Abstract Abstract
This paper reports on a new species of the Baikal endemic sponge (fam. Lubomirskiidae) Swartschewskia khanaevisp. nov. The description of this species is based on morphological and molecular data (ITS and mitochondrial IGRs). Morphologically, S. khanaevisp. nov. differs from S. papyracea by loose tracts arranged in an irregular network as well as the presence on strongyles of compound spines looking like tubercles densely ornamented with simple spines. Moreover, specimens of S. khanaevisp. nov. show a peculiar structure of the aquiferous system at the body surface that may be an adaptive trait for environmental conditions. Phylogenetic analysis has revealed that S. khanaevisp. nov. forms a well-supported (0.99) monophyletic clade with S. papyracea and is allocated as its sister taxa.
Keywords: ITS, mitochondrial IGRs, morphological analysis, Swartschewskia
Introduction
Baikal is the most ancient and deepest lake on the Earth with the huge water volume. The lake is considered to be 25–30 million years old (Mazepova 1995); its maximum depth is 1641 m, and the volume of water body exceeds 23 000 km3 (Mats et al. 2001). Due to these facts, the lake is characterised by minor environmental oscillations. The family Lubomirskiidae represents the most spectacular example of endemic radiation in freshwater sponges under the specific conditions of the great lake. According to molecular phylogeny the endemic family Lubomirskiidae was monophyletic and diverged from Spongillidae (Itskovich et al. 2008; Maikova et al. 2012; Maikova et al. 2017). Existence in a stable environment over a long period of time resulted in the disappearance of gemmulation from the life cycle of Lubomirskiidae (Efremova and Goureeva 1989; Manconi and Pronzato 2002).
At present, 14 species are allocated to the family Lubomirskiidae (Efremova 2001, 2004; Manconi and Pronzato 2019). The actual number of species is most likely underestimated. Comprehensive morphological study of the Baikal sponges revealed at least five forms that showed a constant set of morphological characteristics but could not be related to any known species (Khanaev et al. 2018). In this regard, these forms were suggested to be new species. Additionally, some specimens with uncommon morphology have been described (Weinberg 2005). We also observed several sponge specimens having unusual or transitional traits that interfered with precise species identification in our previous study (Khanaev et al. 2018).
The gaps in our knowledge of Lubomirskiidae morphology and taxonomy concern some aspects in the biology of the Baikal sponges. The absence of gemmules, gemmuloscleres, and parenchymal microscleres, which often contribute to taxonomy, complicates species identification (Manconi and Pronzato 2002). Moreover, the majority of the Lubomirskiidae species were described in the late 19th – early 20th century. The descriptions were limited to the classical taxonomy based on diagnostic morphological characters and were often very brief.
The genus Swartschewskia Makuschok, 1927 is clearly segregated from other Lubomirskiidae genera (Dybowski 1880, Swartschewsky 1901, Makushok 1927a). Only Swartschewskia is characterised by cortex: an ectosomal skeleton, tangential arrangement of primary tracts and stout bent strongyles as megascleres. The genus includes two species: S. papyracea (Dybowski, 1880) and S. irregularis (Swartschewsky, 1902). Swartschewskia papyracea is widely distributed in the depth range of 1–80 m in Baikal. Swartschewskia papyracea morphology was reported in a number of works (Swartschewsky 1902, Makushok 1927a, Rezvoy 1936, Manconi and Pronzato 2002, 2015, 2019; Weinberg 2005, Masuda 2009). On the contrary S. irregularis is extremely rare species inhabiting sublittoral zone of Baikal (70–150 m). The species was described based on the single specimen that was not preserved (Swartschewsky 1902). During the next 120 years only one sponge specimen with a similar morphology was found but no data on its morphology were published (Efremova 2001).
Molecular approach is also limited for phylogenetic studies of the Baikal sponges due to low variability of markers (COI, ITS, rRNA-genes) usually applied for this purpose throughout the world (Itskovich et al. 2008, Harcet et al. 2010). Recently (Maikova et al. 2015, 2016), the protein coding sequences of mitochondrial DNA were used to study the phylogenetic relationship of Baikal sponges only at the genera taxonomic level. This study showed that the nucleotide substitution rate of intergenic regions (IGRs) of the Baikal sponge mtDNA is significantly higher than coding sequences, which makes them very promising for phylogenetic reconstructions of closely related species (Lavrov 2010, Maikova et al. 2012). However, only concatenated nuclear (ITS-regions) and mitochondrial (IGRs) data allowed us to separate closely related species of the family Lubomirskiidae (Maikova et al. 2017). Therefore, in this study we use ITS and mitochondrial IGRs sequences to investigate the phylogenetic position of a new species within the family Lubomirskiidae.
During the 2016 expeditions, unusual sponges were sampled in Olkhonskiye Vorota Strait. These sponges were identified as a separate species based on their morphological and molecular phylogenetic data. The paper describes a new species of Swartschewskia and we present additional data on the morphology of Swartschewskia papyracea (Dybowski, 1880) and provide diagnostic keys for the species belonging to the genus Swartschewskia.
Materials and methods
Study site and sample collection
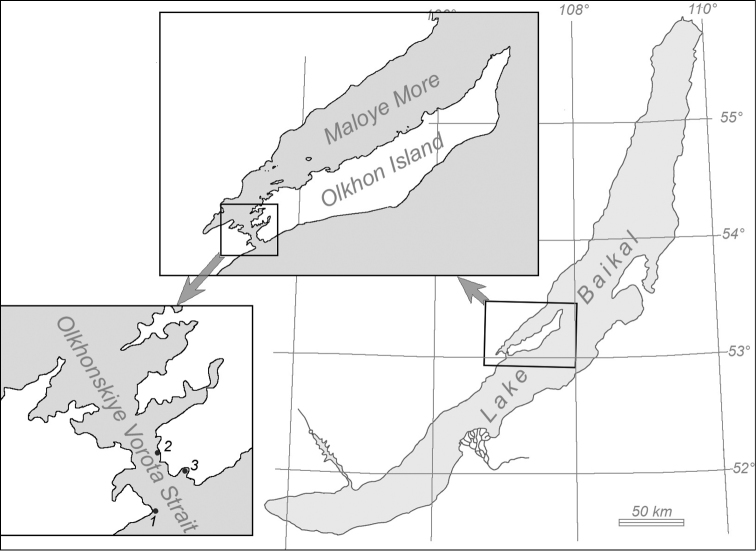
The Olkhonskiye Vorota is a narrow strait that connects the Maloye More Strait with the main part of Baikal. The bottom of the Maloye More and the Olkhonskiye Vorota straits consists of different types of ground: rock debris, boulders, pebbles, various sand fractions, and silt (Kozhov 1947). The samples were collected at the three study sites (Fig. 1). At the study site 1 and 2, an extensive multi-layered bank of rock debris is located along the shore from the shoreline to the depth of 4–10 m. Stone fragments have rather large interstices between each other. The interstices are not filled with smaller fractions of ground (such as gravel, sand or silt); hence, unhampered water movements can take place there. Below 10 m, the bottom is sandy with rare boulders submerged in the sand with their lower side. At the study site 3, the bottom mainly consists of sand with detached boulders and rocks, which can be partially submerged in sand.
Figure 1.
Sampling sites in Lake Baikal: 1, 2 samples of S. khanaevi sp. nov, 3 samples of S. papyracea.
Samples were collected by SCUBA divers. All specimens were photographed and fixed in 96 % ethanol or frozen at -20 °C.
Holotype and four paratypes of the new species (specimens in ethanol and microscopic slides with tissue-free spicules preparations) have been deposited in Zoological Institute of the Russian Academy of Sciences, St Petersburg (ZIN). One paratype has been deposited in Porifera collection of Laboratory of Analytical and Bioorganic Chemistry, Limnological Institute of the Russian Academy of Sciences, Irkutsk (LIN).
DNA extraction, amplification, and sequencing
Total DNA was extracted from approximately 0.1 g of fixed tissue by modified phenol-chloroform method (Maniatis et al. 1984). For molecular analysis, two internal transcribed spacers (ITS1 and ITS2), as well as intergenic regions (IGRs) between tRNATyr and tRNAMet genes of mitochondrial DNA (mtDNA), were sequenced. Amplification of ITS and IGRs sequences was performed using specific primers and parameters described previously (Itskovich et al. 2008; Maikova et al. 2012). Each PCR reaction was purified by electrophoresis in 0.8 % agarose gels and eluted by freezing and thawing. DNA sequencing was carried out using the BigDye Terminator V3.1 Cycle Sequencing Kit (Applied Biosystems, United States) with subsequent analysis of the reaction products on an Applied Biosystems 3130xl Genetic Analyzer sequencer (USA) at Syntol Company (Moscow, Russia).
Sequence alignments and tree reconstructions
PCR-fragments were assembled and aligned using MAFFT (Katoh and Toh 2008) and BioEdit 5.09 (Hall 1999). Bayesian reconstructions were performed using MrBayes v. 3.2.1. (Ronquist and Huelsenbeck 2003). For concatenated data (ITS and IGRs), the nucleotide substitution model GTR+I+G was used for ITS and “mixed” parameter for IGRs. The Markov chain Monte Carlo search was run twice (default parameter) on four chains for 20 000 000 generations. Trees were sampled every 1000 cycles after the first 10 000 burn-in cycles. Genetic distances in pairwise comparisons between all analysed sequences were calculated according to the Kimura’s two-parameter model using MEGA7 (Kumar et al. 2016).
Morphological analysis1
Two variants of skeleton preparations were made for each specimen. In the first case, the small pieces of specimens were saturated with water and frozen. Vertical sections of frozen pieces (0.3–0.5 mm thickness) were made manually to investigate the ectosomal and choanosomal skeleton (Efremova 2004). In the second case, the small piece of the ectosomal skeleton was detached from the choanosomal one, washed with water and oriented upwards with the superior surface. Both kinds of skeleton preparations were mounted on slides and on stubs. Tissue-free spicules preparations were made by dissolving small pieces of sponge in sodium hypochlorite with a subsequent rinse with water and transfer to 96 % ethanol. Clean spicules and skeleton pieces were mounted on slides and aluminium stubs and air-dried. Samples on slides were placed in Canada balsam for the observation with Light Optical Microscope Olympus CX-21. Samples on aluminium stubs were coated with gold for further investigation with a Scanning Electron Microscope Philips SEM 525 (Collective Instrumental Center "Ultramicroanalysis" at LINSBRAS). Digital images of both kinds of skeleton preparations and spicules were made using an integrated camera by SEM. Skeleton preparations were also photographed using the light optical microscope with an ocular camera ToupCam 5.1. Spicules of six samples (length and width; 50 spicules in every sample) were measured in several fields of view using Olympus CX-21 and ocular micrometre. Spicules dimensions were listed as three values: minimum–(mean)–maximum.
Cortex thickness and dermal pores dimensions were measured by Philips SEM 525 digital images in every investigated specimen (N = 9). Dermal pores ranged from rounded (diameter was measured) to ovoid or elliptic (width and length were measured). Dimensions were listed as three values: minimum–(mean)–maximum. Light optical microscope photographs were used for pore fields and oscula measurements. Apertures in the sieve-like osculum of S. papyracea were measured by photograph (N = 1).
The percentage of sponge surface lacking in both oscula and pores was calculated in SpongeArea (original software is available at https://gitlab.com/bukshuk-sci/spongearea). Macro photographs for the analysis were taken using Canon EOS 450D with LPL Copy Stand CS-40.
For the taxonomy of genus and species level and name validity the World Porifera Database was considered as reference (Van Soest et al. 2019).
Results
Phylogenetic analysis
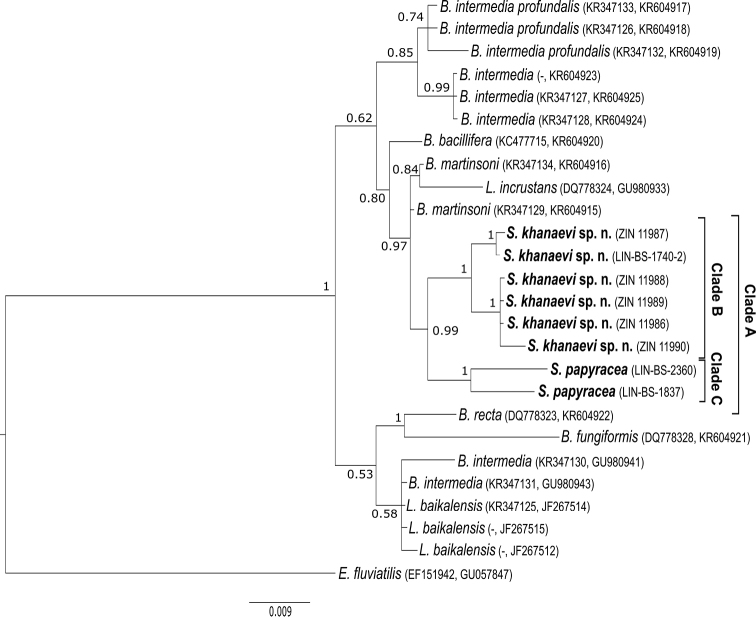
For phylogenetic analysis, the ITS and mitochondrial IGRs sequences were obtained from six specimens of S. khanaevi sp. nov. and two specimens of S. papyracea, which were deposited into GenBank (Table 1). Additionally, we used previously published sequences of Lubomirskiidae and Spongillidae species (Maikova et al. 2017). Ephydatia fluviatilis (Linnaeus, 1759) (fam. Spongillidae) was used as an outgroup (Itskovich et al. 2008). The length of the aligned concatenated sequences was 1266 bp, the ITS and IGRs partitions were 734 bp and 532 bp in the length respectively. The overall variability (K2P) for ITS-regions was 1.9 % and for IGRs – 0.9 %. Within the family Lubomirskiidae the intraspecific genetic distances of the ITS-regions varied from 0 to 0.6 %, while the interspecific ones varied from 0.1 to 4.7 % (average 1.5 %). The intraspecific genetic distances of the IGRs varied from 0 to 0.8 % and the interspecific ones varied from 0 to 4.9 % (average 2.2 %). Based on concatenated data the intraspecific genetic variability was from 0 to 1.6 % and between species ones was from 0.3 to 4.2 % (average 1.7 %). The pairwise genetic distances between the sequences of S. khanaevi sp. nov. varied from 0 to 1.5 % (average 0.7 %), and the ones between the sequences of S. khanaevi sp. nov. and S. papyracea ranged from 1.4 to 2.6 % (average 1.9 %).
Table 1.
Sample numbers in the collection and sequence numbers in GenBank.
| Species | Number in the collection | GenBank number | |
| Sequences of ITS-regions | Sequences of mtDNA intergenic regions (IGRs) | ||
| S. papyracea | LIN-BS-1837 | MH133907 | MH257749 |
| LIN-BS-2360 | MH133908 | MH257750 | |
| S. khanaevi sp. nov. | ZIN 11990 | MH133901 | MH257748 |
| LIN-BS-1740-2 | MH133902 | MH257744 | |
| ZIN 11986 | MH133903 | MH257746 | |
| ZIN 11987 | MH133904 | MH257743 | |
| ZIN 11988 | MH133905 | MH257745 | |
| ZIN 11989 | MH133906 | MH257747 | |
In the phylogenetic tree, the specimens of S. papyracea and S. khanaevi sp. nov. form a well-supported (0.99) monophyletic clade named A (Fig. 2). Within clade A, we recognise two well-supported monophyletic groups named B and C. Clade B contains the specimens of S. khanaevi sp. nov.; S. papyracea was allocated as its sister group (clade C). Thus, S. khanaevi sp. nov. was named as a separate species.
Figure 2.
Phylogenetic tree based on concatenated nuclear (ITS1 and ITS2) and mitochondrial (IGRs) sequences: Bayesian posterior probabilities are shown at the bases of the clusters. Taxon names and collection numbers of sponges analysed in this study are marked in bold. Scale bar denotes substitutions per site.
Systematics
Phylum Porifera Grant, 1836
Class Demospongiae Sollas, 1885
Subclass Heteroscleromorpha Cárdenas, Pérez & Boury-Esnault, 2012
Order Spongillida Manconi & Pronzato, 2002
Family Lubomirskiidae (Weltner, 1895)
Genus. Swartschewskia
Makushok, 1927
6B82B1DF-DBB6-57D8-A41F-FBD66E04B59D
Included species.
Swartschewskia papyracea (Dybowski, 1880), Swartschewskia irregularis (Swartschewsky, 1902).
Type species.
Swartschewskia papyracea (Dybowski, 1880).
Genus diagnosis.
Body shape encrusting to globose or branched. Ectosomal skeleton hard and well developed as more or less regular alveolar network of thick tangential spicular fibres. Choanosomal skeleton sparsely developed with scarce spicules irregularly arranged in few weak fibres. Abundant spongin. Megascleres strongyles, from spiny to smooth (modified from Manconi and Pronzato 2002).
Swartschewskia khanaevi sp. nov.
1826FDDB-C0FD-55F7-997B-E9B98D34028C
http://zoobank.org/40B1B85A-7E8E-4C09-8DD7-15CE4BB56A28
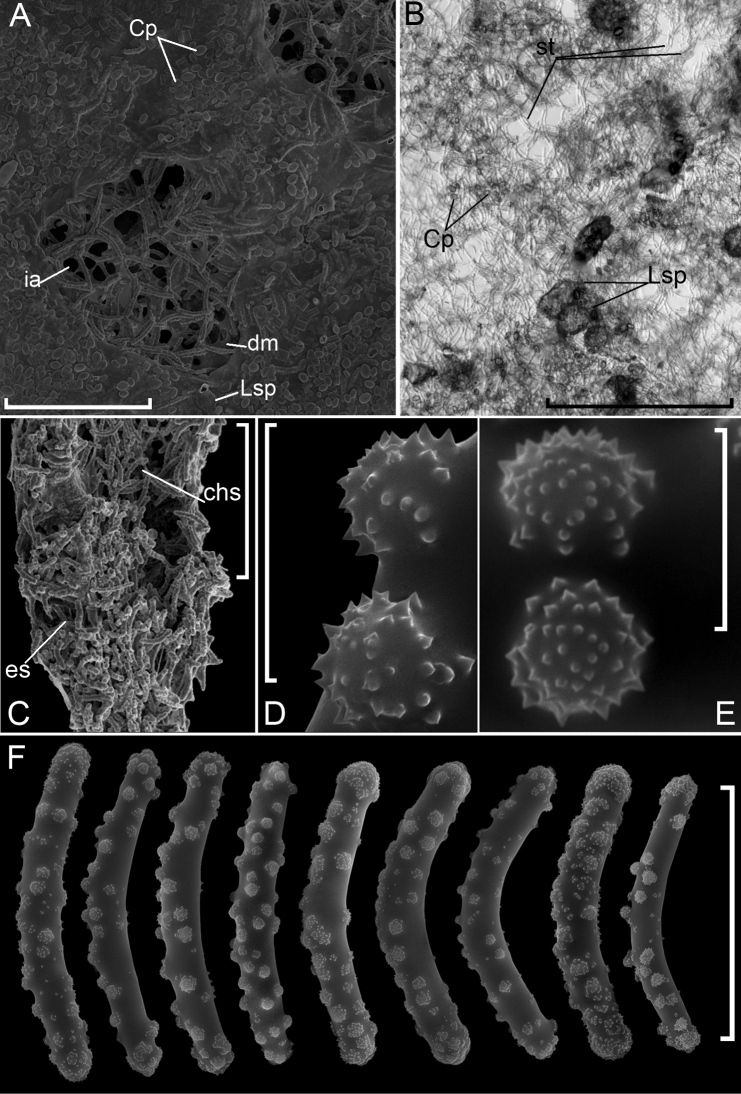
Figure 3.
Swartschewskia khanaevi sp. nov., external view. Abbreviations: osc oscula, pf pore fields. Scale bar: 5 mm.
Figure 4.
Swartschewskia khanaevi sp. nov. A sponge surface B ectosomal skeleton C cross section of skeleton D, E secondarily microspined tuberculated spines on strongyles F strongyles. Abbreviations: chs choanosomal skeleton, CpCocconeis placentula, dm dermal membrane, es ectosomal skeleton, ia inhalant apertures, LspLagenophrys sp., st spicular tracts. Scale bars: 10 μm (D, E), 100 μm (F), 500 μm (A), 1 mm (B).
Table 2.
Swartschewskia khanaevi sp. nov. spicules length and width (N = 50).
| Specimen | Length (µm) | Width (µm) |
| min–(mean)–max | ||
| Holotype | ||
| ZIN 11986 | 111–(128)–141 | 11–(15)–20 |
| Paratypes | ||
| LIN-BS-1740-2 | 99–(127)–146 | 11–(15)–19 |
| ZIN 11987 | 108–(125)–138 | 10–(15)–19 |
| ZIN 11988 | 106–(128)–140 | 11–(14)–18 |
| ZIN 11989 | 104–(123)–138 | 13–(17)–21 |
| ZIN 11990 | 109–(128)–149 | 9–(14)–18 |
Type material.
Holotype: ZIN 11986 (specimen in ethanol), ZIN 11986A (slide), Lake Baikal, the Olkhonskiye Vorota Strait, sampling site 1 (52°59.42'N 106°55.53'E), depth 10 m, SCUBA divers, September 9, 2016, collected by I. V. Khanaev, 1 specimen. Paratypes: ZIN 11987 (specimen in ethanol), ZIN 11987A (slide): ibid, 1 specimen; ZIN 11988 (specimen in ethanol), ZIN 11988A (slide): ibid, 1 specimen; ZIN 11989 (specimen in ethanol), ZIN 11989A (slide): ibid, 1 specimen; LIN-BS-1740-2 (specimen in ethanol, slide): ibid, 1 specimen. ZIN 11990 (specimen in ethanol), ZIN 11990A (slide): the Olkhonskiye Vorota Strait, sampling site 2 (53°01'03.40"N 106°55'47.00"E), depth 2.5 m, SCUBA divers, June 7, 2016, collected by I. V. Khanaev, 1 specimen.
Etymology.
Named after Dr Igor V. Khanaev, scientist and diver who organised a dive program and collected type material.
Description.
Thin encrusting sponge. Sponge thickness is maximal in the centre of the body (0.5–1 mm) and minimal at the edge (0.05–0.3 mm).
The natural colour is yellowish beige and almost white in ethanol with brown areas on the surface. Usually, sponges have from one to three oscula, and only paratype ZIN 11990 has six oscula. Oscula are almost round, deepened, edged with well-developed spicular vallum. Oscula size is 146–(585)–978 × 235–(663)–1148 μm. Dermal pores are non-uniformly distributed on sponge surface. They are mostly aggregated in pore fields. Those are not deepened relatively to sponge surface, diverse in shape and can join to each other. Round or ovoid inhalant apertures of 7–(42)–106 × 7–(54)–140 μm in size perforate dermal membrane. The apertures are located in meshes of ectosomal skeleton network. Pore fields size varies significantly: 0.07–(0.5)–1.5 × 0.09–(0.7)–2 mm. One field usually contains 4–40 pores; the maximum number of pores is 78. There are also isolated pores.
Up to 70–80 % of sponge surface is lacking in both oscula and pores and covered with dense accumulations of Cocconeis placentula Ehrenberg, 1838 and sporadic exemplars of other diatoms (identified by Dr N.A. Bondarenko). Additionally, some ciliated protozoa of genus Lagenophrys von Stein, 1851 (identified by Dr T.Ya. Sitnikova) were observed on all specimen of Swartschewskia khanaevi sp. nov.
Sponge surface is a hard but fragile crust, i.e., ectosomal skeleton; the inner part of the body is soft and can be easily detached from the crust. The ectosomal skeleton has a form of a cortical layer (cortex) of tangentially arranged tracts forming an alveolar network. Meshes are disordered; size and shape vary. In some parts of the cortex, meshes are indistinguishable; tracts cross irregularly. Megascleres in tracts are arranged in loose bundles, 2–8 megascleres in every bundle. The thickness of cortex varies significantly from 44 to 307 μm. The thickest cortex is observed near oscula, the thinnest one in the areas of pore fields. The choanosomal skeleton is weak; it consists of separated spicules and thin disordered fibres.
Megascleres are exclusively strongyles of 99–(127)–149 × 9–(15)–21 µm with different sorts of spines: simple spines, rosette spines, and a peculiar sort of spine, secondarily microspined tuberculated spines. The latter look like tubercles (4–9 µm in diameter and 1–5 µm in height) densely ornamented with simple spines (number 13–58) and these are the most abundant sort of spines. Rosette spines are comparatively rare (0.8–(1.4)–3.2 × 1–(1.6)–3.6 μm in size, contain 3–9 simple spines). The length of isolated simple spines and simple spines in both kinds of complex spines is similar: 0.1–(0.4)–0.9 µm. Microscleres absent.
Swartschewskia papyracea
(Dybowski, 1880)
2E534DAA-8FF0-546F-9239-84BAA6E8CBB7
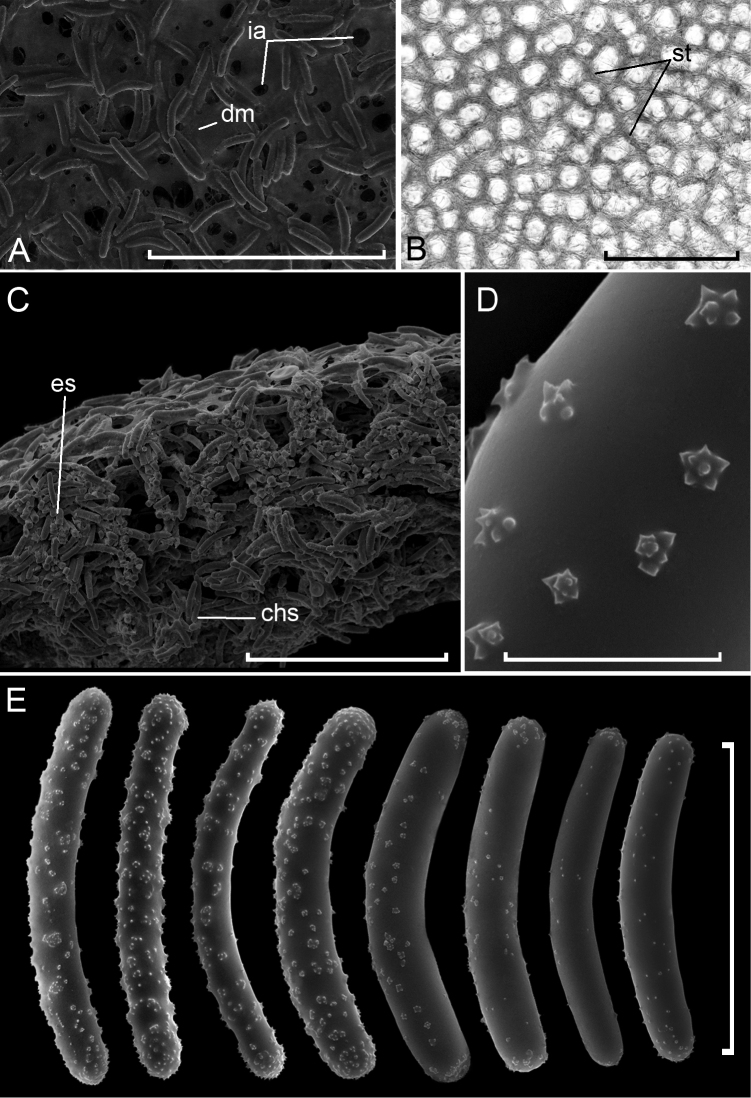
Figure 5.
Swartschewskia papyraceaA sponge surface B ectosomal skeleton C cross section of skeleton D rosette spines on strongyles E strongyles. Abbreviations: chs choanosomal skeleton, dm dermal membrane, es ectosomal skeleton, ia inhalant apertures, st spicular tracts. Scale bars: 10 μm (D), 100 μm (E), 500 μm (A, C), 1 mm (B).
Note.
The morphology of three specimens of S. papyracea sampled in the Olkhonskiye Vorota Strait was examined.
Description.
Body shape is globose. The sponge often has a single osculum but several oscula are also possible. Mostly the oscula look like round pits with 3–5 exhalant apertures on the bottom. One specimen bears a sieve-like osculum that consists of a number of exhalant apertures not deepened relatively to sponge surface. Distribution of dermal pores is uniform. Inhalant apertures are observed almost in every meshes of ectosomal skeleton network. One mesh contains 1–5 round or ovoid apertures, 5–(28)–87 × 6–(35)–102 µm in size. Exhalant apertures in the sieve-like osculum have elongated or round shape, 214–(281)–357 × 178–(219)–286 µm in size.
The ectosomal skeleton is a high ordered alveolar network, mesh shape resembles a convex polygon. There are no parts with a disordered network structure. Megascleres in tracts are arranged in dense bundles, 6–12 megascleres in every bundle.
Megascleres are stout and bent strongyles of 93–(117)–138 × 13–(17)–22 µm. Analysis of the fine morphological structure of S. papyracea spicules indicated the presence of only two sorts of spines: rosette spines and isolated simple spines. Rosette spines are slightly elongated, 0.5–(1.4)–3.2 × 0.6–(1.6)–4.1 µm, and contain 4 – 18 simple spines. Isolated spines and simple spines in rosettes have a similar size of 0.1–(0.4)–0.9 µm.
Discussion
Two species, S. papyracea and S. irregularis, were included in the genus Swartschewskia before the present study. We used the following sources for comparative analysis of diagnostic morphotraits. The original description of S. papyracea was made by W. Dybowski (1880) on several exemplars. Afterwards the type material was lost (Efremova 2001). Due to the impossibility of comparing our data with type material we were guided by the generally accepted recent descriptions of Manconi and Pronzato (2002, 2019). Additionally, we studied the morphology of S. papyracea specimens from the type locality of S. khanaevi sp. nov. Data on S. irregularis morphology are extremely poor. For more than 200 years of studies of the Baikal sponges, only two specimens of S. irregularis were collected. Both specimens are no longer available; therefore, we relied on the original description of the species (Swartschewsky 1902).
Based on molecular data, the new species belongs to the genus Swartschewskia (fam. Lubomirskiidae). The limitations of the molecular approach were previously shown for phylogenetic studies of the Baikal sponges due to low variability of markers (COI, ITS, rRNA-genes) usually applied for this purpose throughout the world (Itskovich et al. 2008, Harcet et al. 2010). The protein coding sequences of mtDNA allowed phylogenetic relationships within Lubomirskiidae to be resolved only at the genera taxonomic level (Maikova et al. 2015, 2016). The mtDNA intergenic regions, as we suggested, could be suitable for the separation of closely related species of Baikal sponges (Maikova et al. 2012) due to their increased rate of substitution accumulation (Lavrov 2010). But on the phylogenetic tree based on mtDNA intergenic regions, the species of Baikal sponges did not form separate clades at the species level, with some exceptions (deep-sea sponges, for example) (Maikova et al. 2012). The concatenated nuclear (ITS-regions) and mitochondrial (IGRs) data were most suitable for studying phylogenetic relationships within the family Lubomirskiidae at the moment (Maikova et al. 2017). The limitation of the molecular approach is apparently related to the low evolutionary rate of both nuclear and mitochondrial DNA of Baikal sponges (Lavrov 2010), which is enhanced by the relatively recent divergence of many species. An exception is the S. papyracea, which shows the acceleration of the accumulation of nucleotide substitutions in mtDNA to be twice relative to other species of the family Lubomirskiidae (Maikova et al. 2015). We hypothesise that this species is one of the most ancient of the existing species. On the phylogenetic tree based on protein-coding mtDNA genes, S. papyracea is closer to a common ancestor than all other species of the Baikal sponges (Maikova et al. 2016). In this study, based on concatenated nuclear (ITS) and mitochondrial (IGRs) data, the maximum interspecific genetic distances were between S. papyracea and other Lubomirskiidae species. Within the genus Swartschewskia the intraspecific and interspecific genetic distances do not overlap. This shows the genetic subdivision of the species within the genus and the genetic isolation of the genus Swartschewskia within the family Lubomirskiidae. The division of the new species into two groups inside the Clade B is not reflected in their morphology.
Swartschewskia khanaevi sp. nov. has skeleton structure and spicules typical for the genus (Manconi and Pronzato 2002, 2019). Swartschewskia khanaevi sp. nov. differs from S. papyracea by the clustering of pores in pore fields, less ordered structure of the ectosomal skeleton and unusual secondarily microspined tuberculated spines on strongyles. In S. papyracea distribution of dermal pores is uniform, ectosomal skeleton is highly ordered alveolar network with polygonal meshes, spicules bear spines grouped in rosettes (Manconi and Pronzato 2002, 2019). Generally, oscula are similar in both species, but S. papyracea has an alternative rather rare kind of osculum, which is sieve-like. It consists of a number of exhalant apertures inside rounded vallum (Manconi and Pronzato 2002). Pore fields of S. khanaevi sp. nov. could hardly be misinterpreted as sieve-like oscula. The total shape of the latter is always roundish; apertures are packed very closely and are noticeably larger than dermal pores (mean size 281 × 219 μm vs 42 × 54 μm). Ectosomal skeleton of S. irregularis even less ordered than of S. khanaevi sp. nov. It lacks polygonal network and looks like randomly arranged spicules. Strongyles of S. irregularis are smooth.
Previous data on the morphology of Swartschewskia species do not contain records of strongyles ornamented with tuberculated spines or pore fields (Dybowski 1880; Swartschewsky 1901, 1902; Makushok 1927a; Rezvoy 1936; Manconi and Pronzato 2002, 2019). Weinberg (2005) mentioned an unusual specimen of S. papyracea as a thin encrusting sponge with numerous oscula and bearing: (a) spicules as stout and bent strongyles thinner than usual in the species and ornamented with massive complex spines; (b) skeleton of clearly divided ectosomal and choanosomal parts, but intensive study of the skeleton was not carried out. The sponge was collected from the Maloye More Strait (the precise locality was unknown). Based on these facts, we suppose that Weinberg met a specimen of S. khanaevi sp. nov. Taking into account sites, where S. khanaevi sp. nov. was collected, the species is most likely a local endemic of the Olkhonskiye Vorota Strait or Maloye More Strait as a whole.
Fossil spicules similar to S. khanaevi sp. nov. were found in the Late Pliocene sediments (interval of 3.2−2.8 Ma) of Lake Baikal (Veynberg 2009). They were described as spicules of extinct species Palaeoswartschewskia sp. 1, they were some thinner and longer than in S. khanaevi sp. nov. and had complex spines. These spines, with a smaller number of simple spines than tuberculated spines and less expressed tubercles (see Veynberg 2009), represent intermediate variant between rosette spines of S. papyracea and tuberculated spines of S. khanaevi sp. nov. In this regard, we cannot ascertain the unambiguous identity of Palaeoswartschewskia sp. 1 and S. khanaevi sp. nov., but these two species are doubtlessly morphologically close.
Non-uniform localisation of pores in S. khanaevi sp. nov. is uncommon amongst the Baikal sponges. Normally, in lubomirskiids pores are evenly distributed throughout the sponge surface. The bottom at the study site consists of stones (a substrate for sponges) and sandy areas located nearby. The latter saturate the water with suspended grains of sand. The number of suspended particles combined with hydrodynamic activity can lead to clogging of the aquiferous system (Bell et al. 2015). The concentration of inhalant pores at restricted areas of the body surface was previously described as an adaptive trait of some sponge species living under the conditions of high sedimentation (Rützler 1974; Werding and Sanchez 1991; Pronzato et al. 1998). A larger size of inhalant apertures of S. khanaevi sp. nov. in comparison with S. papyracea can also prevent clogging.
The presence of sessile ciliates and dense aggregation of diatom algae on the sponge surface is not common for Lubomirskiidae. Isolated diatom algae can be observed sometimes on the lubomirskiids surface. There are no descriptions of mass diatom accumulations on the surface of a number of specimens. Any attached ciliates on sponges in Baikal also have never been mentioned. However, ectosymbiotic sessile ciliates of the Lagenophrys genus were described on Baikal endemic amphipods cuticle (Khalzov et al. 2018). Probably the emergence of a unique epibiotic community on S. khanaevi sp. nov. is possible due to unusual structure of aquiferous system. Permanent exhalant and inhalant water currents are normally presented at the sponge surface and prevent its colonisation. In S. khanaevi sp. nov., exhalant and inhalant apertures are concentrated in restricted areas. Therefore, up to 80 % of the body surface has no currents, which is a favourable substrate for epibiotic organisms.
Key to Swartschewskia species
The key to Lubomirskiidae genera and species was offered by Manconi and Pronzato (2019) and was the basis for the present key.
Spongillida: Lubomirskiidae: Genera
| 1 | Growth form massive (globular) to encrusting with digitiform outgrowths; consistency firm to hard; surface smooth | 2 |
| – | Growth form encrusting to massive, branching; consistency soft; densely conulose, variably long conules | Rezinkovia |
| 2 | Megascleres typically strongyles variably spiny | 3 |
| – | Megascleres typically spiny oxeas | Lubomirskia |
| 3 | Megascleres typically smooth/spiny, stout, bent strongyles with compound spines, rare spiny oxeas | Swartschewskia |
| – | Megascleres typically smooth strongyles with spiny tips; spiny strongyles and/or smooth/spiny oxeas also present | Baikalospongia |
Spongillida: Lubomirskiidae: Swartschewskia : Species
Three species are endemic to Lake Baikal.
| 1 | Massive, rounded or encrusting, bent spiny strongyles; rare oxeas regularly spiny | 2 |
| – | Massive, irregular; bent smooth strongyles | Swartschewskia irregularis (Swartschewsky, 1902) |
| 2 | Strongyles with spines in rosettes | Swartschewskia papyracea (Dybowski, 1880) |
| – | Strongyles with tubercles densely ornamented with simple spines | Swartschewskia khanaevi sp. nov. |
Conclusions
A new species Swartschewskia khanaevi sp. nov. was described based on morphological traits and sequences of nuclear (ITS1 and ITS2) and mitochondrial (IGRs) markers. In the molecular phylogeny the specimens of S. khanaevi sp. nov. are clustered within a well-defined group containing S. papyracea as the most closely related species. Indeed, the specimens’ morphological traits clearly indicate their belonging to Swartschewskia: well-developed ectosomal skeleton of tangential spicular fibres and sparsely developed choanosomal skeleton, stout bent strongyles as megascleres. The major morphological traits that distinguish S. khanaevi sp. nov. from other congeners are the structure of ectosomal skeleton and compound spines on strongyles. Swartschewskia khanaevi sp. nov. was sampled only from the Olkhonskiye Vorota Strait, and we assumed it to be a local endemic of this strait. We suggest the non-uniform localisation of pores on the sponge surface may be an adaptation to biotope conditions.
Supplementary Material
Acknowledgements
This study was performed within the framework of the State Tasks Nos. 0345-2019-0002 and 0345-2019-0009 and supported by the RFBR grant No. 19-04-00787A. The authors are grateful to Dr Igor V. Khanaev and Valery I. Chernykh for samples collection. Authors thank Dr Tatyana Ya. Sitnikova (Limnological Institute SBRAS, Russia), who organised the expedition in 2003 and identified ciliates on the sponge surfaces, and Dr Nina A. Bondarenko (Limnological Institute SBRAS, Russia) for identification of diatom algae on sponges. SEM analyses were carried out in the Center ‘Ultramicroanalysis’ (Limnological Institute SBRAS).
Citation
Bukshuk NA, Maikova OO (2020) A new species of Baikal endemic sponges (Porifera, Demospongiae, Spongillida, Lubomirskiidae). ZooKeys 906: 113–130. https://doi.org/10.3897/zookeys.906.39534
Footnotes
In situ photograph of sponges is provided by Viktor Lyagushkin (scale bar is approximate)
References
- Bell JJ, McGrath E, Biggerstaff A, Bates T, Bennett H, Marlow J, Shaffer M. (2015) Sediment impacts on marine sponges. Marine Pollution Bulletin 94: 5–13. 10.1016/j.marpolbul.2015.03.030 [DOI] [PubMed] [Google Scholar]
- Cárdenas P, Pérez T, Boury-Esnault N. (2012) Sponge systematics facing new challenges. In: Becerro MA, Uriz MJ, Maldonado M, Turon X. (Eds) Advances in Sponge Science: Phylogeny, Systematics, Ecology.Advances in Marine Biology 61: 79–209. 10.1016/B978-0-12-387787-1.00010-6 [DOI] [PubMed]
- Dybowski W. (1880) Studien über die Spongien des russischen Reiches, mit besonderer Berücksichtigung der Spongien-Fauna des Baikal-Sees. Mémoires de l'Académie Impériale des sciences de St. Pétersbourg 7: 1–71. [Google Scholar]
- Efremova SM. (2001) Sponges (Porifera). In: Timoshkin OA. (Ed.) Index of Animal Species Inhabiting Lake Baikal and its Catchment Area.Nauka, Novosibirsk, 182–192.
- Efremova SM. (2004) New genus and new species of sponges from family Lubomirskiidae Rezvoj, 1936. In: Timoshkin OA. (Ed.) Index of Animal Species Inhabiting Lake Baikal and its Catchment Area.Nauka, Novosibirsk, 1261–1278.
- Efremova SM, Goureeva MA. (1989) The problem of the origin and evolution of Baikalian sponges. The 1st Vereshchagin Baikal Conference, Irkutsk. Abstracts, 22–23
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Harcet M, Bilandzija H, Bruvo-Madaric B, Cetkovic H. (2010) Taxonomic position of Eunapius subterraneus (Porifera, Spongillidae) inferred from molecular data – A revised classification needed? Molecular Phylogenetics and Evolution 54: 1021–1027. 10.1016/j.ympev.2009.12.019 [DOI] [PubMed]
- Itskovich V, Gontcharov A, Masuda Y, Nohno T, Belikov S, Efremova S, Meixner M, Janussen D. (2008) Ribosomal ITS sequences allow resolution of freshwater sponge phylogeny with alignments guided by secondary structure prediction. Journal of Molecular Evolution 67: 608–620. 10.1007/s00239-008-9158-5 [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. (2008) Recent developments in the MAFFT multiple sequence alignment program. Briefing on Bioinformatics 9: 286–298. 10.1093/bib/bbn013 [DOI] [PubMed] [Google Scholar]
- Khalzov IA, Mekhanikova IV, Sitnikova TYa. (2018) First data on ectosymbiotic consortia of infusoria and prokaryotes associated with amphipods inhabiting the Frolikha underwater hydrothermal vent, Lake Baikal. Zoological Journal 97: 1525–1530. 10.1134/S0044513418120073 [DOI] [Google Scholar]
- Khamidekh S. (1991) Analysis of anatomic and histological traits of sponges of Lubomirskiidae family. To the question of Baikal sponges taxonomy. PhD Thesis, Zoological Institute, Saint-Petersburg. [In Russian]
- Khanaev IV, Kravtsova LS, Maikova OO, Bukshuk NA, Sakirko MV, Kulakova NV, Butina TV, Nebesnykh IA, Belikov SI. (2018) Current state of the sponge fauna (Porifera: Lubomirskiidae) of Lake Baikal: Sponge disease and the problem of conservation of diversity. Journal of Great Lakes Research 44: 77–85. 10.1016/j.jglr.2017.10.004 [DOI] [Google Scholar]
- Kozhov MM. (1947) Animals of the Lake Baikal. Irkutsk regional Publishers, Irkutsk, 304 pp. [In Russian] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis vertion 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov DV. (2010) Rapid proliferation of repetitive palindromic elements in mtDNA of the endemic Baikalian sponge Lubomirskia baicalensis. Molecular Biology and Evolution 27: 757–760. 10.1093/molbev/msp317 [DOI] [PubMed] [Google Scholar]
- Lavrov DV, Maikova OO, Pett W, Belikov SI. (2012) Small inverted repeats drive mitochondrial genome evolution in Lake Baikal sponges. Gene 505: 91–99. 10.1016/j.gene.2012.05.039 [DOI] [PubMed] [Google Scholar]
- Maikova O, Khanaev I, Belikov S, Sherbakov D. (2015) Two hypotheses of the evolution of endemic sponges in Lake Baikal (Lubomirskiidae). Journal of Zoological Systematics and Evolutionary Research 53: 175–179. 10.1111/.jzs.12086 [DOI] [Google Scholar]
- Maikova O, Sherbakov D, Belikov S. (2016) The complete mitochondrial genome of Baikalospongia intermedia (Lubomirskiidae): description and phylogenetic analysis. Mitochondrial DNA. Part B: Resources 1: 569–570. 10.1080/23802359.2016.1172273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maikova OO, Bukshuk NA, Itskovich VB, Khanaev IV, Nebesnykha IA, Onishchuk NA, Sherbakov DYu. (2017) Transformation of Baikal Sponges (Family Lubomirskiidae) after Penetration into the Angara River. Russian Journal of Genetics 53: 1343–1349. 10.1134/S1022795417120092 [DOI] [Google Scholar]
- Maikova OO, Stepnova GN, Belikov SI. (2012) Variations in noncoding sequences of the mitochondrial DNA in sponges from family Lubomirskiidae. Doklady Biological Sciences 442: 46–48. 10.1134/S1607672912010140 [DOI] [PubMed] [Google Scholar]
- Makushok ME. (1927a) Taxonomy of Baikal sponges. I. Genera Lubomirskia Dyb. and Swartschewskia n. nov. Russian Zoological Journal 7: 79–103. [In Russian] [Google Scholar]
- Makushok ME. (1927b) Taxonomy of Baikal sponges. II. New genus of Baikal sponge fauna Baicalolepis nov. gen. and new species of the genus Baicalolepis fungiformis nov. sp. Russian Zoological Journal 7: 124–131. [In Russian] [Google Scholar]
- Manconi R, Pronzato R. (2002) Suborder Spongillina subord. nov.: Freshwater sponges. In: Hooper JNA, Van Soest RWM. (Eds) Systema Porifera.A guide to the classification of sponges. Kluwer Academic/ Plenum Publishers, New York, Boston, Dordrecht, London, Moscow, 921–1020. 10.1007/978-1-4615-0747-5_97 [DOI]
- Manconi R, Pronzato R. (2015) Phylum Porifera. In: Thorp J, Rogers DC. (Eds) Ecology and general biology: Thorp and Covich’s freshwater invertebrates, 4th ed.Elsevier, Amsterdam, 133–157. 10.1016/B978-0-12-385026-3.00008-5 [DOI]
- Manconi R, Pronzato R. (2019) Phylum Porifera. In: Rogers DC, Thorp J. (Eds) Keys to Palaearctic Fauna: Thorp and Covich’s freshwater invertebrates, 4th ed.Elsevier, Amsterdam, 45–87. 10.1016/B978-0-12-385024-9.00003-4 [DOI]
- Maniatis T, Fritsch EF, Sambrook J. (1984) Molecular Cloning. Mir, Moscow, 480 pp. [Russian Translation] [Google Scholar]
- Masuda Y. (2009) Studies on the Taxonomy and Distribution of Freshwater Sponges in Lake Baikal. In: Müller WEG, Grachev MA. (Eds) Biosilica in Evolution, Morphogenesis, and Nanobiotechnology.Springer, Berlin/Heidelberg, 81–110. 10.1007/978-3-540-88552-8_4 [DOI] [PubMed]
- Mats VD, Ufimtsev GF, Mandelbaum MM. (2001) The Baikal basin in the Cenozoic: Structure and geologic history. Novosibirsk, SBRAS Press, Branch “GEO”, 252 pp. [In Russian] [Google Scholar]
- Mazepova GF. (1995) General characteristics of Lake Baikal. In: Timoshkin OA, Melnik NG, Mazepova GF, Sheveleva NG. (Eds) Guide and key to pelagic animals of Baikal (with ecological notes).Nauka: Siberian Publishing Firm RAS, Novosibirsk, 23–24. [In Russian]
- Pronzato R, Bavestrello G, Cerrano C. (1998) Morpho-functional adaptations of three species of Spongia (Porifera, Demospongiae) from a Mediterranean vertical cliff. Bulletin of Marine Science 63: 317–328. [Google Scholar]
- Rezvoy PD. (1936) Freshwater sponges of the USSR. In: Rezvoy PD. (Ed.) The Fauna of the USSR.AS USSR, Moscow, 1–42.
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rützler K. (1974) The burrowing sponges of Bermuda. Smithsonian contributions to zoology 165: 1–32. 10.5479/si.00810282.165 [DOI] [Google Scholar]
- Sollas WJ. (1885) A Classification of the Sponges. Annals and Magazine of Natural History. 10.1080/00222938509459901 [DOI]
- Swartschewsky BA. (1901) Short communication on Baikal sponge fauna. Mémoires de l'Académie Impériale des sciences de St. Pétersbourg 15: 1–7 [In Russian] [Google Scholar]
- Swartschewsky BA. (1902) Materials on fauna of sponges of Lake Baikal. Memories of Kiev Society of Nature 17: 329–352. [In Russian] [Google Scholar]
- Van Soest RWM, Boury-Esnault N, Hooper JNA, Rützler K, de Voogd NJ, Alvarez B, Hajdu E, Pisera AB, Manconi R, Schönberg C, Klautau M, Kelly M, Vacelet J, Dohrmann M, Díaz M-C, Cárdenas P, Carballo JL, Ríos P, Downey R, Morrow CC. (2019) . World Porifera Database. http://www.marinespecies.org/porifera
- Veynberg E. (2009) Fossil Sponge Fauna in Lake Baikal Region. In: Müller WEG, Grachev MA. (Eds) Biosilica in Evolution, Morphogenesis, and Nanobiotechnology.Springer, Berlin Heidelberg, 185–205. 10.1007/978-3-540-88552-8_8 [DOI]
- Weinberg E, Eckert C, Mehl D, Mueller J, Masuda Y, Efremova S. (1999) Extant and fossil spongiofauna from the underwater Academician ridge of Lake Baikal (SE Siberia). Memories of the Queensland Museum 44: 651–657. [Google Scholar]
- Weinberg EV. (2005) Sponge fauna of Pliocene-Quaternary deposits of Baikal. PhD thesis, Saint-Petersburg: Zoological Institute. [In Russian]
- Weltner W. (1895) Spongilliden studien III. Katalog und Verbreitung der bekannten Süsswasserschwämme. Archiv für Naturgeschichte 61: 114–144. [Google Scholar]
- Werding B, Sanchez H. (1991) Life habits and functional morphology of the sediment infaunal sponges Oceanapia oleracea and Oceanapia peltata (Porifera, Haplosclerida). Zoomorphology 110: 203–208. 10.1007/BF01633004 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.