Abstract
Xenopus tropicalis tadpoles can regenerate an amputated tail, including spinal cord, muscle and notochord, through cell proliferation and differentiation. However, the molecular mechanisms that regulate cell proliferation during tail regeneration are largely unknown. Here we show that JunB plays an important role in tail regeneration by regulating cell proliferation. The expression of junb is rapidly activated and sustained during tail regeneration. Knockout (KO) of junb causes a delay in tail regeneration and tissue differentiation. In junb KO tadpoles, cell proliferation is prevented before tissue differentiation. Furthermore, TGF-β signaling, which is activated just after tail amputation, regulates the induction and maintenance of junb expression. These findings demonstrate that JunB, a downstream component of TGF-β signaling, works as a positive regulator of cell proliferation during Xenopus tail regeneration.
Keywords: JunB, Xenopus tail regeneration, cell proliferation, tissue differentiation, TGF-β signaling
Introduction
After traumatic injury, mammals such as humans and adult mice can heal wounds but not regenerate most of the damaged tissues [1]. In contrast, other vertebrates, such as Xenopus tadpoles, can regenerate lost appendages [2, 3]. Following amputation of the tail of a Xenopus tadpole, the amputation plane is covered by wound epithelium to prevent the leakage of cellular material. After wound healing, a regeneration bud is formed at the amputated site. Subsequently, the tail begins to regrow due to cell proliferation and is completely restored through the differentiation of spinal cord, muscle and notochord [4, 5, 6]. It has been reported that cell proliferation during tail regeneration is regulated by several signaling pathways, such as BMP, Wnt, TGF-β and Hippo [7, 8, 9, 10], and by other factors, such as V-ATPase and small GTPase [11, 12]. However, the detailed molecular mechanisms that regulate cell proliferation during tail regeneration are not well understood.
AP-1 family members (e.g., JunB and c-Jun) are involved in diverse biological processes such as cell proliferation and differentiation [13, 14]. It has been shown that c-Jun is important for mesoderm induction by FGF signaling during Xenopus development [15]. Moreover, we previously reported that over-expression of JunB led to ectopic tail-like structures in Xenopus and induced the expression of differentiation-related genes [16]. Since tail regeneration is caused by the reconstruction of tail structure, in the present study we examined the role of JunB during tail regeneration. We found that immediately after tail amputation junb was expressed in the regenerating tail. Tail regeneration was retarded by knockout (KO) of junb due to a reduction in the number of mitotic cells prior to tissue differentiation. Moreover, we also found that inhibition of TGF-β signaling decreased junb expression during tail regeneration. These results suggest that, downstream of TGF-β signaling, JunB is essential for tail regeneration by regulating cell proliferation.
Materials and Methods
Design and synthesis of sgRNAs
CRISPR direct was used to design sgRNAs against junb (http://crispr.dbcls.jp/). The following primers were used: tyrosinase sgRNA forward 5’-ATT TAG GTG ACA CTA TAG GAA CTG GCC CCT GCA AAC AGT TTT AGA GCT AGA AAT AGC AAG-3’ [17]; junb sgRNA forward [Target 1, 5’-ATT TAG GTG ACA CTA TAG GGG CTG TCG GTA GCA GCT TGT TTT AGA GCT AGA ATA GCA AG-3’; Target 2, 5’-ATT TAG GTG ACA CTA TAG GCA TAA GTG GTC CGA GCG GGT TTT AGA GCT AGA AAT AGC AAG-3’; Target 3, 5’-ATT TAG GTG ACA CTA TAG GAC TGT GCC CGA TAC CGC CGT TTT AGA GCT AGA AAT AGC AAG-3’; Target 4, 5’-ATT TAG GTG ACA CTA TAG GGT GGC AAT GGC ATA ACG GGT TTT AGA GCT AGA AAT AGC AAG-3’; Target 5, 5’-ATT TAG GTG ACA CTA TAG GAA GCT GGA GAG AAT CGC CGT TTT AGA GCT AGA AAT AGC AAG-3’]; reverse 5’-AAA AGC ACC GAC TCG GTG CCA CTT TTT CAA GTT GAT AAC GGA CTA GCC TTA TTT TAA CTT GCT ATT TCT AGC TCT AAA AC-3’. Synthesis of sgRNA was carried out following a previously described protocol [18]. The sgRNA templates were generated by a PCR-based method and in vitro transcription using the MEGA script® SP6 Transcription Kit (Thermo Fisher Scientific).
Growth, manipulation and microinjection of X. tropicalis
Animal experiments were carried out in accordance with the guidelines of the Animal Experimentation Ethics Committee of Hiroshima University and conformed with generally agreed international regulations. X. tropicalis tadpoles were obtained by in vitro fertilization and were cultured in 0.1X Marc’s Modified Ringer’s (MMR) solution containing 50 μg/ml gentamycin on 1% agarose coated dishes. For the KO experiment, 1000 pg (single injection) or 500 pg (combinatorial injection) of sgRNAs and 1 ng of Cas9 protein (Integrated DNA Technologies) were injected into the animal pole at the 1-cell stage in 0.1X MMR containing 6% Ficoll, 50 μg/ml gentamycin and 0.1% BSA within 40 min of fertilization. For the rescue experiment, FLAG-junb mRNA was added to the sgRNAs and Cas9 protein. Tadpoles at stage 41/42 [19] were anesthetized in 0.01% MS-222/0.1X MMR and the tail was surgically amputated at the mid-point. After amputation, tadpoles were kept at 24°C in tap water. The TGF-β receptor inhibitor SB-505124 (Cayman Chemical) was prepared in DMSO (Nacalai) and used at 12.5 μM. Tadpoles were treated with inhibitor-containing medium from 1h before tail amputation and the medium was changed every day until 72 hours post amputation (hpa). F0 junb KO frogs (sg 4 + sg 5 injected males and sg 5 injected females) were confirmed to have germline transmission by direct sequencing of PCR amplicons [20]. These F0 frogs were intercrossed to obtain compound heterozygous F1 junb mutants.
Plasmids
Probes were synthesized using the following plasmids: pDH105-junb [16], pCS-sox2, pCS-myod1, and pCS-shh (gifts from Dr. R. Harland). For the rescue experiment, pDH105-FLAG-junb was generated by subcloning a SalI/XbaI fragment from pDH105-junb into a pDH105-FLAG vector.
Whole-mount immunostaining
Tadpoles were fixed with MEMFA for 30 min at room temperature and stored at −20°C in 100% methanol. After rehydration with 1X PBS, samples were treated with bleaching solution (1% H2O2, 5% formamide, 0.5X SSC). These samples were blocked with 10% normal goat serum (NGS) in PBST (1X PBS, 0.1% Triton X-100, 0.2% BSA) for 3h, and incubated at 4°C overnight with primary antibody (anti-phosphorylated histone H3, Upstate Biotechnology, 1 μg/ml) diluted at 1:500 in PBST plus 10% NGS. A secondary antibody conjugated to Alexa-488 (Molecular Probe, 2 mg/ml) was diluted at 1:500. pH3 positive cells were manually counted in the regenerated tail, and the area of regenerating tissues was measured using Image J software (National Institutes of Health, USA). The number of pH3 positive cells was divided by the individual area (pixels), and normalized by the average of control samples at respective time points.
Whole-mount in situ hybridization
X. tropicalis embryos were fixed in MEMFA for 2h at 24°C. Whole-mount in situ hybridization (WISH) was performed following standard methods [21] with a minor modification as described in Takebayashi-Suzuki et al. [22].
Quantitative RT-PCR (qPCR)
Regenerating tails were digested in lysis solution (0.25 mg/ml proteinase K, 50 mM Tris-HCl (pH7.5), 5 mM EDTA, 50 mM NaCl, 0.5% SDS) and incubated at 42°C overnight. Total RNA was purified by phenol-chloroform extraction and ethanol precipitation, and treated with DNaseI (Roche). cDNA synthesis and qPCR were carried out following previously described protocols [22]. The following primer sequences were used for qPCR analysis: junb, forward 5’-CAT GGA GGA TCA GGA GAG GA-3’ and reverse 5’-CTC TCA CCC TCA CCT TCA GC-3’; rps18, forward 5’-TTC AGC ACA TTT TGC GTG TT-3’ and reverse 5’-GTT CAC CAG CAC GCT TTG TA-3’.
Genotyping
At 72 hpa, individual tadpoles were digested in lysis solution (0.1 mg/ml proteinase K, 10 mM Tris-HCl (pH8.0), 100 mM EDTA, 0.5% SDS) and treated with RNase A (50 μg/ml) to extract genomic DNA. Genomic DNA was purified by phenol-chloroform extraction and ethanol precipitation or using a GenElute mammalian genomic DNA miniprep kit (Sigma-Aldrich). The genomic region of junb was amplified using Q5 High Fidelity DNA polymerase (New England Biolabs). The following primer sequences were used for genotyping of junb KO tadpoles: forward 5’-CCA GCA CCC ACC TAC AAC TT-3’ and reverse 5’-TTC CGA ATG CCT TGG AGT AG-3’. The amplicons were annealed and incubated with T7 Endonuclease I (New England Biolabs). The digested PCR products were separated on a 1.5% agarose gel. For TA cloning, the PCR products were cloned into TOPO vector (Invitrogen). Single colonies were analyzed for each tadpole by Sanger sequencing to determine the mutation types. Sequences of F1 tadpoles were analyzed using poly peak parser software (http://yosttools.genetics.utah.edu/PolyPeakParser/). Compound heterozygous mutants were selected using the genotyping results.
Microscopy and statistics
Fluorescence images were acquired with Axio Zoom V-16 (Zeiss). Tail lengths in regenerating tadpoles were measured using CellSens standard software (Olympus). P values were determined using Student’s t-test. Error bars indicate the standard error of the mean.
Results
junb is expressed during tail regeneration in Xenopus tadpoles
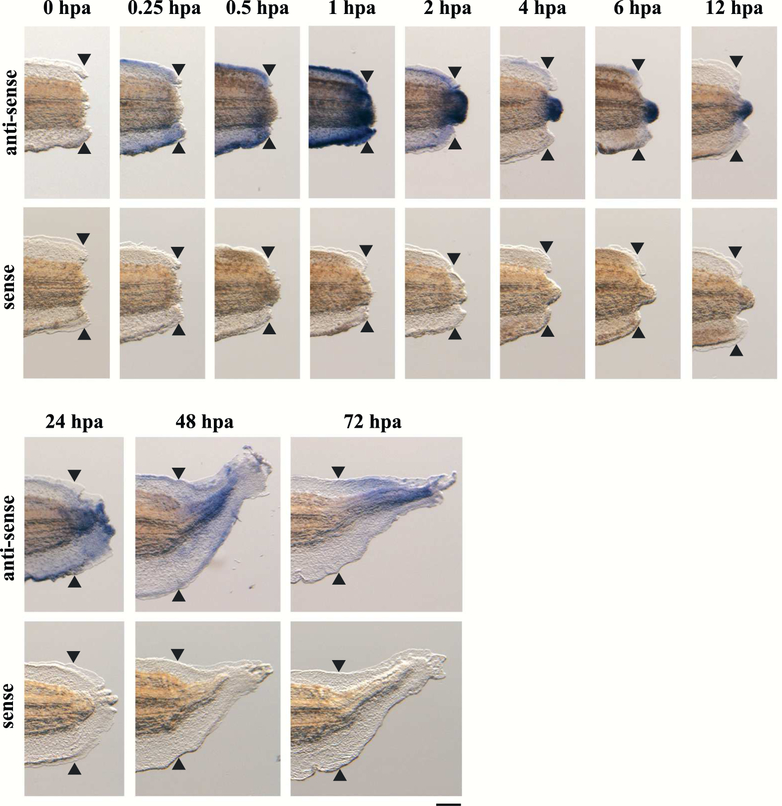
The expression pattern of junb during tail regeneration was analyzed using whole-mount in situ hybridization (WISH) of tadpoles that had undergone tail amputation (Fig. 1). Expression of junb was widely detected near the amputation site from 0.25 hpa (hours post amputation). At 1 hpa, junb was expressed in the tail, including the wound epithelium. The expression of junb intensified in the tip of the regenerating tail at 2–12 hpa and was observed throughout the regenerating tissues at 24–72 hpa. WISH using a junb sense-probe did not show any signals in the regenerating tails. These results demonstrate that junb is expressed during tail regeneration, and that JunB may play an important role in tail regeneration.
Figure 1. junb is expressed during tail regeneration of X. tropicalis tadpoles.
WISH analysis was performed using regenerating tadpoles at 0, 0.25, 0.5, 1, 2, 4, 6, 12, 24, 48 and 72 hpa. The expression of junb is shown in blue/purple. Black arrowheads indicate the amputation plane. Scale bar: 200 μm.
JunB is required for tail regeneration
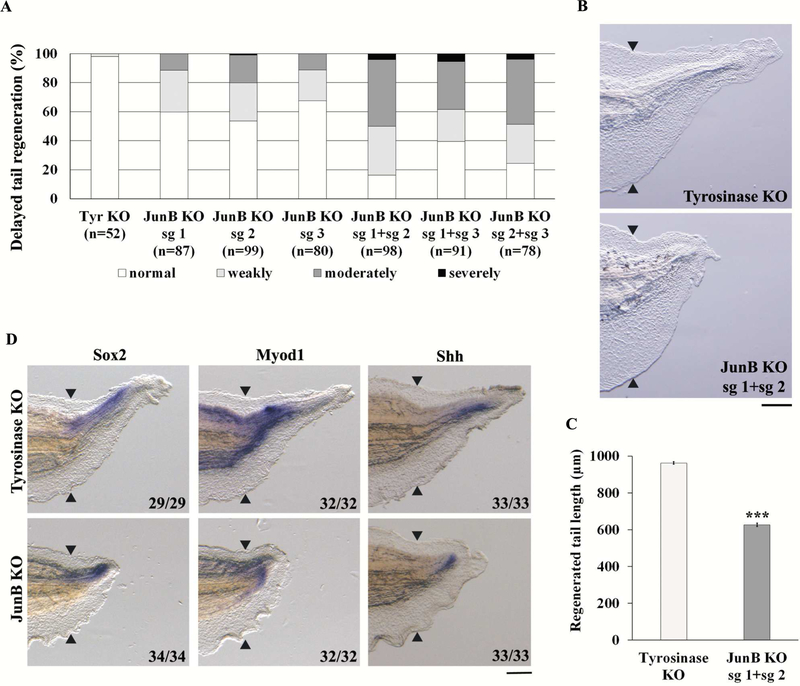
To confirm the role of JunB during tail regeneration, we carried out a KO experiment using the CRISPR-Cas9 system. Three sgRNAs were designed against the genomic region corresponding to the transactivation domain of JunB (Supplementary Fig. 1A); each sgRNA was co-injected with Cas9 protein into 1-cell stage eggs. We amputated the tails of F0 tadpoles at stage 41/42 and graded the degree of tail regeneration at 72 hpa. As shown in Figure 2A, junb KO tadpoles showed a delay in tail regeneration (sg 1, 40%; sg 2, 46%; sg 3, 33%) compared to control tadpoles (tyrosinase KO, 2%). To confirm the induction of mutations in the junb locus, we performed a T7E1 assay of junb KO tadpoles and identified mutations induced by each junb sgRNA, but not by tyrosinase sgRNA (data not shown). Next, we injected combinations of sgRNAs (sg 1 + sg 2, sg 1 + sg 3, sg 2 + sg 3) to increase the mutation rate and induce frame-shift mutations at two sites in junb. The junb KO tadpoles injected with two sgRNAs showed considerable delay in tail regeneration compared to those injected with a single sgRNA, especially for the combination sg 1 + sg 2 (84%, Fig. 2A). In addition, we found that the tail length regenerated in junb KO tadpoles injected with sg 1 + sg 2 was significantly less than in controls (Fig. 2B and C). A sequencing analysis of tadpoles injected with sg 1 + sg 2 showed that all alleles were mutated at both sg 1 and sg 2 target sites (Supplementary Fig. 1B–D). The region of the regenerating tail expressing sox2 (spinal cord marker) [23], myod1 (muscle marker) [23] and sonic hedgehog (shh, notochord marker) [24] was reduced in junb KO tadpoles (100%, Fig. 2D), demonstrating that tissue differentiation was prevented in junb KO at 72 hpa. To assess the specificity of the junb KO, we injected junb mRNA with junb sg 1 + sg 2 into fertilized eggs. The delay in tail regeneration caused by junb KO was partially but significantly rescued by co-injection of junb mRNA, showing that this phenotype results from inactivation of the junb gene (Supplementary Fig. 2A and B). To further explore the phenotype of junb KO, we generated F0 junb KO tadpoles by injecting with a different set of sgRNAs (sg 4 and sg 5, Supplementary Fig. 3A). These F0 tadpoles were raised to adulthood until sexual maturation, and intercrossed to create compound heterozygous F1 junb mutant tadpoles. We identified tadpoles carrying compound heterozygous mutations (Mut/Mut) and found that these tadpoles exhibited a delay in tail regeneration similar to junb KO sg 1 + sg 2 tadpoles (Supplementary Fig. 3B and C). These data strongly indicate that JunB is essential for tail regeneration.
Figure 2. Knockout of junb prevents tail regeneration.
(A) Summary of phenotypes in regenerating tadpoles at 72 hpa. The tadpoles were classified into 4 types: normal tail regeneration; weakly delayed tail regeneration; moderately delayed tail regeneration; and severely delayed tail regeneration. (B) junb KO sg 1 + sg 2 tadpoles show considerably delayed tail regeneration. tyrosinase KO was used as the control. (C) Lengths of regenerating tails. (D) WISH analysis of sox2, myod1 and shh in tyrosinase KO and junb KO tadpoles. Black arrowheads indicate amputation plane. Scale bar: 200 μm. ***P < 0.001, Student’s t-test.
junb KO causes the downregulation of cell proliferation
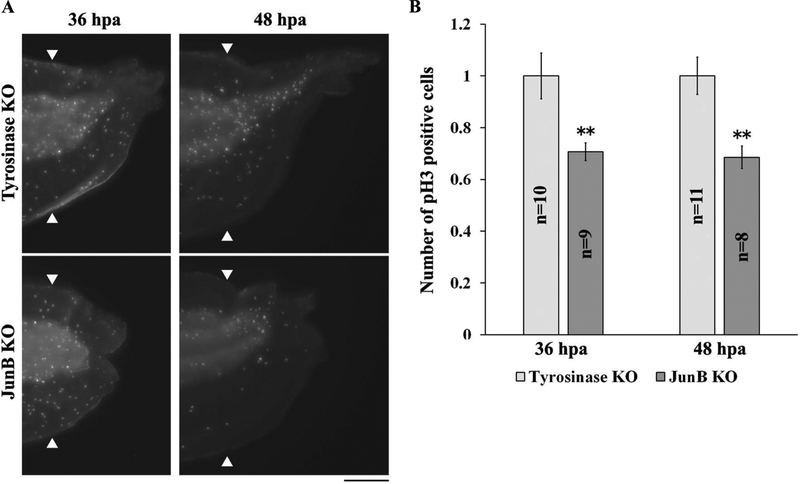
Based on the observations described above and the difficulty of obtaining sufficient numbers of compound heterozygous F1 mutants, we used F0 tadpoles injected with junb sg 1 + sg 2 in the following experiments. Since junb KO tadpoles exhibited a delay in tail regeneration at 72 hpa and because activation of cell proliferation is important for tail regrowth during regeneration, we examined cell proliferation using whole-mount immunostaining with an antibody against phosphorylated histone H3 (pH3). Cell proliferation has been reported to be activated at 24–36 hpa in X. tropicalis regenerating tails [4, 5]. We counted the number of pH3 positive cells in regenerating tails, excluding the fin, at 36 and 48 hpa and found that the number of mitotic cells was significantly reduced at both time points (Fig. 3A and B). Cell proliferation in the fin was not affected by junb KO (data not shown). These results indicate that JunB is important for the regulation of cell proliferation during tail regeneration.
Figure 3. Knockout of junb reduces cell proliferation.
(A) Whole-mount immunostaining of junb KO tadpoles with pH3 antibody at 36 and 48 hpa. (B) Quantification of pH3 positive cells in the region of regenerating tail, excluding the fin. The number of pH3 positive cells was divided by individual area, and normalized against control samples. White arrowheads indicate the amputation plane. Scale bar: 200 μm. **P < 0.01, Student’s t-test.
The expression of junb is regulated by TGF-β signaling
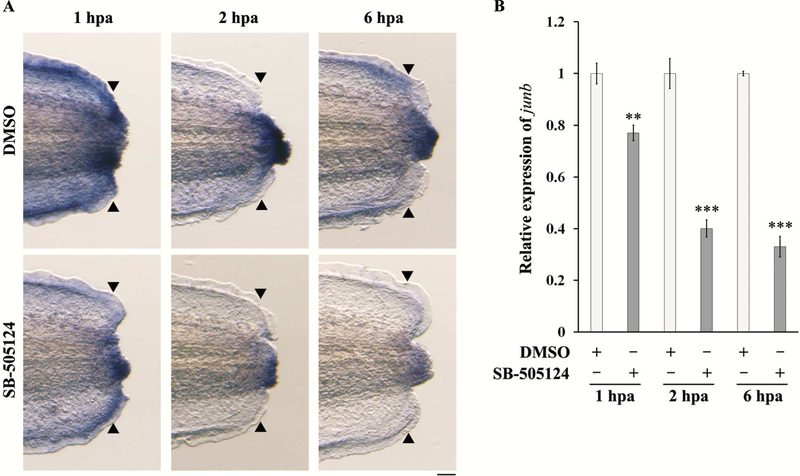
As JunB is important for tail regeneration and its expression is induced just after amputation, we addressed the mechanisms by which the expression of junb is regulated during tail regeneration. Phosphorylated Smad2, a TGF-β signal transducer, is expressed from 0.25 hpa during Xenopus tail regeneration [9]. In addition, Smads bind to the junb promotor in mouse NIH3T3 embryonic fibroblast cells [25]. Therefore, we inhibited TGF-β signaling using the TGF-β receptor inhibitor SB-505124 and examined the expression of junb at 1, 2 and 6 hpa. When TGF-β signaling was blocked by SB-505124, junb expression was downregulated in the regenerating tail (Fig. 4A). qPCR analysis showed that the inhibition of the TGF-β signaling caused a reduction in the amputation-induced level of junb expression at 1 hpa (Fig. 4B). Moreover, junb transcripts were considerably reduced at 2 and 6 hpa by the TGF-β receptor inhibitor treatment (Fig. 4B). These results suggest that the induction and maintenance of junb expression is regulated by TGF-β signaling during tail regeneration.
Figure 4. The expression of junb is downregulated by TGF-β signaling inhibition.
Tadpoles were treated with a medium containing 12.5 μM SB-505124 or DMSO (Control) from 1 h before tail amputation and cultured until 1, 2 and 6 hpa. (A) WISH analysis of junb in SB-treated tadpoles. N ≥ 29 for each sample. (B) qPCR analysis of junb in SB-treated tadpoles. The expression of junb was normalized against rps18 expression, and then against control samples. Scale bar: 50 μm. **P < 0.01, *** < 0.001, Student’s t-test.
Discussion
In this study, we showed that junb was widely expressed in the regenerating tail at 0.25–12 hpa and that expression was sustained at 24–72 hpa except in the fin. In addition, junb KO tadpoles showed a delay in tail regeneration at 72 hpa and a reduction in the number of mitotic cells at 36 and 48 hpa. Cell proliferation is a crucial aspect of tail regeneration after amputation: the number of mitotic cells begins to increase around 24–36 hpa during Xenopus tail regeneration before the activation of tissue differentiation [4, 5, 6]. Our results suggest that the delay in tail regeneration in junb KO tadpoles is caused by the downregulation of cell proliferation, which precedes tissue differentiation. It has been reported that JunB participates in the promotion of cyclinA transcription by binding to its promoter and functions as a positive regulator of cell proliferation [26]. In lymphoma cell lines, JunB knock-down reduces cell proliferation due to a prolongation of the G0/G1 phase [27]. A previous study also showed that JunB may be involved in cell proliferation during fin and finfold regeneration in zebrafish [28]. Therefore, one of the main functions of JunB is the control of cell proliferation, and this function may be conserved among animal species that can undertake regeneration. We also showed here that axial tissues, including the spinal cord, muscle and notochord, could not properly extend in junb KO tadpoles. As these differentiated tissues grow after the activation of cell proliferation at 48–72 hpa during Xenopus tail regeneration [5], it is possible that JunB indirectly contributes to tissue differentiation by regulating cell proliferation.
We found that TGF-β signaling regulates the expression of junb during Xenopus tail regeneration. This observation is consistent with previous reports in cultured cell lines [25, 29]. However, junb expression was not completely eliminated by treatment with a TGF-β receptor inhibitor. These results suggest that, in addition to TGF-β, other factors may regulate the expression of junb during tail regeneration. In our study, we found that cell proliferation, which contributes to tissue differentiation, was perturbed by junb KO. As cell proliferation and differentiation in the regeneration bud are prevented by TGF-β receptor inhibition during Xenopus tail regeneration [9], JunB may be partly involved in events that are promoted by TGF-β signaling.
It is well-known that JunB forms a homodimer or heterodimer with other AP-1 family proteins and regulates the expression of downstream genes. In addition, the binding affinity of JunB to AP-1 elements is higher as a heterodimer with Fos proteins than as a JunB homodimer [30]. Since most AP-1 family genes are expressed during Xenopus tail regeneration [31; data not shown], the relationships between JunB and other AP-1 family proteins and the roles of the AP-1 heterodimer in regeneration will be investigated in future studies.
Supplementary Material
Supplementary Figure 1. Genotyping of junb KO sg 1 + sg 2 tadpoles. (A) Schematic drawing of junb structure and junb sgRNA target sites. Bars, untranslated region; box, coding region; TAD, transactivation domain; DBD, DNA-binding domain; LZD, leucine zipper domain. (B) Sequencing analysis of junb mutations in tyrosinase KO (n = 5) and junb KO (n = 5) tadpoles. Target sites of sg 1 and sg 2 are highlighted in green, and blue boxes indicate the PAM sequence. Deleted sequences are highlighted with red dashes. Insertions and substitutions are shown in blue and red letters, respectively. Mutation types were categorized as wild-type, in-frame, and out-of-frame. (C) Summary of mutation types as in panel B. Sequencing analysis of TA cloning demonstrates that all of alleles contain mutations in both sg 1 and sg 2 target sites. (D) Percentage of mutation types of junb KO sg 1 + sg 2 as in panel B.
Supplementary Figure 2. The delay in tail regeneration in junb KO is rescued by junb mRNA. (A) Co-injection of junb mRNA (250 pg) with junb sg 1 + sg 2 rescues the junb KO phenotype. (B) Lengths of regenerating tails. Black arrowheads indicate the amputation plane. Scale bar: 200 μm. ***P < 0.001, Student’s t-test.
Supplementary Figure 3. Compound heterozygous junb mutants show a delay in tail regeneration. (A) Schematic drawing of junb structure and junb sgRNA target sites. (B) Compound heterozygous junb mutants (Mut/Mut) show considerable delay in tail regeneration compared to wild-type (WT/WT, not sibling) tadpoles. WT/WT tadpoles were used as the control. (C) Lengths of regenerating tails. Black arrowheads indicate the amputation plane. Scale bar: 200 μm. ***P < 0.001, Student’s t-test.
Acknowledgements
We would like to thank the National Xenopus Resource (RRID: SCR_013731) at Marine Biological Laboratory (MBL) for Xenopus animals and care, and the National Bio-Resource Project (NBRP) of the Japan Agency for Medical Research and Development (AMED) under Grant Number JP18km0210085 for providing X. tropicalis. We also thank Dr. R. Harland for marker genes and reagents, and members of the Dr. Y. Kikuchi laboratory at Hiroshima University and our laboratory for helpful discussions. This work was supported by Grant-in-Aids for Scientific Research to K. T.-S. and A. S. from MEXT, Japan Society for the Promotion of Science (JSPS) KAKENHI 25460245, 16K08444, 17K08492 and 19K07247, and by Hiroshima University Natural Science Center for Basic Research and Development, and by NIH (OD010997 and HD084409) and NSF EDGE (1645105) grants to M.E.H..
References
- [1].Stoick-Cooper CL, Moon RT, Weidinger G, Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine, Genes Dev. 21 (2007) 1292–1315. http://www.genesdev.org/cgi/doi/10.1101/gad.1540507. [DOI] [PubMed] [Google Scholar]
- [2].Beck CW, Belmonte JCI, Christen B, Beyond early development: Xenopus as an emerging model for the study of regenerative mechanisms, Dev. Dyn 238 (2009) 1226–1248. 10.1002/dvdy.21890. [DOI] [PubMed] [Google Scholar]
- [3].Mochii M, Taniguchi Y, Shikata I, Tail regeneration in the Xenopus tadpole, Develop. Growth Differ 49 (2007) 155–161. 10.1111/j.1440-169X.2007.00912.x. [DOI] [PubMed] [Google Scholar]
- [4].Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, Koh Y, Gallop JL, Dorey K, Amaya E, Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration, Nat. Cell Biol. 15 (2013) 222–229. 10.1038/ncb2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Love NR, Chen Y, Bonev B, Gilchrist MJ, Fairclough L, Lea R, Mohun TJ, Paredes R, Zeef LA, Amaya E, Genome-wide analysis of gene expression during Xenopus tropicalis tadpole tail regeneration, BMC Dev. Biol 11 (2011) 70 10.1186/1471-213X-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen Y, Love NR, Amaya E, Tadpole tail regeneration in Xenopus, Biochem. Soc. Trans 42 (2014) 617–623. 10.1042/BST20140061. [DOI] [PubMed] [Google Scholar]
- [7].Beck CW, Christen B, Barker D, Slack JMW, Temporal requirement for bone morphogenetic proteins in regeneration of the tail and limb of Xenopus tadpoles, Mech. Dev 123 (2006) 674–688. 10.1016/j.mod.2006.07.001. [DOI] [PubMed] [Google Scholar]
- [8].Lin G, Slack JMW, Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration, Dev. Biol 316 (2008) 323–335. 10.1016/j.ydbio.2008.01.032. [DOI] [PubMed] [Google Scholar]
- [9].Ho DM, Whitman M, TGF-β signaling is required for multiple processes during Xenopus tail regeneration, Dev. Biol 315 (2008) 203–216. 10.1016/j.ydbio.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hayashi S, Ochi H, Ogino H, Kawasumi A, Kamei Y, Tamura K, Yokoyama H, Transcriptional regulators in the Hippo signaling pathway control organ growth in Xenopus tadpole tail regeneration, Dev. Biol 396 (2014) 31–41. 10.1016/j.ydbio.2014.09.018. [DOI] [PubMed] [Google Scholar]
- [11].Adams DS, Masi A, Levin M, H+ pump-dependent changes in membrane voltage are an early mechanism necessary and sufficient to induce Xenopus tail regeneration, Development 134 (2007) 1323–1335. 10.1242/dev.02812. [DOI] [PubMed] [Google Scholar]
- [12].Ivanova AS, Korotkova DD, Ermakova GV, Martynova NY, Zaraisky AG, Tereshina MB, Ras-dva small GTPases lost during evolution of amniotes regulate regeneration in anamniotes, Sci. Rep 8 (2018) 13035 10.1038/s41598-018-30811-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Angel P, Karin M, The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation, Biochim. Biophys. Acta 1072 (1991) 129–157. 10.1016/0304-419X(91)90011-9. [DOI] [PubMed] [Google Scholar]
- [14].Shaulian E, Karin M, AP-1 in cell proliferation and survival, Oncogene 20 (2001) 2390–2400. 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- [15].Dong Z, Xu RH, Kim J, Zhan SN, Ma WY, Colburn NH, Kung HF, AP-1/Jun is required for early Xenopus development and mediates mesoderm induction by fibroblast growth factor but not by activin, J. Biol. Chem 271 (1996) 9942–9946. doi: 10.1074/jbc.271.17.9942. [DOI] [PubMed] [Google Scholar]
- [16].Yoshida H, Okada M, Takebayashi-Suzuki K, Ueno N, Suzuki A, Involvement of JunB proto-oncogene in tail formation during early Xenopus embryogenesis, Zool. Sci 33 (2016) 282–289. 10.2108/zs150136. [DOI] [PubMed] [Google Scholar]
- [17].Blitz IL, Biesinger J, Xie X, Cho KWY, Biallelic genome modification in F0 Xenopus tropicalis embryos using the CRISPR/Cas system, Genesis 51 (2013) 827–834. 10.1002/dvg.22719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nakayama T, Blitz IL, Fish MB, Odeleye AO, Manohar S, Cho KWY, Grainger RM, Cas9-based genome editing in Xenopus tropicalis, Methods Enzymol. 546 (2014) 355–375. 10.1016/B978-0-12-801185-0.00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nieuwkoop PD, Faber J, Normal table of Xenopus laevis (Daudin), North Holland Publishing Company, Amsterdam, 1967. [Google Scholar]
- [20].Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM, Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis, Genesis 51 (2013) 835–843. 10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Harland RM, In situ hybridization: an improved whole-mount method for Xenopus embryos, Methods Cell Biol. 36 (1991) 685–695. DOI: 10.1016/S0091-679X(08)60307-6. [DOI] [PubMed] [Google Scholar]
- [22].Takebayashi-Suzuki K, Kitayama A, Terasaka-Iioka C, Ueno N, Suzuki A, The forkhead transcription factor FoxB1 regu¬lates the dorsal-ventral and anterior-posterior patterning of the ectoderm during early Xenopus embryogenesis, Dev. Biol 360 (2011) 11–29. 10.1016/j.ydbio.2011.09.005. [DOI] [PubMed] [Google Scholar]
- [23].Sugiura T, Taniguchi Y, Tazaki A, Ueno N, Watanabe K, Mochii M, Differential gene expression between the embryonic tail bud and regenerating larval tail in Xenopus laevis, Develop. Growth Differ. 46 (2004) 97–105. 10.1111/j.1440-169X.2004.00727.x. [DOI] [PubMed] [Google Scholar]
- [24].Taniguchi Y, Watanabe K, Mochii M, Notochord-derived hedgehog is essential for tail regeneration in Xenopus tadpole. BMC Dev. Biol 14 (2014) 27 10.1186/1471-213X-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jonk LJC, Itoh S, Heldin CH, ten Dijke P, Kruijer W, Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer, J. Biol. Chem 273 (1998) 21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- [26].Andrecht S, Kolbus A, Hartenstein B, Angel P, Schorpp-Kistner M, Cell cycle promoting activity of JunB through cyclin A activation, J. Biol. Chem 277 (2002) 35961–35968. doi: 10.1074/jbc.M202847200. [DOI] [PubMed] [Google Scholar]
- [27].Zhang J, Wu Z, Savin A, Yang M, Hsu YHR, Jantuan E, Bacani JTC, Ingham RJ, The c-Jun and JunB transcription factors facilitate the transit of classical Hodgkin lymphoma tumour cells through G1, Sci. Rep 8 (2018) 16019 10.1038/s41598-018-34199-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ishida T, Nakajima T, Kudo A, Kawakami A, Phosphorylation of Junb family proteins by the Jun N-terminal kinase supports tissue regeneration in zebrafish, Dev. Biol 340 (2010) 468–479. 10.1016/j.ydbio.2010.01.036. [DOI] [PubMed] [Google Scholar]
- [29].Sundqvist A, Morikawa M, Ren J, Vasilaki E, Kawasaki N, Kobayashi M, Koinuma D, Aburatani H, Miyazono K, Heldin CH, van Dam H, ten Djike P, JUNB governs a feed-forward network of TGFβ signaling that aggravates breast cancer invasion, Nucl. Acids Res 46 (2018) 1180–1195. 10.1093/nar/gkx1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ryseck RP, Bravo R, c-JUN, JUN B, and JUN D differ in their binding affinities to AP-1 and CRE consensus sequences: effect of FOS proteins, Oncogene 6 (1991) 533–542. [PubMed] [Google Scholar]
- [31].Chang J, Baker J, Wills A, Transcriptional dynamics of tail regeneration in Xenopus tropicalis, Genesis 55 (2017) e23015 10.1002/dvg.23015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Genotyping of junb KO sg 1 + sg 2 tadpoles. (A) Schematic drawing of junb structure and junb sgRNA target sites. Bars, untranslated region; box, coding region; TAD, transactivation domain; DBD, DNA-binding domain; LZD, leucine zipper domain. (B) Sequencing analysis of junb mutations in tyrosinase KO (n = 5) and junb KO (n = 5) tadpoles. Target sites of sg 1 and sg 2 are highlighted in green, and blue boxes indicate the PAM sequence. Deleted sequences are highlighted with red dashes. Insertions and substitutions are shown in blue and red letters, respectively. Mutation types were categorized as wild-type, in-frame, and out-of-frame. (C) Summary of mutation types as in panel B. Sequencing analysis of TA cloning demonstrates that all of alleles contain mutations in both sg 1 and sg 2 target sites. (D) Percentage of mutation types of junb KO sg 1 + sg 2 as in panel B.
Supplementary Figure 2. The delay in tail regeneration in junb KO is rescued by junb mRNA. (A) Co-injection of junb mRNA (250 pg) with junb sg 1 + sg 2 rescues the junb KO phenotype. (B) Lengths of regenerating tails. Black arrowheads indicate the amputation plane. Scale bar: 200 μm. ***P < 0.001, Student’s t-test.
Supplementary Figure 3. Compound heterozygous junb mutants show a delay in tail regeneration. (A) Schematic drawing of junb structure and junb sgRNA target sites. (B) Compound heterozygous junb mutants (Mut/Mut) show considerable delay in tail regeneration compared to wild-type (WT/WT, not sibling) tadpoles. WT/WT tadpoles were used as the control. (C) Lengths of regenerating tails. Black arrowheads indicate the amputation plane. Scale bar: 200 μm. ***P < 0.001, Student’s t-test.