocortical connections in the human foetal brain. Acta Paediatr 99, 1119–1127. [DOI] [PubMed] [Google Scholar]
- Kostovic I, and Rakic P (1990). Developmental history of the transient subplate zone in the visual and somatosensory cortex of the macaque monkey and human brain. J Comp Neurol 297, 441–470. [DOI] [PubMed] [Google Scholar]
- Krumm N, O’Roak BJ, Shendure J, and Eichler EE (2014). A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci 37, 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo LC, Karsten SL, Chen J, Levitt P, and Geschwind DH (2007). Genetic analysis of anterior posterior expression gradients in the developing mammalian forebrain. Cereb Cortex 17, 2108–2122. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Johnson MB, Dube U, Shim S, Rasin MR, Sousa AM, Fertuzinhos S, Chen JG, Arellano JI, et al. (2012). Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell 149, 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, et al. (2004). X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet 74, 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FS, Heimer H, Giedd JN, Lein ES, Sestan N, Weinberger DR, and Casey BJ (2014). Mental health. Adolescent mental health--opportunity and obligation. Science 346, 547–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, Clark EP, Kuijer MB, Cushman M, Pommier Y, and Philpot BD (2018). Characterization and structure-activity relationships of indenoisoquinoline-derived topoisomerase I inhibitors in unsilencing the dormant Ube3a gene associated with Angelman syndrome. Mol Autism 9, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Belgard TG, Hawrylycz M, and Molnar Z (2017). Transcriptomic Perspectives on Neocortical Structure, Development, Evolution, and Disease. Annu Rev Neurosci 40, 629–652. [DOI] [PubMed] [Google Scholar]
- Liao BY, and Zhang J (2008). Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci U S A 105, 6987–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, MacArthur DG, Neale BM, Kirby A, Ruderfer DM, Fromer M, et al. (2013). Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron 77, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GN, Corominas R, Lemmens I, Yang X, Tavernier J, Hill DE, Vidal M, Sebat J, and Iakoucheva LM (2015). Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron 85, 742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Lei J, Sanders SJ, Willsey AJ, Kou Y, Cicek AE, Klei L, Lu C, He X, Li M, et al. (2014). DAWN: a framework to identify autism genes and subnetworks using gene expression and genetics. Mol Autism 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S, and Arlotta P (2015). Generating neuronal diversity in the mammalian cerebral cortex. Annu Rev Cell Dev Biol 31, 699–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. (2008). Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 82, 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer A, and Van de Water J (2017). The Role of the Immune System in Autism Spectrum Disorder. Neuropsychopharmacology 42, 284–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW, Lowes L, Alfano L, Berry K, Church K, et al. (2017). Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N Engl J Med 377, 1713–1722. [DOI] [PubMed] [Google Scholar]
- Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, and Rigo F (2015). Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 518, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, Connolly AM, Iannaccone ST, Kirschner J, Kuntz NL, Saito K, et al. (2018). Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy. N Engl J Med 378, 625–635. [DOI] [PubMed] [Google Scholar]
- Mesgarani N, and Chang EF (2012). Selective cortical representation of attended speaker in multi-talker speech perception. Nature 485, 233–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, Fobbs AJ, Sousa AM, Sestan N, Wildman DE, et al. (2012). Prolonged myelination in human neocortical evolution. Proc Natl Acad Sci U S A 109, 16480–16485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. (2014). Transcriptional landscape of the prenatal human brain. Nature 508, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RH (2004). Huntington’s disease genetics. NeuroRx 1, 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. (2012). Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, and Hyman SE (2010). Animal models of neuropsychiatric disorders. Nat Neurosci 13, 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Pattabiraman K, Visel A, and Rubenstein JLR (2015). Genomic perspectives of transcriptional regulation in forebrain development. Neuron 85, 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. (2011). Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet 43, 585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. (2012a). Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. (2012b). Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorati M, Li Z, Liu F, Sousa AMM, Nakagawa N, Li M, Dell’Anno MT, Gulden FO, Pochareddy S, Tebbenkamp ATN, et al. (2016). Zika Virus Disrupts Phospho-TBK1 Localization and Mitosis in Human Neuroepithelial Stem Cells and Radial Glia. Cell Rep 16, 2576–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano G, Ferrara N, Brooks DJ, and Pavese N (2016). Age at onset and Parkinson disease phenotype. Neurology 86, 1400–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes MF, James D, Gil-Perotin S, Kim H, Cotter JA, Ng C, Sandoval K, Rowitch DH, Xu D, McQuillen PS, et al. (2016). Extensive migration of young neurons into the infant human frontal lobe. Science 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, and Geschwind DH (2013). Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z, Judas M, Simic G, Rasin MR, Uylings HB, Rakic P, and Kostovic I (2011). Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci U S A 108, 13281–13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, Thiruvahindrapuram B, Xu X, Ziman R, Wang Z, et al. (2014). Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet 94, 677–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletikos M, Sousa AM, Sedmak G, Meyer KA, Zhu Y, Cheng F, Li M, Kawasawa YI, and Sestan N (2014). Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron 81, 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. (2015). Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, et al. (2014). Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol 32, 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psych EC, Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, et al. (2015). The PsychENCODE project. Nat Neurosci 18, 1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P (2002). Neurogenesis in adult primate neocortex: an evaluation of the evidence. Nat Rev Neurosci 3, 65–71. [DOI] [PubMed] [Google Scholar]
- Rapoport JL, and Gogtay N (2008). Brain neuroplasticity in healthy, hyperactive and psychotic children: insights from neuroimaging. Neuropsychopharmacology 33, 181–197. [DOI] [PubMed] [Google Scholar]
- Riedel A, Klekamp J, Harper C, and Kretschmann HJ (1989). Morphometric study on the postnatal growth of the cerebellum of Australian aborigines and Caucasians. Brain Res 499, 333–343. [DOI] [PubMed] [Google Scholar]
- Robinson EB, St Pourcain B, Anttila V, Kosmicki JA, Bulik-Sullivan B, Grove J, Maller J, Samocha KE, Sanders SJ, Ripke S, et al. (2016). Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat Genet 48, 552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell PE, Fuccillo MV, Maxeiner S, Hayton SJ, Gokce O, Lim BK, Fowler SC, Malenka RC, and Sudhof TC (2014). Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell 158, 198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Campbell AJ, Cottrell JR, Moller RS, Wagner FF, Auldridge AL, Bernier RA, Catterall WA, Chung WK, Empfield JR, et al. (2018). Progress in Understanding and Treating SCN2A-Mediated Disorders. Trends Neurosci 41, 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. (2011). Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70, 863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, et al. (2015). Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 87, 1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. (2012). De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Neale BM, Huang H, Werling DM, An JY, Dong S, Whole Genome Sequencing for Psychiatric, D., Abecasis G, Arguello PA, Blangero J, et al. (2017). Whole genome sequencing in psychiatric disorders: the WGSPD consortium. Nat Neurosci 20, 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, and Caroni P (2011). Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron 71, 35–48. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics, C. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. (2007). Strong association of de novo copy number mutations with autism. Science 316, 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz S, Klin A, and Jones W (2018). Neonatal Transitions in Social Behavior and Their Implications for Autism. Trends Cogn Sci 22, 452–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman RL, and Rakic P (1973). Neuronal migration, with special reference to developing human brain: a review. Brain Res 62, 1–35. [DOI] [PubMed] [Google Scholar]
- Silbereis JC, Pochareddy S, Zhu Y, Li M, and Sestan N (2016). The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron 89, 248–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AMM, Meyer KA, Santpere G, Gulden FO, and Sestan N (2017a). Evolution of the Human Nervous System Function, Structure, and Development. Cell 170, 226–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AMM, Zhu Y, Raghanti MA, Kitchen RR, Onorati M, Tebbenkamp ATN, Stutz B, Meyer KA, Li M, Kawasawa YI, et al. (2017b). Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Pourcain B, Robinson EB, Anttila V, Sullivan BB, Maller J, Golding J, Skuse D, Ring S, Evans DM, Zammit S, et al. (2018). ASD and schizophrenia show distinct developmental profiles in common genetic overlap with population-based social communication difficulties. Mol Psychiatry 23, 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State MW, and Levitt P (2011). The conundrums of understanding genetic risks for autism spectrum disorders. Nat Neurosci 14, 1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State MW, and Sestan N (2012). Neuroscience. The emerging biology of autism spectrum disorders. Science 337, 1301–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur M, and Rubenstein JL (2005). Patterning and plasticity of the cerebral cortex. Science 310, 805–810. [DOI] [PubMed] [Google Scholar]
- Sztainberg Y, Chen HM, Swann JW, Hao S, Tang B, Wu Z, Tang J, Wan YW, Liu Z, Rigo F, et al. (2015). Reversal of phenotypes in MECP2 duplication mice using genetic rescue or antisense oligonucleotides. Nature 528, 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztainberg Y, and Zoghbi HY (2016). Lessons learned from studying syndromic autism spectrum disorders. Nat Neurosci 19, 1408–1417. [DOI] [PubMed] [Google Scholar]
- Thompson BL, and Levitt P (2010). The clinical-basic interface in defining pathogenesis in disorders of neurodevelopmental origin. Neuron 67, 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TN, Coe BP, Dickel DE, Hoekzema K, Nelson BJ, Zody MC, Kronenberg ZN, Hormozdiari F, Raja A, Pennacchio LA, et al. (2017). Genomic Patterns of De Novo Mutation in Simplex Autism. Cell 171, 710–722 e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Tammimies K, Pellecchia G, Alipanahi B, Hu P, Wang Z, Pinto D, Lau L, Nalpathamkalam T, Marshall CR, et al. (2014). Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat Genet 46, 742–747. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Hallstrom BM, Lindskog C, Mardinoglu A, Ponten F, and Nielsen J (2016). Transcriptomics resources of human tissues and organs. Mol Syst Biol 12, 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier WM, Pijnenburg YA, Fox NC, and Scheltens P (2011). Early-onset versus late-onset Alzheimer’s disease: the case of the missing APOE varepsilon4 allele. Lancet Neurol 10, 280–288. [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Janssen B, Verhoef S, Lindhout D, van den Ouweland A, Halley D, Young J, et al. (1997). Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science 277, 805–808. [DOI] [PubMed] [Google Scholar]
- Varghese M, Keshav N, Jacot-Descombes S, Warda T, Wicinski B, Dickstein DL, Harony-Nicolas H, De Rubeis S, Drapeau E, Buxbaum JD, et al. (2017). Autism spectrum disorder: neuropathology and animal models. Acta Neuropathol 134, 537–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. (1991). Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, and Yang J (2017). 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet 101, 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, and Geschwind DH (2011). Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmar F, Siegel M, Woodbury-Smith M, King B, McCracken J, State M, American Academy of, C., and Adolescent Psychiatry Committee on Quality, I. (2014). Practice parameter for the assessment and treatment of children and adolescents with autism spectrum disorder. J Am Acad Child Adolesc Psychiatry 53, 237–257. [DOI] [PubMed] [Google Scholar]
- Volkmar FR, State M, and Klin A (2009). Autism and autism spectrum disorders: diagnostic issues for the coming decade. J Child Psychol Psychiatry 50, 108–115. [DOI] [PubMed] [Google Scholar]
- Vorstman JAS, Parr JR, Moreno-De-Luca D, Anney RJL, Nurnberger JI Jr., and Hallmayer JF (2017). Autism genetics: opportunities and challenges for clinical translation. Nat Rev Genet 18, 362–376. [DOI] [PubMed] [Google Scholar]
- Weiner DJ, Wigdor EM, Ripke S, Walters RK, Kosmicki JA, Grove J, Samocha KE, Goldstein JI, Okbay A, Bybjerg-Grauholm J, et al. (2017). Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet 49, 978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling DM, Brand H, An JY, Stone MR, Zhu L, Glessner JT, Collins RL, Dong S, Layer RM, Markenscoff-Papadimitriou E, et al. (2018). An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat Genet 50, 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling DM, Parikshak NN, and Geschwind DH (2016). Gene expression in human brain implicates sexually dimorphic pathways in autism spectrum disorders. Nat Commun 7, 10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Morris MT, Wang S, Willsey HR, Sun N, Teerikorpi N, Baum TB, Cagney G, Bender KJ, Desai TA, et al. (2018). The Psychiatric Cell Map Initiative: A Convergent Systems Biological Approach to Illuminating Key Molecular Pathways in Neuropsychiatric Disorders. Cell 174, 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. (2013). Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, and State MW (2015). Autism spectrum disorders: from genes to neurobiology. Curr Opin Neurobiol 30, 92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, et al. (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 50, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wells AB, O’Brien DR, Nehorai A, and Dougherty JD (2014). Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci 34, 1420–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev P, and Lecours A (1967). The myelogenetic cycles of regional maturation of the brain (Regional development of the brain in early life) (Philadelphia: F.A. Davis Co; ). [Google Scholar]
- Yeung MS, Zdunek S, Bergmann O, Bernard S, Salehpour M, Alkass K, Perl S, Tisdale J, Possnert G, Brundin L, et al. (2014). Dynamics of oligodendrocyte generation and myelination in the human brain. Cell 159, 766–774. [DOI] [PubMed] [Google Scholar]
- Yuen RK, Thiruvahindrapuram B, Merico D, Walker S, Tammimies K, Hoang N, Chrysler C, Nalpathamkalam T, Pellecchia G, Liu Y, et al. (2015). Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med 21, 185–191. [DOI] [PubMed] [Google Scholar]
- Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, Zhang H, Li L, Sun L, Pan N, et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY (2003). Postnatal neurodevelopmental disorders: meeting at the synapse? Science 302, 826–830. [DOI] [PubMed] [Google Scholar]
- Zwaigenbaum L, and Penner M (2018). Autism spectrum disorder: advances in diagnosis and evaluation. BMJ 361, k1674. [DOI] [PubMed] [Google Scholar]