Mucor is intrinsically resistant to most known antifungals, which makes mucormycosis treatment challenging. Calcineurin is a serine/threonine phosphatase that is widely conserved across eukaryotes. When calcineurin function is inhibited in Mucor, growth shifts to a less virulent yeast growth form, which makes calcineurin an attractive target for development of new antifungal drugs. Previously, we identified two distinct mechanisms through which Mucor can become resistant to calcineurin inhibitors involving Mendelian mutations in the gene for FKBP12, including mechanisms corresponding to calcineurin A or B subunits and epimutations silencing the FKBP12 gene. Here, we identified a third novel mechanism where loss-of-function mutations in the amino acid permease corresponding to the bycA gene contribute to resistance against calcineurin inhibitors. When calcineurin activity is absent, BycA can activate protein kinase A (PKA) to promote yeast growth via a cAMP-independent pathway. Our data also show that calcineurin activity contributes to host-pathogen interactions primarily in the pathogenesis of Mucor.
KEYWORDS: Mucor, mucormycosis, amino acid permease, calcineurin, dimorphism, drug resistance mechanisms, protein kinase A
ABSTRACT
Mucormycosis is an emerging lethal fungal infection in immunocompromised patients. Mucor circinelloides is a causal agent of mucormycosis and serves as a model system to understand genetics in Mucorales. Calcineurin is a conserved virulence factor in many pathogenic fungi, and calcineurin inhibition or deletion of the calcineurin regulatory subunit (CnbR) in Mucor results in a shift from hyphal to yeast growth. We analyzed 36 calcineurin inhibitor-resistant or bypass mutants that exhibited hyphal growth in the presence of calcineurin inhibitors or in the yeast-locked cnbRΔ mutant background without carrying any mutations in known calcineurin components. We found that a majority of the mutants had altered sequence in a gene, named here bycA (bypass of calcineurin). bycA encodes an amino acid permease. We verified that both the bycAΔ single mutant and the bycAΔ cnbRΔ double mutant are resistant to calcineurin inhibitor FK506, thereby demonstrating a novel mechanism of resistance against calcineurin inhibitors. We also found that the level of expression of bycA was significantly higher in the wild-type strain treated with FK506 and in the cnbRΔ mutants but was significantly lower in the wild-type strain without FK506 treatment. These findings suggest that bycA is a negative regulator of hyphal growth and/or a positive regulator of yeast growth in Mucor and that calcineurin suppresses expression of the bycA gene at the mRNA level to promote hyphal growth. BycA is involved in the Mucor hypha-yeast transition as our data demonstrate positive correlations among bycA expression, protein kinase A activity, and Mucor yeast growth. Also, calcineurin, independently of its role in morphogenesis, contributes to virulence traits, including phagosome maturation blockade, host cell damages, and proangiogenic growth factor induction during interactions with hosts.
INTRODUCTION
Mucormycosis is a severe life-threatening infection caused by fungi belonging to the order Mucorales (1). People with weakened immune systems due to diabetes mellitus, neutropenia, hematological disorders, and solid-organ transplantation are at the highest risk of acquiring this infection (2, 3). It is the third most common invasive fungal infection in hematological and allogeneic stem transplantation patients following candidiasis and aspergillosis (1, 4). Over the past decades, there has been a global and ongoing rise in the incidence of mucormycosis, primarily due to the increasing number of diabetic patients and increased use of immunosuppressive drugs (5–10). Mucormycosis is also on the rise in immunocompetent individuals (11–14). Mucorales species grow as molds in the environment and produce sporangiospores that can enter the host via inhalation resulting in pulmonary infection, through the skin due to trauma resulting in cutaneous infections, or through the nasal passages resulting in rhinocerebral infections (15–17). The spores can disseminate within the host, resulting in 95% to 100% mortality even with antifungal drug treatment (18). Mucorales species are intrinsically resistant to most antifungals; thus, Mucorales infections are very difficult to treat, and surgery is often required (19).
Calcineurin is a calcium-calmodulin-dependent phosphatase conserved widely across eukaryotes, including pathogenic fungi (20, 21). Calcineurin is a heterodimer consisting of a catalytic subunit and a regulatory subunit, and both subunits are required for calcineurin function. The role of calcineurin varies depending on the fungal species; for example, calcineurin is required for growth at high temperature (37°C) and at alkaline pH in Cryptococcus neoformans and Cryptococcus gattii (22–24), while calcineurin contributes to azole tolerance and is required for survival in serum, among other functions, in Candida spp. (20, 25). In Aspergillus fumigatus, calcineurin mutants exhibit delayed germination, hyphal growth with irregular branching, and abnormal septa (26). We have previously shown that calcineurin regulates dimorphism in Mucor spp., where the calcineurin inhibitor FK506 (tacrolimus) forces Mucor to grow only as yeast (27).
Hyphal morphology is the predominant growth mode for Mucor spp.; however, by modulation of respiratory conditions, Mucor can be forced to grow as yeast as well (28). While aerobic conditions promote hyphal growth, conditions that include low levels of oxygen and high levels of carbon dioxide enforce yeast growth (29–32). Targeting components involved in mitochondrial or lipid metabolism can also promote yeast growth, even under aerobic conditions (33–36). In addition, previous studies have shown that the addition of cyclic AMP to Mucor in culture results in activation of cAMP-dependent kinase protein kinase A (PKA) and promotes yeast growth (37–40). Wolff et al. also showed that higher levels of expression of PKA regulatory and catalytic subunits are exhibited during anaerobic yeast growth than during aerobic hyphal growth in Mucor (41). Mucor spp. have four isoforms of PKA regulatory subunits, and each is differentially expressed depending on the growth conditions (38, 39). Calcineurin is involved in the genetic regulation of Mucor dimorphism, as deletion of the gene encoding the regulatory subunit of calcineurin (CnbR) resulted in yeast-locked growth, even under aerobic conditions (27). cnbRΔ mutants were previously shown to be avirulent in a wax moth host model (27), thereby indicating that calcineurin is an attractive target for antifungal treatment in mucormycosis. Also, mucormycosis incidence is low in patients receiving FK506 as an immunosuppressant (42). cnbRΔ mutants are also more susceptible to antifungal drugs such as amphotericin B (Ambisome), micafungin, and posaconazole (43).
The cellular receptor for calcineurin is FKBP12, a member of the immunophilin protein family with cis-trans peptide prolyl isomerase activity (44). When FK506 is bound to FKBP12, it inhibits calcineurin phosphatase activity by binding to the calcineurin interface between the catalytic A subunit and the regulatory B subunit, thereby preventing access of substrates to the active site (45, 46). FKBP12 also binds to rapamycin to inhibit the Tor pathway (47), and mutations in the FKBP12 gene confer resistance to both FK506 and rapamycin. Amino acid substitutions in the calcineurin regulatory B and catalytic A subunit surfaces that interact with the FKBP12-FK506 complex can also result in resistance to FK506 (48, 49). Another immunophilin, cyclophilin A (Cyp), serves as a cellular receptor for the drug cyclosporine (CsA). When bound to Cyp, CsA inhibits calcineurin in a manner similar to that seen with FKBP12-FK506 (50). Disruption of the gene encoding Cyp therefore confers resistance to CsA.
In our previous studies, calcineurin inhibitor-resistant Mucor strains, which exhibit hyphal growth instead of yeast growth, were found to have mutations in the FKBP12 gene or the calcineurin catalytic A gene or regulatory B subunit gene (27, 43, 51, 52). In addition, Calo et al. found that Mucor can also silence the FKBP12 gene to become transiently resistant to FK506 and rapamycin via an RNA interference (RNAi)-dependent epimutation pathway (51, 52). In this study, we isolated mutants that do not employ the known calcineurin inhibitor resistance mechanisms. We identified a novel mechanism through which Mucor can become resistant to calcineurin inhibitors. We found that mutations or deletions in a novel gene, bycA (bypass of calcineurin), encoding an amino acid permease that confers resistance to calcineurin inhibitors or loss of calcineurin regulatory B subunit. This gene has not been previously described as involved in the calcineurin pathway, morphogenesis, or virulence in Mucorales. As a result, bycA mutation allowed us to separate the yeast-hypha morphology switch from calcineurin function and to demonstrate that calcineurin, independently of its function in regulating morphology, contributes to Mucor-host interactions.
RESULTS
Isolation of calcineurin bypass mutants in the yeast-locked cnbRΔ mutant background.
The cnbRΔ mutant grows exclusively as a yeast (27). However, we isolated spontaneous mutants that exhibit hyphal growth in the cnbRΔ background. When the mutant (103 cells per spot) was grown in solid yeast extract-peptone-dextrose (YPD) medium at 30°C for a prolonged incubation (exceeding 5 days), we observed that hyphal sectors emerged from the yeast colonies (see Fig. S1A in the supplemental material). After two rounds of single-streak dilutions, 17 independently isolated mutants (calcineurin suppressor [CnSp] mutants) in the cnbRΔ background were isolated (Fig. 1) (Table 1; see also Fig. S1B). The hyphae of the mutants continued to grow, producing aerial hyphae decorated with sporangiophores containing sporangiospores (asexual spores [referred to here as “spores”]). This finding shows that these mutants can complete the entire vegetative cycle without a functional calcineurin. We previously demonstrated that calcineurin function is required for hyphal growth (27). However, these spontaneous mutants do not have a functional calcineurin but exhibit filamentation and produce spores. Thus, the mutants carry a genetic suppressor mutation(s) of the cnbRΔ mutation.
FIG 1.
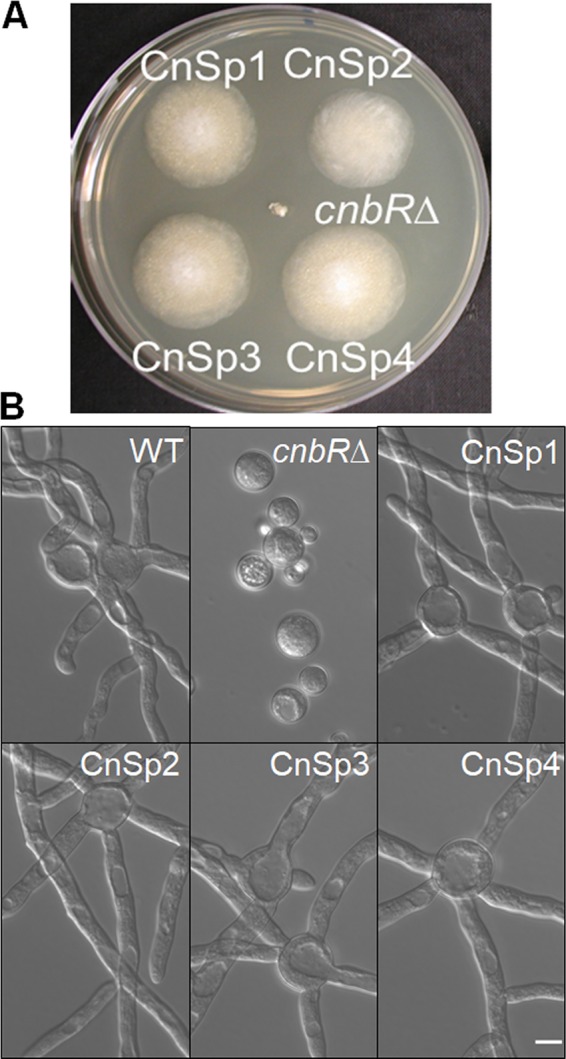
Calcineurin suppressor (CnSp) mutations in the cnbRΔ mutant background restore hyphal growth. (A) Growth of calcineurin suppressor mutants (CnSp1 to CnSp4) and cnbRΔ mutant on a YPD agar plate at 30°C for 4 days postinoculation. While the cnbRΔ mutant shows smaller yeast colonies, the CnSp mutants show larger hyphal colonies. (B) The Mucor WT (R7B), cnbRΔ mutant, or CnSp mutants were grown overnight in liquid YPD medium with shaking at 30°C. Micrographs show that the CnSp mutants exhibit hyphal growth like that seen with the WT (scale bar = 10 μm).
TABLE 1.
Characterization of the bypass mutants isolated in the cnbRΔ mutant background
| Strain | Genotype in the bycA allele | Remark | Type of mutation |
|---|---|---|---|
| CnSp1 | 1350G→A | Nonsense | |
| CnSp2 | Long deletion around bycA | Whole genome sequenced | |
| CnSp3 | 2030C→A | Whole genome sequenced | Nonsense |
| CnSp4 | 361_1579del | Whole genome sequenced | Frameshift |
| CnSp5 | Potential deletion | No bycA PCR product | |
| CnSp6 | No mutation in bycA allele | ||
| CnSp7 | 995C→T | Whole genome sequenced | Missense |
| CnSp8 | 1921_1922insTGACATTGCTTCAGCAG | Whole genome sequenced | Frameshift |
| CnSp9 | 320T→C | Missense | |
| CnSp10 | 2319_2320insC | Whole genome sequenced | Frameshift |
| CnSp11 | 2129_2130insTCACC | Frameshift | |
| CnSp12 | Potential deletion | No bycA PCR product | |
| CnSp13 | 1784_1793delTCAATTTCAT | Frameshift | |
| CnSp14 | 566T→A | Nonsense | |
| CnSp15 | 2238_2239insTC | Frameshift | |
| CnSp16 | No mutation in bycA allele | ||
| CnSp17 | 1732C→T | Nonsense |
Emergence of hyphal sectors from cnbRΔ mutants and isolation of calcineurin suppressor mutants. (A) cnbRΔ mutants were plated on YPD agar for 5 or more days at 30°C. (B) The hyphae emerging from the yeast colonies were transferred to fresh YPD agar for further propagation. (C) Calcineurin suppressor (CnSP) mutants produced hyphae decorated with sporangiophores containing sporangiospores like the WT results (scale bar = 20 μm). Download FIG S1, TIF file, 2.4 MB (2.5MB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Isolation of calcineurin bypass mutants in the cnaBΔ background.
The CsA calcineurin inhibitor does not fully force Mucor to grow as yeast, unlike FK506. Instead, CsA fully inhibits hyphal growth and enforces a yeast-locked phenotype only when Mucor lacks the cnaB gene, which is one of the three calcineurin catalytic subunit genes (cnaA, cnaB, and cnaC) (43). We grew the cnaBΔ mutant on YPD medium containing 2 μg/ml of CsA, and the mutant grew exclusively as yeast because calcineurin was inhibited by CsA. Similarly, we observed that hyphal sectors emerged from yeast colonies as the strains became resistant to CsA after prolonged incubation on CsA medium (Fig. S2). After performing two rounds of single-streak dilutions, we isolated 19 independently derived CsA-resistant (CSR) mutants (Table 2). The mutants exhibited hyphal growth in the presence of either CsA or FK506 (Fig. 2).
TABLE 2.
Characterization of the calcineurin bypass mutants isolated in the cnaBΔ mutant background
| Strain | Genotype in the bycA allele | Remark | Type of mutation |
|---|---|---|---|
| CSR1 | C1240del | Frameshift | |
| CSR2 | Potential deletion | No bycA PCR product | |
| CSR3 | 733 C→A | Missense | |
| CSR4 | 633delG | Whole genome sequenced | Frameshift |
| CSR5 | 407C→T | Whole genome sequenced | Missense |
| CSR6 | 2188delC | Whole genome sequenced | Frameshift |
| CSR7 | 806_825delCCTACCCGCCAGTGGAAGCA | Frameshift | |
| CSR8 | 1711delG | Frameshift | |
| CSR9 | 2188delC | Whole genome sequenced | Frameshift |
| CSR10 | 1649delT | Whole genome sequenced | Frameshift |
| CSR11 | 578G→A | Nonsense | |
| CSR12 | 621delT | Frameshift | |
| CSR13 | 2296_2297insAAT | Frameshift | |
| CSR14 | 218G→A | Nonsense | |
| CSR16 | 2263delC | Whole genome sequenced | Frameshift |
| CSR28 | 215G→A | Missense | |
| CSR29 | No mutation in bycA allele | ||
| CSR35 | No mutation in bycA allele | ||
| CSR38 | No mutation in bycA allele |
FIG 2.
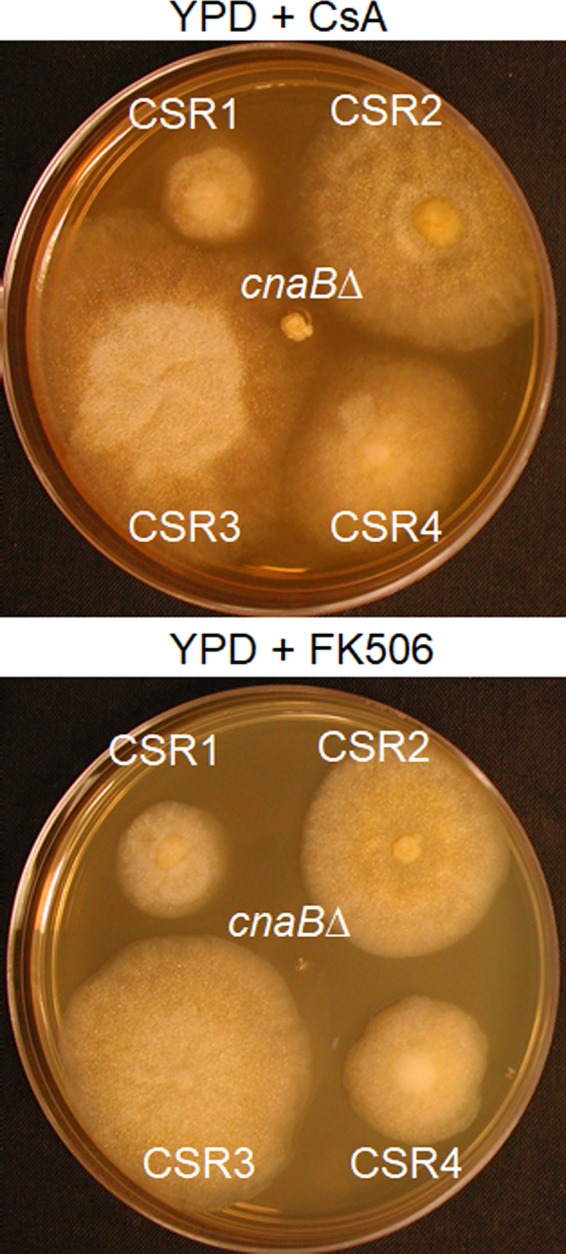
Calcineurin suppressor mutations in the cnaBΔ mutant background confer resistance to calcineurin inhibitors. The images show growth of cyclosporine-resistant mutants (CSR) exhibiting hyphal growth and of cnaBΔ mutants exhibiting yeast growth under conditions of incubation on YPD agar with CsA (100 μg/ml) (top panel) or FK506 (1 μg/ml) (bottom panel) for 4 days at 30°C. CSR1, CSR2, CSR3, and CSR4 are shown. The other CSR mutants exhibited a similar CsA resistance phenotype (data not shown).
Emergence of hyphal sectors from the cnaBΔ mutant grown on CsA-containing medium. cnaBΔ mutants were grown in the presence of CsA (2 μg/ml) for 5 or more days at 30°C. This resulted in the emergence of hyphal sectors from yeast colonies. Download FIG S2, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Surprisingly, none of the mutants carried Mendelian mutations in genes for the calcineurin A subunit or B subunit, FKBP12, or two subtypes of cyclophilin A (cypA and cypB) (data not shown). In addition, the mutants did not carry epimutations in the cypA and cypB genes, and small RNA blots did not reveal any small RNAs derived from the cypA and cypB genes (51) (data not shown). Therefore, we hypothesized that the CSR mutants were also calcineurin bypass mutants that do not require calcineurin activity for hyphal growth.
Spontaneous mutations in the bycA gene result in phenotypes of suppression of the calcineurin mutation.
To characterize the mutation(s) that results in bypass of the calcineurin requirement, we sequenced the whole genomes of 6 CnSp and 6 CSR mutant strains, along with wild-type (WT) strain MU402, which was used for transformation to obtain the cnbR and cnaB mutants with an Illumina HiSeq platform. The whole genomes of each mutant strain were compared to that of the wild-type MU402 strain. Surprisingly, all 12 mutants carried DNA sequence modifications in a single common locus (Table 1 and Table 2). The modifications included an insertion of short sequences, single nucleotide polymorphisms, or deletions of a shorter or longer region in the locus. The gene in the locus was designated bycA (bypass of calcineurin). We further sequenced the bycA gene in the remaining CSR and CnSp mutants and found mutations in the bycA gene in all but 5 mutants (Table 1 and Table 2). Of the 36 suppressor mutants, 31 contained mutations in the bycA gene that could result in suppression of the calcineurin mutation.
bycAΔ mutants are resistant to calcineurin inhibitors, and bycAΔ cnbRΔ double mutants exhibit a hyphal morphology.
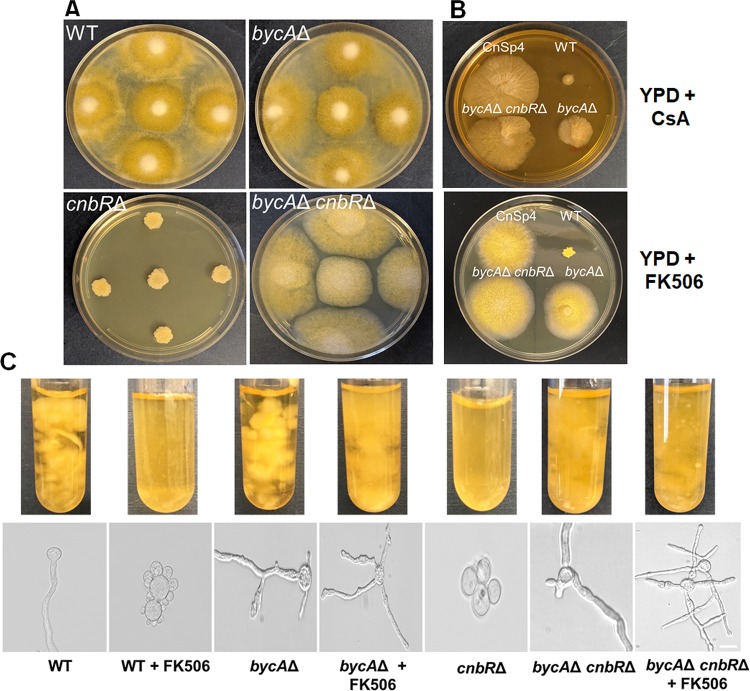
We further verified that bycA is associated with the calcineurin pathway by generating bycA deletion mutants. The bycA gene in wild-type strain MU402 was replaced with the pyrG-dpl237 marker (53), and gene replacement by recombination was confirmed by 5′ and 3′ junction PCR (Fig. S3), open reading frame (ORF)-spanning PCR (Fig. S3), and Southern blotting (not shown). We also performed reverse transcriptase PCR (RT-PCR) to confirm the bycA gene is not expressed in the mutants (see Fig. 4A). Two independent mutants, MSL47.1 and MSL47.2 (bycAΔ::pyrG-dpl237), were obtained. The bycAΔ mutants exhibited hyphal growth and showed no growth defects (Fig. 3A). On solid agar medium containing FK506, a bycAΔ mutant and CnSp4 produce a larger hyphal colony and aerial hyphae, whereas the wild-type strain formed a smaller yeast colony (Fig. 3B). These results indicate that bycAΔ mutants are resistant to FK506.
FIG 4.
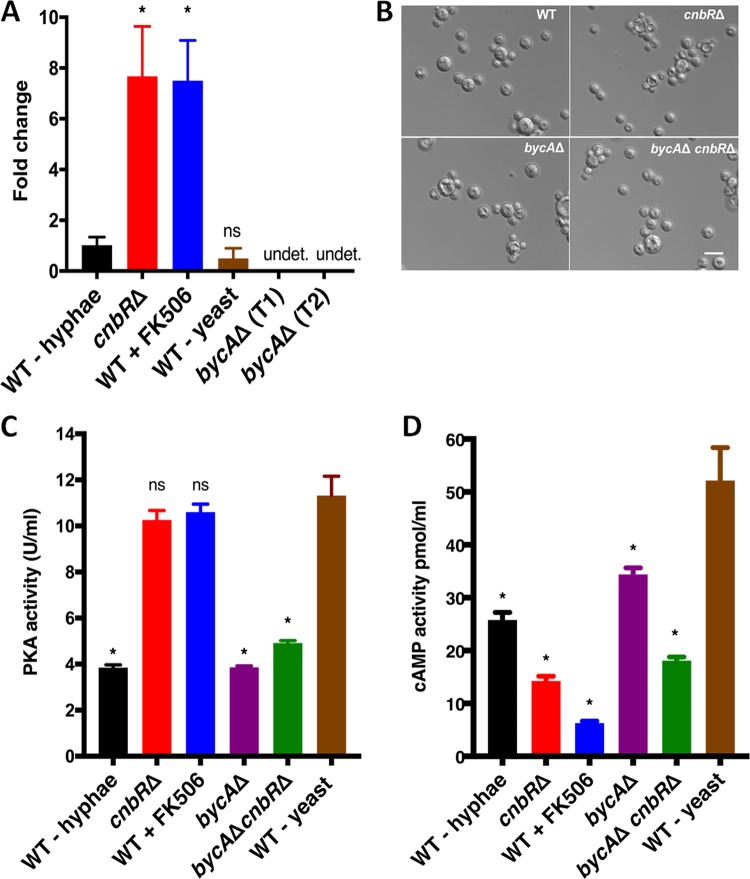
BycA is involved in the Mucor hypha-yeast transition under aerobic conditions. (A) Reverse transcriptase quantitative PCR showed that in the absence of calcineurin function (cnbRΔ and WT + FK506; yeast morphology), bycA expression was 6-fold higher than the level seen with the WT (hyphal morphology) with calcineurin function, suggesting that calcineurin regulates bycA expression at the mRNA level. When the Mucor WT was grown anaerobically under high-CO2 conditions (see Materials and Methods), it grew as yeast; however, there was no significant difference in bycA expression compared to the level seen with the WT-hypha group. One-way analysis of variance (ANOVA) data were significant (P = 0.0001). Dunnett’s post hoc test was used to compare cnbRΔ, WT + FK506, WT-yeast results with those seen with the WT-hypha group (*, P < 0.05; ns, not statistically significant; undet., undetected). As expected, no expression of the bycA gene was detected in either of the bycAΔ mutants. (B) When WT, cnbRΔ, bycAΔ, and bycAΔ cnbRΔ mutants were grown anaerobically overnight under high-CO2 conditions, they all exhibited a yeast morphology, thereby suggesting that neither calcineurin nor BycA has a role in anaerobic morphological pathways (scale bar = 20 μm). (C) Crude protein extracts (0.5 μg) were used to measure overall PKA activity. The WT-hypha and the bycAΔ mutant showed significantly lower PKA activity than WT-yeast (grown anaerobically). In WT + FK506 and the cnbRΔ mutant, PKA activity remained higher; however, the bycAΔ cnbRΔ mutant showed lower PKA activity despite possessing no calcineurin function. One-way ANOVA data were significant (P < 0.0001). Dunnett’s multiple-comparison test was used to compare each group with WT-yeast (*, P < 0.05). (D) Crude extracts from 60 mg biomass were used to measure overall cAMP activity. Yhe WT + FK506 and cnbRΔ strains showed significantly lower cAMP activity than WT-yeast, thereby suggesting that under aerobic conditions, PKA activity is elevated in Mucor yeast in a cAMP-independent manner. One-way ANOVA data were significant (P < 0.0001). Dunnett’s multiple-comparison test was used to compare each group with WT-yeast (*, P < 0.05).
FIG 3.
The bycAΔ single mutant and bycAΔ cnbRΔ double mutant are resistant to calcineurin inhibitors. (A) After 3 days of growth on solid YPD agar, the cnbRΔ mutant grew as yeast; however, despite no calcineurin function, the bycAΔ cnbRΔ double mutant exhibited hyphal growth like that shown by the WT and the bycAΔ mutant. (B) In the presence of CsA (upper panel; 100 μg/ml), the bycAΔ cnbRΔ double mutant and CnSp4 formed hyphal colonies that were larger than those formed by the WT, indicating resistance to CsA. No major difference in colony size was noted between the bycAΔ mutant and the WT. In the presence of FK506 (lower panel; 1 μg/ml), the WT formed a smaller yeast colony whereas the bycAΔ mutant, bycAΔ cnbRΔ double mutant, and CnSp4 each formed a larger hyphal colony. (C) When Mucor was grown overnight in YPD medium containing FK506 (1 μg/ml) at 30°C with shaking, the bycAΔ mutant, bycAΔ cnbRΔ double mutant, and CnSp4 mutant exhibited resistance to FK506 as evidenced by larger biomass and hyphal morphology, whereas the WT cells were sensitive to FK506 as they not only formed less biomass but also grew as yeast. As expected, the cnbRΔ mutant remained in its yeast-locked form (scale bar = 20 μm).
Confirmation of disruption of the bycA gene by junction PCR and ORF-spanning PCR. (A) Illustration of the bycAΔ::pyrG-dpl237 and bycA alleles with ∼1-kb upstream and downstream flanking sequence. P1 (SL3) and P4 (SL8) recognize sequences outside the disruption cassette. P2 (SCL566) and P3 (SCL567) recognize pyrG-dpl237. P5 (SL182) and P6 (SL183) recognize bycA. (B) At the 5′ end, P1 and P2 amplified a 2,203-bp region, and at the 3′ end, P3 and P4 amplified a 1,994-bp region in bycAΔ::pyrG-dpl237. The primer pairs did not produce a fragment in the WT. P5 and P6 amplified a 690-bp region within the bycA gene, so no amplification was noted in the mutants. Images are not to scale. Download FIG S3, TIF file, 0.4 MB (471.6KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Under conditions of growth on solid YPD media containing CsA (50 μg/ml), interestingly, the bycAΔ mutants were not fully resistant to CsA and instead exhibited colony size comparable to that seen with the wild type (R7B). However, CnSp4 exhibited larger colonies on media containing CsA (Fig. 3B). It is possible that the CnaB catalytic A subunit is partially resistant to the inhibition by CsA, during which Cyp-CsA may not interfere with the function of CnaB-CnbR-calmodulin while still inhibiting CnaA-CnbR-calmodulin and CnaC-CnbR-calmodulin. The bycAΔ mutants still harbor an intact cnaB gene, and it is possible that partial inhibition of CnaB could result in a calcineurin-independent phenotype. Therefore, bycAΔ mutants in the cnaBΔ background exhibited full resistance against CsA, representing a result which has yet to be elucidated.
We further disrupted the cnbR gene in the bycAΔ background to generate bycAΔ cnbRΔ double mutants. The deletion and the absence of expression of cnbR were confirmed by PCR and RT-PCR (Fig. S4 and Fig. S5), respectively, and by Southern blotting (data not shown). Two independent mutants, MSL68.1 and MSL68.2 (bycAΔ::pyrG-dpl237 cnbRΔ::leuA), were generated. As shown in Fig. 3A, while the cnbRΔ mutants grow exclusively as yeast, bycAΔ cnbRΔ double mutants exhibited filamentous growth like that exhibited by the wild-type strain and the bycAΔ mutant. bycAΔ cnbRΔ mutants are also completely resistant to either FK506 or CsA (Fig. 3B), as they exhibit normal hyphal growth even in the presence of calcineurin inhibitors (Fig. 3B and C). Taken together, our results genetically validate our hypothesis that bycA mutations bypass the requirement of calcineurin for hyphal growth or suppress the lack of calcineurin.
Confirmation of disruption of the cnbR gene in the bycAΔ background by junction PCR and ORF-spanning PCR. (A) Illustration of the cnbRΔ::leuA deletion in the bycAΔ::pyrG-dpl237 strain and cnbR alleles with ∼1-kb upstream and downstream flanking sequence. P1 (SL243) and P4 (SL244) recognize sequences outside the disruption cassette. P2 (SL391) and P3 (SL392) are specific for leuA. P5 (SCL578) and P6 (SCL579) are specific for cnbR. (B) At the 5′ end, P1 and P2 amplified a 2,388-bp region, and at the 3′ end, P3 and P4 amplified a 1,620-bp region in the cnbRΔ::leuA region. The primer pair did not produce a fragment in the WT. P1 and P4 produced a 5,711-bp product in the mutant and a 3,342-bp product in the WT. P5 and P6 amplified a 170-bp region within the cnbR gene, so no amplification is noted in the mutants. Image is not to scale. Download FIG S4, TIF file, 0.7 MB (745.7KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Confirmation of disruption of the cnbR gene in the bycAΔ background by RT-qPCR. Primers SCL578 and SCL579 were used to perform qPCR on the cDNA obtained from the WT and mutants (see Materials and Methods). Actin (SCL368 and SCL369) served as a control. No cnbR expression was detected in the bycAΔ cnbRΔ double mutant. Download FIG S5, TIF file, 0.1 MB (139.4KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Calcineurin regulates BycA at the mRNA level.
To determine a genetic link between calcineurin and BycA, the expression of bycA was examined under conditions in which calcineurin was either functional or nonfunctional. As shown in Fig. 4A, bycA gene expression was significantly higher than the WT level under conditions in which calcineurin was not functional such as in the cnbRΔ mutant or in the wild type in the presence of FK506. These results suggest that calcineurin negatively regulates the bycA gene at the mRNA level and that BycA expression is positively correlated with Mucor yeast growth.
Under anaerobic (or microaerobic) conditions with high levels of CO2, Mucor also grows as a yeast (54). Interestingly, under those conditions, bycA expression remained low (Fig. 4A). When bycAΔ or bycAΔ cnbRΔ mutants were grown anaerobically with high CO2 levels, they still grew as yeast (Fig. 4B). These results suggest that BycA is involved in aerobic yeast growth but not in anaerobic yeast growth. Whether calcineurin inhibits the expression of the bycA gene transcriptionally or regulates the stability of the bycA mRNA remains to be elucidated.
BycA serves as a link between calcineurin and protein kinase A.
Previous studies by our groups and others have shown that increased levels of bicarbonate ions through addition to Mucor culture or as a consequence of growth under high-carbon-dioxide conditions are sufficient to induce yeast growth (27, 31), as the bicarbonate ions can activate adenylyl cyclase, resulting in the generation of cAMP and, in turn, in the activation of cAMP-dependent kinase, namely, protein kinase A (PKA) (55, 56) (see Fig. 8). Studies have also shown that there is an inverse relationship between PKA and hyphal morphology as the PKA regulatory subunit gene that inhibits the activity of PKA is more highly expressed during hyphal growth (41). Also, high levels of cAMP promote yeast growth, while low levels of cAMP are linked with hyphal morphology (28). Interestingly, even under aerobic conditions when Mucor was forced to grow as yeast by inhibiting calcineurin activity either genetically or by using FK506, the overall cellular PKA activity remained high (27). Therefore, PKA plays pivotal roles in yeast growth. Interestingly, in the wild type when calcineurin is fully functional, the cellular PKA activity is low (27), suggesting an inverse correlation between PKA and calcineurin levels during morphogenesis. However, it was not clear how calcineurin and PKA are linked.
FIG 8.
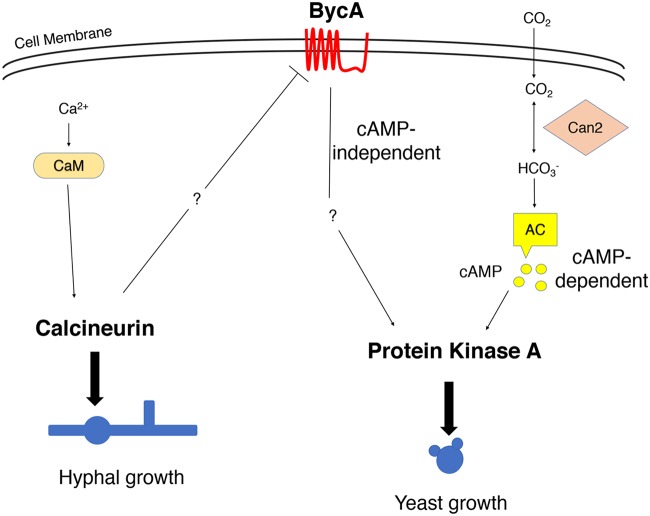
Calcineurin, BycA, and PKA in the morphogenesis of Mucor under aerobic conditions. Calcineurin is the master regulator of Mucor morphology. Active calcineurin positively regulates hyphal growth and negatively regulates yeast growth. This is achieved by suppressing the expression of the bycA gene, thereby preventing an increase in PKA activity. Calcium-calmodulin activates calcineurin to promote hyphal growth under aerobic conditions. However, when calcineurin is not functional, bycA gene expression is significantly elevated. BycA then activates PKA in a cAMP-independent manner to promote yeast growth. Under anaerobic conditions with high CO2 levels, PKA is activated through the CO2-cAMP pathway. Neither BycA nor calcineurin has a defined role in the regulation of Mucor morphology under anaerobic conditions. Can2, carbonyl anhydrase; AC, adenylyl cyclase; CaM, calmodulin.
BycA is a putative amino acid permease and is predicted to have 10 transmembrane domains and 1 pectinesterase domain (Fig. S6). As shown in Table 3, when cnbRΔ and bycAΔ cnbRΔ mutants were grown on YNB medium with methionine or threonine or arginine as a sole nitrogen source, the cnbRΔ mutants exhibited growth levels similar to those seen on YNB complete medium, while bycAΔ cnbRΔ mutants did not produce hyphal mass. This shows that BycA is a bona fide amino acid permease. In Saccharomyces cerevisiae and Candida albicans, the general amino acid permease Gap1 is known to activate PKA in a cAMP-independent manner (57, 58). We hypothesized that in the absence of calcineurin activity, BycA may activate PKA to promote yeast growth. To this end, we measured overall cellular PKA activity in the presence or absence of the bycA gene. As shown in Fig. 4C, PKA activity was significantly higher in WT-yeast, in the cnbRΔ mutant, and in the WT strain subjected to FK506 treatment (WT + FK506) (when bycA expression was significantly higher) than in the WT hyphae (when bycA expression is low). On the other hand, the bycAΔ or bycAΔ cnbRΔ mutants exhibited significantly lower cellular PKA activity when the bycA gene was deleted than were seen under the conditions in which the bycA gene was expressed. No significant differences were observed between the WT hyphae and the bycAΔ or bycAΔ cnbRΔ isolates.
TABLE 3.
Summary of cnbRΔ single mutant and bycAΔ cnbRΔ double mutant growth in the presence of methionine, threonine, and arginine as sole nitrogen source
| Nitrogen source | Growth levela
|
|
|---|---|---|
| cnbRΔ mutant | bycAΔ cnbRΔ mutant | |
| None | − | − |
| Methionine | + | − |
| Threonine | + | − |
| Arginine | + | − |
| Casamino Acids | ++ | ++ |
−, no growth; +, low growth level; ++, high growth level.
Predicted structure of BycA showing 10 transmembrane domains and 1 cytosolic domain. BycA is predicted to have 10 transmembrane domains and 1 cytosolic domain. The transmembrane domains were predicted with the TMHMM 2.0 software package (http://www.cbs.dtu.dk/services/TMHMM/). The predicted cytosolic region contains a pectinesterase domain. Download FIG S6, TIF file, 0.2 MB (219.8KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Interestingly, measurements of cAMP levels showed that the cAMP levels were significantly lower in the cnbRΔ and WT + FK506 yeast groups than in the WT-yeast group despite high PKA activity (Fig. 4D). This suggests that under aerobic conditions when calcineurin is absent, BycA increases PKA activity via a cAMP-independent pathway, a result which has yet to be elucidated.
Phagosome maturation blockade upon phagocytosis by macrophages is dependent on calcineurin rather than morphology.
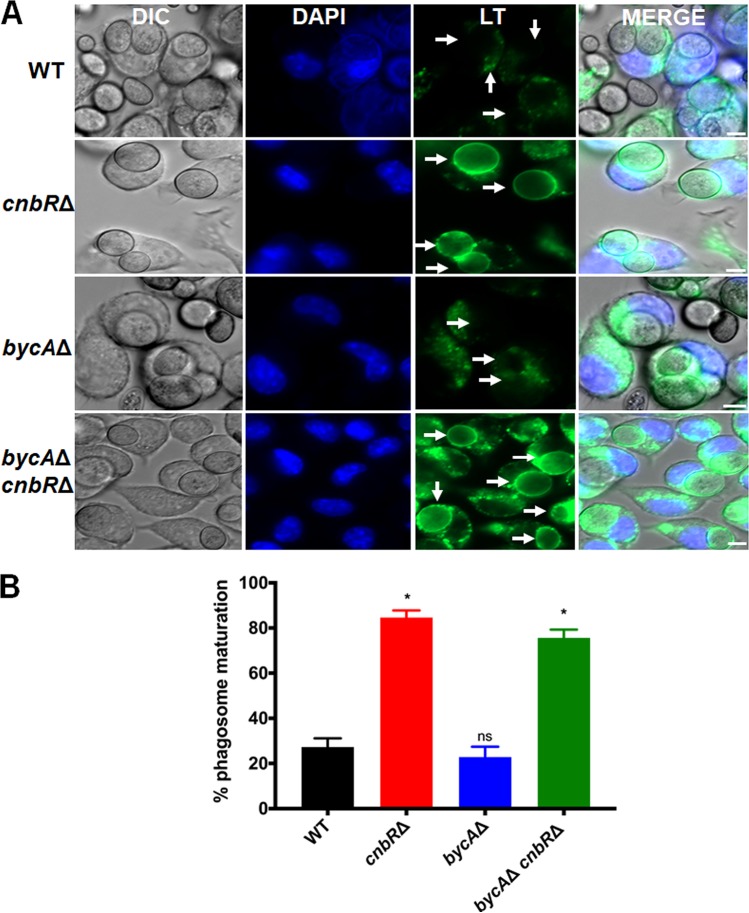
Phagocytosis followed by phagosome maturation or acidification in macrophages is an essential innate immune pathway to control pathogens. We have previously shown that when the macrophage cell line J774.A1 and primary macrophages were challenged with wild-type spores or cnbRΔ yeast, they rapidly phagocytosed both spores and yeast; however, only the macrophages with cnbRΔ yeast underwent phagosome maturation (43). This indicates that Mucor spores escape innate immunity by blocking phagosome maturation as spores survive better than cnbRΔ yeast during coculture with macrophages (Fig. S9). However, it was not clear if the blockade of phagosome maturation by Mucor is dependent on its morphology or on the presence of functional calcineurin because yeast cells lack calcineurin function. To address this issue, we cocultured WT spores, cnbRΔ yeast, and bycAΔ cnbRΔ double mutant spores with macrophages to monitor phagosome maturation. Macrophages containing Mucor spores or yeast cells were stained with Lysotracker Green DND-26 (Thermofisher). Lysotracker stains only acidic organelles in cells such as lysosomes or mature phagosomes and therefore can be used to determine whether phagosomes containing Mucor cells are acidic, an indication of phagosome maturation. As shown in Fig. 5, only about 20% of macrophages challenged with either wild-type (n = 480) or bycAΔ (n = 290) spores underwent phagosomal maturation, which is significantly lower than the proportion seen with macrophages challenged with the cnbRΔ mutant (n = 279; yeast-locked). Interestingly, ∼80% of phagosomes containing bycAΔ cnbRΔ double mutants (n = 413) spores underwent maturation. Our data suggest that a novel downstream function of calcineurin, independent of its function in governing morphology, is involved in the inhibition of phagosome maturation.
FIG 5.
Phagosome maturation in macrophages containing Mucor is dependent on pathogen calcineurin function and not morphology. (A) J774.A1 macrophages (5 × 105) were challenged with Mucor spores or yeast at an MOI of 1 along with LysoTracker Green DND-26 and Hoechst 33342 stain (blue). White arrows indicate where phagosome maturation should be observed in the field (scale bar = 5 μm). DIC, differential interference contrast; DAPI, 4′,6-diamidino-2-phenylindole; LT, LysoTracker. (B) The macrophages containing the cnbRΔ single mutant and the bycAΔ cnbRΔ double mutant underwent significantly higher phagosome maturation than those containing the WT. Data are shown as percent maturation. The numbers (n) of macrophages containing Mucor counted for each group were as follows: WT = 480; bycAΔ mutant = 290; cnbRΔ mutant = 279; bycAΔ cnbRΔ mutant = 413. One-way ANOVA data were significant (P < 0.0001). Dunnett’s multiple-comparison test was used to compare each group to the WT (*, P < 0.05).
Mucor without functional calcineurin causes reduced cell damage and induces lower levels of FGF-2 expression in endothelial cells.
Mucormycosis is an angioinvasive disease, and hence Mucorales interaction and the subsequent damage to the endothelium lining the blood vessels are important steps in disease pathology (59). We have previously shown that Mucor hyphae but not yeast cells (cnbRΔ mutant) induce fibroblast growth factor-2 (FGF-2) protein expression in lymphoblastoid cell lines and bone marrow macrophages (43, 60). We wanted to determine the extent to which calcineurin contributes to the endothelial damage and FGF-2 protein response by Mucor. To this end, we used human umbilical vein endothelial cells (HUVECs) and challenged cells with the wild type, the bycAΔ single mutant, the cnbRΔ single mutant, or the bycAΔ cnbRΔ double mutant for a period of 24 h. HUVECs challenged with the cnbRΔ single mutant and the bycAΔ cnbRΔ double mutant showed 70% less cytotoxicity than HUVECs challenged with wild-type or bycAΔ spores as quantified by measuring lactate dehydrogenase (LDH) levels (Fig. 6A) and significantly less FGF-2 protein production (Fig. 6B). These data suggest a morphology-independent function of calcineurin in regulating Mucor interaction with endothelial cells.
FIG 6.
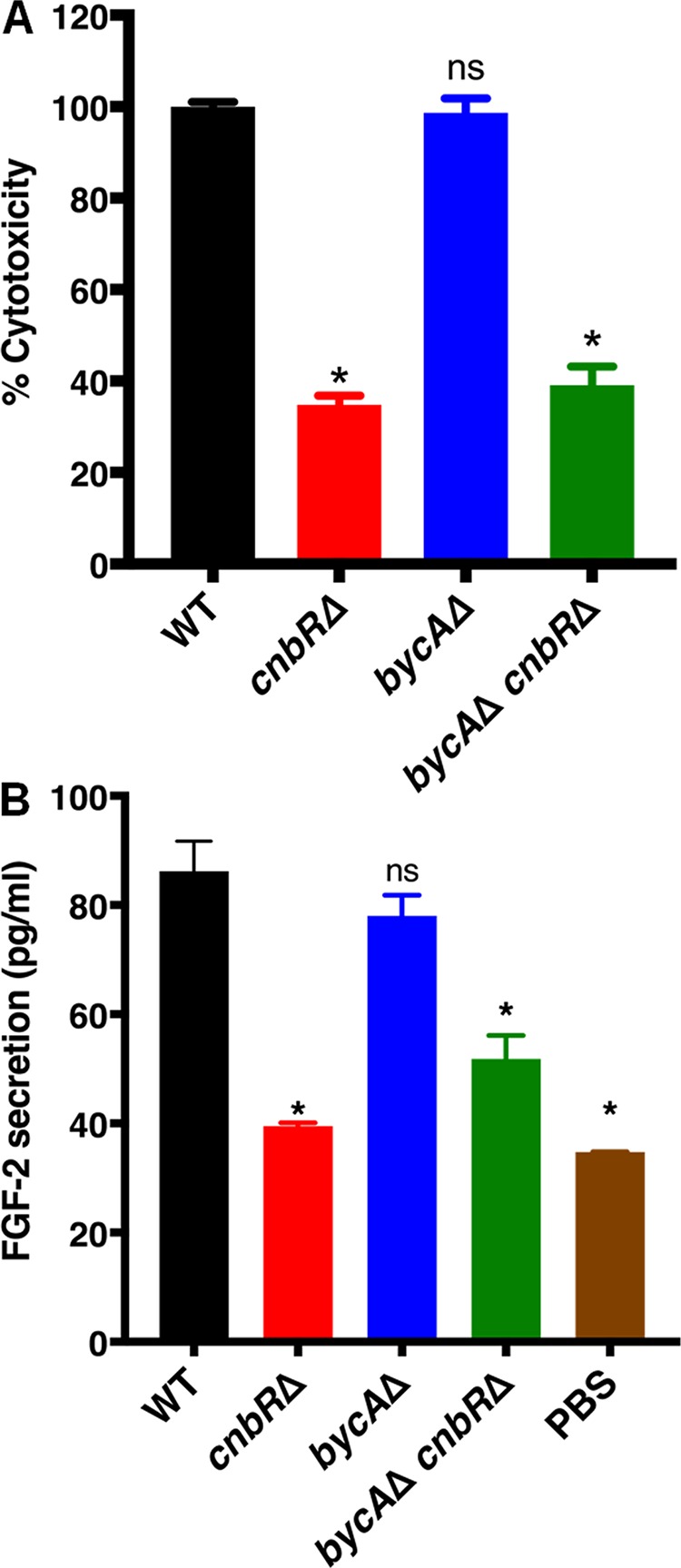
Calcineurin mutants cause less endothelial cell damage and FGF-2 protein secretion. A total of 5 × 103 HUVECs were challenged with Mucor spores or yeast at an MOI of 10. After 24 h, the supernatant was collected. (A) Levels of LDH (indicating cytotoxicity) were quantified. Calcineurin mutants caused less damage than the WT. One-way ANOVA data were significant (P < 0.0001). Dunnett’s multiple-comparison test was used to compare each group to the WT (*, P < 0.05). (B) FGF-2 levels were quantified using ELISA. The WT strain induced significantly higher FGF-2 protein secretion than the calcineurin mutants. One-way ANOVA data were significant (P < 0.0001). Dunnett’s multiple-comparison test was used to compare each group to the WT (*, P < 0.05).
bycAΔ cnbRΔ double mutants are less virulent in a Galleria mellonella host model of mucormycosis.
The yeast-locked cnbRΔ mutant is less virulent than the wild type in a G. mellonella (wax moth) host (27). To test if calcineurin is involved in virulence, we injected wax moths (n = 15/group) with 10,000 spores of the wild type, the bycAΔ single mutant, the cnbRΔ single mutant, or the bycAΔ cnbRΔ double mutant or with phosphate-buffered saline (PBS) (mock) and monitored survival for 7 days. As shown in Fig. 7, all of the wax moth larvae challenged with wild-type or bycAΔ spores succumbed to infection within a week, while the cnbRΔ and bycAΔ cnbRΔ mutants exhibited a significantly higher survival rate. Interestingly, only about 65% of the wax moth infected with the CnSp4 mutant (spontaneous double mutant) or with the bycAΔ cnbRΔ mutant survived, while 100% of the wax moth infected with the cnbRΔ mutant survived. We also injected wax moth larvae with 10,000 (1×), 20,000 (2×), or 30,000 spores (3×) of the bycAΔ cnbRΔ double mutant; as shown in Fig. S7, it was only the 3× spore inoculum that achieved 100% mortality.
FIG 7.
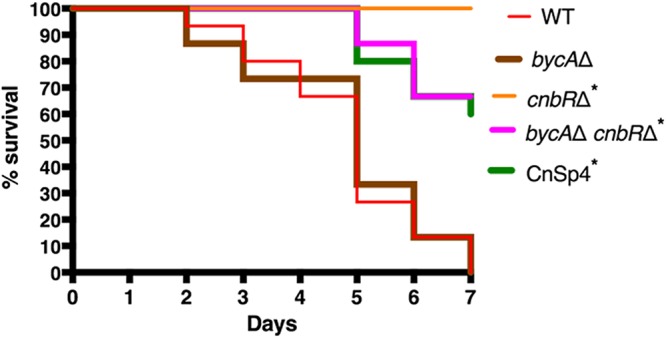
Calcineurin mutants were less virulent in a Galleria mellonella (wax moth) model of mucormycosis. Wax moth larvae (n = 15/group) were inoculated with 1 × 104 Mucor spores or yeast in a mixture containing 2 μl PBS via injection into the last left proleg and were monitored for survival. All wax moth larvae challenged with the WT or the bycAΔ mutant succumbed to mortality within a week. The cnbRΔ mutant was avirulent, whereas about 65% of wax moth larvae inoculated with the bycAΔ cnbRΔ mutant or CnSp4 mutant survived. Results of a log rank (Mantel-Cox) test were statistically significant (P < 0.0001). A pairwise comparison was also performed with the following groups: WT versus bycAΔ mutant (P = 0.82) and WT versus cnbRΔ mutant or bycAΔ cnbRΔ mutant or CnSp4 mutant (*, P < 0.0001).
A bycAΔ cnbRΔ double mutant is lethal in a wax moth larva host only at higher inocula. The wax moth larvae (n = 15/group) were inoculated with 1 × 104 (1×) to 3 × 104 (3×) bycAΔ cnbRΔ double mutant or WT spores (1×) in a mixture containing 2 μl PBS via the last left proleg and were monitored for survival. All wax moth challenged with the WT or the bycAΔ cnbRΔ mutant (3×) succumbed to mortality within a week. A total of 65% of the wax moths inoculated with the bycAΔ cnbRΔ mutant (1×) and 45% of the wax moths inoculated with the bycAΔ cnbRΔ mutant (2×) survived. Results obtained with a log rank (Mantel-Cox) test were statistically significant (P < 0.0001). A pairwise comparison was also performed with the following groups: WT versus bycAΔ cnbRΔ mutant (1×) (*, P < 0.0001); WT versus bycAΔ cnbRΔ mutant (2×) (*, P = 0.0014); WT versus bycAΔ cnbRΔ (3×) (P = 0.9460). Download FIG S7, TIF file, 0.1 MB (91.8KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To determine virulence in an immunocompromised murine host, inoculations were performed with the wild-type strain (R7B), the bycAΔ single mutant, the cnbRΔ single mutant, the bycAΔ cnbRΔ double mutant, or PBS (mock) via the intratracheal or intravenous route. However, there was no significant difference in survival between the infected groups (Fig. S8). It is interesting that calcineurin mutants were significantly less virulent than the WT in the wax moth larval host but were as virulent as the WT in the murine model.
Mucor virulence in the murine model of systemic and pulmonary mucormycosis. (A) Cyclophosphamide- and cortisone acetate-treated CF1 mice were challenged with 1.5 × 106 spores or yeast cells in a mixture containing PBS via lateral tail vein (systemic mucormycosis model). The animals (n = 5/group) were monitored for survival. Results from a log rank (Mantel-Cox) test were not statistically significant (P = 0.0585), suggesting no major difference in survival between the WT and calcineurin mutant groups. (B) For the pulmonary mucormycosis model (n = 5/group), the same inocula were introduced via intratracheal route and the animals were monitored for survival. Results of a log rank (Mantel-Cox) test was not statistically significant (P = 0.1188). Download FIG S8, TIF file, 0.2 MB (197.4KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Calcineurin is conserved widely across pathogenic fungi and is involved in cell wall integrity, morphogenesis, and virulence (20, 21, 61). We have previously shown that in the presence of a calcineurin inhibitor (FK506), Mucor exhibits yeast morphology even under aerobic conditions; however, in strains that are resistant to FK506, Mucor grows as hyphae (27). FKBP12 is the drug receptor for FK506, and when bound, the FKBP12-FK506 complex inhibits the phosphatase activity of calcineurin by binding to the interface of the calcineurin regulatory and catalytic subunits (45). One of the common ways through which most pathogenic microbes become resistant to drugs is that of having mutations in the drug receptor, in this case, FKBP12 or the catalytic A subunit or regulatory B subunit of calcineurin (27). Calo et al. have shown that Mucor can exhibit transient resistance to FK506 by triggering RNA interference specific to the gene encoding FKBP12, resulting in silencing of the drug target gene (51, 52). In this study, we identified a novel resistance mechanism through which Mucor can also become resistant to calcineurin inhibitors. Whole-genome sequencing (WGS) and targeted sequencing of the genomes of spontaneous mutants (CnSp and CSR mutants) revealed DNA sequence alterations in the bycA locus in 31 of 36 resistant isolates (Table 1 and Table 2).
A novel link between calcineurin and BycA, an amino acid permease in Mucor.
On the basis of our sequencing analysis, we hypothesized that deletion of bycA should result in stable resistance to calcineurin inhibitors. Indeed, bycAΔ mutants are resistant to FK506. Unlike cnbRΔ mutants, which grow as yeast, cells of the bycAΔ cnbRΔ mutant fully grow as hyphae such as were seen with the WT (Fig. 3). This verifies that Mucor can become resistant to calcineurin inhibitors as a consequence of the presence of a second mutation in the bycA gene. Mucor has a secondary conserved mechanism that promotes hyphal growth even in the absence of a functional calcineurin. Interestingly, however, we did not observe Mendelian DNA mutations in the bycA locus in 5 (of 36) of the mutants on which we performed sequencing even though they exhibited the same phenotype as the remaining suppressors; one possibility is that Mucor may also produce small RNAi targeting bycA or that there could be another mechanism(s) that confers Mucor resistance against calcineurin inhibitors. Future studies are required to elucidate these mechanisms further.
How is BycA linked to calcineurin and morphogenesis?
It is not known if BycA is a direct posttranslation modification target of calcineurin. However, our data suggest that calcineurin negatively regulates the expression of BycA at the mRNA level. It is also possible that calcineurin is involved in the stability of the bycA mRNA. Further investigation is ongoing to elucidate how calcineurin regulates expression of BycA. Interestingly, Chow et al. revealed that, in Cryptococcus neoformans, a gene encoding a putative amino acid permease (CNAG_01118) is overexpressed when calcineurin is deleted and that this is independent of the presence of Crz1, a well-known target of calcineurin (62). Whether deletion of the amino acid gene would result in suppression of calcineurin mutant phenotypes remains to be tested in C. neoformans.
BycA as a missing link between calcineurin and PKA.
Studies by us and others have shown that Mucor morphology is primarily dependent on (i) respiratory conditions and (ii) calcineurin and PKA activity (27–30, 37–39, 41). Mucor exhibits hyphal growth under aerobic conditions. Under anaerobic conditions with high CO2, Mucor grows as yeast and has PKA activity that is increased via the bicarbonate-cAMP pathway (27, 28, 30). We have previously shown that even under aerobic conditions when calcineurin function is inhibited by FK506 or absent in the cnbRΔ mutant, Mucor grows as a yeast and PKA activity remained high (27). The bicarbonate-cAMP pathway is likely not involved under aerobic conditions as the carbon dioxide levels are very low. It has been suggested that there is an antagonistic relationship between calcineurin and PKA in fungal systems in Ustilago maydis and S. cerevisiae (63, 64).
In this study, we found that calcineurin and PKA are inversely related through BycA and that the overall PKA activity was lower in wild-type hyphae with lower bycA expression but higher in yeast with higher bycA expression (Fig. 4A and C). PKA activity also remained lower in the bycAΔ cnbRΔ double mutant because even though calcineurin function was suppressed, the double mutant did not have BycA function to activate PKA and promote yeast growth. For the same reason, the bycAΔ cnbRΔ double mutants exhibited hyphal morphology. We also found that the amino acid permease BycA was able to activate PKA without a requirement for cAMP. Our finding that BycA can activate PKA is congruent with previous findings in C. albicans and S. cerevisiae, where the general amino acid permease Gap1 can activate PKA in a cAMP-independent manner (57, 58). It is possible that this is achieved through an imbalance between the catalytic and regulatory subunits of PKA. It was previously reported that kelch repeat homolog proteins 1 and 2 (Krh1 and Krh2) promote the association between PKA catalytic and regulatory subunits in S. cerevisiae and that deletion of krh1/2 leads to a lower cAMP requirement for PKA activation (65). Since Krh proteins are evolutionarily conserved in eukaryotes, it is possible that such a mechanism may also exist in Mucor. Our future studies will further focus on determining how BycA activates PKA in Mucor.
Our previous studies showed that cellular PKA activity levels were elevated when calcineurin was inhibited in C. neoformans and Rhizopus delemar (27). How this link is achieved in these fungi remains to be elucidated. There is evidence that higher expression of an amino acid permease in C. neoformans is correlated with the phenotypes regulated by PKA; treatment with urea resulted in a 27-fold increase in the level of an amino acid permease (CNAG_01118) and in increased capsule production, known to be regulated by PKA (66–68). Our current study investigated the links between calcineurin, amino acid permease, and PKA which could also exist in C. neoformans and possibly other fungi.
Calcineurin governs virulence and plays important roles in host-pathogen interactions in Mucor.
Studies of several pathogenic fungi have shown that morphology is linked with expression of virulence factors, colonization of the host, evasion of host immune responses, etc. (69, 70). The cnbRΔ mutants were shown to be significantly less virulent in a heterologous wax moth larva host model (27); however, it was not clear if the diminished virulence potential of cnbRΔ mutants was due to morphology or loss of the calcineurin or both. In the current study, using a hyphal growth strain lacking calcineurin (bycAΔ cnbRΔ double mutant), we identified a novel downstream function of calcineurin that is independent of its function in governing morphology and that contributes to Mucor-host interactions and virulence. For example, macrophages challenged with mutants that lack calcineurin displayed significantly higher phagosome maturation than macrophages challenged with the wild type, irrespective of their morphology (Fig. 5). The ability of Mucor to cause damage to the endothelium was dependent upon the presence of calcineurin (Fig. 6A).
Angiogenesis is the process by which new blood vessels arise from preformed blood vessels. Proteins such as fibroblast growth factor-2 (FGF-2) and vascular endothelial growth factor (VEGF) promote angiogenesis (71). We have previously shown that the ability of C. albicans to induce FGF-2 is morphology dependent, as nonfilamentous strains of C. albicans failed to induce FGF-2 (72). We have also shown that only the WT and not cnbRΔ mutants induced a host FGF-2 response in Mucor (43). In this study, we further found that a bycAΔ cnbRΔ double mutant also failed to induce FGF-2 (Fig. 6B). Our data suggest that the ability of Mucor to induce host FGF-2 is dependent on a novel downstream function of calcineurin which is independent of its function in regulating morphology. Studies by us and others have shown that candidalysin from C. albicans and gliotoxins from Aspergillus fumigatus regulate the host FGF-2 response (72, 73). Hence, it is possible that toxins from Mucor (74, 75) also regulate host FGF-2 response. We are currently working on identifying the Mucor factor(s) that facilitates Mucor interactions with endothelial cells to induce FGF-2 response.
Larvae of Galleria mellonella have largely been used to study virulence and for testing antifungal drugs (27, 47, 74). Wax moth larvae possess an innate immune system that is both structurally and functionally similar to that of mammals at the humoral and cellular levels (76). We have previously shown that cnbRΔ mutants are avirulent in wax moth larva host (27). In this study, we found that bycAΔ cnbRΔ double mutants are also less virulent (Fig. 7). It is congruent that more wild-type spores survived than cnbRΔ yeast during interactions with bone marrow murine macrophages (see Fig. S9 in the supplemental material).
Survival of wild-type spores and cnbRΔ yeast during cocultures with bone marrow macrophages. The spores survived the macrophages at significantly higher levels than were seen with the yeast-locked mutant. The experiment was performed as described previously (K.-W. Jung, Y. Lee, E. Y. Huh, S. C. Lee, et al., mBio 10:e01726-18, 2019, https://doi.org/10.1128/mBio.01726-18). Download FIG S9, TIF file, 0.1 MB (62.2KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Interestingly, unlike in the wax moth host system, the cnbRΔ mutant also caused significant mortality like that seen with the wild type in the pulmonary and systemic murine model of mucormycosis. This observation may be due to the neutropenic host conditions caused by treatment with cyclophosphamide and cortisone. Calcineurin is required to escape innate immune cells by blocking phagosome maturation; however, lack or significant lower numbers of phagocytic cells resulted in higher mortality by the yeast-locked cnbRΔ mutant. In addition, treatment with cyclophosphamide and cortisone acetate could impact the phagocytic potential of the innate immune cells, as it was previously shown that treatment with cortisone acetate resulted in failure of alveolar macrophages to inhibit germination of Rhizopus oryzae (77). Alternatively, Mucor yeast is more immunogenic than Mucor spores and therefore the yeast might have caused mortality in a different manner than the spores (43). This could represent an analogy to the Cryptococcus rim101 mutants (78). The rim101 mutant lacks virulence traits and yet is more immunogenic than the wild type; nevertheless, the mutants still caused mortality in a murine infection model via a substantial host immune response. Further study is required to determine if Mucor cnbRΔ yeast would cause hyperimmune responses in a murine lung infection model.
In summary, we identified a novel mechanism through which Mucor can become resistant to calcineurin inhibitors, where loss-of-function mutations in the bycA gene confer resistance against calcineurin inhibitors. This resistance mechanism is achieved by BycA regulating the activity of protein kinase A via a cAMP-independent pathway that has yet to be elucidated (Fig. 8). As calcineurin is a major virulence factor in many pathogenic fungi, it is worth investigating if this relationship between calcineurin, BycA, and PKA is also conserved in other pathogenic fungal systems. Calcineurin also governs key Mucor-host interactions and is an attractive target for developing antifungals to treat mucormycosis.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were conducted at the University of Texas at San Antonio (UTSA) in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines and in full compliance with the United States Animal Welfare Act (Public Law 98-198) and National Institutes of Health (NIH) guidelines. The animal protocol (MU104) used in this study was approved by the UTSA IACUC. The experiments were conducted in the Division of Laboratory Animal Resources (DLAR) facilities, which are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Fungal strains and growth conditions.
All fungal strains and plasmids used in this study are listed in Table S1A in the supplemental material. For spore production, Mucor strains were inoculated and maintained on yeast extract-peptone-glucose (YPG; 3 g/liter yeast extract, 10 g/liter peptone, 20 g/liter glucose, 2% agar, pH 4.5) or yeast extract-peptone dextrose (YPD; 10 g/liter yeast extract, 20 g/liter peptone, 20 g/liter glucose, 2% agar, pH 6.5) agar at 26°C or 30°C for 4 days. cnbRΔ mutants were maintained on YPD agar at 30°C. For in vitro Mucor-host interaction studies or in vivo survival studies, cnbRΔ mutants were grown overnight in liquid YPD at 30°C with shaking. For high-CO2 conditions, the flasks were entirely filled with YPD broth, and the wild type (R7B) was inoculated on the bottom of the flasks. The flasks were sealed with Parafilm and were left at room temperature overnight without being disturbed. For testing with calcineurin inhibitors, Mucor strains were grown in liquid YPD or on YPD agar plates supplemented with FK506 (Astellas Pharma Inc.) (1 μg/ml) or CsA (LC Laboratories) (2 or 100 μg/ml) at 30°C for 2 to 5 days.
(A) List of strains used in the study. (B) List of primers used in the study. Download Table S1, DOCX file, 0.03 MB (27.2KB, docx) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Generation of suppressor mutants CnSp and CSR.
For generation of CnSp mutants, 103 cells of cnbRΔ mutants were spotted on YPD agar and grown at 30°C for 5 or more days. Hyphal sectors emerging from yeast colonies were propagated on fresh YPD plates. For CSR mutant generation, cnaBΔ mutants were grown on YPD agar containing CsA (2 μg/ml) for 5 days or longer, and the hyphal colonies were propagated further.
Whole-genome sequencing and targeted sequencing of CnsP and CSR mutants.
Genomic DNA of the 12 spontaneous mutant strains and the MU402 strain was used for sequencing. A paired-ended Truseq library was constructed with the genomic DNA and sequenced on an Illumina HiSeq 2000 platform at the University of North Carolina at Chapel Hill School of Medicine. The reads were mapped to the genome of CBS277.49 (79) using the short-read component of the Burrows-Wheeler Aligner (BWA) (80). SNP calling was performed using the Genome Analysis Toolkit (GATK ver. 2.4-9) pipeline and the Unified Genotyper with the haploid setting (81). Whole-genome seqeunce data were deposited at the NCBI Sequence Read Archive (SRA) under accession number PRJNA597331.
Disruption of genes.
The primers used in the study are listed in Table S1B. To disrupt the bycA gene, nearly 1-kb upstream and downstream sequences flanking the bycA gene were PCR amplified (Phusion High-Fidelity DNA polymerase; NEB) using primers SL3 and SL4 and primers SL7 and SL8, respectively. The pyrG-dpl237 marker (53) was amplified using SL5 and SL6 primers. All three fragments were gel excised, and 75 ng of each fragment was amplified using an overlap PCR strategy with primers SL9 and SL10 such that the marker was placed between the flanking regions. The product was cloned into a TOPO vector (pSL26), represent minus signs, please specify what the which was then linearized using SmaI enzyme and transformed into Mucor strain Mu402 (pyrG leuA double mutant) via electroporation (82). The transformants were selected on minimal media at pH = 3.2 with Casamino Acids (MMC; 10 g Casamino Acids, 0.5 g yeast nitrogen base without amino acids and ammonium sulfate, 20 g glucose, 1 mg niacin, 1 mg thiamine, 15 g agar, 1 liter of distilled water [dH2O]) containing 0.5 M sorbitol as described previously (82). Two independent transformants (MSL47.1 and MSL47.2; Table S1A) of 12 were positive for disruption of bycA.
For disruption of the cnbR gene, 5′ and 3′ flanking regions were amplified using SL243 and SL281 and using SL282 and SL244, respectively. A leuA marker was amplified using SCL737 and SCL738. Overlap PCR was performed using primers SCL286 and SCL287. The pSL58 plasmid containing the disruption cassette was linearized and used to transform MSL47.2. The transformants were selected on YNB medium (1.5 g ammonium sulfate, 1.5 g glutamic acid, 0.5 g yeast nitrogen base without amino acids and ammonium sulfate, 10 g glucose, 20 g agar, 1 liter dH2O). Two of 18 independent transformants (MSL68.1 and MSL68.2) were positive for disruption of cnbR.
Reverse transcriptase quantitative PCR (RT-qPCR).
Based on the ORF sequences, we designed primers for bycA (SL346 and SL347) and cnbR (SCL 578 and SCL 579) that span across two exons. Mucor strains were grown overnight in YPD liquid medium, and total RNA was isolated the next day using TRIzol reagent according to the manufacturer’s instructions. 1 μg of RNA from each sample was used for cDNA synthesis (High Capacity cDNA reverse transcription kit with RNase inhibitor; Applied Biosystems), and qPCR was performed using SYBR green QPCR master mix (Thermo Scientific). The actin gene (primers SCL368 and SCL369) served as an internal control. Three independent replications were performed for each experiment with RNA obtained from two independent preparations.
Protein kinase A activity assay.
Mucor strains were grown under appropriate conditions overnight, and collection of crude protein extracts and overall PKA activity measurement were performed using a DetectX PKA (protein kinase A) activity kit according to the manufacturer’s instructions. Briefly, crude protein extracts were obtained by subjecting the samples to activated cell lysis buffer with protease inhibitor cocktail, phenylmethylsulfonyl fluoride (PMSF), and sodium orthovanadate. Each sample was vigorously shaken in a bead beater (five times for 1 min each time with 1 min of cooling) followed by centrifugation to collect supernatants containing crude extracts. Total protein was quantified using the Bradford method (Bio-Rad). A 0.5-μg volume of each sample was used to determine overall PKA activity according to the manufacturer’s instructions.
cAMP determination.
Mucor strains were grown under appropriate conditions overnight, and cells corresponding to 60 mg (wet weight) were flash frozen and immersed in 0.1 M HCl the next day. Crude extracts were obtained as described above. cAMP levels were measured according to the manufacturer’s instructions (Direct cAMP enzyme-linked immunosorbent assay [ELISA] kit; Enzo).
Phagosome maturation assay.
Murine macrophage cell line J774.A1 was maintained in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and a 200 U/ml mixture of penicillin and streptomycin antibiotics at 37°C and 5% CO2. Upon confluence, a cell scraper was used to detach cells, and 5 × 105 cells were plated in each well of a 24-well glass plate. After 24 h, Mucor spores or yeast cells were added at a multiplicity of infection (MOI) of 1 along with 10 μM LysoTracker Green DND-26 and 1 μg/ml Hoechst 33342 stain. A Zeiss Axio Observer microscope was used 30 min later to image fields containing macrophages and Mucor cells. The experiment was performed in triplicate and was repeated on two different occasions.
LDH assay and FGF-2 ELISA.
Primary human umbilical vein endothelial cells (HUVECs) were purchased from Lonza and were maintained in endothelial basal medium (EBM) containing hydrocortisone, ascorbic acid, insulin growth factor, heparin, and FBS at 37°C and 5% CO2 according to the manufacturer’s instructions. Confluent cells were trypsinized, and 5 × 103 cells/well were seeded in a 96-well plate. After 24 h, 5 × 104 of the appropriate fungal cells in a mixture containing PBS (MOI =10) or an equal volume of PBS was added to each well. At 24 h postinfection, the supernatant was collected to quantify LDH levels (CytoTox 96 nonradioactive cytotoxicity assay; Promega) or FGF-2 protein levels using ELISA (R&D Systems). The level of LDH release was calculated as described previously (83).
Virulence test.
For production of Mucor spores, appropriate strains were grown on YPG agar at 26°C for 4 days under light. The cnbRΔ mutant was grown overnight in YPD broth at 30°C with aeration. Both the spores and yeast were washed two times with PBS, and different inoculums ranging from 1 × 104 to 3 × 104 cells in a mixture with 2 μl PBS were injected into the wax moth host through the last left proleg. Differences between the survival curves were evaluated for significance using the Kaplan-Meier test. The experiment was performed on two different occasions with n = 15 animals for each group.
Six-week-old CF1 mice were immunocompromised with cyclophosphamide (250 mg/kg of body weight via the intraperitoneal route) and cortisone acetate (500 mg/kg via the subcutaneous route) every 5 days starting 2 days before inoculation (84). On the day of inoculation, the mice were anaesthetized using isoflurane and 1.5 × 106 spores or yeast cells in a mixture containing PBS were introduced via the intratracheal route (pulmonary), followed by routine monitoring. Two independent experiments were performed with n = 5 for each group. Data represent results from a single experiment.
Statistics.
Prism (Version 7; GraphPad Software Inc.) was used to perform statistical analysis. A P value of ≤0.05 was considered significant.
Data availability.
Whole-genome seqeunce data were deposited at the NCBI Sequence Read Archive (SRA) under accession number PRJNA597331.
ACKNOWLEDGMENTS
We are indebted to Jose Lopez-Ribot, Chiung-Yu Hung, Astrid Cardona, Stephen Saville, Ashraf Ibrahim, Floyd Wormley Jr., and Praveen Juvvadi for valuable discussions. We also thank Anna Averette, Gloria Adedoyin, and Alexis Garcia for technical support for this study.
This work was supported by NIH/NIAID R03 AI119617, a Korean Food Research Institution (KFRI) grant, and UTSA Research funds to S.C.L. S.C.L. holds a Voelcker Fund Young Investigator Pilot Award from the Max and Minnie Tomerlin Voelcker Fund. J.H. was supported by NIH/NIAID R37 AI39115-21, R01 AI50113-15, and P01 AI104533-05. J.H. is codirector and a fellow of the Canadian Institute for Advanced Research Program Fungal Kingdom: Threats & Opportunities.
Footnotes
Citation Vellanki S, Billmyre RB, Lorenzen A, Campbell M, Turner B, Huh EY, Heitman J, Lee SC. 2020. A novel resistance pathway for calcineurin inhibitors in the human-pathogenic Mucorales Mucor circinelloides. mBio 11:e02949-19. https://doi.org/10.1128/mBio.02949-19.
REFERENCES
- 1.Petrikkos G, Skiada A, Lortholary O, Roilides E, Walsh TJ, Kontoyiannis DP. 2012. Epidemiology and clinical manifestations of mucormycosis. Clin Infect Dis 54(Suppl 1):S23–S34. doi: 10.1093/cid/cir866. [DOI] [PubMed] [Google Scholar]
- 2.Baldin C, Ibrahim AS. 2017. Molecular mechanisms of mucormycosis—the bitter and the sweet. PLoS Pathog 13:e1006408. doi: 10.1371/journal.ppat.1006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prabhu RM, Patel R. 2004. Mucormycosis and entomophthoramycosis: a review of the clinical manifestations, diagnosis and treatment. Clin Microbiol Infect 10:31–47. doi: 10.1111/j.1470-9465.2004.00843.x. [DOI] [PubMed] [Google Scholar]
- 4.Eucker J, Sezer O, Graf B, Possinger K. 2001. Mucormycoses. Mycoses 44:253–260. doi: 10.1111/j.1439-0507.2001.00656.x. [DOI] [PubMed] [Google Scholar]
- 5.Ambrosioni J, Bouchuiguir-Wafa K, Garbino J. 2010. Emerging invasive zygomycosis in a tertiary care center: epidemiology and associated risk factors. Int J Infect Dis 14(Suppl 3):e100–e103. doi: 10.1016/j.ijid.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 6.Prakash H, Chakrabarti A. 21 March 2019, posting date Global epidemiology of mucormycosis. J Fungi (Basel) doi: 10.3390/jof5010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saegeman V, Maertens J, Meersseman W, Spriet I, Verbeken E, Lagrou K. 2010. Increasing incidence of mucormycosis in University hospital, Belgium. Emerg Infect Dis 16:1456–1458. doi: 10.3201/eid1609.100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kontoyiannis DP, Yang H, Song J, Kelkar SS, Yang X, Azie N, Harrington R, Fan A, Lee E, Spalding JR. 2016. Prevalence, clinical and economic burden of mucormycosis-related hospitalizations in the United States: a retrospective study. BMC Infect Dis 16:730. doi: 10.1186/s12879-016-2023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres-Narbona M, MYCOMED Zygomycosis Study Group, Guinea J, Martínez-Alarcón J, Muñoz P, Gadea I, Bouza E. 2007. Impact of zygomycosis on microbiology workload: a survey study in Spain. J Clin Microbiol 45:2051–2053. doi: 10.1128/JCM.02473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrikkos G, Skiada A, Drogari-Apiranthitou M. 2014. Epidemiology of mucormycosis in Europe. Clin Microbiol Infect 20(Suppl 6):67–73. doi: 10.1111/1469-0691.12563. [DOI] [PubMed] [Google Scholar]
- 11.Mignogna MD, Fortuna G, Leuci S, Adamo D, Ruoppo E, Siano M, Mariani U. 2011. Mucormycosis in immunocompetent patients: a case-series of patients with maxillary sinus involvement and a critical review of the literature. Int J Infect Dis 15:e533–e540. doi: 10.1016/j.ijid.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Venkatesh D, Dandagi S, Chandrappa PR, Hema KN. 2018. Mucormycosis in immunocompetent patient resulting in extensive maxillary sequestration. J Oral Maxillofac Pathol 22:S112–S116. doi: 10.4103/jomfp.JOMFP_163_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shatriah I, Mohd-Amin N, Tuan-Jaafar TN, Khanna RK, Yunus R, Madhavan M. 2012. Rhino-orbito-cerebral mucormycosis in an immunocompetent patient: case report and review of literature. Middle East Afr J Ophthalmol 19:258–261. doi: 10.4103/0974-9233.95269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pozo Laderas JC, Pontes Moreno A, Pozo Salido C, Robles Arista JC, Linares Sicilia MJ. 2015. Mucormicosis diseminadas en pacientes sin inmunodeficiencias: una enfermedad que también existe. Rev Iberoam Micol 32:63–70. doi: 10.1016/j.riam.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Fanfair R, Benedict K, Bos J, Bennett SD, Lo Y-C, Adebanjo T, Etienne K, Deak E, Derado G, Shieh W-J, Drew C, Zaki S, Sugerman D, Gade L, Thompson EH, Sutton D, Engelthaler D, Schupp J, Brandt ME, Park BJ. 2012. Necrotizing cutaneous mucormycosis after a tornado in Joplin, Missouri, in 2011. N Engl J Med 367:2214–2225. doi: 10.1056/NEJMoa1204781. [DOI] [PubMed] [Google Scholar]
- 16.Muqeetadnan M, Rahman A, Amer S, Nusrat S, Hassan S, Hashmi S. 2012. Pulmonary mucormycosis: an emerging infection. Case Rep Pulmonol 2012:120809. doi: 10.1155/2012/120809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tribble DR, Rodriguez CJ. 2014. Combat-related invasive fungal wound infections. Curr Fungal Infect Rep 8:277–286. doi: 10.1007/s12281-014-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katragkou A, Walsh TJ, Roilides E. 2014. Why is mucormycosis more difficult to cure than more common mycoses? Clin Microbiol Infect 20:74–81. doi: 10.1111/1469-0691.12466. [DOI] [PubMed] [Google Scholar]
- 19.Spellberg B, Edwards J Jr, Ibrahim A. 2005. Novel perspectives on mucormycosis: pathophysiology, presentation, and management. Clin Microbiol Rev 18:556–569. doi: 10.1128/CMR.18.3.556-569.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juvvadi PR, Lee SC, Heitman J, Steinbach WJ. 2017. Calcineurin in fungal virulence and drug resistance: prospects for harnessing targeted inhibition of calcineurin for an antifungal therapeutic approach. Virulence 8:186–197. doi: 10.1080/21505594.2016.1201250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park H, Lee SC, Cardenas ME, Heitman J. 2019. Calcium-calmodulin-calcineurin signalling: a globally conserved virulence cascade in eukaryotic microbial pathogens. Cell Host Microbe 26(4):453–462. doi: 10.1016/j.chom.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J 16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen YL, Lehman VN, Lewit Y, Averette AF, Heitman J. 2013. Calcineurin governs thermotolerance and virulence of Cryptococcus gattii. G3 (Bethesda) 3:527–539. doi: 10.1534/g3.112.004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Del Poeta M, Cruz MC, Cardenas ME, Perfect JR, Heitman J. 2000. Synergistic antifungal activities of bafilomycin A(1), fluconazole, and the pneumocandin MK-0991/caspofungin acetate (L-743,873) with calcineurin inhibitors FK506 and L-685,818 against Cryptococcus neoformans. Antimicrob Agents Chemother 44:739–746. doi: 10.1128/aac.44.3.739-746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blankenship JR, Wormley FL, Boyce MK, Schell WA, Filler SG, Perfect JR, Heitman J. 2003. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot Cell 2:422–430. doi: 10.1128/ec.2.3.422-430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juvvadi PR, Lamoth F, Steinbach WJ. 2014. Calcineurin-mediated regulation of hyphal growth, septation, and virulence in Aspergillus fumigatus. Mycopathologia 178:341–348. doi: 10.1007/s11046-014-9794-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SC, Li A, Calo S, Heitman J. 2013. Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog 9:e1003625. doi: 10.1371/journal.ppat.1003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orlowski M. 1991. Mucor dimorphism. Microbiol Rev 55:234–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartnicki-Garcia S. 1963. Symposium on biochemical bases of morphogenesis in fungi. III. Mold-yeast dimorphism of Mucor. Bacteriol Rev 27:293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartnicki-Garcia S, Nickerson WJ. 1962. Induction of yeast-like development in Mucor by carbon dioxide. J Bacteriol 84:829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartnicki-Garcia S, Nickerson WJ. 1962. Assimilation of carbon dioxide and morphogenesis Mucor rouxii. Biochim Biophys Acta 64:548–551. doi: 10.1016/0006-3002(62)90314-1. [DOI] [PubMed] [Google Scholar]
- 32.Bartnicki-Garcia S, Nickerson WJ. 1962. Nutrition, growth, and morphogenesis of Mucor rouxii. J Bacteriol 84:841–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zorzopulos J, Jobbagy AJ, Terenzi HF. 1973. Effects of ethylenediaminetetraacetate and chloramphenicol on mitochondrial activity and morphogenesis in Mucor rouxii. J Bacteriol 115:1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark-Walker GD. 1973. Relationship between dimorphology and respiration in Mucor genevensis studied with chloramphenicol. J Bacteriol 116:972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito ET, Cihlar RL, Inderlied CB. 1982. Lipid synthesis during morphogenesis of Mucor racemosus. J Bacteriol 152:880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia JR, Hiatt WR, Peters J, Sypherd PS. 1980. S-Adenosylmethionine levels and protein methylation during morphogenesis of Mucor racemosus. J Bacteriol 142:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lubbehusen T, Gonzalez Polo V, Rossi S, Nielsen J, Moreno S, McIntyre M, Arnau J. 2004. Protein kinase A is involved in the control of morphology and branching during aerobic growth of Mucor circinelloides. Microbiology 150:143–150. doi: 10.1099/mic.0.26708-0. [DOI] [PubMed] [Google Scholar]
- 38.Ocampo J, McCormack B, Navarro E, Moreno S, Garre V, Rossi S. 2012. Protein kinase A regulatory subunit isoforms regulate growth and differentiation in Mucor circinelloides: essential role of PKAR4. Eukaryot Cell 11:989–1002. doi: 10.1128/EC.00017-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ocampo J, Fernandez Nuñez L, Silva F, Pereyra E, Moreno S, Garre V, Rossi S. 2009. A subunit of protein kinase A regulates growth and differentiation in the fungus Mucor circinelloides. Eukaryot Cell 8:933–944. doi: 10.1128/EC.00026-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaremberg V, Donella-Deana A, Moreno S. 2000. Mechanism of activation of cAMP-dependent protein kinase: in Mucor rouxii the apparent specific activity of the cAMP-activated holoenzyme is different than that of its free catalytic subunit. Arch Biochem Biophys 381:74–82. doi: 10.1006/abbi.2000.1948. [DOI] [PubMed] [Google Scholar]
- 41.Wolff AM, Appel KF, Petersen JB, Poulsen U, Arnau J. 2002. Identification and analysis of genes involved in the control of dimorphism in Mucor circinelloides (syn. racemosus). FEMS Yeast Res 2:203–213. doi: 10.1111/j.1567-1364.2002.tb00085.x. [DOI] [PubMed] [Google Scholar]
- 42.Singh N, Aguado JM, Bonatti H, Forrest G, Gupta KL, Safdar N, John GT, Pursell KJ, Munoz P, Patel R, Fortun J, Martin-Davila P, Philippe B, Philit F, Tabah A, Terzi N, Chatelet V, Kusne S, Clark N, Blumberg E, Julia MB, Humar A, Houston S, Lass-Florl C, Johnson L, Dubberke ER, Barron MA, Lortholary O. 2009. Zygomycosis in solid organ transplant recipients: a prospective, matched case-control study to assess risks for disease and outcome. J Infect Dis 200:1002–1011. doi: 10.1086/605445. [DOI] [PubMed] [Google Scholar]
- 43.Lee SC, Li A, Calo S, Inoue M, Tonthat NK, Bain JM, Louw J, Shinohara ML, Erwig LP, Schumacher MA, Ko DC, Heitman J. 2015. Calcineurin orchestrates dimorphic transitions, antifungal drug responses and host-pathogen interactions of the pathogenic mucoralean fungus Mucor circinelloides. Mol Microbiol 97:844–865. doi: 10.1111/mmi.13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siekierka JJ, Hung SH, Poe M, Lin CS, Sigal NH. 1989. A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature 341:755–757. doi: 10.1038/341755a0. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Farmer JD Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66:807–815. doi: 10.1016/0092-8674(91)90124-H. [DOI] [PubMed] [Google Scholar]
- 46.Juvvadi PR, Fox D, Bobay BG, Hoy MJ, Gobeil SMC, Venters RA, Chang Z, Lin JJ, Averette AF, Cole DC, Barrington BC, Wheaton JD, Ciofani M, Trzoss M, Li X, Lee SC, Chen Y-L, Mutz M, Spicer LD, Schumacher MA, Heitman J, Steinbach WJ. 2019. Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents. Nat Commun 10:4275. doi: 10.1038/s41467-019-12199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bastidas RJ, Shertz CA, Lee SC, Heitman J, Cardenas ME. 2012. Rapamycin exerts antifungal activity in vitro and in vivo against Mucor circinelloides via FKBP12-dependent inhibition of Tor. Eukaryot Cell 11:270–281. doi: 10.1128/EC.05284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cardenas ME, Muir RS, Breuder T, Heitman J. 1995. Targets of immunophilin-immunosuppressant complexes are distinct highly conserved regions of calcineurin A. EMBO J 14:2772–2783. doi: 10.1002/j.1460-2075.1995.tb07277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fox DS, Cruz MC, Sia RA, Ke H, Cox GM, Cardenas ME, Heitman J. 2001. Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12-FK506 in Cryptococcus neoformans. Mol Microbiol 39:835–849. doi: 10.1046/j.1365-2958.2001.02295.x. [DOI] [PubMed] [Google Scholar]
- 50.Jin L, Harrison SC. 2002. Crystal structure of human calcineurin complexed with cyclosporin A and human cyclophilin. Proc Natl Acad Sci U S A 99:13522–13526. doi: 10.1073/pnas.212504399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calo S, Nicolas FE, Lee SC, Vila A, Cervantes M, Torres-Martinez S, Ruiz-Vazquez RM, Cardenas ME, Heitman J. 2017. A non-canonical RNA degradation pathway suppresses RNAi-dependent epimutations in the human fungal pathogen Mucor circinelloides. PLoS Genet 13:e1006686. doi: 10.1371/journal.pgen.1006686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calo S, Shertz-Wall C, Lee SC, Bastidas RJ, Nicolás FE, Granek JA, Mieczkowski P, Torres-Martínez S, Ruiz-Vázquez RM, Cardenas ME, Heitman J. 2014. Antifungal drug resistance evoked via RNAi-dependent epimutations. Nature 513:555–558. doi: 10.1038/nature13575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia A, Adedoyin G, Heitman J, Lee SC. 2017. Construction of a recyclable genetic marker and serial gene deletions in the human pathogenic Mucorales Mucor circinelloides. G3 (Bethesda) 7:2047–2054. doi: 10.1534/g3.117.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lubbehusen TL, Nielsen J, McIntyre M. 2003. Morphology and physiology of the dimorphic fungus Mucor circinelloides (syn. M. racemosus) during anaerobic growth. Mycol Res 107:223–230. doi: 10.1017/s0953756203007299. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y, Cann MJ, Litvin TN, Iourgenko V, Sinclair ML, Levin LR, Buck J. 2000. Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science 289:625–628. doi: 10.1126/science.289.5479.625. [DOI] [PubMed] [Google Scholar]
- 56.Turnham RE, Scott JD. 2016. Protein kinase A catalytic subunit isoform PRKACA; history, function and physiology. Gene 577:101–108. doi: 10.1016/j.gene.2015.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Donaton MC, Holsbeeks I, Lagatie O, Van Zeebroeck G, Crauwels M, Winderickx J, Thevelein JM. 2003. The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol Microbiol 50:911–929. doi: 10.1046/j.1365-2958.2003.03732.x. [DOI] [PubMed] [Google Scholar]
- 58.Kraidlova L, Van Zeebroeck G, Van Dijck P, Sychrová H. 2011. The Candida albicans GAP gene family encodes permeases involved in general and specific amino acid uptake and sensing. Eukaryot Cell 10:1219–1229. doi: 10.1128/EC.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ibrahim AS, Spellberg B, Walsh TJ, Kontoyiannis DP. 2012. Pathogenesis of mucormycosis. Clin Infect Dis 54:S16–S22. doi: 10.1093/cid/cir865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L, eMERGE Network, Pittman KJ, Barker JR, Salinas RE, Stanaway IB, Williams GD, Carroll RJ, Balmat T, Ingham A, Gopalakrishnan AM, Gibbs KD, Antonia AL, Heitman J, Lee SC, Jarvik GP, Denny JC, Horner SM, DeLong MR, Valdivia RH, Crosslin DR, Ko DC. 2018. An atlas of genetic variation linking pathogen-induced cellular traits to human disease. Cell Host Microbe 24:308–323.e6. doi: 10.1016/j.chom.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Juvvadi PR, Lamoth F, Steinbach WJ. 2014. Calcineurin as a multifunctional regulator: unraveling novel functions in fungal stress responses, hyphal growth, drug resistance, and pathogenesis. Fungal Biol Rev 28:56–69. doi: 10.1016/j.fbr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chow EW, Clancey SA, Billmyre RB, Averette AF, Granek JA, Mieczkowski P, Cardenas ME, Heitman J. 2017. Elucidation of the calcineurin-Crz1 stress response transcriptional network in the human fungal pathogen Cryptococcus neoformans. PLoS Genet 13:e1006667. doi: 10.1371/journal.pgen.1006667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Egan JD, Garcia-Pedrajas MD, Andrews DL, Gold SE. 2009. Calcineurin is an antagonist to PKA protein phosphorylation required for postmating filamentation and virulence, while PP2A is required for viability in Ustilago maydis. Mol Plant Microbe Interact 22:1293–1301. doi: 10.1094/MPMI-22-10-1293. [DOI] [PubMed] [Google Scholar]
- 64.Kafadar KA, Cyert MS. 2004. Integration of stress responses: modulation of calcineurin signaling in Saccharomyces cerevisiae by protein kinase A. Eukaryot Cell 3:1147–1153. doi: 10.1128/EC.3.5.1147-1153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peeters T, Louwet W, Geladé R, Nauwelaers D, Thevelein JM, Versele M. 2006. Kelch-repeat proteins interacting with the Gα protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc Natl Acad Sci U S A 103:13034–13039. doi: 10.1073/pnas.0509644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frazzitta AE, Vora H, Price MS, Tenor JL, Betancourt-Quiroz M, Toffaletti DL, Cheng N, Perfect JR. 2013. Nitrogen source-dependent capsule induction in human-pathogenic Cryptococcus species. Eukaryot Cell 12:1439–1450. doi: 10.1128/EC.00169-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, Alspaugh JA. 2010. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog 6:e1000776. doi: 10.1371/journal.ppat.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alspaugh JA. 2016. Unveiling protein kinase A targets in Cryptococcus neoformans capsule formation. mBio 7:e00021-16. doi: 10.1128/mBio.00021-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boyce KJ, Andrianopoulos A. 2015. Fungal dimorphism: the switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host. FEMS Microbiol Rev 39:797–811. doi: 10.1093/femsre/fuv035. [DOI] [PubMed] [Google Scholar]
- 70.Gauthier GM. 2017. Fungal dimorphism and virulence: molecular mechanisms for temperature adaptation, immune evasion, and in vivo survival. Mediators Inflamm 2017:8491383. doi: 10.1155/2017/8491383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoo SY, Kwon SM. 2013. Angiogenesis and its therapeutic opportunities. Mediators Inflamm 2013:127170. doi: 10.1155/2013/127170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vellanki S, Huh EY, Saville SP, Lee SC. 5 March 2019, posting date Candida albicans morphology-dependent host FGF-2 response as a potential therapeutic target. J Fungi (Basel) doi: 10.3390/jof5010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ben-Ami R, Lewis RE, Leventakos K, Kontoyiannis DP. 2009. Aspergillus fumigatus inhibits angiogenesis through the production of gliotoxin and other secondary metabolites. Blood 114:5393–5399. doi: 10.1182/blood-2009-07-231209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee SC, Billmyre RB, Li A, Carson S, Sykes SM, Huh EY, Mieczkowski P, Ko DC, Cuomo CA, Heitman J. 2014. Analysis of a food-borne fungal pathogen outbreak: virulence and genome of a Mucor circinelloides isolate from yogurt. mBio 5:e01390-14. doi: 10.1128/mBio.01390-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mueller KD, Zhang H, Serrano CR, Billmyre RB, Huh EY, Wiemann P, Keller NP, Wang Y, Heitman J, Lee SC. 2019. Gastrointestinal microbiota alteration induced by Mucor circinelloides in a murine model. J Microbiol 57:509–520. doi: 10.1007/s12275-019-8682-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Desbois AP, Coote PJ. 2012. Utility of greater wax moth larva (Galleria mellonella) for evaluating the toxicity and efficacy of new antimicrobial agents. Adv Appl Microbiol 78:25–53. doi: 10.1016/B978-0-12-394805-2.00002-6. [DOI] [PubMed] [Google Scholar]
- 77.Waldorf AR, Levitz SM, Diamond RD. 1984. In vivo bronchoalveolar macrophage defense against Rhizopus oryzae and Aspergillus fumigatus. J Infect Dis 150:752–760. doi: 10.1093/infdis/150.5.752. [DOI] [PubMed] [Google Scholar]
- 78.O’Meara TR, Holmer SM, Selvig K, Dietrich F, Alspaugh JA. 2013. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. mBio 4:e00522-12. doi: 10.1128/mBio.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corrochano LM, Kuo A, Marcet-Houben M, Polaino S, Salamov A, Villalobos-Escobedo JM, Grimwood J, Álvarez MI, Avalos J, Bauer D, Benito EP, Benoit I, Burger G, Camino LP, Cánovas D, Cerdá-Olmedo E, Cheng J-F, Domínguez A, Eliáš M, Eslava AP, Glaser F, Gutiérrez G, Heitman J, Henrissat B, Iturriaga EA, Lang BF, Lavín JL, Lee SC, Li W, Lindquist E, López-García S, Luque EM, Marcos AT, Martin J, McCluskey K, Medina HR, Miralles-Durán A, Miyazaki A, Muñoz-Torres E, Oguiza JA, Ohm RA, Olmedo M, Orejas M, Ortiz-Castellanos L, Pisabarro AG, Rodríguez-Romero J, Ruiz-Herrera J, Ruiz-Vázquez R, Sanz C, Schackwitz W, et al. 2016. Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr Biol 26:1577–1584. doi: 10.1016/j.cub.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lopman B, Vennema H, Kohli E, Pothier P, Sanchez A, Negredo A, Buesa J, Schreier E, Reacher M, Brown D, Gray J, Iturriza M, Gallimore C, Bottiger B, Hedlund KO, Torven M, von Bonsdorff CH, Maunula L, Poljsak-Prijatelj M, Zimsek J, Reuter G, Szucs G, Melegh B, Svennson L, van Duijnhoven Y, Koopmans M. 2004. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363:682–688. doi: 10.1016/S0140-6736(04)15641-9. [DOI] [PubMed] [Google Scholar]
- 81.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vellanki S, Navarro-Mendoza MI, Garcia A, Murcia L, Perez-Arques C, Garre V, Nicolas FE, Lee SC. 2018. Mucor circinelloides: growth, maintenance, and genetic manipulation. Curr Protoc Microbiol 49:e53. doi: 10.1002/cpmc.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watkins TN, Gebremariam T, Swidergall M, Shetty AC, Graf KT, Alqarihi A, Alkhazraji S, Alsaadi AI, Edwards VL, Filler SG, Ibrahim AS, Bruno VM, Watkins TN, Gebremariam T, Swidergall M, Shetty AC, Graf KT, Alqarihi A, Alkhazraji S, Alsaadi AI, Edwards VL, Filler SG, Ibrahim AS, Bruno VM. 2018. Inhibition of EGFR signaling protects from mucormycosis. mBio 9:e01384-18. doi: 10.1128/mBio.01384-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luo G, Gebremariam T, Lee H, French SW, Wiederhold NP, Patterson TF, Filler SG, Ibrahim AS. 2013. Efficacy of liposomal amphotericin B and posaconazole in intratracheal models of murine mucormycosis. Antimicrob Agents Chemother 57:3340–3347. doi: 10.1128/AAC.00313-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Emergence of hyphal sectors from cnbRΔ mutants and isolation of calcineurin suppressor mutants. (A) cnbRΔ mutants were plated on YPD agar for 5 or more days at 30°C. (B) The hyphae emerging from the yeast colonies were transferred to fresh YPD agar for further propagation. (C) Calcineurin suppressor (CnSP) mutants produced hyphae decorated with sporangiophores containing sporangiospores like the WT results (scale bar = 20 μm). Download FIG S1, TIF file, 2.4 MB (2.5MB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Emergence of hyphal sectors from the cnaBΔ mutant grown on CsA-containing medium. cnaBΔ mutants were grown in the presence of CsA (2 μg/ml) for 5 or more days at 30°C. This resulted in the emergence of hyphal sectors from yeast colonies. Download FIG S2, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Confirmation of disruption of the bycA gene by junction PCR and ORF-spanning PCR. (A) Illustration of the bycAΔ::pyrG-dpl237 and bycA alleles with ∼1-kb upstream and downstream flanking sequence. P1 (SL3) and P4 (SL8) recognize sequences outside the disruption cassette. P2 (SCL566) and P3 (SCL567) recognize pyrG-dpl237. P5 (SL182) and P6 (SL183) recognize bycA. (B) At the 5′ end, P1 and P2 amplified a 2,203-bp region, and at the 3′ end, P3 and P4 amplified a 1,994-bp region in bycAΔ::pyrG-dpl237. The primer pairs did not produce a fragment in the WT. P5 and P6 amplified a 690-bp region within the bycA gene, so no amplification was noted in the mutants. Images are not to scale. Download FIG S3, TIF file, 0.4 MB (471.6KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Confirmation of disruption of the cnbR gene in the bycAΔ background by junction PCR and ORF-spanning PCR. (A) Illustration of the cnbRΔ::leuA deletion in the bycAΔ::pyrG-dpl237 strain and cnbR alleles with ∼1-kb upstream and downstream flanking sequence. P1 (SL243) and P4 (SL244) recognize sequences outside the disruption cassette. P2 (SL391) and P3 (SL392) are specific for leuA. P5 (SCL578) and P6 (SCL579) are specific for cnbR. (B) At the 5′ end, P1 and P2 amplified a 2,388-bp region, and at the 3′ end, P3 and P4 amplified a 1,620-bp region in the cnbRΔ::leuA region. The primer pair did not produce a fragment in the WT. P1 and P4 produced a 5,711-bp product in the mutant and a 3,342-bp product in the WT. P5 and P6 amplified a 170-bp region within the cnbR gene, so no amplification is noted in the mutants. Image is not to scale. Download FIG S4, TIF file, 0.7 MB (745.7KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Confirmation of disruption of the cnbR gene in the bycAΔ background by RT-qPCR. Primers SCL578 and SCL579 were used to perform qPCR on the cDNA obtained from the WT and mutants (see Materials and Methods). Actin (SCL368 and SCL369) served as a control. No cnbR expression was detected in the bycAΔ cnbRΔ double mutant. Download FIG S5, TIF file, 0.1 MB (139.4KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predicted structure of BycA showing 10 transmembrane domains and 1 cytosolic domain. BycA is predicted to have 10 transmembrane domains and 1 cytosolic domain. The transmembrane domains were predicted with the TMHMM 2.0 software package (http://www.cbs.dtu.dk/services/TMHMM/). The predicted cytosolic region contains a pectinesterase domain. Download FIG S6, TIF file, 0.2 MB (219.8KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A bycAΔ cnbRΔ double mutant is lethal in a wax moth larva host only at higher inocula. The wax moth larvae (n = 15/group) were inoculated with 1 × 104 (1×) to 3 × 104 (3×) bycAΔ cnbRΔ double mutant or WT spores (1×) in a mixture containing 2 μl PBS via the last left proleg and were monitored for survival. All wax moth challenged with the WT or the bycAΔ cnbRΔ mutant (3×) succumbed to mortality within a week. A total of 65% of the wax moths inoculated with the bycAΔ cnbRΔ mutant (1×) and 45% of the wax moths inoculated with the bycAΔ cnbRΔ mutant (2×) survived. Results obtained with a log rank (Mantel-Cox) test were statistically significant (P < 0.0001). A pairwise comparison was also performed with the following groups: WT versus bycAΔ cnbRΔ mutant (1×) (*, P < 0.0001); WT versus bycAΔ cnbRΔ mutant (2×) (*, P = 0.0014); WT versus bycAΔ cnbRΔ (3×) (P = 0.9460). Download FIG S7, TIF file, 0.1 MB (91.8KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mucor virulence in the murine model of systemic and pulmonary mucormycosis. (A) Cyclophosphamide- and cortisone acetate-treated CF1 mice were challenged with 1.5 × 106 spores or yeast cells in a mixture containing PBS via lateral tail vein (systemic mucormycosis model). The animals (n = 5/group) were monitored for survival. Results from a log rank (Mantel-Cox) test were not statistically significant (P = 0.0585), suggesting no major difference in survival between the WT and calcineurin mutant groups. (B) For the pulmonary mucormycosis model (n = 5/group), the same inocula were introduced via intratracheal route and the animals were monitored for survival. Results of a log rank (Mantel-Cox) test was not statistically significant (P = 0.1188). Download FIG S8, TIF file, 0.2 MB (197.4KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Survival of wild-type spores and cnbRΔ yeast during cocultures with bone marrow macrophages. The spores survived the macrophages at significantly higher levels than were seen with the yeast-locked mutant. The experiment was performed as described previously (K.-W. Jung, Y. Lee, E. Y. Huh, S. C. Lee, et al., mBio 10:e01726-18, 2019, https://doi.org/10.1128/mBio.01726-18). Download FIG S9, TIF file, 0.1 MB (62.2KB, tif) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) List of strains used in the study. (B) List of primers used in the study. Download Table S1, DOCX file, 0.03 MB (27.2KB, docx) .
Copyright © 2020 Vellanki et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Whole-genome seqeunce data were deposited at the NCBI Sequence Read Archive (SRA) under accession number PRJNA597331.