Abstract
In Western countries, about 5% of the population is affected by an autoimmune disease; in the United States, up to 23.5 million Americans suffer from autoimmune disorders1. Women comprise over 80% of the affected individuals for many autoimmune conditions, including Sjögren’s syndrome, systemic sclerosis, Addison’s disease, primary biliary cirrhosis, systemic lupus erythematosus (SLE), and autoimmune thyroid diseases like Grave’s disease and Hashimoto’s thyroiditis2. Additionally, the prevalence of rheumatoid arthritis, multiple sclerosis, and Sjögren’s syndrome is also skewed towards women (3:1, 2:1, and 9:1, respectively)3. Though the reason for this sex-based disparity remains unknown, it has been speculated that sex hormone-dependent signaling, acting either in conjunction with or independently from sex-specific and X-linked gene expression, may modulate the immune system in women such that it predisposes them to autoimmunity4. Sex hormones clearly have a critical role in autoimmunity, as exemplified by the 15-fold reduction in risk of developing SLE in men compared to premenopausal women, though this risk drops by only 2-fold after menopause5. The focus of this brief review, however, will be to explore the potential link between X-chromosome inactivation, a critical biological event that occurs only in female cells, and the predisposition of women to autoimmunity.
Keywords: X-chromosome inactivation, autoimmune disease, female predilection, sex disparity
Introduction
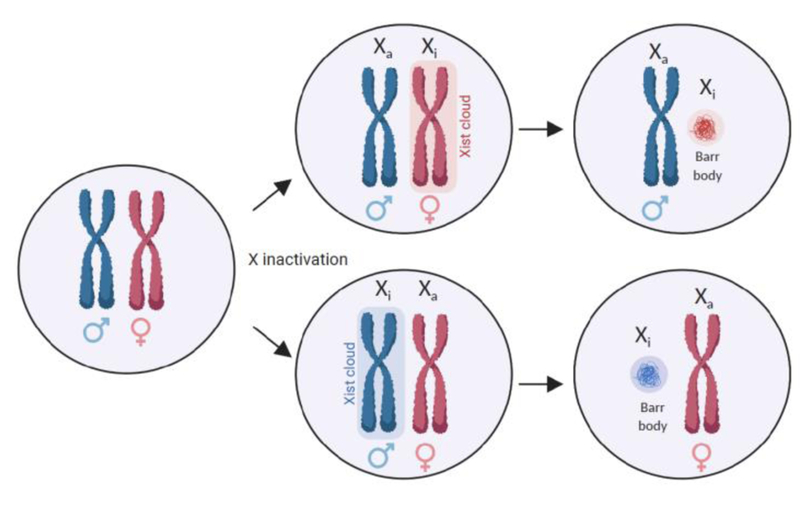
The human X-chromosome encodes more than one thousand genes and one could imagine the undesirable consequences of a two-fold difference in the expression of hundreds of X-linked genes between male cells that carry one X chromosome and female cells bearing two X chromosomes. This raises the question of how sex chromosome gene dosage is regulated. Dosage compensation is the process by which female mammals equalize the dosage of X-encoded genes between XX female and XY male cells. The solution to the sex chromosome gene dosage problem that has evolved in mammals is called X-chromosome inactivation (XCI), also known as lyonization or silencing (Figure 1). XCI involves the transcriptional silencing of one of the X-chromosomes early in female embryonic development. This silenced state of the X-chromosome is epigenetically inherited during cell division. Silencing of the X chromosome is controlled by the X-inactivation center on the X chromosome, and requires the action of Xist, a long non-coding RNAs that acts in cis to recruit the polycomb repressive complex 2 (PRC2). Xist spreads across the X chromosome forming an “Xist cloud” and maintains the epigenetically silenced state across the entire X chromosome with a few exceptions that are discussed later in this review [6]. The inactivated X chromosome (Xi) condenses to form a heterochromatic “Barr Body”, and all progeny cells inherit the epigenetic state of the inactivated X-chromosome. At the same time, the lncRNA Tsix, an antisense transcript of Xist, interferes with Xist transcription, and thwarts the heterochromatinization of the active X-chromosome (Xa) [7]. In most somatic cells, silencing of the Xi appears to be highly stable, typically only reversing in the female germline.
Figure 1. X chromosome inactivation and X-linked immune genes.
During development, in female cells, X-chromosome inactivation occurs randomly for one of the two X chromosomes, which equalizes the dosage of gene products from the X chromosome between females (XX) and males (XY). Each X chromosome has an equal probability of being silenced. Once established, inactivation of the X chromosome is stable, and perpetuates throughout subsequent division of the daughter cells. Through this process, each female is a mosaic of cells, composed of cells where either the maternally or paternally inherited X is expressed, and the other inherited X is silenced.
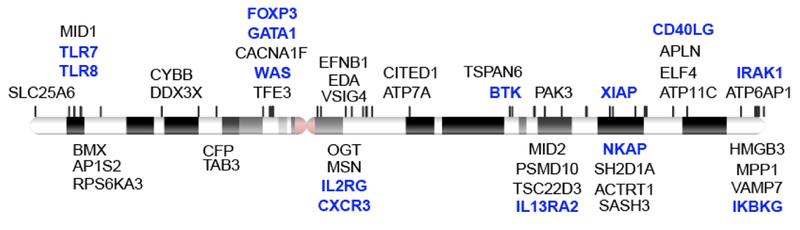
The X chromosome has a high density of immunity-related genes [8,9], which has led to the speculation that aberrant XCI may contribute to immune pathologies including autoimmunity (Figure 2). It is speculated that even the inherent randomness in initiating the XCI process may impact the immune system. Furthermore, reactivation of the Xi in immune cells, either at specific genes or the whole chromosome level, may predispose an individual to disease. The theoretical immune ramifications of physiological XCI or pathologically dysregulated XCI will be explored in the next sections.
Figure 2. The X chromosome has a high density of immunity-related genes.
Genes from the gene ontology terms GO_REGULATION_OF_IMMUNE_SYSTEM_PROCESS and GO_IMMUNE_RESPONSE that are located on the X chromosome are shown. The genes highlighted in blue have been linked to inherited immune disorders including immunodeficiencies.
Consequences of random or skewed X-inactivation
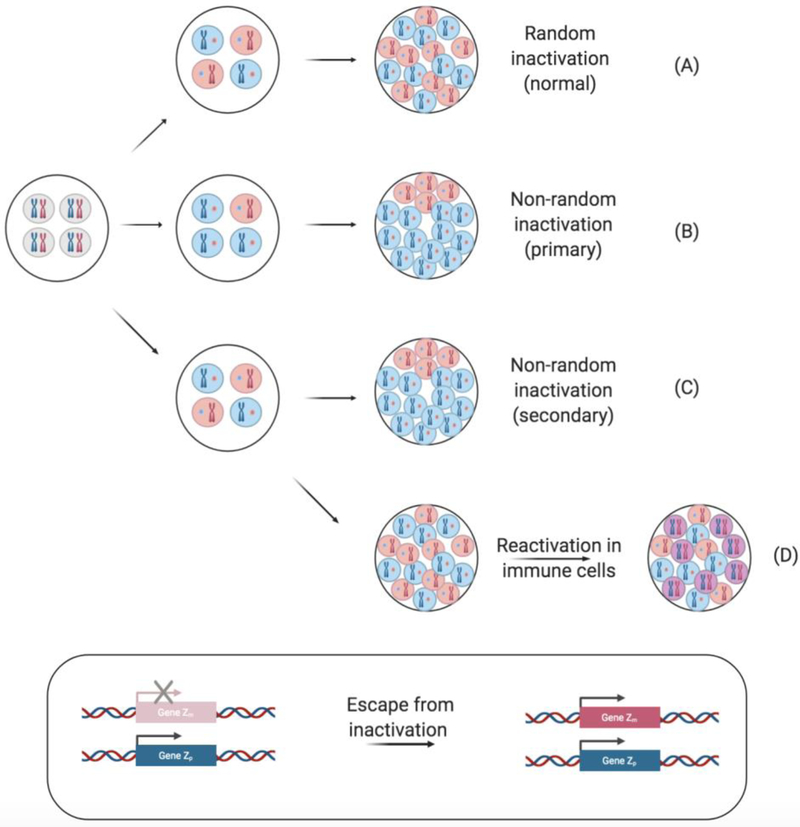
As random XCI occurs very early in development and Xi is stably inherited across somatic cell divisions, bias in XCI mosaicism may itself contribute to disease heterogeneity. The potential for biased X-inactivation lies at the heart of this theory, also termed the “loss of mosaicism” hypothesis (Figure 3). It has been reported that in approximately 10% of women, X inactivation is skewed to the point where 95% of their cells express X-encoded genes from one parent [10]. Given the stochastic nature of the XCI process and that the embryo contains a very small number of cells (estimated to be between 10 and 100) at the onset of XCI [11–13], it is possible that a majority of embryonic progenitor cells may inactivate the same parental X-chromosome by chance. It is also possible that inactivation of the X chromosome from a particular parent may offer a selective fitness advantage to a cell, if biased XCI has an impact on their ability to proliferate, migrate into tissues, use particular metabolic nutrients, and so forth. In the immune compartment, the impacts of biased XCI on cellular physiology may contribute to autoimmunity. Indeed, biased XCI has been linked to some autoimmune diseases [14–18].
Figure 3. Models of skewed X-chromosome inactivation and reactivation.
In female cells, either the maternal (pink) or paternal (blue) X chromosome is randomly inactivated. Early in development, each cell has two active X chromosomes (gray). (A) Normal XCI is random, res ulting in a 1:1 ratio of cells expressing a maternal or paternal X chromosome. (B) Nonrandom X-inactivation can be primary and arise simply by chance or due to a genetic predisposition, resulting in the preferential inactivation of either the maternal or paternal X. (C) Secondary nonrandom X-inactivation occurs when an X-linked mutation affects cell fitness and inactivation of the mutated allele results in a fitness cost. Though random XCI may have initially occurred, one may find a greater proportion of the cells inactivating the X chromosome associated with the disease allele, through a form of selection in some tissues. (D) Additionally, following random X-inactivation, some cells, notably naive lymphocytes, have been found to reactivate the Xi, expressing both the maternal and paternal X chromosome (purple). A number of genes have also been described to escape X-inactivation and are thus biallelically expressed (inset).
It has long been speculated that proteins expressed from the Xi can serve as autoantigens if immune cells are incompletely tolerized to them [2]. This may be particularly relevant in individuals with highly skewed XCI. As one hypothetical example, thymocytes, reactive to an antigen encoded by one of the parental X-encoded alleles, may fail to be negatively selected due to skewed silencing of that allele in the medullary thymic epithelial cells, only to encounter that antigen in the periphery. An analogous scenario may arise in developing B cells, although we believe that it may only be applicable to alleles encoding localized or membrane-bound self-antigens present in the developing B cell niche. This theory of autoimmunity is relies upon the assumption that an immunogenic X-linked variant is present in the heterozygous state [19]. While provocative, this theory does not easily lend itself to being tested. It also follows from this theory that female predilection to autoimmunity should be seen in all mammals with the potential for random and skewed XCI; it is not known if this is indeed the case.
While some autoimmune diseases (autoimmune thyroid diseases [16,17], systemic sclerosis [16,20], and rheumatoid arthritis [20]) have been associated with XCI skewing, it has not been observed in Sjögren’s or primary biliary cirrhosis, suggesting that this hypothesis may not be applicable to all autoimmune conditions. Additionally, assessment of skewed XCI has been carried out using PBMCs, which is convenient and useful, but not directly informative about the degree of skewing in the tissues, which may be more relevant to disease [21]. Furthermore, it is unclear whether loss of mosaicism in XCI truly reflects skewed initiation of XCI or a differential loss of cells expressing a deleterious X-linked allele. In autoimmune individuals with skewed XCI, is the pattern of skewed XCI a cause or consequence of autoimmunity? If skewed XCI is a cause of the disease, when and in what cell types does it occur? Is it possible to measure the cellular fitness costs of X-linked alleles in individuals with highly skewed patterns of XCI? As suggested previously by others, the association between skewed XCI and certain autoimmune diseases may not be causal, but rather an epiphenomenon, related to the inheritance of X-linked susceptibility genes [18,22] or related to age, as any selective pressure that can contribute to XCI skewing is afforded time to act [23].
Skewed XCI in lymphocyte populations can also arise in the context of clonal expansions. In one interesting case study, the biased XCI pattern was best explained by selection of expanded clones rather than the skewed onset of XCI [24]. This study examined two patients, a boy and his mother, who both had a CD40LG duplication linked to autoimmune disease. While the young patient was affected with multilineage autoimmune cytopenias that were refractory to glucocorticoids, rapamycin, intravenous immunoglobulin, and rituximab, his mother had been diagnosed with mixed connective tissue disease and autoimmune thyroiditis, which spontaneously resolved after 8 years, during her pregnancy. Flow cytometry revealed that though the CD40LG duplication was present in both patients, only the son (XY) exhibited functional disomy, as the induction of CD40LG on the mother’s CD4+ T cells (XX) was indistinguishable from unaffected relatives. A CpG-methylation dependent PCR assay for the X-linked human androgen receptor (HUMARA) revealed nonrandom XCI in the mother’s CD4+ T cells, suggesting that cells bearing the duplicated CD40LG on the Xa experienced a survival disadvantage. It is speculated that the eventual resolution of her disease was a result of the selective survival disadvantage of T cells expressing a higher level of CD40LG encoded by a duplication on their Xa. In contrast, her young son required pharmacological intervention with cyclosporine, as his sole X-chromosome, which does not undergo XCI, contained the pathogenic CD40LG duplication. This example shows how a deleterious mutation in an important X-linked immune gene can be more severe in a male than a female, as a result of epigenetic silencing of the affected X-linked allele.
Physiological escape and pathological reactivation of X inactivation
A small section of the X chromosome that is highly homologous to the Y chromosome is called the pseudoautosomal region and is the site of meiotic crossovers. Genes in this region are present on both the X and Y chromosomes and are biallelically expressed from both chromosomes [25,26]. It is now known that some genes outside the pseudoautosomal region, can also escape XCI despite the biological importance of XCI in mediating gene dosage (Figure 3). It is estimated that about 10–15% of genes on the Xi escape silencing [27–29]. Indeed, escape from XCI, even for pseudoautosomal genes, is generally partial and incomplete, in the sense that the level of expression of the escapee genes on the Xi is lower than their counterparts on the Xa [28]. Interestingly, tissue-specific patterns of escape from XCI have been noted, particularly in immune cells. In both humans and in mice, naive T and B cells from female individuals do not exhibit the canonical XIST RNA clouds that enshroud the inactivated X chromosome in other somatic cells [8]. However, upon activation, XIST returns to the Xi in B cells and T cells. Using a combination of RNA FISH, DNA FISH, and immunofluorescence, the authors revealed that the Xi chromatin in female T cells is much more euchromatic than those of fibroblasts, given than the Xi was missing H3K27me3, H4K20me, macroH2A, and H2AUb enrichment [8]. This partial reactivation of the Xi in female resting lymphocytes appeared to predispose X-linked genes, many of which are immune related, to be biallelically expressed at a higher level than in male cells.
Compared to healthy age-matched controls, B cells derived from female SLE patients exhibit abnormal XIST RNA localization that is associated with increased biallelic expression. It was also found that B cells from female SLE patients biallelically express a higher level of CD40LG and CXCR3. Recently, a study using a bicistronic CXCR3 dual-reporter mouse, wherein each CXCR3 allele was linked to a green or red fluorescent reporter, demonstrated that CXCR3 escapes XCI, and that cells that biallelically express CXCR3 produce more CXCR3 than monoallelically-expressing cells, in turn also expressing more IFN-γ, IL-2, and CD69 following Leishmania infection [30]. Previous studies examining the mechanisms of CD40LG overexpression in SLE have shown that the CD40LG core promoter and enhancer are unmethylated in men, while healthy women have one methylated and one unmethylated gene. However, in CD4+ T cells from women with lupus, CD40LG escapes from XCI and is biallelically expressed, as indicated by its demethylation and overexpression [31]. In contrast, the levels of CD40LG on CD4+ T cells in men with lupus were not elevated. These results suggest that biallelic expression of key X-linked immune genes in female subjects can result in pathological overexpression. Using an allele-specific PCR, Souyris et al. showed that TLR7 escapes XCI in a substantial proportion B cells, monocytes, and pDCs[32]. Biallelic expression of TLR7 was associated with increased TLR7 protein levels and imparted biallelically expressing B cells with a greater propensity to proliferate in response to TLR7 agonists, and switch to IgG. Biallelic expression of TLR7 was also noted in men with Kleinfelter’s Syndrome with a supernumerary X chromosome (47, XXY), who are also prone to lupus. Although pathological biallelic expression of immune genes resulting in higher immune gene dosage may be contribute to autoimmunity in women, its contribution relative to other predisposing factors such genetics (including the role of sex hormones) and environment remains to be ascertained.
There is ample evidence for the escape from XCI for a number of X-linked, immune-related genes. Heterogeneity in monoallelic or biallelic expression of these genes can result in differential responsiveness of the cells to TLR7 ligands, CD40, and CXCR3 ligands. If the differential responsiveness ultimately offers a fitness advantage to these cells, it could skew the lymphocyte and myeloid cell populations towards monoallelic or biallelic expression. As alluded to by Wang et al., it is possible that immunity-related genes like CD40LG, CXCR3, and TLR7 may be more susceptible to reactivation from the Xi in lymphocytes, possibly because of their proximity to parts of the Xi that have euchromatin characteristics.
A recent study using highly sensitive approaches to measure allele-specific gene expression, found that no X chromosome was 100% inactive in any of the female cells examined. Furthermore, the degree of XCI was heterogeneous between cells [33]. XCI heterogeneity was found to be dictated the level and localization of XIST RNA expression. As XIST is expressed at higher levels in the resting G0 phase than the G1 phase of the cell cycle, the degree of XCI also varies with the cell growth stage. Additional heterogeneity in XCI among individuals and among cell types results in cells with a unique expression profile of the Xi escapee genes. This is likely to contribute to the clinical heterogeneity among patients, as well as the likelihood that a certain cell type or tissue is the target or aggressor in a particular autoimmune disease.
The question of “why”
Why have immune cells have evolved a mechanism to evade the silencing of the Xi, at least for some genes with immune-related functions? One theory is to increase the potential degree of expression of the immunity-related X-linked genes in the event of an infection, with one adverse consequence being aberrant overexpression of genes leading to autoimmunity [13]. If this offers a fitness advantage to women, particularly in the protection against infectious diseases, it may help explain the accumulation of genes with an immune function on the X-chromosome [34]. It is also possible that biallelic expression and relaxed silencing of immune-related genes on the Xi have little to do with immunity itself. Leakiness of XCI may be an adaptive response to deleterious X-linked alleles with severe loss-of-function mutations that reduces the viability of males (e.g., male fetuses dying in utero), but serves to rescue cellular function in females due to skewed or incomplete XCI, as in incontinentia pigmenti and Rett’s syndrome. In these diseases, females benefit from mosaicism, skewed towards cells bearing the disease-causing mutation on the Xi or exhibiting biallelic expression. This has been clearly observed in the case of Rett syndrome, where the survival of heterozygous female patients depends upon cells with the wild-type allele of MECP2 on Xa, resulting in dramatic skewing of XCI [35]. In other conditions, like incontinentia pigmenti (OMIM: 308300) and oral-facial-digital syndrome type I (OMIM: 311200), it appears that the wild-type alleles of the affected genes do escape XCI, but the escape is incomplete and tissue-specific. Some X-linked immunodeficiencies may also fall into this pattern. It is important to note that the escape of a particular gene from XCI may also be driven by the biallelic expression of a closely linked gene that has even more stark ramifications if it is not biallelically expressed. Indeed, in neural progenitor cells, genes that escape XCI were shown to be present in epigenetically defined clusters called topologically-associated domains (TADs) [36]. Understanding the gene-specific effects of biallelic expression on particular cells will require the development of new genetic tools.
Open questions
Genes in the pseudoautosomal region that escape XCI are excluded from XCI from its onset, as they contain (epi)genetic features that exclude them from being silenced during propagation of XCI [37,38]. The epigenetic features of loci outside the pseudoautosomal region that escape from XCI once it is established are poorly understood. What are the epigenetic characteristics of non-pseudoautosomal regions that escape XCI? What factors control escape of these loci from XCI and their subsequent reversion to XCI? Gene clusters that escape XCI in neural progenitors were shown to form TAD-like structures [36]. Do polymorphisms at these genes have an impact on epigenetic features such as TADs in a manner that predisposes an individual to autoimmunity? What are the cell types and contexts in which genes escape from XCI? Why do immune-related genes escape XCI in naive lymphocytes and why does lymphocyte activation restore XCI at these loci? These are important open questions that hold the key to understanding the epigenetic basis for the female predilection of human autoimmunity
Highlights.
Over three-fourths of individuals affected by autoimmune conditions are women.
Perturbations in X-chromosome silencing may contribute to an autoimmune diathesis in women.
The X-chromosome harbors several genes critical to immune function.
The silenced X-chromosome undergoes physiological and pathological reactivation in immune cells.
Acknowledgments
The author was supported by a training grant on systemic autoimmunity from the NIH (AI074549). The author thanks Shiv Pillai and Vinay Mahajan for their critical reading of this review. Figures were created using BioRender.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].The Autoimmune Diseases Coordinating Committee, Progress in autoimmune diseases research: Report to Congress, 2005. https://www.niaid.nih.gov/sites/default/files/adccfinal.pdf.
- [2].Libert C, Dejager L, Pinheiro I, The X chromosome in immune functions: when a chromosome makes the difference, Nat. Rev. Immunol 10 (2010) 594–604. [DOI] [PubMed] [Google Scholar]
- [3].Desai MK, Brinton RD, Autoimmune Disease in Women: Endocrine Transition and Risk Across the Lifespan, Front. Endocrinol 10 (2019) xix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Moulton VR, Sex Hormones in Acquired Immunity and Autoimmune Disease, Front. Immunol 9 (2018) 2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lockshin MD, Sex differences in autoimmune disease, Lupus. 15 (2006) 753–756. [DOI] [PubMed] [Google Scholar]
- [6].Clemson CM, McNeil JA, Willard HF, Lawrence JB, XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure, J. Cell Biol. 132 (1996) 259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Caley DP, Pink RC, Trujillano D, Carter DRF, Long noncoding RNAs, chromatin, and development, ScientificWorldJournal. 10 (2010) 90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang J, Syrett CM, Kramer MC, Basu A, Atchison ML, Anguera MC, Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X, Proc. Natl. Acad. Sci. U. S. A 113 (2016) E2029–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, Frankish A, Lovell FL, Howe KL, Ashurst JL, Fulton RS, Sudbrak R, Wen G, Jones MC, Hurles ME, Andrews TD, Scott CE, Searle S, Ramser J, Whittaker A, Deadman R, Carter NP, Hunt SE, Chen R, Cree A, Gunaratne P, Havlak P, Hodgson A, Metzker ML, Richards S, Scott G, Steffen D, Sodergren E, Wheeler DA, Worley KC, Ainscough R, Ambrose KD, Ansari-Lari MA, Aradhya S, Ashwell RIS, Babbage AK, Bagguley CL, Ballabio A, Banerjee R, Barker GE, Barlow KF, Barrett IP, Bates KN, Beare DM, Beasley H, Beasley O, Beck A, Bethel G, Blechschmidt K, Brady N, Bray-Allen S, Bridgeman AM, Brown AJ, Brown MJ, Bonnin D, Bruford EA, Buhay C, Burch P, Burford D, Burgess J, Burrill W, Burton J, Bye JM, Carder C, Carrel L, Chako J, Chapman JC, Chavez D, Chen E, Chen G, Chen Y, Chen Z, Chinault C, Ciccodicola A, Clark SY, Clarke G, Clee CM, Clegg S, Clerc-Blankenburg K, Clifford K, Cobley V, Cole CG, Conquer JS, Corby N, Connor RE, David R, Davies J, Davis C, Davis J, Delgado O, DeShazo D, Dhami P, Ding Y, Dinh H, Dodsworth S, Draper H, Dugan-Rocha S, Dunham A, Dunn M, Durbin KJ, Dutta I, Eades T, Ellwood M, Emery-Cohen A, Errington H, Evans KL, Faulkner L, Francis F, Frankland J, Fraser AE, Galgoczy P, Gilbert J, Gill R, Glöckner G, Gregory SG, Gribble S, Griffiths C, Grocock R, Gu Y, Gwilliam R, Hamilton C, Hart EA, Hawes A, Heath PD, Heitmann K, Hennig S, Hernandez J, Hinzmann B, Ho S, Hoffs M, Howden PJ, Huckle EJ, Hume J, Hunt PJ, Hunt AR, Isherwood J, Jacob L, Johnson D, Jones S, de Jong PJ, Joseph SS, Keenan S, Kelly S, Kershaw JK, Khan Z, Kioschis P, Klages S, Knights AJ, Kosiura A, Kovar-Smith C, Laird GK, Langford C, Lawlor S, Leversha M, Lewis L, Liu W, Lloyd C, Lloyd DM, Loulseged H, Loveland JE, Lovell JD, Lozado R, Lu J, Lyne R, Ma J, Maheshwari M, Matthews LH, McDowall J, McLaren S, McMurray A, Meidl P, Meitinger T, Milne S, Miner G, Mistry SL, Morgan M, Morris S, Müller I, Mullikin JC, Nguyen N, Nordsiek G, Nyakatura G, O’Dell CN, Okwuonu G, Palmer S, Pandian R, Parker D, Parrish J, Pasternak S, Patel D, Pearce AV, Pearson DM, Pelan SE, Perez L, Porter KM, Ramsey Y, Reichwald K, Rhodes S, Ridler KA, Schlessinger D, Schueler MG, Sehra HK, Shaw-Smith C, Shen H, Sheridan EM, Shownkeen R, Skuce CD, Smith ML, Sotheran EC, Steingruber HE, Steward CA, Storey R, Swann RM, Swarbreck D, Tabor PE, Taudien S, Taylor T, Teague B, Thomas K, Thorpe A, Timms K, Tracey A, Trevanion S, Tromans AC, d’Urso M, Verduzco D, Villasana D, Waldron L, Wall M, Wang Q, Warren J, Warry GL, Wei X, West A, Whitehead SL, Whiteley MN, Wilkinson JE, Willey DL, Williams G, Williams L, Williamson A, Williamson H, Wilming L, Woodmansey RL, Wray PW, Yen J, Zhang J, Zhou J, Zoghbi H, Zorilla S, Buck D, Reinhardt R, Poustka A, Rosenthal A, Lehrach H, Meindl A, Minx PJ, Hillier LW, Willard HF, Wilson RK, Waterston RH, Rice CM, Vaudin M, Coulson A, Nelson DL, Weinstock G, Sulston JE, Durbin R, Hubbard T, Gibbs RA, Beck S, Rogers J, Bentley DR, The DNA sequence of the human X chromosome, Nature. 434 (2005) 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Migeon BR, The role of X inactivation and cellular mosaicism in women’s health and sex -specific diseases, JAMA. 295 (2006) 1428–1433. [DOI] [PubMed] [Google Scholar]
- [11].Teklenburg G, Weimar CHE, Fauser BCJM, Macklon N, Geijsen N, Heijnen CJ, Chuva SM Sousa Lopes de, Kuijk EW, Cell lineage specific distribution of H3K27 trimethylation accumulation in an in vitro model for human implantation, PLoS One. 7 (2012) e32701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Monteiro J, Derom C, Vlietinck R, Kohn N, Lesser M, Gregersen PK, Commitment to X inactivation precedes the twinning event in monochorionic MZ twins, Am. J. Hum. Genet 63 (1998) 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gribnau J, Barakat TS, X-chromosome inactivation and its implications for human disease, bioRxiV. (2017). doi: 10.1101/076950. [DOI] [Google Scholar]
- [14].Lleo A, Battezzati PM, Selmi C, Gershwin ME, Podda M, Is autoimmunity a matter of sex?, Autoimmun. Rev 7 (2008) 626–630. [DOI] [PubMed] [Google Scholar]
- [15].Rubtsova K, Marrack P, Rubtsov AV, Sexual dimorphism in autoimmunity, J. Clin. Invest 125 (2015) 2187–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ozbalkan Z, Bagişlar S, Kiraz S, Akyerli CB, Ozer HTE, Yavuz S, Birlik AM, Calgüneri M, Ozçelik T, Skewed X chromosome inactivation in blood cells of women with scleroderma, Arthritis Rheum. 52 (2005) 1564–1570. [DOI] [PubMed] [Google Scholar]
- [17].Ozcelik T, Uz E, Akyerli CB, Bagislar S, Mustafa CA, Gursoy A, Akarsu N, Toruner G, Kamel N, Gullu S, Evidence from autoimmune thyroiditis of skewed X-chromosome inactivation in female predisposition to autoimmunity, Eur. J. Hum. Genet 14 (2006) 791–797. [DOI] [PubMed] [Google Scholar]
- [18].Brix TH, Knudsen GPS, Kristiansen M, Kyvik KO, Orstavik KH, Hegedüs L, High frequency of skewed X-chromosome inactivation in females with autoimmune thyroid disease: a possible explanation for the female predisposition to thyroid autoimmunity, J. Clin. Endocrinol. Metab 90 (2005) 5949–5953. [DOI] [PubMed] [Google Scholar]
- [19].Lambert NC, The price of silence, Arthritis Rheum. 60 (2009) 3164–3167. [DOI] [PubMed] [Google Scholar]
- [20].Kanaan SB, Onat OE, Balandraud N, Martin GV, Nelson JL, Azzouz DF, Auger I, Arnoux F, Martin M, Roudier J, Ozcelik T, Lambert NC, Evaluation of X Chromosome Inactivation with Respect to HLA Genetic Susceptibility in Rheumatoid Arthritis and Systemic Sclerosis, PLoS One. 11 (2016) e0158550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brooks WH, X chromosome inactivation and autoimmunity, Clin. Rev. Allergy Immunol. 39 (2010) 20–29. [DOI] [PubMed] [Google Scholar]
- [22].Brix TH, Hansen PS, Kyvik KO, Hegedüs L, Preliminary evidence of a noncausal association between the X-chromosome inactivation pattern and thyroid autoimmunity: a twin study, Eur. J. Hum. Genet 18 (2009) 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Busque L, Mio R, Mattioli J, Brais E, Blais N, Lalonde Y, Maragh M, Gilliland DG, Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age, Blood. 88 (1996) 59–65. [PubMed] [Google Scholar]
- [24].Le Coz C, Trofa M, Syrett CM, Martin A, Jyonouchi H, Jyonouchi S, Anguera MC, Romberg N, CD40LG duplication-associated autoimmune disease is silenced by nonrandom X-chromosome inactivation, J. Allergy Clin. Immunol 141 (2018) 2308–2311.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Helena Mangs A, Morris BJ, The Human Pseudoautosomal Region (PAR): Origin, Function and Future, Curr. Genomics 8 (2007) 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Disteche CM, Dosage compensation of the sex chromosomes, Annu. Rev. Genet 46 (2012) 537–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Calabrese JM, Sun W, Song L, Mugford JW, Williams L, Yee D, Starmer J, Mieczkowski P, Crawford GE, Magnuson T, Site-specific silencing of regulatory elements as a mechanism of X inactivation, Cell. 151 (2012) 951–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Carrel L, Willard HF, X-inactivation profile reveals extensive variability in X-linked gene expression in females, Nature. 434 (2005) 400–404. [DOI] [PubMed] [Google Scholar]
- [29].Zhang Y, Castillo-Morales A, Jiang M, Zhu Y, Hu L, Urrutia AO, Kong X, Hurst LD, Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving, Mol. Biol. Evol 30 (2013) 2588–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Oghumu S, Varikuti S, Stock JC, Volpedo G, Saljoughian N, Terrazas CA, Satoskar AR, Cutting Edge: CXCR3 Escapes X Chromosome Inactivation in T Cells during Infection: Potential Implications for Sex Differences in Immune Responses, J. Immunol 203 (2019) 789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B, Demethylation of CD40LG on the Inactive X in T Cells from Women with Lupus, The Journal of Immunology. 179 (2007) 6352–6358. [DOI] [PubMed] [Google Scholar]
- [32].Souyris M, Mejía JE, Chaumeil J, Guéry J-C, Female predisposition to TLR7-driven autoimmunity: gene dosage and the escape from X chromosome inactivation, Semin. Immunopathol (2018). doi: 10.1007/s00281-018-0712-y. [DOI] [PubMed] [Google Scholar]
- [33].Garieri M, Stamoulis G, Blanc X, Falconnet E, Ribaux P, Borel C, Santoni F, Antonarakis SE, Extensive cellular heterogeneity of X inactivation revealed by single-cell allele-specific expression in human fibroblasts, Proc. Natl. Acad. Sci. U. S. A 115 (2018) 13015–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schurz H, Salie M, Tromp G, Hoal EG, Kinnear CJ, Möller M, The X chromosome and sex-specific effects in infectious disease susceptibility, Hum. Genomics 13 (2019) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cheung AYL, Horvath LM, Carrel L, Ellis J, X-chromosome inactivation in rett syndrome human induced pluripotent stem cells, Front. Psychiatry 3 (2012) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, Dekker J, Structural organization of the inactive X chromosome in the mouse, Nature. 535 (2016) 575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Blaschke RJ, Rappold G, The pseudoautosomal regions SHOX and disease, Curr. Opin. Genet. Dev 16 (2006) 233–239. [DOI] [PubMed] [Google Scholar]
- [38].Singh NP, Madabhushi SR, Srivastava S, Senthilkumar R, Neeraja C, Khosla S, Mishra RK, Epigenetic profile of the euchromatic region of human Y chromosome, Nucleic Acids Res. 39 (2011) 3594–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]