Legionella pneumophila is an intracellular human pathogen that utilizes amoebae as its environmental host. The adaptation of L. pneumophila to the intracellular environment requires coordination of expression of its multicomponent pathogenesis system, which is composed of a secretion system and effector proteins. However, the regulatory factors controlling the expression of this pathogenesis system are only partially uncovered. Here, we discovered a novel regulatory system that is activated by copper and controls the expression of a single effector protein. The genes encoding both the regulatory system and the effector protein are located on a genomic island that undergoes horizontal gene transfer within the Legionella genus. This regulator-effector genomic island represents the first reported case of local regulation of effectors in Legionella. The discovery of this regulatory mechanism is an important step forward in the understanding of how the regulatory network of effectors functions and evolves in the Legionella genus.
KEYWORDS: Legionella, effectors, copper, signal, horizontal gene transfer, two-component system
ABSTRACT
The intracellular pathogen Legionella pneumophila utilizes the Icm/Dot type IV secretion system to translocate >300 effector proteins into host cells during infection. The regulation of some of these effector-encoding genes was previously shown to be coordinated by several global regulators, including three two-component systems (TCSs) found in all the Legionella species examined. Here, we describe the first Legionella genomic island encoding a single Icm/Dot effector and a dedicated TCS, which regulates its expression. This genomic island, which we named Lci, undergoes horizontal gene transfer in the Legionella genus, and the TCS encoded from this island (LciRS) is homologous to TCSs that control the expression of various metal resistance systems found in other bacteria. We found that the L. pneumophila sensor histidine kinase LciS is specifically activated by copper via a unique, small periplasmic sensing domain. Upon activation by LciS, the response regulator LciR directly binds to a conserved regulatory element and activates the expression of the adjacently located lciE effector-encoding gene. Thus, LciR represents the first local regulator of effectors identified in L. pneumophila. Moreover, we found that the expression of the lciRS operon is repressed by the Fis1 and Fis3 regulators, leading to Fis-mediated effects on copper induction of LciE and silencing of the expression of this genomic island in the absence of copper. This island represents a novel type of effector regulation in Legionella, shedding new light on the ways by which the Legionella pathogenesis system evolves its effector repertoire and expands its activating signals.
INTRODUCTION
Legionella pneumophila is an intracellular human pathogen that multiplies within alveolar macrophages and causes a severe pneumonia known as Legionnaires’ disease (1–3). In the environment, L. pneumophila thrives in many different protozoan cells (4–6), which serve as its training grounds for pathogenesis (7). Inside its eukaryotic host cells, the bacterium remodels its phagosome to generate the Legionella-containing vacuole (LCV) (8, 9). Establishment of the LCV depends on the Icm/Dot type IV secretion system, which delivers more than 300 effector proteins, which modulate numerous host-cell functions during infection (10–14). The enormous number of effectors that participate in LCV establishment and the various host cell pathways manipulated by L. pneumophila effectors (15–17) imply that a successful infection will require different levels of coordination among the effectors, including on the level of gene expression.
To date, five regulatory systems have been shown to directly regulate the expression of effector-encoding genes (EEGs) in L. pneumophila: (i) the PmrAB two-component system (TCS) activates the expression of about 40 EEGs (18, 19); (ii) the CpxRA TCS activates or represses the expression of about 30 EEGs and also regulates the expression of several icm/dot genes (20–23); (iii) the LetAS-RsmYZ-CsrA regulatory cascade represses the expression of about 40 EEGs during exponential phase (24–31); (iv) two Fis regulators repress the expression of about 20 EEGs during exponential phase (32); and (v) the Fur regulator controls the expression of a single EEG (mavN) as well as several other proteins involved in iron acquisition (33–37). In addition, these regulatory systems have been shown to assemble into an interconnected regulatory network using accessory components, such as modulators (AckA-Pta and PTSNtr [22, 38]) and connectors (LetE and LerC [22, 39, 40]). All the direct regulators of EEGs described above (PmrA, CpxR, CsrA, Fis1, Fis3, and Fur) were found to be present in all the Legionella species examined (41, 42), and they function as global regulators that regulate the expression of a large number of genes, including EEGs, scattered throughout the L. pneumophila genome (18–20, 22). Besides global regulators, local regulators that regulate the expression of a small number of adjacent genes are also common in many bacterial systems (43, 44), including pathogenesis-related genes in Vibrio cholerae and Salmonella enterica (45). Local regulators were shown to function as either activators or repressors, and in many cases, they are found in genomic islands together with the genes they regulate (46–49).
Genomic islands are genetic elements acquired via horizontal gene transfer (HGT) that include sets of genes that encode proteins that may be beneficial for the bacteria under certain conditions (50, 51). Many pathogenicity islands have been described in bacteria, some of which contain a large number of genes and encode complete pathogenesis systems (50, 52). For example, the Salmonella enterica pathogenicity island 2 (SPI-2) encodes the complex components of a type III system, effector proteins, and the SsrAB TCS, which functions as a local regulator and coordinates their expression (53). Smaller pathogenicity islands that encode a few proteins are sometimes referred to as islets (50). For example, the Streptococcus pneumoniae RlrA pathogenicity islet encodes the RlrA local transcriptional regulator, which controls the expression of genes located on the same islet that are essential for lung infection (54). Another example is the Listeria monocytogenes virulence gene cluster that encodes phospholipases, listeriolysin, metalloprotease, the ActA protein, and the PrfA transcriptional regulator, which controls their expression as well as the expression of genes outside the island (55).
All known L. pneumophila TCSs that regulate the expression of EEGs are global regulators found in all the Legionella species sequenced. They are not part of genomic islands, and the signal sensed by their cognate sensor histidine kinases (SHKs) is unknown. Here, we describe a novel L. pneumophila TCS effector island. The TCS is composed of the LciS SHK, which specifically senses copper and activates the cognate LciR response regulator (RR). LciR functions as a local regulator, activating the expression of a single adjacent EEG (lciE). The LciRS-LciE genomic island undergoes HGT throughout the Legionella genus and represents a novel type of effector regulation in Legionella.
RESULTS
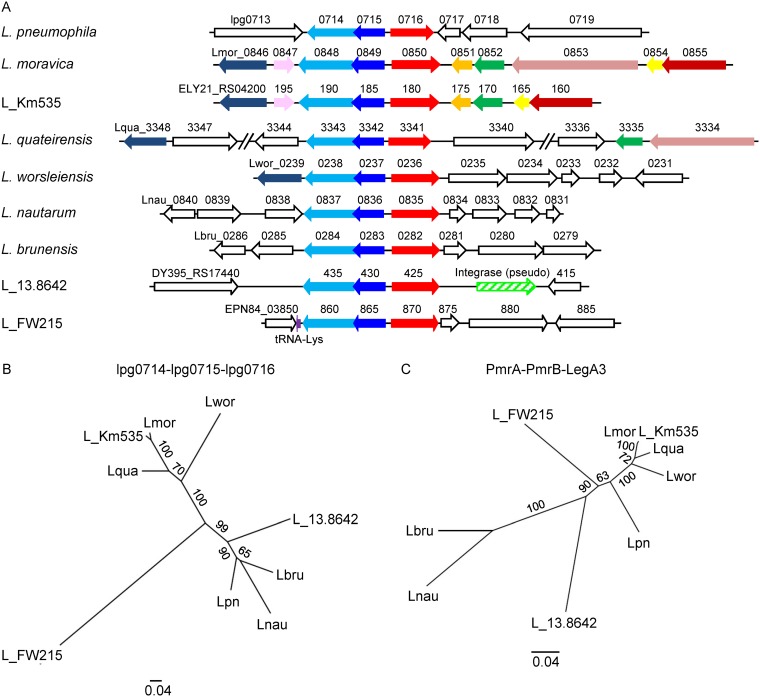
The CpxR and PmrA direct regulators of effector-encoding genes (EEGs) belong to the winged helix-turn-helix (wHTH) family of response regulators (RRs), which function as part of the CpxRA and PmrAB two-component systems (TCSs) (18–20, 22, 23). L. pneumophila harbors a third TCS from the same family, consisting of lpg0714, which encodes a sensor-histidine kinase (SHK), and lpg0715, which encodes a wHTH-type RR. This TCS is found in L. pneumophila, in five other characterized Legionella species, and in three uncharacterized Legionella species. In all these species, a gene, which was shown in L. pneumophila to encode an effector protein (56) (lpg0716 in L. pneumophila), is located next to it, forming a regulator-effector island (Fig. 1A; see also Fig. S1 in the supplemental material).
FIG 1.
Regulator-effector island undergoes HGT in the Legionella genus. (A) Schematic presentation of the genes located in the genomic region near the lpg0714-lpg0715-lpg0716 orthologs in six characterized Legionella species, L. pneumophila (Lpg), L. moravica (Lmor), L. quateirensis (Lqua), L. worsleiensis (Lwor), L. nautarum (Lnau), and L. brunensis (Lbru), and three uncharacterized Legionella species, Legionella sp. strain 13.8642 (L_13.8642), Legionella sp. strain FW215 (L_FW215), and Legionella sp. strain Km535 (L_Km535), harboring the regulator-effector island. Homologous genes are marked by the same color, and nonhomologous genes are marked in white. The genes are indicated by their locus tag number. The integrase pseudogene is marked with a hatched arrow, and the Lys tRNA gene is marked by a purple arrow. (B) A maximum-likelihood phylogeny tree of nine Legionella species harboring the regulator-effector island reconstructed on the basis of concatenated amino acid alignment of the lpg0714-lpg0715-lpg0716 orthologous open reading frames. (C) Similarly constructed phylogeny tree of the PmrA-PmrB and LegA3 orthologous ORFs. Bootstrap values are presented on the branches.
Presence and absence of lpg0714-lpg0715-lpg0716 in the Legionella genus. A maximum likelihood tree of 41 Legionella species was reconstructed on the basis of concatenated amino acid alignment of 78 orthologous open reading frames (41). For each species, the presence (grey) or absence (white) of lpg0714, lpg0715, and lpg0716 genes is indicated. Download FIG S1, TIF file, 0.9 MB (907.2KB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The lpg0714-lpg0715-lpg0716 island undergoes horizontal gene transfer in the Legionella genus.
Comparison of the genomic location of the regulator-effector island in the nine Legionella species in which it was found indicated that it is usually located in different positions (Fig. 1A). However, in genomes of the four closely related species (L. worsleiensis, L. moravica, L. quateirensis, and Legionella sp. strain Km535), it is positioned in a similar genomic region, which underwent additional changes in its gene content (Fig. 1A). Comparison of the genomic region of this regulator-effector island is found in L. pneumophila and in the other eight Legionella species revealed that it contains highly variable genes (Fig. S2A), indicating that it is prone to changes in gene content. To examine whether this regulator-effector island undergoes HGT in the Legionella genus, we reconstructed the phylogenetic tree of these nine species based on the protein sequences encoded by the lpg0714-lpg0715 TCS and the lpg0716 effector and compared it to the phylogenetic tree reconstructed using a TCS (PmrAB) and a core effector (LegA3) present in all the Legionella species. While the analysis using PmrAB-LegA3 resulted in a tree structure similar to that of the known Legionella phylogenetic tree (as far as the characterized Legionella species are concerned [41]), the tree based on the regulator-effector island resulted in a different topology (Fig. 1B). Moreover, while the GC content of the pmrAB-legA3 DNA sequence was similar to the genomic GC content in all nine species (Fig. S2B), the GC content of the regulator-effector island in three of the species (L. pneumophila, L. brunensis, and L. nautarum) was considerably lower, suggesting a recent event of HGT in these species. The genomic position, the GC content, and the tree structure of L. worsleiensis, L. moravica, L. quateirensis, and Legionella sp. strain Km535 imply a single HGT event, which occurred before their speciation. Collectively, these results suggest that this regulator-effector island undergoes HGT in the Legionella genus as a unit.
Regulator-effector island undergoes HGT in the Legionella genus. (A) Schematic presentation of the genomic region where the lpg0714-lpg0715-lpg0716 L. pneumophila orthologs are located in the six characterized Legionella species, L. pneumophila (Lpn), L. moravica (Lmor), L. quateirensis (Lqua), L. worsleiensis (Lwor), L. nautarum (Lnau), and L. brunensis (Lbru), and the three uncharacterized Legionella species, Legionella sp. strain 13.8642 (L_13.8642), Legionella sp. strain FW215 (L_FW215), and Legionella sp. strain Km535 (L_Km535), harboring the regulator-effector island. Homologous genes are indicated by the same color, and nonhomologous genes are white. The regions between lpg0713, encoding an oligopeptide transporter, and lpg0719, encoding a valyl-tRNA synthetase in the nine species, are presented. The genes are indicated by their locus tag number. (B) Comparison of the GC content of the lpg0714-lpg0715-lpg0716 genes and the pmrA-pmrB-legA3 genes to the genomic GC content of the nine Legionella species harboring the regulator-effector island. Download FIG S2, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
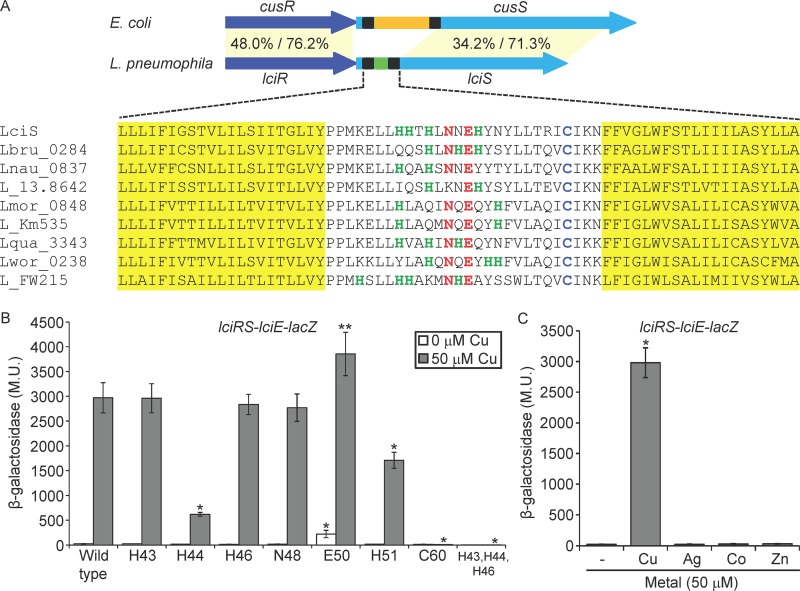
The L. pneumophila lpg0714-lpg0715 TCS is homologous to TCSs involved in copper sensing in other bacteria.
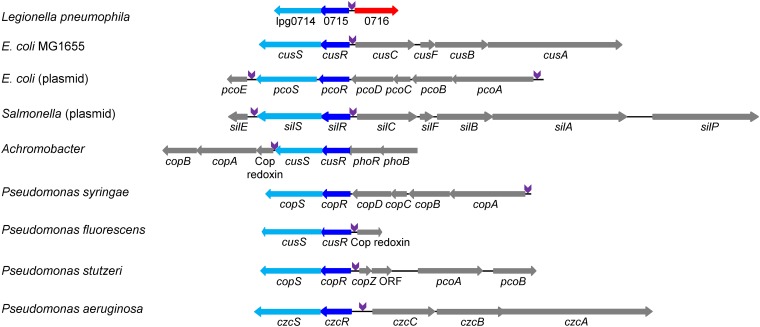
Examination of the proteins encoded by lpg0714 and lpg0715 indicated that homologous TCSs are present in many bacteria. In most of them, the genes located next to it encode different proteins and systems involved in copper resistance (Fig. 2); however, cases of genes encoding resistance to silver and zinc were also described (57, 58). A few of these systems were studied before, including the CusRS TCS of Escherichia coli, which is located next to the CusCFBA copper transport system (59, 60). Examination of the lpg0716 EEG and of all other genes located next to the homologous TCSs from the different bacteria indicated that a conserved sequence is found at a precise distance from the predicted or validated −10 promoter elements in one or two of the surrounding genes (Fig. 2 and Fig. S3).
FIG 2.
Two-component systems homologous to lpg0714-lpg0715 are present in many bacteria and are located next to genes encoding metal resistance systems. Schematic representation of genomic regions containing TCSs homologous to lpg0714-lpg0715 found in different bacteria. The homologous response regulators (RRs) are shown in dark blue, the homologous sensor histidine kinases (SHKs) are shown in light blue, the effector lpg0716 is shown in red, and the genes involved in copper resistance, or in resistance to other metals, are shown in gray (most of these genes are not homologous to one another). The position of the conserved regulatory element predicted to be recognized by the lpg0715 RR is shown in purple (Fig. S3). The different designations of the homologous RRs and SHKs are indicated.
Consensus regulatory element of genes found next to the LciRS homologous TCSs in different bacteria. The regulatory region of genes located near the genes encoding the LciRS homologous TCS in different bacteria is shown: L. pneumophila (Lpg), L. moravica (Lmor), L. quateirensis (Lqua), L. worsleiensis (Lwor), L. nautarum (Lnau), L. brunensis (Lbru), Legionella sp. strain 13.8642 (L_13.8642), Legionella sp. strain FW215 (L_FW215), Legionella sp. strain Km535 (L_Km535), Escherichia coli (Esc), Salmonella enterica (Sal), Enterobacter cloacae (Ent), Achromobacter species (Ach), Pseudomonas aeruginosa (Pa), Pseudomonas fluorescens (Pf), Pseudomonas putida (Pp), Pseudomonas stutzeri (Ps), and Bordetella pertussis (Bper). In purple are the putative LciR inverted-repeat regulatory elements, in light blue are the two additional regulatory elements found between the inverted-repeat and the −10 promoter element, in dark blue are the −10 promoter elements, and the experimentally determined transcription start sites (TSSs) are underlined and marked in blue. Download FIG S3, TIF file, 1.5 MB (1.6MB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Due to the homology of this TCS and its predicted regulatory element to systems involved in copper sensing and detoxification, we named the L. pneumophila lpg0714-lpg0716 genes lci, for Legionella copper island. The SHK lpg0714 was named LciS, the RR lpg0715 was named LciR, and the effector lpg0716 was named LciE.
lciE is induced by copper in L. pneumophila in an LciRS-dependent manner.
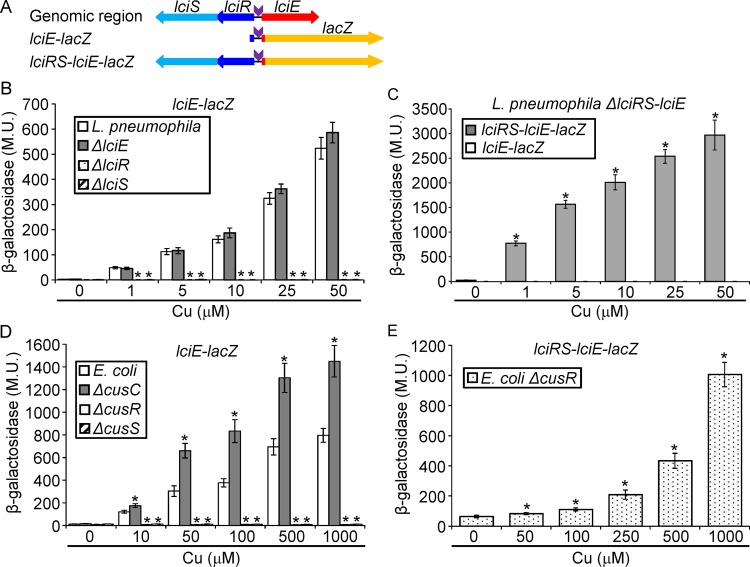
To determine whether lciE is induced in response to copper exposure, two fusions were constructed (Fig. 3A). The first fusion contains the 300-bp regulatory region of lciE fused to lacZ (designated lciE-lacZ). The second construct contains the same fusion as well as the lciRS genes in their original genomic organization (designated lciRS-lciE-lacZ) (Fig. 3A). To examine the expression of these constructs in response to copper exposure, we first determined the concentrations of copper that L. pneumophila can tolerate (Fig. S4A) and examined the effect of copper on the expression of lciE using a range of concentrations, from 1 to 50 μM. Using the lciE-lacZ fusion, the level of expression of lciE increased gradually as the concentration of copper was elevated, reaching more than 100-fold induction at the highest copper concentration (Fig. 3B). To determine the importance of the LciRS TCS for LciE expression, four deletion mutants were constructed, lciR, lciS, and lciE single deletion mutants and an lciRS-lciE triple deletion mutant. None of these mutants had an intracellular growth phenotype when examined in the amoeba host Acanthamoeba castellanii and in HL-60-derived human macrophages, as well as in a competition assay in amoeba with and without the addition of copper (Fig. S5). The copper induction of lciE was found to be completely dependent on the lciR and lciS genes, and as expected, the lciE deletion did not affect the levels of expression of the lciE-lacZ fusion (Fig. 3B). To further substantiate these results, copper induction was examined in an L. pneumophila strain deleted for the entire lciRS-lciE region (Fig. 3C). Using the lciE-lacZ fusion (Fig. 3A), no induction by copper was obtained, but when the lciRS-lciE-lacZ fusion (Fig. 3A) was examined, high levels of expression were obtained due to exposure to copper, which increased gradually as the concentrations of copper increased (Fig. 3C). In addition, the expression of lciE was found to be completely dependent on the presence of a functional LciRS TCS, since no induction was obtained when the conserved histidine of the LciS SHK and conserved aspartic acid of the LciR RR were mutated (Fig. S6A). Collectively, these results indicate that the expression of the Icm/Dot effector LciE is activated by copper, and its activation is completely dependent on the presence and functionality of the LciRS TCS.
FIG 3.
lciE is induced by copper in L. pneumophila and E. coli in a TCS-dependent manner. (A) Schematic representation of the L. pneumophila lciRS-lciE genomic region and the two lacZ fusions constructed (lciE-lacZ and lciRS-lciE-lacZ) to determine the expression of lciE. (B) The expression of the lciE-lacZ fusion was examined in the wild-type strain (white bars), in the lciE deletion mutant (gray bars), in the lciR deletion mutant (dotted bars), and in the lciS deletion mutant (hatched bars) at the stationary phase under different copper concentrations (indicated below the bars). (C) The expression of the lciE-lacZ fusion (white bars) and the lciRS-lciE-lacZ fusion (gray bars) was examined in the L. pneumophila triple lciRS-lciE deletion mutant at the stationary phase under different copper concentrations (indicated below the bars). (D) The expression of the lciE-lacZ fusion was examined in E. coli (white bars), in the E. coli cusC deletion mutant (gray bars), in the E. coli cusR deletion mutant (dotted bars), and in the cusS deletion mutant (hatched bars) at the stationary phase under different copper concentrations (indicated below the bars). (E) The expression of the lciRS-lciE-lacZ fusion was examined in the E. coli cusR deletion mutant at the stationary phase under different copper concentrations (indicated below the bars). The levels of expression of the lacZ fusions were found to be significantly different (*, P < 10−5, paired Student's t test) between expression of the same lacZ fusion under the same copper concentrations between the mutants and the one in the wild-type strain (panels B and D) or between the expression of the same strain grown without copper and the one grown under different copper concentrations (panels C and E). β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results from at least three different experiments.
Effect of copper on L. pneumophila and E. coli growth in media. L. pneumophila (A) and E. coli (B) were grown with different concentrations of copper (indicated on the right), and their growth (OD600) was examined in intervals of 1 h for the time indicated. Download FIG S4, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LciRS TCS and the effector LciE are dispensable for intracellular growth. (A and B) The intracellular growth of the lciR, lciS, lciE, and lciRS-lciE deletion mutants was examined in A. castellanii (A) and HL-60-derived human macrophages (B). Symbols: diamonds, L. pneumophila wild-type strain JR32; squares, icmT mutant; triangles, lciS deletion mutant; black circles, lciR deletion mutant; star, lciE deletion mutant; white circles, lciRS-lciE triple deletion mutant. The experiments were performed three times, and similar results were obtained. (C and D) Intracellular competition assay between the L. pneumophila lciRS-lciE triple deletion mutant (white circles) and the JR32 wild-type strain (diamonds) in A. castellanii. The experiment was performed without (C) and with (D) 200 μM copper. Download FIG S5, TIF file, 0.8 MB (804.7KB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effect of mutations and copper on the expression of lciE and lciR. (A) The effect of mutations on the amino acids predicted to be required for phosphorylation of LciR (LciRD51A) and LciS (LciSH148A) on lciE induction by copper. The levels of expression of wild-type lciRS-lciE-lacZ fusion and the lciRS-lciE-lacZ fusion containing a mutation in the amino acids, with and without 50 μM copper, are indicated below the bars. The levels of expression of the lacZ fusions were found to be significantly different (*, P < 10−5, paired Student’s t test) between mutated fusions and the wild-type fusion after exposure to copper. (B) lciR expression is activated neither by copper nor by the LciR regulator. The expression of the lciR-lacZ fusion was examined in the wild-type strain and in the lciR deletion mutant with (grey bars) and without (white bars) the addition of 50 μM copper. Download FIG S6, TIF file, 0.7 MB (776.2KB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
lciE is induced in E. coli after copper exposure in a CusRS- or LciRS-dependent manner.
As indicated above, the LciRS homologous TCSs as well as the LciR predicted regulatory element are conserved in many bacteria. To examine if these TCSs function similarly, we examined the expression of the lciE-lacZ construct (Fig. 3A) in E. coli. To this end, we determined the maximal copper concentration that E. coli tolerates without any effect on growth to be 1 mM (Fig. S4B), which is much higher than the one found for L. pneumophila (Fig. S4A). Analysis of the lciE-lacZ construct in E. coli resulted in gradual induction of lciE expression as the concentration of copper increased (Fig. 3D). Moreover, this induction was completely dependent on the E. coli CusRS TCS (Fig. 3D). Interestingly, when lciE-lacZ expression was examined in the cusC deletion mutant (CusC together with CusFBA form a copper transporter located in the E. coli envelope [59, 61]), higher levels of expression of lciE were observed at all copper concentrations examined (Fig. 3D). This result was obtained probably because E. coli lacking a functional CusCFBA system is exposed to higher concentrations of copper, which led to higher expression of lciE-lacZ. Comparing this result to the one obtained with the L. pneumophila lciE deletion mutant (compare Fig. 3B and D), which was induced similarly to wild-type L. pneumophila at all copper concentrations, indicates that in L. pneumophila, LciE is not involved in copper transport when the bacteria are grown in media, as expected from an Icm/Dot effector protein that functions during infection of host cells. Furthermore, the higher concentrations of copper tolerated by E. coli led us to examine the copper induction of the L. pneumophila LciRS TCS using E. coli as an in vivo heterologous system. Examination of the lciRS-lciE-lacZ construct in an E. coli cusR deletion mutant indicated that the L. pneumophila LciRS functions in E. coli and can respond to higher concentrations of copper than those tolerated by L. pneumophila (Fig. 3E). Collectively, these results indicate that the CusRS and LciRS TCSs function similarly, and both activate the expression of lciE in response to copper exposure.
LciR recognizes a conserved regulatory element located upstream of lciE.
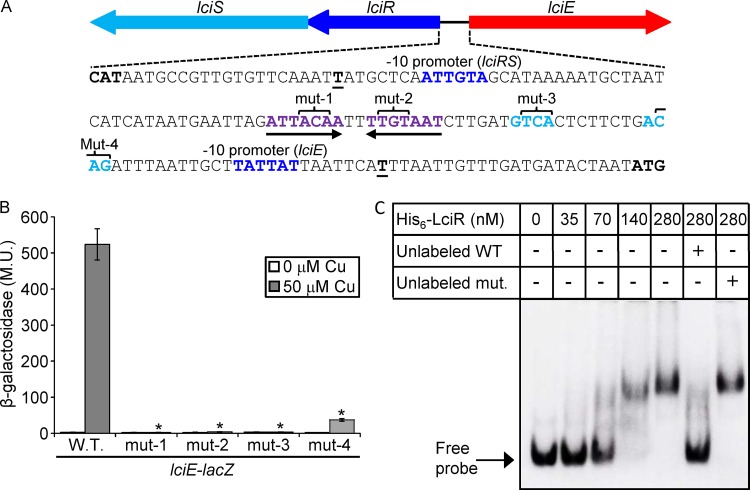
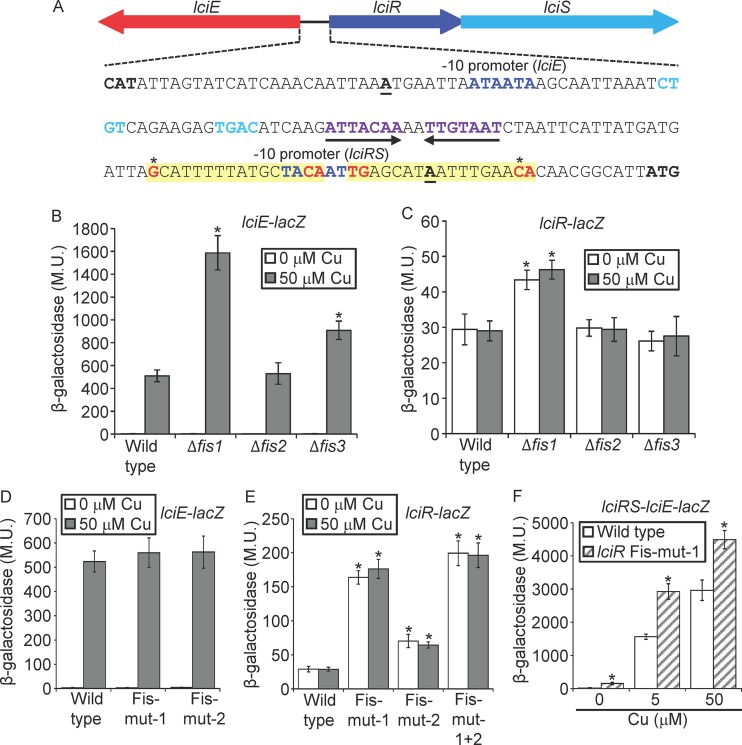
We identified a conserved regulatory element located upstream of lciE and other genes known or expected to be regulated by LciRS homologous TCSs (Fig. 2 and Fig. S3). This regulatory element constitutes a 7-bp inverted repeat (ATTACAAnnTTGTAAT) as well as two shorter (4 bp each) conserved sequences located between the inverted repeat and the lciE −10 promoter element (Fig. 4A and Fig. S3). Examination of the L. pneumophila genomic sequence for the presence of additional such regulatory elements revealed that this site is not present elsewhere in the genome, which is in line with our observation that the LciRS-LciE genomic island was acquired by HGT. To determine the importance of each of the putative lciE regulatory elements for the copper induction by LciR, they were mutagenized and examined for their levels of expression before and after copper induction. Three of these mutated lciE-lacZ fusions completely lost their ability to be induced by copper, and the fourth mutant retained a very limited ability to respond to copper (Fig. 4B).
FIG 4.
LciR directly regulates the expression of lciE using a conserved regulatory element. (A) The intergenic DNA sequence located between lciR and lciE. The lciR and lciE −10 promoter elements are in dark blue, and the nucleotides representing the putative LciR consensus are in purple (the inverted-repeat sequence) or light blue (the two sequences located between the inverted repeat and the lciE −10 promoter); the inverted repeat is also marked with arrows. The transcription start sites are boldface and underlined. The four triplets of base pairs mutated in each part of the suspected regulatory element are indicated. (B) The effect of mutations in the putative LciR regulatory element. The levels of expression of wild-type lciE-lacZ fusion and the lciE-lacZ fusions containing mutations in the nucleotides marked in panel A were examined with and without 50 μM copper. The levels of expression of the lacZ fusions were found to be significantly different (*, P < 10−5, paired Student's t test) between the fusions containing the wild-type regulatory region and the mutated regulatory region under the same copper concentrations. β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results from at least three different experiments. (C) L. pneumophila His6-LciR protein binds to the lciE regulatory region. Gel mobility shift assay was performed with purified His6-LciR protein and the DIG-labeled lciE regulatory region. The first lane did not contain any protein. The rest of the lanes contained increasing amounts of the His6-LciR protein in 2-fold increments, starting from 35 nM. Competition was performed using unlabeled probe as a specific competitor (unlabeled WT) or a probe containing a mutation in the LciR regulatory element (unlabeled mut.).
Since the lciR gene that encodes the RR and the lciE EEG share an intergenic region (Fig. 4A), we examined whether the lciR gene is activated by copper and whether it is an autoregulator. The lciR gene was neither induced by copper nor affected by the lciR deletion mutant (Fig. S6B). These results indicate that the LciRS TCS specifically activates the expression of lciE in response to copper and suggest that the two conserved short regulatory elements located between the inverted repeat and the −10 promoter element of lciE play a critical role in directing the activation by LciR to lciE and not to lciR.
The L. pneumophila LciR protein directly binds to the regulatory region of lciE.
To further support the results presented, the L. pneumophila LciR protein was His tagged, overexpressed, purified, and used for gel mobility shift assays with the lciE regulatory region. The L. pneumophila His6-LciR protein was found to bind to the regulatory region of the lciE gene, as evidenced by a shift in the migration of the DNA probe (Fig. 4C). The band shift degree as well as the amount of the shifted probe correlated with the increasing amounts of His6-LciR (Fig. 4C). In addition, competition with an unlabeled probe reduced the band shift (Fig. 4C, compare lanes 5 and 6). To further validate the specificity of the binding, we performed additional competition assays with an unlabeled probe containing a mutation in the LciR binding site. When the unlabeled mutated probe was used, a dramatic decrease in competition was observed compared to that of the unlabeled wild-type probe (Fig. 4C, compare lanes 6 and 7).
The mobility shift assay (Fig. 4C), together with the examination of lciE gene expression in the lciR and lciS deletion mutants (Fig. 3) and the analysis of the mutations in the LciR consensus sequence (Fig. 4B), establish LciR as a direct regulator of the lciE EEG in L. pneumophila.
Identification of amino acids required for LciS copper sensing.
Even though the E. coli CusRS and the L. pneumophila LciRS TCSs are homologous and both respond to elevated copper concentrations (Fig. 3), the periplasmic sensing domain of CusS and LciS are considerably different in size (150 amino acids in E. coli compared to 28 amino acids in L. pneumophila) and show no sequence homology (Fig. 5A). To identify amino acids required for copper induction in the L. pneumophila LciS periplasmic sensor domain, we aligned the sequences of LciS from the nine Legionella species harboring it. Since the amino acids histidine, cysteine, and methionine were previously shown to be involved in copper binding (62, 63), we specifically looked for these residues. Several histidine residues and a single cysteine were found in the short LciS periplasmic domain, together with other conserved amino acids (Fig. 5A). To examine the importance of these conserved amino acids for copper sensing by LciS, seven amino acid residues in the periplasmic domain of LciS (H43, H44, H46, N48, E50, H51, and C60) were changed separately to alanine residues. These mutations were introduced into the lciRS-lciE-lacZ fusion and used to determine the levels of copper induction (Fig. 5B). The most striking result was obtained with the mutation of the cysteine at position 60, which completely abolished the induction by copper. In addition, two of the histidine residues mutated (H44 and H51) significantly reduced induction by copper. We noticed that the number of histidine residues varies between the periplasmic sensor domains of the different Legionella LciS proteins, but at least two histidine residues were present in each LciS periplasmic domain. Thus, we constructed a triple mutation in which the three adjacent histidine residues (H43, H44, and H46) all were changed to alanine residues. This combined mutation resulted in a complete lack of induction by copper (Fig. 5B). To determine the specificity of LciS, we also exposed the bacteria to other similar metals (57, 64), and as seen in Fig. 5C, the LciRS TCS was found to be specific to copper and no induction was obtained with any of the other metals examined. Collectively, these results indicate that the Legionella LciS small periplasmic domain contains few histidine residues and a single cysteine residue specifically required for copper sensing.
FIG 5.
Identification of amino acids required for copper induction located in the LciS periplasmic sensor domain. (A) Schematic illustration of the L. pneumophila lciRS and the E. coli cusRS genes. The periplasmic domain of the nine Legionella SHKs is much smaller than the periplasmic domain found in other bacteria, such as E. coli (28 amino acids compared to about 150 amino acids, respectively). In addition, protein sequence alignment of the LciS transmembrane regions and periplasmic domains from nine Legionella species harboring the LciRS TCS is shown. The amino acids that compose the SHK predicted transmembrane domains are colored yellow. Amino acids found in the periplasmic domain which might be involved in copper-binding are marked by different colors: histidine residues, green; cysteine, light blue; and asparagine and glutamic acid, red. (B) The effect of mutations in the periplasmic domain of the L. pneumophila LciS on copper induction. The levels of expression of wild-type lciRS-lciE-lacZ fusion and the lciRS-lciE-lacZ fusions containing mutations in the amino acids indicated below the bars, with and without 50 μM copper. The levels of expression of the lacZ fusions were found to be significantly different (**, P < 10−4; *, P < 10−5; both by paired Student's t test) between the mutated fusions and the wild-type fusion with or without copper. (C) The effect of different metals on lciE expression. The levels of expression of wild-type lciRS-lciE-lacZ fusion after exposure to the metals indicated below the bars (50 μM). The level of expression of the lacZ fusion was found to be significantly different (P < 10−5, paired Student's t test) between expression with metals and the one without metal exposure. β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results from at least three different experiments.
The LciRS-LciE genomic island is regulated by the Fis repressors.
It was previously shown in S. enterica as well as in other bacteria that genomic islands that undergo HGT are silenced by nucleoid-associated proteins (NAPs), such as H-NS and Hha (65–67). Since the LciRS-LciE island undergoes HGT in the Legionella genus (Fig. 1 and Fig. S1 and S2), we were interested in examining whether this island is silenced by NAPs. The H-NS NAP is not present in Legionella, but three Fis paralogs (Fis1, Fis2, and Fis3), which are also NAPs, were previously shown to directly repress the expression of EEGs (32). Examination of the lciRS-lciE intergenic region led to the identification of four potential Fis regulatory elements (TG-N13-C), two close to or overlapping the −10 promoter element of lciE (Fig. S7) and two others close to or overlapping the −10 promoter element of lciR (Fig. 6A and Fig. S7). To determine whether the Fis repressors are involved in the regulation of the LciRS-LciE island, the expression of the lciE-lacZ fusion was examined in deletion mutants of each of the three fis genes. In the absence of copper, the expression levels of the lciE-lacZ fusion in the three fis deletion mutants were similar to those in the wild-type strain (Fig. 6B). However, in the presence of copper, the expression of the lciE-lacZ fusion was significantly higher in the fis1 and fis3 deletion mutants (Fig. 6B). In addition, when the expression of the lciR-lacZ fusion was examined in the same mutants under the same conditions, there was an increase in the level of expression of the lciR-lacZ fusion in the fis1 deletion mutant that was independent of the presence of copper (Fig. 6C). These results indicate a direct repression of Fis on lciE, on lciR, or on both genes.
FIG 6.
lciRS-lciE island is repressed by Fis. (A) The intergenic DNA sequence located between lciE and lciR. The lciR and lciE −10 promoter elements are in dark blue, and the nucleotides representing the LciR consensus are in purple (the inverted-repeat sequence) or light blue (the two sequences located between the inverted repeat and the −10 promoter of lciE); the inverted repeat is also marked with arrows. The transcription start sites are boldface and underlined. The putative lciR Fis regulatory elements are shaded in yellow, conserved nucleotides of the Fis consensus are marked in red, and the nucleotides mutated are marked by asterisks. (B and C) The expression levels of the lciE-lacZ fusion (B) and the lciR-lacZ fusion (C) were examined in the wild-type strain and in the three fis deletion mutants at the stationary phase. Expression was examined with (gray bars) and without (white bars) 50 μM copper. The levels of expression of the lacZ fusions were found to be significantly different (*, P < 10−5, paired Student's t test) between expression of the wild-type strain and each fis deletion mutant under the same conditions. (D and E) The levels of expression of wild-type lciE-lacZ fusion and the two lciE-lacZ fusions containing mutations (mut-1 and mut-2) in the putative Fis regulatory elements (D) and wild-type lciR-lacZ fusion and the three lciR-lacZ fusions containing mutations (mut-1, mut-2, and mut-1 + 2) in the putative Fis regulatory elements (E) were examined with (gray bars) and without (white bars) 50 μM copper. The levels of expression of the lacZ fusions were found to be significantly different (*, P < 10−5, paired Student's t test) between fusions containing the wild-type regulatory region and the mutated regulatory region under the same copper concentrations. (F) The levels of expression of wild-type lciRS-lciE-lacZ fusion (white bars) and the same fusion containing a mutation in the downstream Fis regulatory element of lciR (hatched gray bars) were examined in the lciRS-lciE deletion mutant without copper and with 5 μM and 50 μM copper. The levels of expression of the lacZ fusions were found to be significantly different (*, P < 10−5, paired Student's t test) between the wild-type fusion and the mutated fusion under the same copper concentrations. β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results from at least three different experiments.
Predicted Fis regulatory elements in the regulatory regions of lciE and lciR homologs in different Legionella species. The promoter region of the lciE (A) and lciR (B) genes in the nine Legionella species harboring the island, L. pneumophila (lciE and lciR), L. moravica (Lmor), L. quateirensis (Lqua), L. worsleiensis (Lwor), L. nautarum (Lnau), L. brunensis (Lbru), Legionella sp. strain 13.8642 (L_13.8642), Legionella sp. strain FW215 (L_FW215), and Legionella sp. strain Km535 (L_Km535). In purple are the putative LciR inverted-repeat regulatory elements, in light blue are the two additional regulatory elements found between the inverted repeat and the −10 promoter element of lciE, in dark blue are the −10 promoter elements, and the experimentally determined transcription start sites (TSSs) are underlined and marked in blue. The predicted Fis sites in both promoter regions are marked: the site overlapping or close to the −10 promoter element is colored yellow, and a second predicted Fis site (when present) is underlined. In both predicted Fis sites, the conserved TG and CA consensus nucleotides are in red and the nucleotides mutated are marked by asterisks. Download FIG S7, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fis1 and Fis3 repress the expression of lciR and affect the copper induction of lciE.
To distinguish between the three possibilities described above, we constructed site-directed mutations in the four described putative Fis regulatory elements (Fig. 6A and Fig. S7). The two mutations constructed in the putative Fis regulatory elements of lciE did not affect its level of expression, with or without copper (Fig. 6D). However, both mutations constructed in the putative Fis regulatory element of lciR (Fig. 6A) showed a relief of repression (Fig. 6E). The mutation in the Fis regulatory element located immediately downstream to the −10 promoter element of lciR (Fig. 6A) showed a 6-fold relief of repression, and the mutation in the upstream Fis site showed 2-fold relief of repression. A combined mutation in both Fis regulatory elements led to a level of expression similar to that of the mutation in the downstream Fis site (Fig. 6E). It is important to note that the Fis regulatory element located downstream to the lciR −10 promoter element is conserved in all nine Legionella species harboring the LciRS-LciE genomic island, while the three other putative Fis regulatory elements examined are not conserved (Fig. S7). The effect observed on the level of expression of the lciR-lacZ fusion containing the mutations in the Fis regulatory elements was considerably stronger than the effect observed on the wild-type lciR-lacZ fusion in the fis1 deletion mutant. Since a double deletion of both fis1 and fis3 is not viable in L. pneumophila (32), only the mutation in the Fis regulatory element can fully expose the degree of repression mediated by both Fis1 and Fis3 on the expression of LciR. To determine the effect of the Fis repression of LciR on the expression of LciE, we used the lciRS-lciE-lacZ fusion to construct a single-base-pair mutation in the downstream Fis regulatory element of LciR and examined the expression of LciE from this fusion. As can be seen in Fig. 6F, the level of expression of the lciRS-lciE-lacZ fusion containing the mutation in the lciR regulatory element was much higher in the absence of copper (6-fold) as well as in the presence of copper compared to that of the wild-type fusion. Collectively, these results indicate that the repression mediated by Fis on the LciR regulator affects the level of expression of the LciE effector with and without copper, showing that Fis proteins silence this genomic island by repressing the expression of the positive regulator LciR.
DISCUSSION
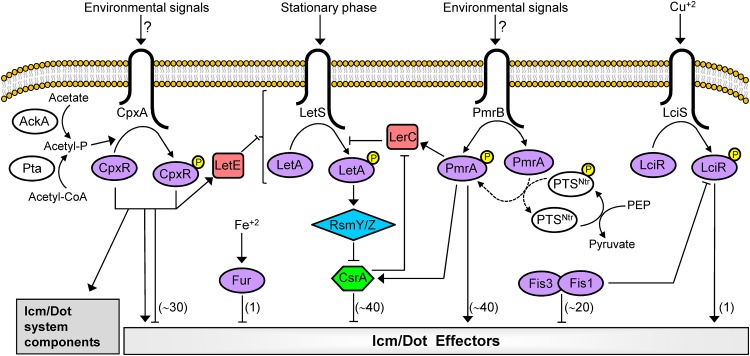
The L. pneumophila Icm/Dot secretion system translocates into host cells the largest number of effectors known in a single bacterium, and these effectors manipulate numerous host cell processes for the benefit of the bacteria (10–14). One of the challenges encountered by bacteria using such a multicomponent pathogenesis system is the coordination of the expression of the pathogenesis genes encoding these components to result in a successful infection. Thus far, several regulatory systems that control the expression of EEGs were identified in L. pneumophila (Fig. 7), including a pair of TCSs (PmrAB and CpxRA) that belong to the wHTH family of transcriptional regulators (68). Here, we described a third L. pneumophila TCS (LciRS) that belongs to the wHTH family of RRs, which directly regulates the expression of a single EEG (Fig. 7). In contrast to PmrA and CpxR, LciR is present only in several Legionella species and undergoes HGT as part of a genomic island together with the single EEG it regulates, representing the first case of local regulation of Icm/Dot effectors in Legionella.
FIG 7.
Model of the regulatory network of direct regulators that regulate the expression of L. pneumophila EEGs. The four TCSs, CpxRA, PmrAB, LciRS, and LetAS, as well as the components of the LetAS-RsmYZ-CsrA regulatory cascade, which were found to be involved in the regulation of EEGs, are shown. The four SHKs (CpxA, LetS, PmrB, and LciS) are drawn inside the bacterial inner membrane. The four RRs (CpxR, LetA, PmrA, and LciR), as well as the other DNA binding regulators (Fur, Fis1, and Fis3), are colored purple. The RRs are phosphorylated (small yellow circle) and/or dephosphorylated by their cognate SHKs. The CsrA RNA binding regulator is colored green, and the RsmY/RsmZ small RNAs are colored blue. Connector proteins that participate in the regulation of EEGs are colored red, and modulators are colored white. Acetyl-P, acetyl phosphate; PEP, phosphoenolpyruvate; PTS, phosphotransferase system. The numbers of EEGs that were shown to be regulated by each of the regulatory systems are indicated in parentheses. Arrows and T-shaped symbols indicate activation and repression, respectively. Solid lines indicate direct regulation and broken lines indirect regulation.
One of the most interesting findings regarding the LciRS-LciE genomic island is that it undergoes HGT in the Legionella genus (Fig. 1; see also Fig. S1 and S2 in the supplemental material). Two aspects regarding this genomic island were left unresolved, namely, (i) the way by which it is transferred between Legionella species and (ii) the way by which it integrates into the bacterial genome. A hint regarding these two issues was obtained when we analyzed the LciRS-LciE genomic island found in two uncharacterized Legionella species (Legionella sp. strain 13.8642 and Legionella sp. strain FW215). In Legionella sp. strain 13.8642, a pseudogene was found to be located next to the LciRS-LciE genomic island (Fig. 1). This pseudogene contains a deletion of a single nucleotide after nucleotide 41, making the protein it used to encode nonfunctional. However, prior to its pseudogenization, this gene encoded a protein with a high degree of homology to a phage integrase. Homologous integrase-encoding genes were found intact in several Legionella species (but not next to the LciRS-LciE genomic island) and in Legionella sp. strain 13.8642 it is completely intact, excluding the single-nucleotide deletion described above. It is possible that this integrase was involved in the integration of the LciRS-LciE genomic island in this Legionella species. Integrase-encoding genes, as well as mutated integrase-encoding genes, were previously shown to be located near genomic islands in other bacteria (69, 70). Another known feature of genomic islands is that they sometimes integrate into bacterial genomes next to tRNA genes (50, 71). In this case too, in one of the uncharacterized Legionella species, Legionella sp. strain FW215, the LciRS-LciE genomic island was found in proximity to a Lys tRNA gene (Fig. 1). This indicates that tRNA genes are also an entry site for the LciRS-LciE genomic island in Legionella. Although the precise mechanism by which the LciRS-LciE genomic island undergoes HGT in bacteria is not known, the above-mentioned findings suggest that it utilizes transfer and integration mechanisms similar to the ones previously described for genomic islands in other bacteria.
The LciRS TCS was found to be activated in the presence of copper (Fig. 3), and homologous TCSs present in other bacteria are also activated by copper and are usually located next to genes involved in metal resistance (mainly copper) (59). Comparison of the Legionella LciRS TCS to the homolog TCSs present in other bacteria resulted in the identification of a major difference: the Legionella SHK LciS was found to contain a small (28 amino acids long) (Fig. 5A) periplasmic sensor domain, which is completely nonhomologous to the periplasmic sensing domains found in the homologous SHKs present in other bacteria, including the ones that sense copper. This finding is intriguing, since SHKs were previously shown to change their sensing domain by recombination or mutations and, in this way, change the signal they respond to and deviate from other SHKs (72). However, in the case of LciS, the sensing domain was completely changed but the signal recognized by the SHK remained the same. One can speculate on the evolutionary driving forces that can lead a sensing domain to be replaced without changing the signal it responds to. For example, an advantage can be achieved if the new sensing domain alters the sensitivity or specificity to the signal. However, examination of these two aspects, by comparing the E. coli CusS and L. pneumophila LciS SHKs, did not result in differences in the sensitivity to copper (compare Fig. 3B and D) or in the specificity to copper (Fig. 5C and data not shown). Even though the LciS periplasmic sensor domain is not homologous to other sensing domains that sense copper, it is important to mention that we did recognize specific amino acids critical for the sensing of copper by the L. pneumophila LciS, and the same amino acids were previously shown to be involved in copper binding in different proteins (62, 63).
Genes regulated by regulatory systems that sense a specific signal were shown to encode proteins with functions directly related to the signal sensed by their regulators. This phenomenon was demonstrated in many systems, including regulators of amino acids biosynthetic pathways and regulators of metal resistance systems (59, 73). In addition, the L. pneumophila mavN EEG, which encodes an iron transporter localized to the LCV, is regulated by the iron-specific repressor Fur (33, 34). However, in many cases the signal sensed by a regulator is not directly related to the function mediated by the genes it regulates. In these cases, the signal sensed indicates a change in the bacterial environment. Such cases were described in many pathogenesis systems, such as the S. enterica PhoPQ TCS, which senses Mg2+ and cationic antimicrobial peptides (74, 75), and the S. enterica PmrAB TCS, which senses acidic pH and high Fe3+ concentrations (76, 77). Both of these TCSs regulate the expression of numerous genes encoding diverse functions (78, 79). The function of LciE, which is activated by the LciRS TCS in response to copper, is currently unknown, and it contains no known protein domains. However, we identified two other L. pneumophila effectors with unknown functions that show a significant degree of homology to LciE (Fig. S8). The core effector CetLp1 (lpg0140), and another effector (lpg2888) that is found in most of the Legionella species examined, harbor a protein domain homologous to LciE (Fig. S8). In addition, these three effectors contain four predicted transmembrane domains located at the C-terminal half of the protein (Fig. S8). These similarities among the three effectors suggest that they perform a related function. Since both copper and zinc are used by eukaryotic cells to kill invading bacteria (80, 81), we examined the copper- or zinc-dependent induction of CetLp1 and lpg2888, or an effect of LciR on their levels of expression, but their expression was unaffected (Fig. S9A and B). Moreover, we generated a triple deletion mutant of cetLp1, lpg288, and lciE and examined this mutant using a competition assay in amoeba with and without the addition of copper, and no intracellular growth phenotype was observed (Fig. S9C to F).
L. pneumophila contains two effectors with a domain homologous to LciE. Schematic representation of the effectors LciE (lpg0716), CetLp1 (lpg0140), and lpg2888. The domain homologous among the three effectors is marked in light blue, and putative transmembrane domains are marked in black. In addition, protein sequence alignment of the homologous domain found in the three effectors is shown from the six characterized Legionella species in which the LciRS-LciE genomic island was found, L. pneumophila (LciE/lpg2888/CetLp1), L. moravica (Lmor), L. quateirensis (Lqua), L. worsleiensis (Lwor), L. nautarum (Lnau), and L. brunensis (Lbru). Download FIG S8, TIF file, 2.1 MB (2.1MB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Examination of the connection of CetLp1 and lpg2888 to copper and LciE. The levels of expression of cetLp1 (A) and lpg2888 (B) lacZ fusions were examined without the addition of metals and after exposure to 50 μM copper (grey bars) or zinc (hatched bars). The same fusions were also examined in the lciR deletion mutant under the same conditions. (C and D) Intracellular competition assay between the L. pneumophila cetLp1-lpg2888 double deletion mutant (circles) and the JR32 wild-type strain (diamonds) in A. castellanii. (E and F) Intracellular competition assay between the L. pneumophila cetLp1-lpg2888-lciE triple deletion mutant (circles) and the JR32 wild-type strain (diamonds) in A. castellanii. The experiment was performed without (C and E) and with (D and F) 200 μM copper. Download FIG S9, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Unlike all the regulatory systems described thus far for L. pneumophila effectors, the LciRS-LciE genomic island represents a new type of effector regulation, unprecedented in the Legionella genus. In this system, a single effector is locally regulated by a dedicated regulatory system in response to a specific signal. Moreover, both the regulatory system and the effector form a unit that undergoes HGT in the Legionella genus. Uncovering this novel type of regulation sheds new light on the ways the effector repertoire of Legionella evolves and the activating signals of effectors expand.
MATERIALS AND METHODS
Bacteria strains and media.
The L. pneumophila wild-type strain used in this work was JR32, a streptomycin-resistant, restriction-negative mutant of L. pneumophila Philadelphia-1, which is a wild-type strain in terms of intracellular growth (82). In addition, mutant strains derived from JR32 that were used in this study are listed in Data Set S1 in the supplemental material. The E. coli strains used in this work are also listed in Data Set S1. Bacterial media, plates, and antibiotics were as previously described (83).
Strains, plasmids, and primers used in this study. Download Data Set S1, XLSX file, 0.03 MB (33.9KB, xlsx) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmid construction.
To construct lacZ translational fusions (Data Set S1), the 300-bp regulatory regions of the lciE and lciR genes were amplified by PCR using the primers listed in Data Set S1. The PCR products were then digested with BamHI and EcoRI, cloned into pGS-lac-02, and sequenced. The lciE lacZ fusion, which does not contain the lciRS genes, was designated lciE-lacZ. In addition, a second lciE lacZ fusion was constructed containing the lciRS genes in an organization similar to that in the genome. To construct this fusion, an internal EcoRI site present in the lciR gene was mutagenized in a way that does not change the LciR amino acid sequence. A 1,950-bp region was amplified by PCR using the four primers listed in Data Set S1, digested with BamHI and EcoRI cloned into pGS-lac-02, and sequenced. This lciE lacZ fusion, which contains the lciRS genes, was designated lciRS-lciE-lacZ.
To construct a substitution mutation in the putative LciR binding site in the regulatory region of the lciE gene, a substitution mutation in the putative Fis binding site in the regulatory region of the lciE and lciR genes, and substitution mutations in the lciR and lciS coding sequence, site-directed mutagenesis was performed by regular PCR or the PCR overlap extension approach (84), as previously described (19). The primers used for the mutagenesis are listed in Data Set S1, and the plasmids resulting from the site-directed mutagenesis are listed in Data Set S1.
To construct deletion substitution mutants in the L. pneumophila lciE and lciS genes, a 1-kb DNA fragment located on each side of the planned deletion was amplified by PCR using the primers listed in Data Set S1. The resulting plasmids were digested with the suitable enzymes, and the inserts were used for a four-way ligation containing the Km resistance cassette (Pharmacia). The plasmids generated, pAA-lpg0714-Km and pMLpUC18+0716Up-Km-Dw (Data Set S1), were digested with PvuII, and the resulting fragment was cloned into the pLAW344 allelic exchange vector digested with EcoRV to generate the plasmids pAA-lpg0714-pLAW and pMLpLAW344-0716-Up-Km-Dw (Data Set S1). In addition, the insert of pMLpUC18+0716Up-Km-Dw was also cloned into the pGY100 allelic exchange vector, digested with XmnI, to generate the plasmid pMLpGY100+lpg0716-Up-Km-Dw, which was later digested with SalI to take out the Km resistance cassette and generate the plasmid pMLpGY100+lpg0176-UP+Dw (Data Set S1). The latter plasmid was used to generate a clean lciE deletion.
To generate the triple lciRS-lciE deletion mutant, the insert containing the 1-kb upstream region of lciS and the insert containing the 1-kb downstream region of lciE were used for a four-way ligation, as described above, to generated plasmid pMLpUC18+0714Dw-Km-0716Dw (Data Set S1). This plasmid was digested with PvuII, and the resulting fragment was cloned into the pLAW344 allelic exchange vector digested with EcoRV to generate the plasmid pMLpLAW344-0714Dw-Km-0716Dw (Data Set S1).
To generate a clean deletion mutant in lpg2888, a 1-kb DNA fragment located on each side of the planned deletion was amplified by PCR using the primers listed in Data Set S1. The resulting plasmids were digested with the suitable enzymes, and the inserts were used for a three-way ligation into pUC-18 to generate pMW-18-Δlpg2888-3W (Data Set S1). This plasmid was digested with PvuII, and the resulting fragment was cloned into the pGY100 allelic exchange vector digested with XmnI to generate the plasmid pMW-100-Δlpg2888 (Data Set S1). The clean and marked allelic exchange deletion mutants were constructed as previously described (83, 85).
For the construction of the plasmid expressing the His-tagged LciR, the lciR gene was amplified by PCR using the primers listed in Data Set S1, cloned into pET-15b, and sequenced to generate the plasmid pML-pET15b+lpg0715 (Data Set S1).
Bacterial growth in the presence of copper.
To determine the copper concentrations to be used in the β-galactosidase assays with both L. pneumophila and E. coli, the bacteria were grown in fresh AYE lacking Fe(NO3)3 or LB, respectively, with a wide range of copper concentrations, and the optical density at 600 nm (OD600) was determined in intervals of 1 h until reaching stationary phase. The same analysis was also performed with the other metals examined.
β-Galactosidase assay.
β-Galactosidase assays were performed as previously described (19). L. pneumophila strains were grown for 48 h on charcoal-yeast extract (CYE) plates containing chloramphenicol (Cm). The bacteria were scraped off the plate and suspended in ACES-yeast extract (AYE) broth, and the bacterial OD600 was calibrated to 0.1 in fresh AYE lacking Fe(NO3)3, containing different concentrations of copper (or other metals, when indicated) and Cm. When other metals were used, 2 μM bathocuproine sulfonate (BCS) was added to the medium to adsorb any copper traces present in the stocks of the other metals. This concentration of BCS was also included when copper was used, and it did not affect the induction by copper. The resulting cultures were grown on a roller drum for about 18 h, until reaching an OD600 of about 3.2 (early stationary phase), and used for β-galactosidase assay.
β-Galactosidase assays in E. coli were performed similarly, but the E. coli strains were grown for about 6 h in LB containing different concentrations of copper, until reaching an OD600 of about 2.5 (early stationary phase), and used for β-galactosidase assay. The assays were done for 20, 50, or 100 μl of culture, and the substrate for β-galactosidase hydrolysis was o-nitrophenyl-β-d-galactopyranoside.
Protein purification and gel mobility shift assay.
His6-LciR was purified from E. coli BL21(DE3) using nickel bead columns (Qiagen) according to the manufacturer’s instructions. After purification, the fractions containing the protein were dialyzed overnight against a buffer containing 10 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 50 mM KCl, 0.1 mM EDTA, and 0.1 mM dithiothreitol. Glycerol was added to a concentration of 50%, and the purified protein was then stored at –20°C. A gel mobility shift assay was performed as previously described (32), with a few modifications. The regulatory region lciE (176 bp) was amplified by PCR using the primers listed in Data Set S1 and 3′ end labeled with digoxigenin (DIG) by using DIG-11-ddUTP (Roche). Increasing amounts of the purified His6-LciR protein (between 35 and 280 nM) were mixed with 0.75 nM the DIG-labeled probe in buffer containing 10 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 50 mM KCl, 0.1 mM EDTA, 0.1 mM dithiothreitol, 250 μg/ml bovine serum albumin, and 50 μg/ml herring sperm DNA. For the competition experiments, a 100-fold excess of the unlabeled probe or mutated unlabeled probe was allowed to bind the His6-LciR protein for 30 min before addition of the DIG-labeled probe. The binding reaction was carried out for 30 min at room temperature, and samples then were loaded onto a 5% polyacrylamide–0.25× Tris-acetate-EDTA gel in 0.5× Tris-acetate-EDTA running buffer. Following electrophoresis, the gel was transferred to a nylon membrane and fixed by UV cross-linking. The DIG-labeled DNA fragments were detected by following the manufacturer’s instructions (Roche).
Intracellular growth assays.
Intracellular growth assays of L. pneumophila strains in A. castellanii and HL-60-derived human macrophages were performed as previously described (86). Intracellular competition assays of L. pneumophila strains in A. castellanii also were performed as previously described (22).
Reconstruction of phylogenetic trees.
Trees were reconstructed on the basis of concatenated alignments of the three proteins indicated for each tree. The trees were reconstructed using RAxML (87) under the LG + GAMMA evolutionary model with 100 bootstrap resampling.
ACKNOWLEDGMENTS
We thank Yaron S. Feldheim for plasmid construction and for reading the manuscript. We thank Ziv Lifshitz and Michael Wexler for plasmids and strains construction. We thank Anna Pasechnek for plasmid construction. We thank Tal Zusman for her ideas and help throughout this study and for reading the manuscript. We thank David Burstein for his help with the bioinformatic analyses and for reading the manuscript.
This research was supported by Israeli Science Foundation grant 877/15 (to G.S.).
Footnotes
This article is a direct contribution from Gil Segal, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Ralph Isberg, Tufts Medical School, and Craig Roy, Yale University School of Medicine.
Citation Linsky M, Vitkin Y, Segal G. 2020. A novel Legionella genomic island encodes a copper-responsive regulatory system and a single Icm/Dot effector protein transcriptionally activated by copper. mBio 11:e03232-19. https://doi.org/10.1128/mBio.03232-19.
REFERENCES
- 1.Cunha BA, Burillo A, Bouza E. 2016. Legionnaires’ disease. Lancet 387:376–385. doi: 10.1016/S0140-6736(15)60078-2. [DOI] [PubMed] [Google Scholar]
- 2.Newton HJ, Ang DK, van Driel IR, Hartland EL. 2010. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev 23:274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burillo A, Pedro-Botet ML, Bouza E. 2017. Microbiology and epidemiology of Legionnaire’s disease. Infect Dis Clin North Am 31:7–27. doi: 10.1016/j.idc.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Franco IS, Shuman HA, Charpentier X. 2009. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol 11:1435–1443. doi: 10.1111/j.1462-5822.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 5.Gimenez G, Bertelli C, Moliner C, Robert C, Raoult D, Fournier PE, Greub G. 2011. Insight into cross-talk between intra-amoebal pathogens. BMC Genomics 12:542. doi: 10.1186/1471-2164-12-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boamah DK, Zhou G, Ensminger AW, O'Connor TJ. 2017. From many hosts, one accidental pathogen: the diverse protozoan hosts of Legionella. Front Cell Infect Microbiol 7:477. doi: 10.3389/fcimb.2017.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moliner C, Fournier PE, Raoult D. 2010. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol Rev 34:281–294. doi: 10.1111/j.1574-6976.2010.00209.x. [DOI] [PubMed] [Google Scholar]
- 8.Horwitz MA. 1983. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med 158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR. 2001. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J Cell Sci 114:4637–4650. [DOI] [PubMed] [Google Scholar]
- 10.Qiu J, Luo ZQ. 2017. Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol 15:591–605. doi: 10.1038/nrmicro.2017.67. [DOI] [PubMed] [Google Scholar]
- 11.Finsel I, Hilbi H. 2015. Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell Microbiol 17:935–950. doi: 10.1111/cmi.12450. [DOI] [PubMed] [Google Scholar]
- 12.Isaac DT, Isberg R. 2014. Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future Microbiol 9:343–359. doi: 10.2217/fmb.13.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubber A, Roy CR. 2010. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 14.Sherwood RK, Roy CR. 2016. Autophagy evasion and endoplasmic reticulum subversion: the yin and yang of Legionella intracellular infection. Annu Rev Microbiol 70:413–433. doi: 10.1146/annurev-micro-102215-095557. [DOI] [PubMed] [Google Scholar]
- 15.Dorer MS, Kirton D, Bader JS, Isberg RR. 2006. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog 2:e34. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Connor TJ, Boyd D, Dorer MS, Isberg RR. 2012. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science 338:1440–1444. doi: 10.1126/science.1229556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubori T, Shinzawa N, Kanuka H, Nagai H. 2010. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog 6:e1001216. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Khodor S, Kalachikov S, Morozova I, Price CT, Abu Kwaik Y. 2009. The PmrA/PmrB two-component system of Legionella pneumophila is a global regulator required for intracellular replication within macrophages and protozoa. Infect Immun 77:374–786. doi: 10.1128/IAI.01081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol 63:1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]
- 20.Altman E, Segal G. 2008. The response regulator CpxR directly regulates the expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J Bacteriol 190:1985–1996. doi: 10.1128/JB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gal-Mor O, Segal G. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J Bacteriol 185:4908–4919. doi: 10.1128/jb.185.16.4908-4919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldheim YS, Zusman T, Speiser Y, Segal G. 2016. The Legionella pneumophila CpxRA two-component regulatory system: new insights into CpxR’s function as a dual regulator and its connection to the effectors regulatory network. Mol Microbiol 99:1059–1079. doi: 10.1111/mmi.13290. [DOI] [PubMed] [Google Scholar]
- 23.Tanner JR, Li L, Faucher SP, Brassinga AK. 2016. The CpxRA two-component system contributes to Legionella pneumophila virulence. Mol Microbiol 100:1017–1038. doi: 10.1111/mmi.13365. [DOI] [PubMed] [Google Scholar]
- 24.Hammer BK, Tateda ES, Swanson MS. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol Microbiol 44:107–118. doi: 10.1046/j.1365-2958.2002.02884.x. [DOI] [PubMed] [Google Scholar]
- 25.Molofsky AB, Swanson MS. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol Microbiol 50:445–461. doi: 10.1046/j.1365-2958.2003.03706.x. [DOI] [PubMed] [Google Scholar]
- 26.Nevo O, Zusman T, Rasis M, Lifshitz Z, Segal G. 2014. Identification of Legionella pneumophila effectors regulated by the LetAS-RsmYZ-CsrA regulatory cascade, many of which modulate vesicular trafficking. J Bacteriol 196:681–692. doi: 10.1128/JB.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasis M, Segal G. 2009. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol Microbiol 72:995–1010. doi: 10.1111/j.1365-2958.2009.06705.x. [DOI] [PubMed] [Google Scholar]
- 28.Sahr T, Bruggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C, Buchrieser C. 2009. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol 72:741–762. doi: 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch D, Fieser N, Gloggler K, Forsbach-Birk V, Marre R. 2003. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol Lett 219:241–248. doi: 10.1016/S0378-1097(03)00050-8. [DOI] [PubMed] [Google Scholar]
- 30.Forsbach-Birk V, McNealy T, Shi C, Lynch D, Marre R. 2004. Reduced expression of the global regulator protein CsrA in Legionella pneumophila affects virulence-associated regulators and growth in Acanthamoeba castellanii. Int J Med Microbiol 294:15–25. doi: 10.1016/j.ijmm.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Sahr T, Rusniok C, Impens F, Oliva G, Sismeiro O, Coppee JY, Buchrieser C. 2017. The Legionella pneumophila genome evolved to accommodate multiple regulatory mechanisms controlled by the CsrA-system. PLoS Genet 13:e1006629. doi: 10.1371/journal.pgen.1006629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zusman T, Speiser Y, Segal G. 2014. Two Fis regulators directly repress the expression of numerous effector-encoding genes in Legionella pneumophila. J Bacteriol 196:4172–4183. doi: 10.1128/JB.02017-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isaac DT, Laguna RK, Valtz N, Isberg RR. 2015. MavN is a Legionella pneumophila vacuole-associated protein required for efficient iron acquisition during intracellular growth. Proc Natl Acad Sci U S A 112:E5208–E5217. doi: 10.1073/pnas.1511389112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portier E, Zheng H, Sahr T, Burnside DM, Mallama C, Buchrieser C, Cianciotto NP, Hechard Y. 2015. IroT/mavN, a new iron-regulated gene involved in Legionella pneumophila virulence against amoebae and macrophages. Environ Microbiol 17:1338–1350. doi: 10.1111/1462-2920.12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hickey EK, Cianciotto NP. 1997. An iron- and fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect Immun 65:133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chatfield CH, Mulhern BJ, Viswanathan VK, Cianciotto NP. 2012. The major facilitator superfamily-type protein LbtC promotes the utilization of the legiobactin siderophore by Legionella pneumophila. Microbiology 158:721–735. doi: 10.1099/mic.0.055533-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robey M, Cianciotto NP. 2002. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect Immun 70:5659–5669. doi: 10.1128/iai.70.10.5659-5669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Speiser Y, Zusman T, Pasechnek A, Segal G. 2017. The Legionella pneumophila incomplete phosphotransferase system is required for optimal intracellular growth and maximal expression of PmrA-regulated effectors. Infect Immun 85:e00121-17. doi: 10.1128/IAI.00121-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feldheim YS, Zusman T, Kapach A, Segal G. 2018. The single-domain response regulator LerC functions as a connector protein in the Legionella pneumophila effectors regulatory network. Mol Microbiol 110:741–760. doi: 10.1111/mmi.14101. [DOI] [PubMed] [Google Scholar]
- 40.Bachman MA, Swanson MS. 2004. The LetE protein enhances expression of multiple LetA/LetS-dependent transmission traits by Legionella pneumophila. Infect Immun 72:3284–3293. doi: 10.1128/IAI.72.6.3284-3293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burstein D, Amaro F, Zusman T, Lifshitz Z, Cohen O, Gilbert AJ, Pupko T, Shuman HA, Segal G. 2016. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet 48:167–175. doi: 10.1038/ng.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomez-Valero L, Rusniok C, Carson D, Mondino S, Perez-Cobas AE, Rolando M, Pasricha S, Reuter S, Demirtas J, Crumbach J, Descorps-Declere S, Hartland EL, Jarraud S, Dougan G, Schroeder GN, Frankel G, Buchrieser C. 2019. More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells. Proc Natl Acad Sci U S A 116:2265–2273. doi: 10.1073/pnas.1808016116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Browning DF, Busby SJ. 2016. Local and global regulation of transcription initiation in bacteria. Nat Rev Microbiol 14:638–650. doi: 10.1038/nrmicro.2016.103. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto K. 2014. The hierarchic network of metal-response transcription factors in Escherichia coli. Biosci Biotechnol Biochem 78:737–747. doi: 10.1080/09168451.2014.915731. [DOI] [PubMed] [Google Scholar]
- 45.Dorman CJ, Dorman MJ. 2017. Control of virulence gene transcription by indirect readout in Vibrio cholerae and Salmonella enterica serovar Typhimurium. Environ Microbiol 19:3834–3845. doi: 10.1111/1462-2920.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown NL, Stoyanov JV, Kidd SP, Hobman JL. 2003. The MerR family of transcriptional regulators. FEMS Microbiol Rev 27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 47.Wilkinson SP, Grove A. 2006. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol 8:51–62. [PubMed] [Google Scholar]
- 48.Maddocks SE, Oyston PC. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 49.Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hacker J, Kaper JB. 2000. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 51.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobrindt U, Hochhut B, Hentschel U, Hacker J. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 53.Fabrega A, Vila J. 2013. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev 26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hava DL, Hemsley CJ, Camilli A. 2003. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J Bacteriol 185:413–421. doi: 10.1128/jb.185.2.413-421.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de las Heras A, Cain RJ, Bielecka MK, Vázquez-Boland JA. 2011. Regulation of Listeria virulence: PrfA master and commander. Curr Opin Microbiol 14:118–127. doi: 10.1016/j.mib.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 56.Zhu W, Banga S, Tan Y, Zheng C, Stephenson R, Gately J, Luo ZQ. 2011. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS One 6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silver S. 2003. Bacterial silver resistance: molecular biology and uses and misuses of silver compounds. FEMS Microbiol Rev 27:341–353. doi: 10.1016/S0168-6445(03)00047-0. [DOI] [PubMed] [Google Scholar]
- 58.Perron K, Caille O, Rossier C, Van Delden C, Dumas J-L, Köhler T. 2004. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J Biol Chem 279:8761–8768. doi: 10.1074/jbc.M312080200. [DOI] [PubMed] [Google Scholar]
- 59.Kim EH, Nies DH, McEvoy MM, Rensing C. 2011. Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J Bacteriol 193:2381–2387. doi: 10.1128/JB.01323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munson GP, Lam DL, Outten FW, O'Halloran TV. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol 182:5864–5671. doi: 10.1128/jb.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rensing C, Franke S, Rensing C, Franke S. 11 January 2007. Copper homeostasis in Escherichia coli and other Enterobacteriaceae. EcoSal Plus 2007 doi: 10.1128/ecosalplus.5.4.4.1. [DOI] [PubMed] [Google Scholar]
- 62.Lu CH, Lin YF, Lin JJ, Yu CS. 2012. Prediction of metal ion-binding sites in proteins using the fragment transformation method. PLoS One 7:e39252. doi: 10.1371/journal.pone.0039252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krishna Deepak RNV, Chandrakar B, Sankararamakrishnan R. 2017. Comparison of metal-binding strength between methionine and cysteine residues: implications for the design of metal-binding motifs in proteins. Biophys Chem 224:32–39. doi: 10.1016/j.bpc.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 64.Garber ME, Rajeev L, Kazakov AE, Trinh J, Masuno D, Thompson MG, Kaplan N, Luk J, Novichkov PS, Mukhopadhyay A. 2018. Multiple signaling systems target a core set of transition metal homeostasis genes using similar binding motifs. Mol Microbiol 107:704–717. doi: 10.1111/mmi.13909. [DOI] [PubMed] [Google Scholar]
- 65.Banos RC, Vivero A, Aznar S, Garcia J, Pons M, Madrid C, Juarez A. 2009. Differential regulation of horizontally acquired and core genome genes by the bacterial modulator H-NS. PLoS Genet 5:e1000513. doi: 10.1371/journal.pgen.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 67.Silphaduang U, Mascarenhas M, Karmali M, Coombes BK. 2007. Repression of intracellular virulence factors in Salmonella by the Hha and YdgT nucleoid-associated proteins. J Bacteriol 189:3669–3673. doi: 10.1128/JB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 69.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun 66:480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manson JM, Gilmore MS. 2006. Pathogenicity island integrase cross-talk: a potential new tool for virulence modulation. Mol Microbiol 61:555–559. doi: 10.1111/j.1365-2958.2006.05262.x. [DOI] [PubMed] [Google Scholar]
- 71.Dobrindt U, Blum-Oehler G, Hartsch T, Gottschalk G, Ron EZ, Fünfstück R, Hacker J. 2001. S-Fimbria-encoding determinant sfa(I) is located on pathogenicity island III(536) of uropathogenic Escherichia coli strain 536. Infect Immun 69:4248–4256. doi: 10.1128/IAI.69.7.4248-4256.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Capra EJ, Laub MT. 2012. Evolution of two-component signal transduction systems. Annu Rev Microbiol 66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rademacher C, Masepohl B. 2012. Copper-responsive gene regulation in bacteria. Microbiology 158:2451–2464. doi: 10.1099/mic.0.058487-0. [DOI] [PubMed] [Google Scholar]
- 74.Garcia Vescovi E, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 75.Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol 50:219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- 76.Wosten MM, Groisman EA. 1999. Molecular characterization of the PmrA regulon. J Biol Chem 274:27185–27190. doi: 10.1074/jbc.274.38.27185. [DOI] [PubMed] [Google Scholar]
- 77.Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67:83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dalebroux ZD, Miller SI. 2014. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr Opin Microbiol 17:106–113. doi: 10.1016/j.mib.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gunn JS. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol 16:284–290. doi: 10.1016/j.tim.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 80.Chandrangsu P, Rensing C, Helmann JD. 2017. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol 15:338–350. doi: 10.1038/nrmicro.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.German N, Doyscher D, Rensing C. 2013. Bacterial killing in macrophages and amoeba: do they all use a brass dagger? Future Microbiol 8:1257–1264. doi: 10.2217/fmb.13.100. [DOI] [PubMed] [Google Scholar]
- 82.Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun 61:5361–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Segal G, Shuman HA. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect Immun 65:5057–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 85.Zusman T, Yerushalmi G, Segal G. 2003. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect Immun 71:3714–3723. doi: 10.1128/iai.71.7.3714-3723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Segal G, Shuman HA. 1999. Legionella pneumophila utilize the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun 67:2117–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Presence and absence of lpg0714-lpg0715-lpg0716 in the Legionella genus. A maximum likelihood tree of 41 Legionella species was reconstructed on the basis of concatenated amino acid alignment of 78 orthologous open reading frames (41). For each species, the presence (grey) or absence (white) of lpg0714, lpg0715, and lpg0716 genes is indicated. Download FIG S1, TIF file, 0.9 MB (907.2KB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Regulator-effector island undergoes HGT in the Legionella genus. (A) Schematic presentation of the genomic region where the lpg0714-lpg0715-lpg0716 L. pneumophila orthologs are located in the six characterized Legionella species, L. pneumophila (Lpn), L. moravica (Lmor), L. quateirensis (Lqua), L. worsleiensis (Lwor), L. nautarum (Lnau), and L. brunensis (Lbru), and the three uncharacterized Legionella species, Legionella sp. strain 13.8642 (L_13.8642), Legionella sp. strain FW215 (L_FW215), and Legionella sp. strain Km535 (L_Km535), harboring the regulator-effector island. Homologous genes are indicated by the same color, and nonhomologous genes are white. The regions between lpg0713, encoding an oligopeptide transporter, and lpg0719, encoding a valyl-tRNA synthetase in the nine species, are presented. The genes are indicated by their locus tag number. (B) Comparison of the GC content of the lpg0714-lpg0715-lpg0716 genes and the pmrA-pmrB-legA3 genes to the genomic GC content of the nine Legionella species harboring the regulator-effector island. Download FIG S2, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Consensus regulatory element of genes found next to the LciRS homologous TCSs in different bacteria. The regulatory region of genes located near the genes encoding the LciRS homologous TCS in different bacteria is shown: L. pneumophila (Lpg), L. moravica (Lmor), L. quateirensis (Lqua), L. worsleiensis (Lwor), L. nautarum (Lnau), L. brunensis (Lbru), Legionella sp. strain 13.8642 (L_13.8642), Legionella sp. strain FW215 (L_FW215), Legionella sp. strain Km535 (L_Km535), Escherichia coli (Esc), Salmonella enterica (Sal), Enterobacter cloacae (Ent), Achromobacter species (Ach), Pseudomonas aeruginosa (Pa), Pseudomonas fluorescens (Pf), Pseudomonas putida (Pp), Pseudomonas stutzeri (Ps), and Bordetella pertussis (Bper). In purple are the putative LciR inverted-repeat regulatory elements, in light blue are the two additional regulatory elements found between the inverted-repeat and the −10 promoter element, in dark blue are the −10 promoter elements, and the experimentally determined transcription start sites (TSSs) are underlined and marked in blue. Download FIG S3, TIF file, 1.5 MB (1.6MB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effect of copper on L. pneumophila and E. coli growth in media. L. pneumophila (A) and E. coli (B) were grown with different concentrations of copper (indicated on the right), and their growth (OD600) was examined in intervals of 1 h for the time indicated. Download FIG S4, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LciRS TCS and the effector LciE are dispensable for intracellular growth. (A and B) The intracellular growth of the lciR, lciS, lciE, and lciRS-lciE deletion mutants was examined in A. castellanii (A) and HL-60-derived human macrophages (B). Symbols: diamonds, L. pneumophila wild-type strain JR32; squares, icmT mutant; triangles, lciS deletion mutant; black circles, lciR deletion mutant; star, lciE deletion mutant; white circles, lciRS-lciE triple deletion mutant. The experiments were performed three times, and similar results were obtained. (C and D) Intracellular competition assay between the L. pneumophila lciRS-lciE triple deletion mutant (white circles) and the JR32 wild-type strain (diamonds) in A. castellanii. The experiment was performed without (C) and with (D) 200 μM copper. Download FIG S5, TIF file, 0.8 MB (804.7KB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effect of mutations and copper on the expression of lciE and lciR. (A) The effect of mutations on the amino acids predicted to be required for phosphorylation of LciR (LciRD51A) and LciS (LciSH148A) on lciE induction by copper. The levels of expression of wild-type lciRS-lciE-lacZ fusion and the lciRS-lciE-lacZ fusion containing a mutation in the amino acids, with and without 50 μM copper, are indicated below the bars. The levels of expression of the lacZ fusions were found to be significantly different (*, P < 10−5, paired Student’s t test) between mutated fusions and the wild-type fusion after exposure to copper. (B) lciR expression is activated neither by copper nor by the LciR regulator. The expression of the lciR-lacZ fusion was examined in the wild-type strain and in the lciR deletion mutant with (grey bars) and without (white bars) the addition of 50 μM copper. Download FIG S6, TIF file, 0.7 MB (776.2KB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predicted Fis regulatory elements in the regulatory regions of lciE and lciR homologs in different Legionella species. The promoter region of the lciE (A) and lciR (B) genes in the nine Legionella species harboring the island, L. pneumophila (lciE and lciR), L. moravica (Lmor), L. quateirensis (Lqua), L. worsleiensis (Lwor), L. nautarum (Lnau), L. brunensis (Lbru), Legionella sp. strain 13.8642 (L_13.8642), Legionella sp. strain FW215 (L_FW215), and Legionella sp. strain Km535 (L_Km535). In purple are the putative LciR inverted-repeat regulatory elements, in light blue are the two additional regulatory elements found between the inverted repeat and the −10 promoter element of lciE, in dark blue are the −10 promoter elements, and the experimentally determined transcription start sites (TSSs) are underlined and marked in blue. The predicted Fis sites in both promoter regions are marked: the site overlapping or close to the −10 promoter element is colored yellow, and a second predicted Fis site (when present) is underlined. In both predicted Fis sites, the conserved TG and CA consensus nucleotides are in red and the nucleotides mutated are marked by asterisks. Download FIG S7, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
L. pneumophila contains two effectors with a domain homologous to LciE. Schematic representation of the effectors LciE (lpg0716), CetLp1 (lpg0140), and lpg2888. The domain homologous among the three effectors is marked in light blue, and putative transmembrane domains are marked in black. In addition, protein sequence alignment of the homologous domain found in the three effectors is shown from the six characterized Legionella species in which the LciRS-LciE genomic island was found, L. pneumophila (LciE/lpg2888/CetLp1), L. moravica (Lmor), L. quateirensis (Lqua), L. worsleiensis (Lwor), L. nautarum (Lnau), and L. brunensis (Lbru). Download FIG S8, TIF file, 2.1 MB (2.1MB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Examination of the connection of CetLp1 and lpg2888 to copper and LciE. The levels of expression of cetLp1 (A) and lpg2888 (B) lacZ fusions were examined without the addition of metals and after exposure to 50 μM copper (grey bars) or zinc (hatched bars). The same fusions were also examined in the lciR deletion mutant under the same conditions. (C and D) Intracellular competition assay between the L. pneumophila cetLp1-lpg2888 double deletion mutant (circles) and the JR32 wild-type strain (diamonds) in A. castellanii. (E and F) Intracellular competition assay between the L. pneumophila cetLp1-lpg2888-lciE triple deletion mutant (circles) and the JR32 wild-type strain (diamonds) in A. castellanii. The experiment was performed without (C and E) and with (D and F) 200 μM copper. Download FIG S9, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains, plasmids, and primers used in this study. Download Data Set S1, XLSX file, 0.03 MB (33.9KB, xlsx) .
Copyright © 2020 Linsky et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.