Abstract
Noroviruses are the leading cause of acute gastroenteritis and foodborne disease in the United States (U.S.). About 1 in 5 reported norovirus outbreaks are spread through foodborne transmission, presenting opportunities for prevention. We describe the epidemiology of U.S. foodborne norovirus outbreaks reported to national surveillance systems, including differences between genotypes. Foodborne outbreaks that occurred during August 2009–July 2015 with norovirus reported as a single confirmed etiology to the National Outbreak Reporting System (NORS) were matched with outbreaks reported to CaliciNet, a U.S. laboratory norovirus outbreak surveillance network. We analyzed these matched outbreaks stratified by genotype for epidemiologic characteristics, including setting, size and duration, health outcomes of case-patients, implicated food, and outbreak contributing factors. Four hundred ninety-three confirmed foodborne norovirus outbreaks were reported in both NORS and CaliciNet. The most common norovirus genotypes reported were GII.4 (52%), GII.6 (9%), and GI.3 (8%). Compared to non-GII.4 outbreaks, GII.4 outbreaks had higher hospitalization rates (12.8 vs. 4.8 per 1,000 cases, P < 0.01). While contaminated foods were identified and reported in only 35% of outbreaks, molluscan shellfish (4% overall) were more often implicated in non-GII.4 outbreaks than in GII.4 outbreaks (7% vs. 1%, P = 0.04). Of the 240 outbreaks reporting at least one contributing factor, food workers were implicated as the source of contamination in 182 (76%), with no difference between GII.4 and non-GII.4 (73% vs 79%, P = 0.3). Foodborne norovirus outbreaks are frequently reported in the U.S., most of which are caused by GII.4 noroviruses. Viruses of this genotype are associated with higher rates of hospitalization; non-GII.4 noroviruses are more frequently associated with contaminated molluscan shellfish. These surveillance data highlight the diversity of noroviruses causing foodborne disease and can help guide appropriate food safety interventions, including worker hygiene, improved food handling and preparation, and further development of norovirus vaccines.
Key words: : norovirus, food, surveillance, outbreaks, genotypes
1. Introduction
Noroviruses are the leading cause of acute gastroenteritis globally1,2). Each year in the United States (U.S.), norovirus infections result in an estimated 19–21 million illnesses, 56,000–71,000 hospitalizations, and 570–800 deaths3). Although a generally mild and self-limiting infection that affects all ages, noroviruses can cause severe health outcomes including hospitalization and death, particularly in the young (< 5 years), the elderly (≥ 65 years), and those with underlying conditions3). Noroviruses are genetically diverse and can be classified into at least seven known genogroups (GI-GVII), of which three (GI, GII, and GIV) cause disease in humans4). Norovirus genogroups can be further subdivided into at least 32 genotypes, with GII.4 being the most commonly reported4).
Noroviruses are transmitted primarily via the fecal-oral route, specifically through direct person-to-person contact, consumption of contaminated food or water, or contact with contaminated environmental surfaces. Noroviruses can also be spread through the vomitus-oral route, specifically through aerosolization of virions bound to vomitus droplets. Among the reported modes of transmission, about 1 in 5 norovirus outbreaks in the U.S. result from foodborne transmission5,6). Moreover, noroviruses are the most common of 31 major foodborne pathogens in the U.S., and the WHO Foodborne Disease Burden Epidemiology Reference Group recently reported that norovirus is also the leading cause of foodborne illness worldwide1,7). In the U.S., noroviruses are estimated to cause 58% of all domestically-acquired foodborne illnesses from known agents, including 26% of foodborne illness-associated hospitalizations and 11% of such deaths7).
With more widespread use of molecular diagnostics, including sequence-based genotyping, differences in the epidemiologic and clinical characteristics of norovirus genotypes have been identified6,8). For example, a summary of norovirus outbreaks reported to CaliciNet found non-GII.4 norovirus outbreaks were more often associated with foodborne transmission compared to outbreaks caused by GII.4 noroviruses6). Additionally, a systematic literature review of published norovirus outbreaks reported that GII.4 outbreaks more frequently result in severe health outcomes, specifically hospitalization and death8). As these studies demonstrate, pairing of laboratory and epidemiologic data from outbreak investigations provides greater insight into the mechanisms by which noroviruses spread and can inform the development of more effective prevention and control measures. This report describes epidemiologic and genotypic characteristics of U.S. foodborne norovirus outbreaks to help guide development of effective food safety interventions.
2. Methods
In 2009, the Centers for Disease Control and Prevention (CDC) launched the National Outbreak Reporting System (NORS), a web-based platform used by local, state, and territorial health departments in the U.S. to report all waterborne and foodborne disease outbreaks and enteric disease outbreaks transmitted by contact with environmental sources, infected persons or animals, or unknown modes of transmission to CDC2). Also in 2009, CDC launched CaliciNet, a norovirus outbreak laboratory surveillance network of federal, state, and local public health laboratories in the U.S., to collect information on norovirus including genotype information6). Outbreaks reported to NORS and/or CaliciNet are defined as two or more cases of a similar illness epidemiologically linked to a common exposure, such as a setting or food item. Primary mode of transmission was determined by the reporting agency using information obtained during the investigation and by referencing CDC guidance documents9). Foodborne outbreaks that occurred during August 2009–July 2015 with norovirus listed as the single confirmed etiology reported to NORS were matched with outbreaks reported to CaliciNet using an algorithm based on outbreak identification numbers and a proximity match based on reporting state and date of first illness for multiple identification number matches.
Matched outbreaks were stratified into two groups based on genotype (i.e., GII.4 vs non-GII.4 outbreaks) for all analyses. Chi-square or Fisher’s exact tests were used to perform bivariate analyses on health outcomes of case-patients, setting, food type, food contamination during processing and preparation, outbreak contributing factors, level of preparation, and point of contamination. Mann Whitney U-test was used to compare outbreak size and duration. Food contamination during processing and preparation and outbreak contributing factors were analyzed in categories pre-defined in NORS10,11). Briefly, outbreak contributing factors include factors that led to contamination of food by an etiologic agent, proliferation or amplification of these etiologic agents, or the survival of these etiologic agents11). Implicated food items were classified using a categorization scheme developed by the Interagency Food Safety Analytics Collaboration12).
3. Results
During August 2009–July 2015, foodborne transmission was reported in 975 (17%) of 5,734 confirmed norovirus outbreaks reported to NORS and in 978 (14%) of 7,059 norovirus outbreaks reported to CaliciNet. A total of 493 foodborne norovirus outbreaks were matched in both NORS and CaliciNet and had complete information on norovirus genogroup and genotype. Ninety-three (19%) outbreaks were caused by norovirus genogroup I (GI) and 400 (81%) were caused by genogroup II (GII). The most common genotypes reported were GII.4 (258, 52%), GII.6 (45, 9%), and GI.3 (38, 8%). Among the 258 GII.4 outbreaks, GII.4 Sydney (163, 63%) and GII.4 New Orleans (86, 33%) were most often detected. A complete list of GII.4 and non-GII.4 outbreaks can be found in the footnote of Table 1.
Table 1. Foodborne norovirus outbreak characteristics reported to the National Outbreak Reporting System and CaliciNet, United States, August 2009–July 2015.
| Overall | GII.4* outbreaks | non-GII.4† outbreaks | P-value‡ | |
|---|---|---|---|---|
| n = 493 | n = 258 | n = 235 | ||
| Outbreak characteristics | ||||
| Number of primary cases, median (range) | 13 (2-294) | 13 (2-210) | 14 (2-294) | 0.7587 |
| Duration (in days), median (range) | 3 (1-33) | 3 (1-19) | 3 (1-33) | 0.3443 |
| Case-patient health outcomes | ||||
| Healthcare visit, n (per 100 case-patients) | 492 (5.8) | 278 (6.2) | 214 (5.4) | 0.1349 |
| Emergency department visit, n (per 100 case-patients) | 319 (2.5) | 164 (3.7) | 155 (4.0) | 0.4781 |
| Hospitalization, n (per 1,000 case-patients) | 87 (8.8) | 63 (12.8) | 24 (4.8) | < 0.0001 |
| Death, n (per 10,000 case-patients) | 4 (3.8) | 4 (7.5) | 0 (0.0) | 0.0651 |
*GII.4 Sydney (163, 33.1%), GII.4 New Orleans (86, 17.4%), GII.4 Den Haag (8, 1.6%), GII.4 Osaka (1, 0.2%)
†GII.6 (45, 9.1%), GI.3 (38, 7.7%), GI.6 (29, 5.9%), GII.1 (22, 4.5%), GII.2 (18, 3.7%), GII.12 (16, 3.3%), GII.7 (14, 2.8%), GII.3 (12, 2.4%), GI.2 (11, 2.2%), GI.4 (7, 1.4%), GI.5 (4, 0.8%), GII.13 (4, 0.8%), GII.14 (4, 0.8%), GI.7 (3, 0.6%), GII.5 (3, 0.6%), GII.17 (3, 0.6%), GI.1 (1, 0.2%), GII.25 (1, 0.2%)
‡Statistical comparisons made between GII.4 and non-GII.4 outbreaks. Mann-Whitney U-test was used to compare the median number of primary cases and median outbreak duration; Fisher's exact test was used to compare the case-patient health outcomes
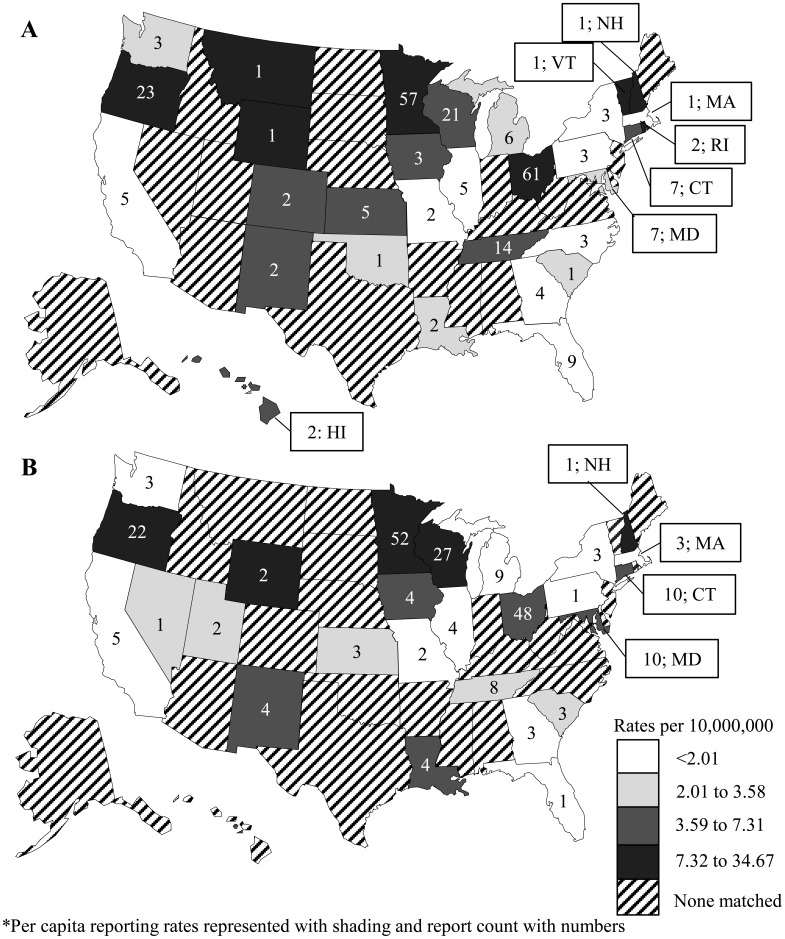
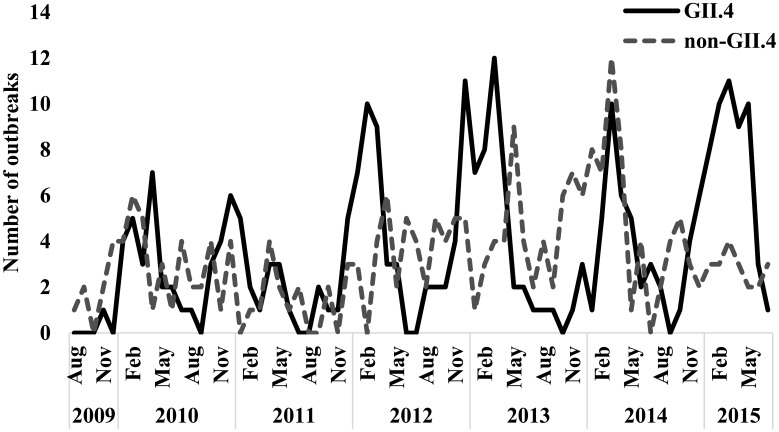
Overall, 33 states identified and reported foodborne norovirus outbreaks that could be matched between NORS and CaliciNet (Fig. 1). Among these states, per capita reporting rates of GII.4 (Fig. 1A) and non-GII.4 (Fig. 1B) norovirus outbreaks did not significantly differ within the same state (e.g., 4.9 GII.4 vs 7.0 non-GII.4 outbreaks per 10,000,000 residents in Connecticut, P = 0.5). However, GII.4 outbreaks were reported slightly more often during cooler months, with 171 of 258 (66%) occurring during October–March compared to non-GII.4 outbreaks (132 of 235 [56%], P = 0.03; Fig. 2). Outbreak duration and size did not differ between the two genotype categories. The 493 outbreaks lasted a median of three (range: 1-33) days, and had a median outbreak size of 13 (range: 2-294) primary cases (Table 1). There were no differences between GII.4 and non-GII.4 outbreaks with regard to most health outcomes, with at least 492 healthcare visits (5.8 per 100 case-patients), 319 emergency department visits (2.5 per 100 case-patients), and 4 deaths (3.8 per 10,000 case-patients; Table 1) reported during the 493 outbreaks. However, hospitalizations were significantly more frequent among GII.4 outbreaks (12.8 per 1,000 case-patients) compared to non-GII.4 outbreaks (4.8 per 1,000 case-patients, P < 0.0001).
Fig. 1.
Number and per capita rate of foodborne norovirus GII.4 (n = 258; A) and non-GII.4 (n = 235; B) outbreaks reported to the National Outbreak Reporting System and CaliciNet by state, United States, August 2009–July 2015.
Fig. 2.
Monthly distribution of foodborne norovirus GII.4 (n = 258, black) and non-GII.4 (n = 235, dashed gray) outbreaks reported to the National Outbreak Reporting System and CaliciNet, United States, August 2009–July 2015.
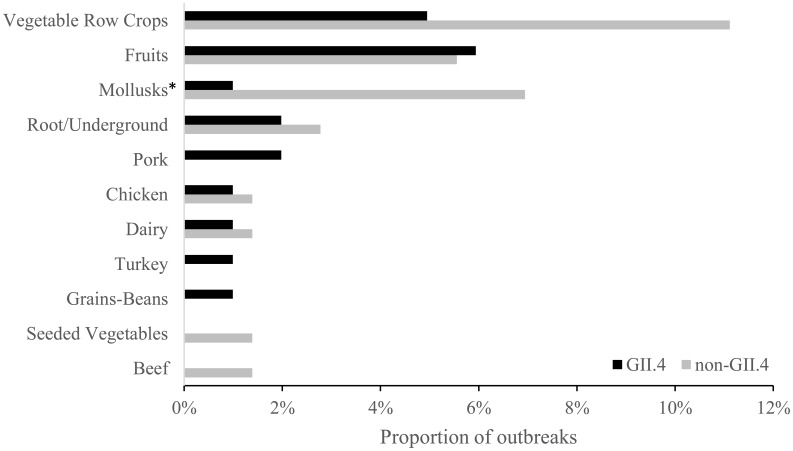
Specific food items were implicated in only 173 (35%) of outbreaks, of which 43 (25%) could be classified into a single food commodity and the remaining 130 (75%) outbreaks were either complex foods (i.e., multiple commodity foods) or unable to be classified. The only single food commodity implicated significantly more often in non-GII.4 outbreaks compared to GII.4 outbreaks was molluscan shellfish (7% vs 1%, P = 0.04; Fig. 3). Although not significantly different, vegetable row crops were also implicated more frequently in non-GII.4 outbreaks (11%) than in GII.4 outbreaks (5%, P = 0.1). Of the 173 outbreaks with a known food item, 140 (81%) had information on the level of preparation (Table 2). Of the 140, 108 (77%) outbreaks reported food was consumed raw and 47 (34%) reported the food was heat processed. Both raw and heat processed level of preparation could be reported in the same outbreak due to multiple implicated food items. There was no difference in level of preparation between GII.4 and non-GII.4 outbreaks. GII.4 and non-GII.4 outbreaks were similar with regard to outbreak setting (e.g., restaurants: 68% vs 66%, P = 0.4) and implication of food workers as the source of contamination among outbreaks reporting contributing factor information (73% vs 79%, P = 0.3; Table 2). Among the 182 outbreaks implicating food workers, bare-hand contact with ready-to-eat food was reported in 54 (57%) of GII.4 outbreaks and 45 (51%) of non-GII.4 outbreaks (P = 0.5) and glove-hand contact in 23 (24%) of GII.4 outbreaks and 29 (33%) of non-GII.4 outbreaks (P = 0.3, Table 2). Two hundred seventy-five outbreaks (56%) identified a point of contamination, of which almost all stated contamination occurred during preparation (264, 96%). Of the 275 outbreaks, 6% of non-GII.4 outbreaks involved foods contaminated before preparation compared to 2% of GII.4 outbreaks (P = 0.2), and 98% of GII.4 outbreaks were contaminated during preparation compared to 94% of non-GII.4 outbreaks (P = 0.2, Table 2).
Fig. 3.
Proportion of implicated single food commodities among 101 GII.4 and 72 non-GII.4 foodborne norovirus outbreaks reported to the National Outbreak Reporting System and CaliciNet, United States, August 2009–July 2015. One hundred thirty (81 GII.4 and 49 non-GII.4) outbreaks were either complex foods (i.e., multiple commodity foods) or unable to be classified and are not shown on the figure. Food commodities are grouped according to the categorization scheme developed by the Interagency Food Safety Analytics Collaboration12.
*The proportion of non-GII.4 outbreaks implicating molluscan shellfish was significantly greater than that among GII.4 outbreaks (P = 0.04).
Table 2. Contamination factors of foodborne norovirus outbreaks reported to the National Outbreak Reporting System and CaliciNet, United States, August 2009–July 2015.
| GII.4 outbreaks | non-GII.4 outbreaks | P-value‡ | |
|---|---|---|---|
| n = 258 | n = 235 | ||
| Outbreak contributing factor known, n (%)1 | 129 (50%) | 111 (47%) | |
| Contaminated raw product, n (%) | 10 (8%) | 8 (7%) | 1.0 |
| Cross-contamination, n (%) | 6 (5%) | 6 (5%) | 1.0 |
| Food worker implicated, n (%)2 | 94 (73%) | 88 (79%) | 0.3 |
| Bare hand contact, n (%) | 54 (57%) | 45 (51%) | 0.5 |
| Gloved hand contact, n (%) | 23 (24%) | 29 (33%) | 0.3 |
| Other hand contact, n (%) | 30 (32%) | 24 (27%) | 0.5 |
| Level of preparation, n (%)3 | 82 (81%) | 58 (81%) | |
| Eaten raw, n (%) | 61 (60%) | 47 (65%) | 0.5 |
| Heat processed, n (%) | 32 (32%) | 15 (21%) | 0.1 |
| Point of contamination, n (%)4 | 135 (49%) | 140 (51%) | |
| Before preparation, n (%) | 3 (2%) | 8 (6%) | 0.2 |
| Preparation, n (%) | 132 (98%) | 132 (94%) | 0.2 |
1At least one contributing factor (not C-N/A, P-N/A, or S-N/A listed) is selected. Multiple contributing factors may be selected for each outbreak; therefore, the sum of contributing factors may be larger than the number of outbreaks with a known contributing factor.
2Among the outbreaks with at least one known contributing factor (not C-N/A, P-N/A, or S-N/A listed) is selected (n = 244).
3Among the outbreaks with at least one implicated food item listed (n = 173). Multiple levels of preparation may be selected for each outbreak; therefore, the sum of level of preparation methods may be larger than the number of outbreaks with a known level of preparation
4Among the outbreaks where point of contamination listed as either before preparation or preparation (n = 275).
‡Statistical comparisons made between GII.4 and non-GII.4 outbreaks. Chi-square test was used to compare the outbreak contributing factors, level of preparation, and point of contamination.
4. Discussion
This analysis highlights key features of foodborne norovirus outbreaks caused by different genotypes in the United States to help guide the development of effective norovirus prevention and control strategies. Overall, a majority of outbreaks were caused by GII.4 noroviruses. GII.4 noroviruses have been well documented as the most common norovirus genotype for all modes of transmission; however, GII.4 is most frequently identified among person-to-person outbreaks6). After stratifying by genotype, GII.4 outbreaks were found to result in more severe health outcomes, specifically hospitalizations, compared to non-GII.4 outbreaks. This association was similarly reported in a systematic literature review of published norovirus outbreaks8). Since GII.4 noroviruses have been associated with longer viral shedding, more vomiting, and have different environmental stability and disinfectant resistance, outbreaks of this genotype may be more challenging to prevent and control13). Such insights from combined epidemiology and laboratory outbreak surveillance may also help inform vaccine development efforts, including formulation of antigenic targets (e.g., inclusion of both GII.4 and non-GII.4) and impacts likely protective of public health (e.g., decreasing disease severity or viral shedding).
Another association observed was between molluscan shellfish and non-GII.4 norovirus outbreaks. A nine-year study of norovirus contamination of shellfish in Italy also detected non-GII.4 (36 samples, 53%) genotypes more often than GII.4 (32 samples, 47%) in mollusks14). In the U.S., a GII.12 norovirus outbreak implicating insufficiently steamed oysters resulted in over 200 illnesses among both restaurant patrons and their household members15). Therefore, protecting against contamination of molluscan growing beds and ensuring proper processing before consumption, with methods such as high hydrostatic pressure, may prevent the occurrence of large foodborne norovirus outbreaks16). Interestingly, vegetable row crops were also more commonly implicated in non-GII.4 outbreaks. The higher number of non-GII.4 norovirus outbreaks implicating these specific single food categories may indicate contamination of these products before preparation. As reported in a Danish investigation, a single lot of norovirus GI-contaminated lettuce from a supplier resulted in 23 outbreaks17). Moreover, raspberries are frequently implicated in foodborne norovirus outbreaks with the source of contamination frequently occurring during production18). A study in the European Union found three of 26 raspberry batches were contaminated with an average of 4.3 log genomic equivalent copies of norovirus GI per 20 grams19). Such sampling of fresh produce, like berries and leafy greens, may provide an early indication of unsanitary production or processing conditions and an opportunity to intervene.
Contamination during preparation accounted for the vast majority of reported foodborne norovirus outbreaks and occurred somewhat more often with GII.4 outbreaks than with outbreaks caused by non-GII.4. Moreover, the most frequently implicated food vehicles were those containing multiple food categories, particularly among GII.4 outbreaks, which usually require handling by a food worker immediately before consumption. This summary also found a majority of implicated foods were consumed raw, which would indicate no viral kill step, such as heat processing, occurred before consumption. These observations are consistent with previous accounts of GII.4 contamination during preparation by a food worker since GII.4 is more commonly associated with person-to-person transmission6). For example, guests at a wedding fell ill after eating a mushroom dish prepared by a GII.4-positive food worker who reported working while ill with diarrhea20). Additionally, a series of four norovirus outbreaks occurred after event attendees consumed food requiring manual preparation by a GII.4-positive food worker who reported returning to work 45 hours after symptoms subsided21).
The prominent role of infected food workers identified in this analysis suggests that implementation of food safety interventions focused on worker health and hygiene have the potential to substantially reduce the burden of foodborne norovirus outbreaks. First, hiring a certified kitchen manager to supervise kitchen operations and staff has been demonstrated to reduce the number of norovirus outbreaks22). Currently, the U.S. Food and Drug Administration (FDA) recommends as part of its 2013 model Food Code that restaurants have a kitchen manager certified in food safety23). Second, excluding food workers until at least 48 hours after symptoms of norovirus have subsided is frequently recommended to prevent further spread13,23). This recommendation is based on the principle that isolating an individual during the acute phase of illness, which is also the peak of viral shedding, reduces the opportunity for additional transmission to occur24). As outbreaks involving post-symptomatic food workers have demonstrated, returning to work sooner than 48 hours after symptoms have subsided poses a risk of norovirus transmission21). Unfortunately, a survey of restaurant workers determined that one in five reported working while ill with vomiting or diarrhea at least once during a single year25). Interviewed workers stated they feared they would lose their job or place unnecessary burden on co-workers by remaining at home while ill. Therefore, implementing management practices to allay fears of repercussions and encourage reporting of illness by food workers have been recommended13). Specifically, food establishments should consider establishing measures to ensure paid sick leave and adequate staffing to cover the ill food worker are in place to encourage timely notification of illness and adherence to work exclusion13). Finally, several studies have demonstrated washing hands frequently with soap and water is effective at removing norovirus from contaminated hands26). Moreover, avoiding bare-hand contact with ready-to-eat foods has been cited in the U.S. FDA Food Code as a key infection control measure, specifically in food service settings23). Although all states in the U.S. require food workers to wash their hands, an observational study found that only 27% of food workers do so when it is recommended and this decreases to 16% when food workers wear gloves27,28). Thus, additional efforts are needed to encourage compliance with and enforcement of worker exclusion and hand hygiene measures.
Though this summary advances the understanding of foodborne norovirus outbreaks, the data are subject to at least four limitations. First, outbreaks reported to NORS and CaliciNet were incompletely matched. Although 493 outbreaks were matched, this represents only a fraction of all foodborne norovirus outbreaks in NORS and in CaliciNet. As such, additional risk factors may be assessed if remaining NORS and CaliciNet reports could be matched. To improve NORS and CaliciNet report matching, updates to the surveillance systems are routinely implemented based on discussions with reporting agencies about the ways they detect, store, and report outbreaks. Moreover, an effort is underway to link outbreak reports in NORS and the National Environmental Assessment Reporting System (NEARS) to gain additional data on environmental contributing factors and illness prevention policies, such as food worker illness reporting and exclusion29). Second, potential collinearity and confounding may exist between the identified risk factors and outcomes of interest. However, a systematic literature review found that among foodborne norovirus outbreaks in healthcare settings, GII.4 noroviruses were associated with higher hospitalization rates than non-GII.4 viruses after controlling for potential confounding factors8). Third, additional associations between food commodities and norovirus genotype may exist, yet only 35% of reported outbreaks were able to identify a food commodity. Identifying and implicating a food item is often challenging due to the inability of an investigator to identify a common food exposure epidemiologically, the limited availability of validated norovirus detection assays for food matrices, and the rapid use or discarding of potentially contaminated food products at the implicated location. Therefore, reporting agencies should strive to improve their ability to rapidly detect and respond to a potential foodborne outbreak to minimize case-patient recall bias and increase the likelihood of collecting potentially contaminated food samples. Presently, a validated norovirus detection assay for shellfish is available through U.S. Food and Drug Administration laboratories, and norovirus detection assays for select produce commodities (e.g., berries) are under development30,31). However, additional research into developing and validating norovirus detection assays for other food commodities would greatly enhance the ability of an investigator to link an implicated food product and ill case-patients. Finally, variability exists in the frequency and completeness of reporting to NORS and CaliciNet among state and local health departments. Not all states reported foodborne norovirus outbreaks, yet this may not indicate a lack of outbreaks, rather different outbreak detection capacities in state and local surveillance platforms. As a result, the true number of foodborne norovirus outbreaks is likely higher than those reported to NORS and CaliciNet, which indicates a need to continue to build capacity of state and local health departments to detect, investigate, and report outbreaks. A recent evaluation of Norovirus Sentinel Testing and Tracking (NoroSTAT), a collaborative network with selected state health departments that report specific epidemiologic and laboratory data on norovirus outbreaks, found norovirus outbreak reporting and report completeness has improved among both NoroSTAT and non-NoroSTAT states since the implementation of NORS in 200932).
This analysis provides some important insights that can help inform efforts to prevent foodborne norovirus outbreaks in the United States. First, GII.4 noroviruses were detected in a majority of foodborne norovirus outbreaks. Moreover, food workers were a significant source of norovirus contamination among the matched outbreaks. Finally, GII.4 noroviruses resulted in more hospitalizations and non-GII.4 noroviruses were detected more often in outbreaks implicating molluscan shellfish. These findings suggest that interventions targeting food worker hygiene should be implemented, molluscan growing waters should be protected from fecal contamination, and further development of effective vaccines against both GII.4 and non-GII.4 noroviruses seems warranted to help reduce norovirus circulation in human populations and thereby prevent future foodborne norovirus outbreaks.
Acknowledgments
This study was supported in part by appointment to the Research Participation Program at the CDC (to ZM) administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the CDC.
Abbreviations: CDC: Centers for Disease Control and Prevention, FDA: Food and Drug Administration, GI: genogroup I, GII: genogroup II, NEARS: National Environmental Assessment Reporting System, NoroSTAT: Norovirus Sentinel Testing and Tracking, NORS: National Outbreak Reporting System, WHO: World Health Organization
Footnotes
Conflict of interest: The authors have no conflict of interest.
References
- 1.Pires SM, Fischer-Walker CL, Lanata CF, et al. Aetiology-Specific Estimates of the Global and Regional Incidence and Mortality of Diarrhoeal Diseases Commonly Transmitted through Food. PLOS ONE. 2015; 10: e0142927. 10.1371/journal.pone.0142927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall AJ, Wikswo ME, Manikonda K, Roberts VA, Yoder JS, Gould LH. Acute gastroenteritis surveillance through the National Outbreak Reporting System, United States. Emerging Infectious Diseases. 2013; 19: 1305–1309. 10.3201/eid1908.130482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall AJ, Lopman BA, Payne DC, et al. Norovirus Disease in the United States. Emerging Infectious Diseases. 2013; 19: 1198–1205. 10.3201/eid1908.130465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinjé J. Advances in laboratory methods for detection and typing of norovirus. Journal of Clinical Microbiology. 2015; 53: 373–381. 10.1128/JCM.01535-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall AJ, Wikswo ME, Pringle K, Gould LH, Parashar UD, Division of Viral Diseases, National Center for Immunization and Respiratory Diseases, CDC Vital signs: foodborne norovirus outbreaks - United States, 2009-2012. MMWR Morb Mortal Wkly Rep. 2014; 63: 491–495. [PMC free article] [PubMed] [Google Scholar]
- 6.Vega E, Barclay L, Gregoricus N, Shirley SH, Lee D, Vinjé J. Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013. Journal of Clinical Microbiology. 2014; 52: 147–155. 10.1128/JCM.02680-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United States--major pathogens. Emerging Infectious Diseases. 2011; 17: 7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai R, Hembree CD, Handel A, et al. Severe outcomes are associated with genogroup 2 genotype 4 norovirus outbreaks: a systematic literature review. Clinical Infectious Diseases. 2012; 55: 189–193. 10.1093/cid/cis372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.United States Centers for Disease Control and Prevention. National Outbreak Reporting System (NORS) Guidance. Atlanta, GA.
- 10.United States Centers for Disease Control and Prevention. Appendix E: Available Picklist Values. Atlanta, GA.
- 11.United States Centers for Disease Control and Prevention. Appendix D: NORS Guidance for Contributing Factors (CF) in Foodborne Outbreak Reports. Atlanta, GA.
- 12.United States Centers for Disease Control and Prevention. Interagency Food Safety Analytics Collaboration (IFSAC): Food Categorization Scheme. Atlanta, GA.
- 13.Barclay L, Park GW, Vega E, et al. Infection control for norovirus. Clinical Microbiology and Infection. 2014; 20: 731–740. 10.1111/1469-0691.12674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavoni E, Consoli M, Suffredini E, et al. Noroviruses in seafood: a 9-year monitoring in Italy. Foodborne Pathogens and Disease. 2013; 10: 533–539. 10.1089/fpd.2012.1399 [DOI] [PubMed] [Google Scholar]
- 15.Alfano-Sobsey E, Sweat D, Hall A, et al. Norovirus outbreak associated with undercooked oysters and secondary household transmission. Epidemiology and Infection. 2012; 140: 276–282. 10.1017/S0950268811000665 [DOI] [PubMed] [Google Scholar]
- 16.Ye M, Li X, Kingsley DH, Jiang X, Chen H. Inactivation of human norovirus in contaminated oysters and clams by high hydrostatic pressure. Applied and Environmental Microbiology. 2014; 80: 2248–2253. 10.1128/AEM.04260-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller L, Rasmussen LD, Jensen T, et al. Series of Norovirus Outbreaks Caused by Consumption of Green Coral Lettuce, Denmark, April 2016. PLoS Curr. 2016; 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarvikivi E, Roivainen M, Maunula L, et al. Multiple norovirus outbreaks linked to imported frozen raspberries. Epidemiology and Infection. 2012; 140: 260–267. 10.1017/S0950268811000379 [DOI] [PubMed] [Google Scholar]
- 19.De Keuckelaere A, Li D, Deliens B, Stals A, Uyttendaele M. Batch testing for noroviruses in frozen raspberries. International Journal of Food Microbiology. 2015; 192: 43–50. 10.1016/j.ijfoodmicro.2014.09.024 [DOI] [PubMed] [Google Scholar]
- 20.Maritschnik S, Kanitz EE, Simons E, et al. A Food Handler-Associated, Foodborne Norovirus GII.4 Sydney 2012-Outbreak Following a Wedding Dinner, Austria, October 2012. Food Environ Virol. 2013; ▪▪▪: 12. PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thornley CN, Hewitt J, Perumal L, et al. Multiple outbreaks of a novel norovirus GII.4 linked to an infected post-symptomatic food handler. Epidemiology and Infection. 2013; 141: 1585–1597. 10.1017/S0950268813000095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hedberg CW, Smith SJ, Kirkland E, Radke V, Jones TF, Selman CA, EHS-Net Working Group Systematic environmental evaluations to identify food safety differences between outbreak and nonoutbreak restaurants. Journal of Food Protection. 2006; 69: 2697–2702. 10.4315/0362-028X-69.11.2697 [DOI] [PubMed] [Google Scholar]
- 23.United States Food and Drug Administration. FDA Food Code. Silver Spring, MD.
- 24.Division of Viral Diseases, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention Updated norovirus outbreak management and disease prevention guidelines. MMWR Recomm Rep. 2011; 60 (RR-3): 1–18. [PubMed] [Google Scholar]
- 25.Carpenter LR, Green AL, Norton DM, et al. Food worker experiences with and beliefs about working while ill. Journal of Food Protection. 2013; 76: 2146–2154. 10.4315/0362-028X.JFP-13-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P, Yuen Y, Hsiao HM, Jaykus LA, Moe C. Effectiveness of liquid soap and hand sanitizer against Norwalk virus on contaminated hands. Applied and Environmental Microbiology. 2010; 76: 394–399. 10.1128/AEM.01729-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kambhampati A, Shioda K, Gould LH, et al. A State-by-State Assessment of Food Service Regulations for Prevention of Norovirus Outbreaks. Journal of Food Protection. 2016; 79: 1527–1536. 10.4315/0362-028X.JFP-16-088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green LR, Selman CA, Radke V, et al. Food worker hand washing practices: an observation study. Journal of Food Protection. 2006; 69: 2417–2423. 10.4315/0362-028X-69.10.2417 [DOI] [PubMed] [Google Scholar]
- 29.Brown LG, Hoover ER, Selman CA, Coleman EW, Schurz Rogers H. Outbreak characteristics associated with identification of contributing factors to foodborne illness outbreaks. Epidemiology and Infection. 2017; 145: 2254–2262. 10.1017/S0950268817001406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woods JW, Calci KR, Marchant-Tambone JG, Burkhardt W, III. Detection and molecular characterization of norovirus from oysters implicated in outbreaks in the US. Food Microbiology. 2016; 59: 76–84. 10.1016/j.fm.2016.05.009 [DOI] [PubMed] [Google Scholar]
- 31.DePaola A, Jones JL, Woods J, et al. Bacterial and viral pathogens in live oysters: 2007 United States market survey. Applied and Environmental Microbiology. 2010; 76: 2754–2768. 10.1128/AEM.02590-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah MP, Wikswo ME, Barclay L, et al. Near Real-Time Surveillance of U.S. Norovirus Outbreaks by the Norovirus Sentinel Testing and Tracking Network — United States, August 2009–July 2015. MMWR. Morbidity and Mortality Weekly Report. 2017; 66: 185–189. 10.15585/mmwr.mm6607a1 [DOI] [PMC free article] [PubMed] [Google Scholar]