Abstract
Recent studies have highlighted the critical role of angiogenesis and sympathetic innervation in adipose tissue remodeling during the development of obesity. Therefore, developing an easy and efficient method to document the dynamic changes in adipose tissue is necessary. Here, we describe a modified immunofluorescent approach that efficiently co-stains blood vessels and nerve fibers in adipose tissues. Compared to traditional and recently developed methods, our approach is relatively easy to follow and more efficient in labeling the blood vessels and nerve fibers with higher densities and less background. Moreover, the higher resolution of the images further allows us to accurately measure the area of the vessels, amount of branching, and length of the fibers by open source software. As a demonstration using our method, we show that brown adipose tissue (BAT) contains higher amounts of blood vessels and nerve fibers compared to white adipose tissue (WAT). We further find that among the WATs, subcutaneous WAT (sWAT) has more blood vessels and nerve fibers compared to epididymal WAT (eWAT). Our method thus provides a useful tool for investigating adipose tissue remodeling.
Keywords: Immunology and Infection, Issue 144, Adipose tissue, blood vessels, angiogenesis, nerve fibers, sympathetic innervation, white adipose tissue, WAT, brown adipose tissue, BAT
Introduction
Adipose tissue has key metabolic and endocrine functions1. It dynamically expands or shrinks in response to different nutrient stresses2. The active tissue remodeling process consists of multiple physiological paths/steps including angiogenesis, fibrosis, and shaping of local inflammatory microenvironments2,3,4. Some physical stimuli, such as cold exposure and exercise, may trigger sympathetic activation, which ultimately leads to new blood vessel formation and sympathetic innervation in adipose tissues5,6. These remodeling processes are linked tightly to systemic metabolic outcomes including insulin sensitivity, the hallmark of type 2 diabetes2. Thus, visualization of these pathological changes is very important to understanding the healthy status of whole adipose tissues.
Angiogenesis is the process of new blood vessel formation. Since blood vessels provide oxygen, nutrients, hormones, and growth factors to tissue, angiogenesis has been considered a key step in adipose tissue remodeling, which has been documented with different techniques6,7,8,9,10,11,12,13. However, there remain questions about the resolution of the images, efficiency of immunostaining, and methods for quantification of vessel density. Compared to new blood vessel formation, innervation in adipose tissue has been underestimated for a long time. Recently, Zeng et al.14 used advanced intravital two-photon microscopy and demonstrated that adipocytes are surrounded by layers of nerve fibers14. Since then, researchers have started to appreciate the pivotal role of sympathetic innervation in regulation of adipose tissue physiology. Thus, developing an easy and practical approach to document adipose nerve innervation is important.
Here, we report an optimized method for the co-staining of blood vessels and nerve fibers based on our previous protocols. With this method, we can achieve clear images of blood vessels and nerve fibers without noisy background. Moreover, we obtain a resolution that is high enough for performing quantitative measurement of densities with open source software. By using this new approach, we can successfully compare the structures and densities of blood vessels and nerve fibers in different adipose depots.
Protocol
All procedures containing animal subjects have been approved by the Animal Welfare Committee of University of Texas Health Science Center at Houston (animal protocol number: AWC-18–0057).
1. Reagent Preparation
1× phosphate-buffered saline (PBS, pH 7.4): to make 1 L of 1× PBS, dissolve 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 in 800 mL of distilled water. Adjust the pH to 7.4 and fill with distilled water to achieve a final volume of 1 L.
-
1% paraformaldehyde in 1× PBS (1% PFA, wt/vol): to achieve a final volume of 50 mL, add 3.125 mL of 16% PFA (see Table of Materials) to 46.875 mL of 1× PBS. Mix well and store at 4 °C for later use.
CAUTION: Paraformaldehyde is toxic. All procedures should be carried out under a fume hood to avoid inhalation and skin contact.
0.1% polysorbate 20 (see Table of Materials) in 1× PBS (1× PBST, pH 7.4, vol/vol): to achieve a final volume of 50 mL, add 0.05 mL of polysorbate 20 to 49.95 mL of 1× PBS. Vortex and store at 4 °C for up to 1 month.
1% octoxynol-9 (see Table of Materials) in 1× PBS (1× PBS-TX, pH 7.4, vol/vol): to achieve a final volume of 10 mL, add 0.1 mL of octoxynol-9 to 9.9 mL of 1× PBS. Vortex and store at 4 °C for up to 1 month.
2% sodium azide (wt/vol): to produce 10 mL of 2% sodium azide, add 0.2 g of sodium azide to 10 mL of 1× PBS. Mix well and store at 4 °C for later use.
90% glycerol in 1× PBS (vol/vol): to achieve a final volume of 10 mL, add 1 mL of 1× PBS to 9 mL of glycerol. Vortex and store at room temperature (RT) for later use.
2. Animal Ddissection and Adipose Tissue Collection
Use 6-week-old C57BL/6J male mice. Anesthetize the mice with isoflurane (4%–5% for induction then 1%–2% for maintenance). Once anesthetized, perform a toe-pinch reflex to determine the anesthetic depth, judging by the lack of pedal reflexes. Once the mice are deeply anesthetized, dissect the mice following the protocol as previously published15.
Sterilize the blunt scissors and surgical area of the chest (no need to shave the hair). Open the chest by cutting the diaphragm and ribs along the lateral surface with a size of ~2 cm using blunt scissors. Clamp the chest flag with hemostat and reflect it by putting the hemostat over the head.
Make a small incision into the left ventricle (~0.5 cm) with iris scissors. Insert an olive-tipped perfusion needle through the incision site and clamp the needle tip in place with the hemostat. Attach the needle to a 50 mL syringe containing paraformaldehyde fixation solution (1% PFA, pH 7.4).
-
Open the right atrium by section cutting with iris scissors as the perfusion outlet. Start the perfusion by gently and continuously pushing the syringe at a perfusion rate close to 10 mL/min. Stop pushing when the fluid exiting the outlet is clear of any blood. The whole process should require ~5 min.
CAUTION: Perform these steps under a fume hood to prevent toxicity from PFA.
Dissect white and brown adipose tissues from the mice. Use scissors to break the tissue samples into small pieces of chunks approximately 5 × 5 × 3 mm3. Transfer the small chunks with the sterilized tweezers into the tissue embedding cassettes with proper labeling of the tissue samples on the cassettes.
-
Immerse the embedding cassettes containing tissue samples into the fixation solution (1% PFA in 1× PBS, pH 7.4) at 4 °C for 24–48 h.
NOTE: The fixation solution and fixation time vary based on sizes of the tissues16,17,18.
-
Wash the samples with fresh 1× PBS buffer three times (0.5 mL/wash).
CAUTION: Perform this step under a fume hood. The paraformaldehyde waste should be properly handled according to hazardous waste disposal procedures.
NOTE: The tissues can be stored in 1× PBS at 4 °C for further processing.
3. Antibody Incubation
Cut the fixed tissue samples into approximately 2 mm3 cubes with the sterilized scissors. For permeabilization, transfer the samples into a 1.5 mL tube containing 1 mL of 1× PBS-TX (1% octoxynol-9 in 1× PBS, see Table of Materials, pH 7.4) and gently rotate the tubes at RT for 1 h at 18 rpm.
Carefully remove the 1× PBS-TX by aspiration. Wash the samples 3× by adding the 1× PBS directly into the same tubes (no need to change the tubes). During each wash, invert the tubes several times.
For blocking, add 0.5 mL of blocking buffer (Table of Materials) to the samples and incubate at RT for 2 h with gentle rotation.
To prepare 0.4 mL of primary antibody solution, dilute 2 μL of anti - endomucin (the marker of blood vessels)5 (1:200) and 2 μL of anti — tyrosine hydroxylase (TH, the marker of nerve fibers)5 (1:200, 1 μg/mL) antibodies (Table of Materials for antibody information) into 396 μL of blocking buffer. Vortex and spin down to recover the volume.
-
For the first antibody incubation, carefully remove the blocking buffer from the tissue chunks. Add 100 μL of primary antibody solution prepared in step 3.4 into tubes and incubate at 4 °C overnight.
NOTE: The antibody dilution and incubation time may vary among different antibodies. It is recommended to add 0.02% sodium azide to the antibody solution to prevent microbial growth. To prepare 0.4 mL of primary antibody solution with sodium azide, add 4 μL of antibodies (1 μL of each) and 4 μL of 2% sodium azide into 392 μL of blocking buffer.
Carefully collect the primary antibody solution for reuse if desired. Wash the samples with 1× PBST (pH 7.4, 100 μL/wash) three times (30 min each) with gentle rotation at 18 rpm.
-
For the preparation of secondary antibody solution, dilute 2 μL of fluorophore (wave length 495 nm) conjugated anti-goat IgG (1:200) and 2 μL of fluorophore (wave length 650 nm) conjugated anti-rabbit IgG (1:200) (see Table of Materials) into 396 μL of blocking buffer. Vortex and spin briefly to collect all the liquid.
NOTE: Protect the samples from light during the following procedures.
For the second antibody incubation, remove the last wash buffer from samples. Add 100 μL of secondary antibody solution into tubes and incubate at RT for 2 h with gentle rotation at 18 rpm.
-
Carefully remove the secondary antibody solution. Wash the samples 3× with 1× PBST (pH 7.4, 30 min each) with gentle rotation at 18 rpm.
NOTE: Do not reuse this secondary antibody solution, as it may be contaminated with the trace amounts of primary antibodies brought by the tissues.
-
For optical clearance, immerse the samples in 1 mL of 90% glycerol and keep the samples at 4 °C in darkness until they become transparent.
NOTE: The immersion duration depends on the tissue type and size. Typically, a 2 mm3 cube of white adipose tissue needs overnight incubation, while the same size brown adipose tissue sample needs longer.
-
Adhere a silicone isolator to the slide to create a well for volume imaging. Alternatively, attach several layers of transparent tape to the slides and cut a square out of the middle to create a well (Figure 2).
NOTE: Adjust the well depth to fit the sample size. In this protocol, a 1 mm well depth is used.
-
Carefully transfer the samples into the well and fill it with the mounting medium (see Table of Materials). Lay a cover slip on the surface and seal the corners of the cover slip with high viscosity medium (see Table of Materials). Let the mounting medium cure for 24 h at RT in darkness.
NOTE: Ensure that no space is left between the sample and cover slip. Avoid bubbles under the cover slip. Avoid placing excessive pressure on the samples.
Figure 2: Diagrams of the wells for volume imaging.
(A) Diagram of a slide with a silicone isolator. (B) Diagram of a well with multiple layers of tape made by our lab.
4. Image Acquisition
Acquire Z-stack images (refer to steps 4.2–4.7) with the 20× objective of a confocal microscope/software.
Start the system. Click on the <Configuration> tab and activate the argon laser and HeNe 633 laser. Click on the <Acquire> tab. In the <Visible> laser lines panel, move the corresponding intensity sliders up to choose the laser lines of 488 nm and 633 nm. Initially, the intensities can be set to 20%–30%. Select the 488/561/633 triple dichoric mirror. Activate the PMTs by clicking on the <Active> checkboxes, choosing PMT1 for the emission excited by the 488 nm laser and PMT3 for that of the 633 nm laser. Set the wavelength range to 500–550 nm for PMT1 and 650–750nm for PMT3. Select the psudocolor to be used for image display by double clicking the colored rectangle beside each PMT and choosing a color from the pop-up menu.
-
Click on <Live> to check the image of samples. Use the z-Position knob to select a plane of focus within the XY region of interest. Adjust the brightness of the images by tuning the laser intensity, smart gain and offset. For each fluorescence channel, perform this adjustment under the <QLUT> (Quick Look Up Table) display mode. Click on the <QLUT> button to enter the QLUT mode, in which saturated pixels are displayed in blue to aid the setting of appropriate brightness level. Click twice on the <QLUT> button to go back to the pseudocolor display mode.
NOTE: The numerical values of the setting may vary among each experiment.
-
While scanning, turn the Z-Position knob counterclockwise to move the plan of focus to one end of the volume of interest and then click on the <End> arrowhead to set the scanning end position. Then turn the Z-Position knob clockwise to move the plane of focus through the specimen to the other end of the volume of interest and then click on the <Begin> arrowhead to set the scanning begin position.
NOTE: For the comparison between different samples, define the same Z-volume for different samples.
Adjust the z-step size to 3 μm. Change the image quality by selecting a format of 1024 × 1024, speed of 100 Hz, and line average of 2.
Select <Start> to initiate the z-stack image acquisition.
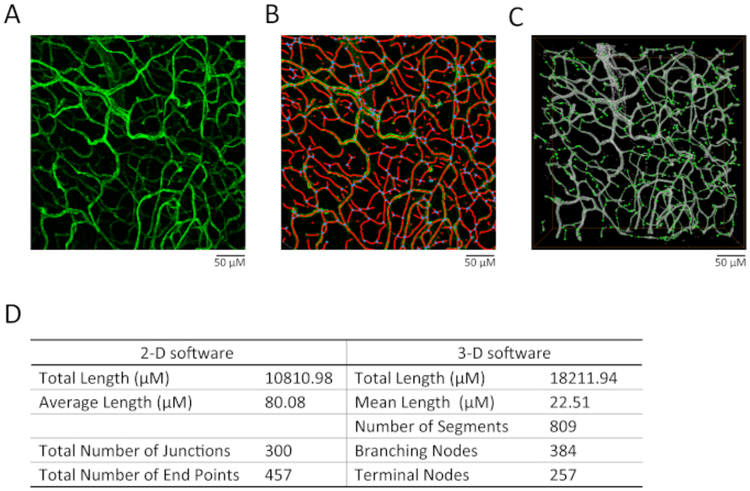
For maximum projection of the acquired image stack, click on <Process> tab and then <Tool>, select <3D Projection> and enter <Maximum> in the method list without changes to X, Y and Z. Set <Threshold> to 0. Click on <Apply>. The maximum intensity of the Z-volume will be stacked into a 2D image which will be shown (Figure 4A).
Figure 4: Comparison of 2D and 3D analyses on vessel structure.
(A) The effect of maximum projection on the images. (B) The analytic results by the 2D software (see Table of Materials). Particularly, the red lines indicate selected skeleton, while the blue dots indicate branding points. (C) The analytic results by the 3D software (see Table of Materials). The gray area is the selected region for analysis. The green line indicates the measured segments and green dots indicate the measured nodes. (D) The comparison of vessel length, segment numbers, branching node numbers, and terminal node numbers analyzed by 2D or 3D methods.
5. Analysis of Blood Vessel and Nerve Fiber Networks
1. Analysis of 2D images via open source software (see Table of Materials)19
Open the stacked images in the software. Adjust Vessel diameter and intensity until all the vessel structures can be properly selected (Figure 4B).
Select Run analysis and export the data following guidance of the software.
2. Analysis of 3D images via the licensed software (see Table of Materials)
Open the raw data of images with the software. Adjust the threshold and other parameters until the vessel structures in each layer of the z-volume are properly selected (refer to the user guide for details in the software) (Figure 4C).
Analyze the selected segments characters and export the data following the guidance of the software.
Representative Results
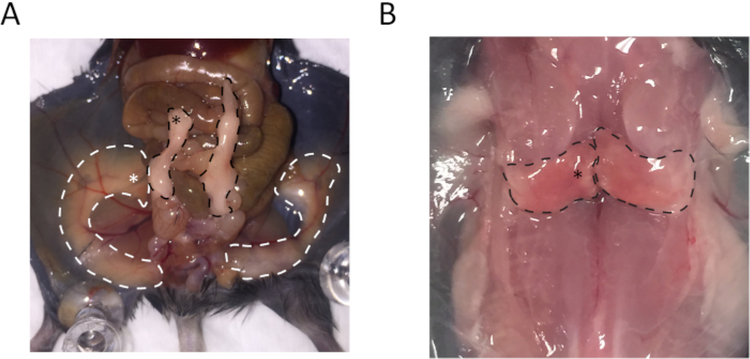
The distal region of the epididymal white adipose tissue (eWAT), medial region of the dorsolumbar subcutaneous white adipose tissue (sWAT), and medial region of the interscapular brown adipose tissue (BAT) were collected. The locations for collecting these tissues are indicated in Figure 1.
Figure 1: Anatomy of subcutaneous white adipose tissue (sWAT), epididymal white adipose tissue (eWAT), and brown adipose tissue (BAT) indicates regions used for collecting the samples.
(A) The regions outlined by white dashed lines represent sWAT, and the white asterisk highlighted location is the site for collecting sWAT. Regions outlined by black dashed lines are eWAT, and the black asterisk highlighted location is the site for collecting eWAT. (B) The regions outlined by black dashed lines are BAT, and the tissue collection region is highlighted by the asterisk.
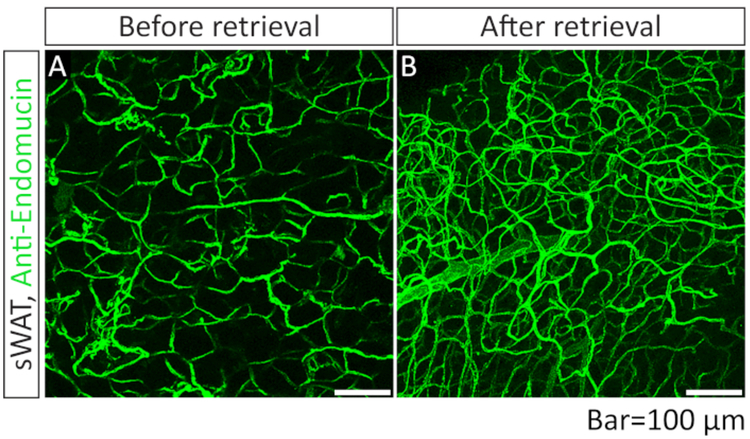
After whole-mount staining, the tissue chunks were mounted in a well of 1 mm depth (Figure 2) and imaged with the confocal microscope. We first tested the effect of the clearing step with 90% glycerol incubation on quality of the images. We found that more blood vessels were positively stained with α-endomucin antibody in the glycerol-incubated sWAT, suggesting that the clearing step is critical for complete staining of the blood vessels (Figure 3, compare panels A and B).
Figure 3: Comparison of IF images of blood vessels acquiredwith or without optical clearing step with 90% glycerol.
(A) Whole-mount immunofluorescence (IF) staining with anti-endomucin antibody (green) in sWAT. The sample was not subjected to the optical clearing step (step 3.10). (B) Whole-mount IF staining with anti-endomucin antibody in sWAT. The sample was subjected to the optical clearing step (step 3.10) before the mounting steps (steps 3.11 and 3.12).
However, the clearing step with glycerol did not affect the integrity or shape of the blood vessels (Figure 3). Given these beneficial effects, in the following experiments, the clearing step was performed.
We further compared the results of 2D and 3D analyses using different software (see Table of Materials). We found that, while the 3D images apparently provided more detailed structural information, the two analytic methods eventually did not show significant differences in terms of the length and branch number of the vessels (Figure 4).
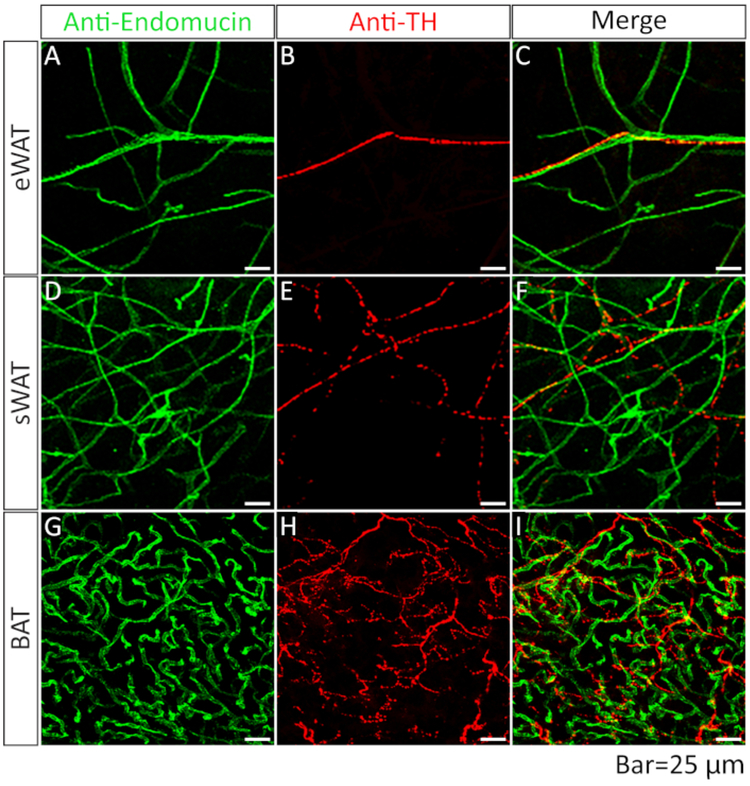
Previous publications have demonstrated that different adipose depots possess heterogeneous properties1,18. Here we sought to determine whether blood vessels and nerve fibers exhibit different patterns among the depots using our newly developed method. To achieve this, we performed co-IF staining with α-endomucin (for blood vessels) and α-TH (for nerve fibers) antibodies in adipose tissue. Interestingly, results showed that there were significantly more blood vessels and nerve fibers in BAT compared to WATs (Figure 5, compare bottom lanes to top and middle lanes)
Figure 5: Comparison of blood vessels and nerve fibers acquired from different adipose depots.
Whole-mount IF staining with anti-endomucin (green) and anti-tyrosine hydroxylase (TH) (red) antibodies in eWAT (A, B, merged in C), sWAT (D, E, merged in F) and BAT (G, H, merged in I).
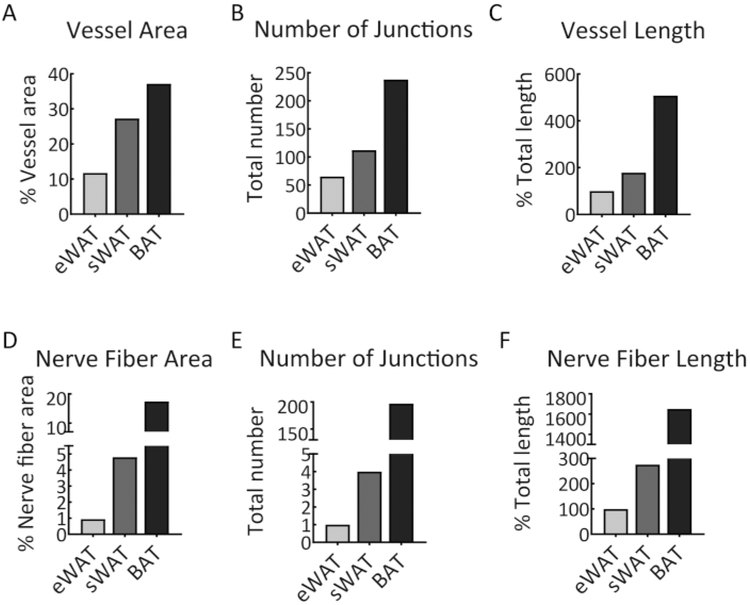
Among the WATs, the sWAT exhibited higher blood vessel density than the eWAT (Figure 5, compare the middle to top lane). Of note, while the nerve fibers expanded in parallel with the blood vessels, they did not show significant co-localization (Figure 5, merged lanes). We further quantitatively measured the vessel area, number of junctions, and tube length with the 2D method and found similar results as described above (Figure 6).
Figure 6: Quantitative analysis of vascular and nerve fiber networks.
(A) The percentage of blood vessel area in the samples of eWAT, sWAT, and BAT. (B) The number of junctions of the vascular networks in eWAT, sWAT, and BAT. (C) The total vessel length of vascular networks in eWAT, sWAT, and BAT. (D) The percentage of neve fiber area in the samples of eWAT, sWAT and BAT. (E) The number of junctions of the nerve fibers in eWAT, sWAT, and BAT. (F) The total length of nerve fibers in eWAT, sWAT, and BAT. The analyses were performed with 2D software. The results were achieved from the images presented in Figure 5.
In summary, this approach successfully co-stained blood vessels and nerve fibers in different adipose tissues.
Discussion
Adipose tissue remodeling is directly linked to metabolic dysregulation during obesity development1,2. Angiogenesis and sympathetic innervation are both essential for the dynamic remodeling process2,12. Therefore, developing an applicable approach to visualize the new blood vessels as well as nerve fibers are of great importance. Previous methods have been reported for documenting angiogenesis in adipose tissue. However, some issues remain with these approaches, including low efficiency, fussy resolution, and noisy background. Meanwhile, sympathetic innervation has only been recognized recently as a critical step in adipose tissue pathology14. Even though the related findings are interesting, applied methods require advanced microscopy tools and are time-consuming. Here, we report an easy-to-follow approach modified from our previous protocol for co-staining blood vessels and sympathetic nerve fibers. Interestingly, we successfully stained the blood vessels and nerve fibers with high resolution, and the staining has qualities comparable to previously reported images18.
We previously stained the blood vessels in adipose tissues by immunohistochemistry or immunofluorescence with the α-CD31 antibody, a marker of endothelial cells, on paraffin-embedded slides8,13,20. While the method allows us to distinguish blood vessel density between different groups, the microvascular vessels that were positively stained were not complete. Here, we chose a whole-mount method and performed IF staining with a α-endomucin antibody, another marker for blood vessels. With this optimized approach, we were able to achieve staining with more blood vessels, especially microblood vessels. Moreover, in this method, since we avoided steps such as paraffin embedding and formalin fixation, which have the potential to impair the integrity of blood vessels, we obtained images with higher resolution and more intact vessels. The unimpaired structure of the vessels further allowed us to measure and compare their lengths, branching, and areas. Of note, we added a clearing step with glycerol, so the tissue chunks could become more transparent, and this simple but critical step significantly enhanced efficiency of the staining18,21.
Due to high levels of lipid contents in the adipose tissue, which might limit the accessibility of antibodies to the inner regions of the tissue, we cut the depots into small pieces to ensure adequate antibody binding to the target proteins. Therefore, our method successfully stains more blood vessels and nerve fibers than staining on whole tissues. However, it may lose spatial information and hence affect the integrity of the nerve fibers. To resolve this apparent issue and ensure the images are comparable among individual mice, it is suggested to collect the small chunks from the same regions in the adipose depots. If more spatial information is further needed, larger chunks should be obtained for staining. For larger samples, longer fixation times with higher doses of FPA, higher concentrations of detergents for permeabilization, and longer periods of antibody incubation may be needed.
Adipose tissue innervation has been investigated recently. Multiple advanced techniques have been applied to visualize nerve fibers14,17. These methods are either expensive or time-consuming. Here, we simply performed an IF stain with α-TH (a sympathetic nerve marker) and successfully stained the sympathetic nerve fibers at a high quality. More importantly, we performed the co-staining with α-endomucin and α-TH to investigate how the blood vessels interplay with sympathetic nerve fibers during adipose tissue remodeling.
Of note, we compared the results from 2D and 3D analyses using different software. It was found that, while the 3D images provided more detailed structural information, the two analytic methods did not show significant difference in terms of the length and branching of the vessels. Eventually, when analyzing with the 3D method, the software itself may detect some false signals that need to be manually adjusted. Given that the 2D software is purposely designed for vessel analysis, it is thus suggested to use this method for quantitatively measuring the structure of vessels.
With this method, blood vessels and nerve fibers in different adipose tissue were co-stained, and their densities, branching, and lengths were compared19. It was found that both blood vessels and nerve fibers are much thicker in BAT compared to WAT, suggesting that BAT is a more metabolically active adipose depot. Moreover, the blood vessel and nerve densities in sWAT were higher than in the eWAT, indicating that different WATs have different remodeling profiles.
In conclusion, this method efficiently co-stains blood vessels and nerve fibers in adipose tissue5. Furthermore, it serves as a useful tool for studying the dynamic changes in adipose tissue during obesity development.
Supplementary Material
Acknowledgments
This study was supported by the National Institute of Health (NIH) grant R01DK109001 (to K.S.).
Footnotes
Video Link
The video component of this article can be found at https://www.jove.com/video/59266/
Disclosures
The authors declare that they have no competing financial interests.
References
- 1.Rosen ED, Spiegelman BM What we talk about when we talk about fat. Cell. 156 (1–2), 20–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun K, Kusminski CM, Scherer PE Adipose tissue remodeling and obesity. Journal of Clinical Investigations. 121 (6), 2094–2101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun K et al. Endotrophin triggers adipose tissue fibrosis and metabolic dysfunction. Nature Communication. 5, 3485(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y et al. Divergent functions of endotrophin on different cell populations in adipose tissue. American Journal of Physiology-Endocrinology and Metabolism. 311 (6), E952–E963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y et al. Transient Overexpression of VEGF-A in Adipose Tissue Promotes Energy Expenditure via Activation of the Sympathetic Nervous System. Molecular and Cellular Biology. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue Y et al. Hypoxia-independent angiogenesis in adipose tissues during cold acclimation. Cell Metabolism. 9 (1), 99–109 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Chen S et al. LncRNA TDRG1 enhances tumorigenicity in endometrial carcinoma by binding and targeting VEGF-A protein. BBA Molecular Basis of Disease. 1864 (9 Pt B), 3013–3021 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Sun K et al. Dichotomous effects of VEGF-A on adipose tissue dysfunction. Proceedings of the National Academy of Sciences of the United States of America. 109 (15), 5874–5879 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.During MJ et al. Adipose VEGF Links the White-to-Brown Fat Switch With Environmental, Genetic, and Pharmacological Stimuli in Male Mice. Endocrinology. 156 (6), 2059–2073 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elias I et al. Adipose tissue overexpression of vascular endothelial growth factor protects against diet-induced obesity and insulin resistance. Diabetes. 61 (7), 1801–1813 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung HK et al. Adipose vascular endothelial growth factor regulates metabolic homeostasis through angiogenesis. Cell Metabolism. 17 (1), 61–72 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Cao Y Angiogenesis and vascular functions in modulation of obesity, adipose metabolism, and insulin sensitivity. Cell Metabolism. 18 (4), 478–489 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Sun K et al. Brown adipose tissue derived VEGF-A modulates cold tolerance and energy expenditure. Molecular Metabolism. 3 (4), 474–483 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng W et al. Sympathetic neuro-adipose connections mediate leptin-driven lipolysis. Cell. 163 (1), 84–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gage GJ, Kipke DR, Shain W Whole animal perfusion fixation for rodents. Journal of Visualized Experiments. (65), (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry R et al. Imaging of adipose tissue. Methods in Enzymology. 537, 47–73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang H, Ding X, Cao Y, Wang H, Zeng W Dense Intra-adipose Sympathetic Arborizations Are Essential for Cold-Induced Beiging of Mouse White Adipose Tissue. Cell Metabolism. 26 (4), 686–692 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Chi J et al. Three-Dimensional Adipose Tissue Imaging Reveals Regional Variation in Beige Fat Biogenesis and PRDM16-Dependent Sympathetic Neurite Density. Cell Metabolism. 27 (1), 226–236 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Zudaire E, Gambardella L, Kurcz C, Vermeren S A computational tool for quantitative analysis of vascular networks. PLoS One. 6 (11), e27385(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An YA et al. Angiopoietin-2 in white adipose tissue improves metabolic homeostasis through enhanced angiogenesis. eLife. 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chi J, Crane A, Wu Z, Cohen P Adipo-Clear: A Tissue Clearing Method for Three-Dimensional Imaging of Adipose Tissue. Journal of Visualized Experiments. (137), (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.