Abstract
Metabolomics, the study of metabolic profiles in a biological sample, has seen rapid growth due to advances in measurement technologies such as mass spectrometry (MS). While MS metabolite reference libraries have been generated for metabolomics applications, mass spectra alone are unable to unambiguously identify many metabolites in a sample; these unidentified compounds are typically annotated as “features”. Surface-enhanced Raman spectroscopy (SERS) is an interesting technology for metabolite identification based on vibrational spectra. However, no reports have been published that present SERS metabolite spectra from chemical libraries. In this paper, we demonstrate that an untargeted approach utilizing citrate-capped silver nanoparticles yields SERS spectra for ~20% of 80 compounds chosen randomly from a commercial metabolite library. Furthermore, prescreening of the metabolites according to chemical functionality allowed for the efficient identification of samples within the library that yield distinctive SERS spectra under our experimental conditions. Last, we present a reference database of 63 metabolite SERS spectra for use tools for metabolite identification; this set includes 30 metabolites that have not had previously published SERS spectra.
Keywords: Surface-enhanced Raman spectroscopy, SERS, Metabolite, Reference library
1. Introduction
Metabolites are low-molecular weight compounds that contribute to a cell’s ability to utilize energy, grow, reproduce, excrete waste, and respond to environmental stimuli. The Human Metabolome Database currently includes over 114,000 metabolites classified as amino acids, nucleotides, lipids, carbohydrates, etc. [1]. To begin to address the challenges in analyzing this complex set of molecules, the Metabolomics Standards Initiative has proposed standardized reporting format for metabolomics studies [2]. That group defines Level 1 identifications as being based on co-characterization with authentic standards; identification at level 1 is laborious and time consuming. Level 2 identifications are based on comparison with spectral libraries.
Mass spectrometry (MS), coupled with gas or liquid chromatography, and nuclear magnetic resonance (NMR) spectroscopy are the most widely used methods for metabolite studies [2-5]. However, NMR suffers from limited sensitivity, and mass spectra are often insufficient to identify metabolites with high confidence. Vibrational spectroscopies, such as Fourier-transform infrared spectroscopy (FTIR), provide a cost-effective and sensitive alternative, but its use is limited by the strong IR activity of water.
We propose the use of Raman spectroscopy as an alternative to NMR or MS for metabolite identification. Raman measures inelastically scattered photons characteristic of a molecule’s vibrational modes. Raman does not suffer from significant water interferences due to water’s relatively sparse spectrum [6, 7]. While the Raman effect is extremely weak [8], higher sensitivity may be achieved by adsorbing the analyte to plasmonic nanoparticles [9-11]. When using noble metal nanoparticles, routine surface-enhanced Raman spectroscopy (SERS) enhancement factors range from 106 to 108 compared to their unenhanced Raman counterparts [10]. Consequently, SERS has emerged as an intriguing bioanalytical method due to its excellent selectivity, its high sensitivity, and its compatibility with aqueous media [12, 13]. Despite a long-standing interest in the SERS detection of metabolites, previous studies have only targeted specific metabolites or metabolite classes, such as amino acids [14-17], rather than comprehensively analyzing a complex metabolite library. We are not aware of a published SERS spectral database for metabolites. In this paper, we present SERS spectra obtained from a commercially available set of standards that can be used as Level 2 metabolite identifications.
2. Materials & Methods
2.1. Materials
The Mass Spectrometry Metabolite Library (IROA Technologies) was purchased from MilliporeSigma. Methanol was purchased from Honeywell Burdick & Jackson. Nitric acid, hydrochloric acid, acetonitrile, phenethylamine (Alfa Aesar), methyl indole-3-acetate (Alfa Aesar), 1-napthylamine (Tokyo Chemical Industry), N,N-dimethyl-1,4-phenylenediamine (Acros Organics), dethiobiotin (IBA Lifesciences), and 3-methyladenine (MP Biomedicals) were purchased from VWR Analytical. Silver nitrate was purchased from Aldrich. Sodium citrate tribasic dihydrate and toluene were purchased from Sigma-Aldrich. Sodium bromide (NaBr) was purchased from EMD Millipore. Metabolites were used as received without further purification.
2.2. SERS Sample Preparation
First, we conducted preliminary studies to optimize the metabolite SERS response while maintaining a method that can be used in future high-throughput experiments. L-cysteine, spermidine, and L-histidine were used to evaluate the following standard SERS substrates: Ag and Au colloids prepared with sodium citrate, Ag colloids prepared with sodium borohydride (NaBH4), and Ag and Au film-over-nanospheres (FONs). Published procedures were used for preparation of these substrates [18-20].
Our prescreen demonstrated several advantages of the Ag colloids prepared with sodium citrate: (1) these colloids produced superior signal enhancement compared to the FONs substrates, consistent with previous reports [21-23], (2) common functional groups found in biological molecules (thiols and amines) have a strong affinity to silver [11, 24, 25], and (3) the colloid substrate prepared with sodium citrate produced a smaller background signal compared to NaBH4 particles and the FONs substrates (Figure S65).
The silver colloids were prepared by the common Lee and Meisel method [19], which involves the slow reduction of silver ions by trisodium citrate to yield quasispherical nanoparticles. Trisodium citrate serves as both a reducing agent and a stabilizing ligand for the colloids. Specifically, 90.6 mg of silver nitrate was added to 500 mL of water and brought to a rolling boil. 10 mL of 1% trisodium citrate (109.5 mg) was added dropwise. The solution continued boiling for an additional 20 min. The resulting nanoparticles exhibited a strong plasmon band at 403 nm and were approximately 42 nm in diameter, verified by UV-Vis (UV-3100PC, VWR) and dynamic light scattering (NanoBrook Omni, Brookhaven Instruments), respectively (Figure S67).
Each metabolite from the library was reconstituted in 5% methanol to yield a 0.1 mg/mL concentration. Ammonium formate buffer (10 mM final concentration, pH 2.7) was added to prepare the samples for characterization using the sequential injection capillary electrophoresis – mass spectrometry method described in the companion paper [26]. Prior to analysis, the analytes were stored at −20 °C according to the manufacturer’s recommendation.
To prepare the metabolite samples for SERS analysis, approximately 15 μL of metabolite solution was mixed with 250 μL of suspended Ag colloids in a Greiner Bio-One Glass-Bottom Microplate (Thomas Scientific) for 35 min to promote analyte adsorption to the nanoparticle surface. 50 μL of NaBr (1 M) was then added to induce aggregation for SERS measurements. The final analyte concentration was ~0.005 mg/mL. Each metabolite was placed in a single well of a glass-bottom microtiter plate.
2.3. SERS Instrumentation
SERS measurements were performed with a custom-built Raman spectrometer using a 633 nm HeNe laser (Thor Labs). The laser was focused onto the sample using an inverted microscope objective (Nikon, 20x, NA = 0.5) with approximately 600 μW of power at the objective. The backscattered radiation passed through a Rayleigh rejection filter (Semrock) before dispersion in the spectrometer (Acton SP2300, Princeton Instruments, 600 g mm−1). The photons were detected using a back-illuminated deep depletion CCD (PIXIS, Spec-10, Princeton Instruments) and recorded using Winspec32 software (Princeton Instruments) with a 3 min acquisition time. The wavenumber was calibrated with a toluene/acetonitrile (1:1) solution using nine calibration points. The spectra were background subtracted (Multipeak Fitting 2.0 Package) and plotted in IGOR Pro (WaveMetrics). Each spectrum presented is an average of three scans obtained from different positions in the well of the glass-bottom microtiter plate.
3. Results & Discussion
3.1. Success Rate Quantification in an Untargeted SERS Metabolite Study
There is not a large literature on the diversity of compounds that generate strong SERS signals. Therefore, we began by quantifying the percentage of analytes in a reference library that researchers could expect to be successful without targeted optimization. We began with a metabolite library that includes 460 metabolites that were characterized by mass spectrometry (data in companion paper) [26].
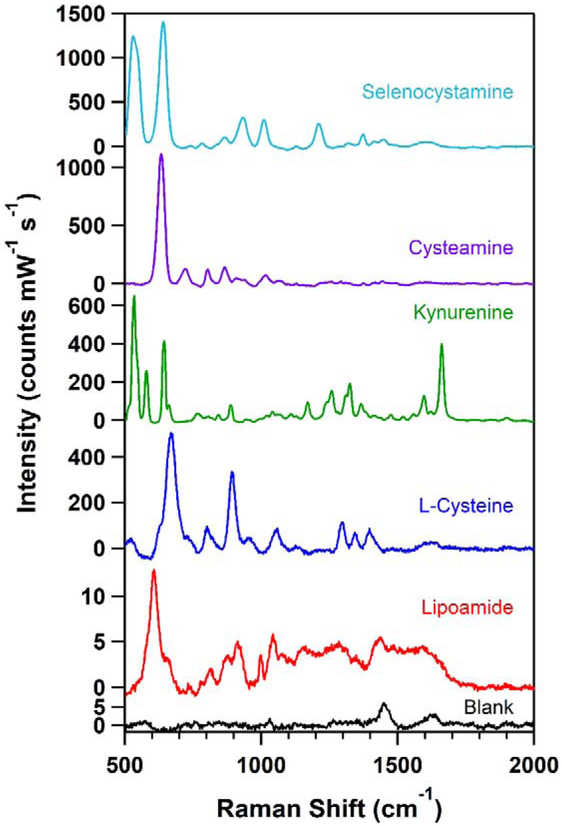
To establish a benchmark success rate, each metabolite in the library was denoted by an integer. Eighty integer values were randomly selected from a discrete uniform distribution (MatLab, MathWorks) and the corresponding metabolites were subjected to SERS analysis with the same experimental conditions. Not all spectra were useful, and inclusion of a compound in our SERS database, the spectra had to meet the following criteria: (1) a maximum intensity higher than the intensity observed in the blank and (2) distinct signatures including at least three peaks not attributed to the blank (Figure S66), and a “failure” was a metabolite that did not meet those conditions. Figure S66 provides an example for distinguishing a metabolite that met all conditions versus a metabolite that did not.
The blank spectrum contained two peaks at 1440 cm−1 and 1635 cm−1, attributed to sodium citrate and the ammonium formate buffer, respectively (Figure S64). Given that the sodium citrate is weakly bound to the silver nanoparticles, we expect the metabolite, in many cases, to displace the citrate at the surface of the particle, decreasing the intensity of the background citrate peak.
In this context, the success rate () was calculated by dividing the number of successes (i.e., the sum of metabolites that met all criteria for inclusion in the database) by the sample size (n). The standard error (σ) is the standard deviation of a binomial distribution and was calculated using the following equation:
where the sample statistic () can be extrapolated to the population (i.e., the 460 component metabolite library) [27]. The unbiased success rate and standard error using this method were determined to be 19 ± 4%. Therefore, in an untargeted library SERS study, we predict approximately 20% of the samples will generate useful SERS signatures under our experimental conditions. As it is well known that the SERS effect relies on analyte-substrate proximity [28] and surface-binding properties [29], a targeted approach is expected to greatly improve the identification of metabolites with a strong SERS response.
3.2. Targeted SERS Approach
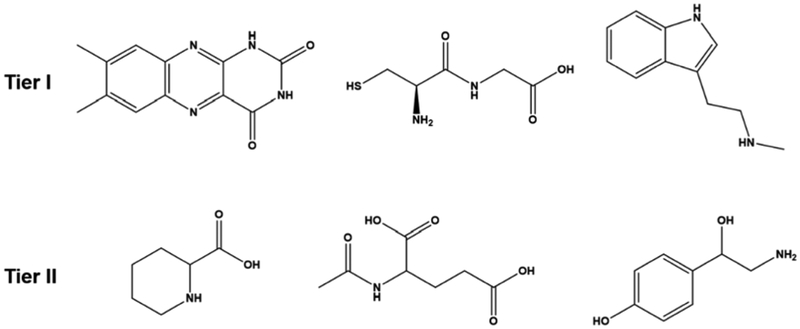
Hierarchical tiers of the library were generated based on two primary criteria: the presence of functional groups with an affinity for silver and analytes with high polarizability. To be identified as a Tier I analyte, the molecule’s structure needed to contain multiple amine or thiol functional groups, which have an affinity to silver. In addition, the presence of aromatic groups was noted due to their high polarizability. Tier II metabolites contained either a single amine or thiol group, carboxylates that bind weakly to silver, and/or carbon chains with few (if any) aromatic groups. Figure 1 illustrates representative structures for metabolites categorized in each tier.
Figure 1.
Representative chemical structures for metabolites categorized as Tier I or II.
The overall success rates for SERS analysis of the Tier I and Tier II metabolites were 44% and 18%, respectively. The Tier I success rate is more than twice the unbiased counterpart, demonstrating the power of a targeted approach. Upon analyzing a second tier, the success rate decreased as expected and was comparable to the unbiased rate.
3.3. SERS Reference Database Generation
In total, 52% (237/460) of the metabolite library was analyzed. 63 of the 237 metabolites were considered successful and these spectra constitute the first SERS metabolite reference database (Figures S1-S63). Using our unbiased success rate (Section 3.1), we calculate that only 22 of the remaining 223 samples are expected to yield SERS spectra. By employing a targeted approach, we were able to identify the majority of SERS-active analytes in a high throughput, time efficient manner. It is important to note that 30 of the 63 metabolites do not have previously published SERS spectra.
The following metabolites are presented in our SERS spectral database: 1-methylnicotinamide, 1-napthylamine, 2-quinolinecarboxylic acid, 3’, 5’-cyclic AMP, 3-methoxytyramine, 3-methyladenine, 4-imidazoleacetic acid, 5-oxo-L-proline, agmatine sulfate, biliverdin, bis(3-aminopropyl)amine, caffeine, carbamoyl phosphate, Cys-Gly, cysteamine, cytochrome C, deoxyadenosine monophosphate, dethiobiotin, dihydrofolate, dopamine, glutathione, histamine, homocysteine, homocystine, indole-3-acetic acid, kynurenine, L-arginine, L-asparagine, L-cystathionine, L-cysteic acid, L-cysteine, L-cystine, leucine, L-histidine, lipoamide, L-lysine, L-methionine sulfoximine, L-tryptophan, L-tryptophanamide, lumichrome, mandelic acid, methyl indole-3-acetate, methylguanidine, N,N-dimethyl-1,4-phenylenediamine, N-acetyl-DL-glutamic acid, N-acetyl-L-cysteine, nicotinamide, N-methyl-D-aspartic acid, N-methyltryptamine, octopamine, phenethylamine, pipecolate, pterin, riboflavin, selenocystamine, selenomethionine, spermidine, tetrahydrofolate, thiamine, thyrotropin releasing hormone, tryptamine, tyramine, vitamin B12. Spectra are graphed in supporting information, and tabulated in an Excel spreadsheet.
Figure 2 presents representative metabolite spectra spanning a wide range of chemical classifications. The spectra clearly have higher intensities than the blank spectrum in addition to unique peaks. Spectra in the referenced publications in Table 1 are comparable to our data. Therefore, SERS is a viable method for the chemical identification of metabolites with extensive diversity in molecular structure and biological function. Table 1 provides additional information about the metabolites discussed in Figures 2-3. The Human Metabolome Database was utilized to assign the biological and chemical classifications [1]. Table S1 contains a master classification list for all of the successful metabolites in the library.
Figure 2.
Representative SERS spectra of successful metabolites in the library. Additional information is included in Table 1.
Table 1.
Classifications, chemical structures, and previous literature reports (if any) for the metabolites in Figures 2-3.
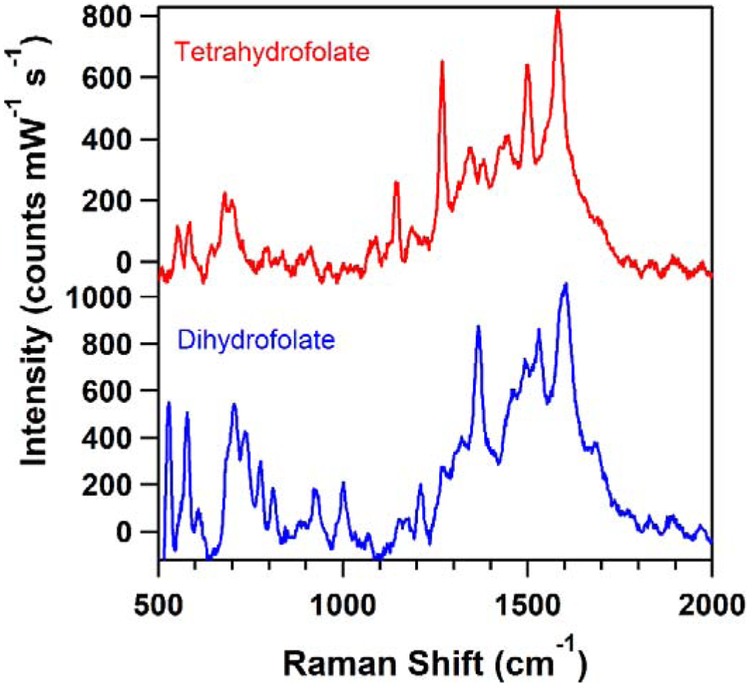
Figure 3.
SERS spectra comparing tetrahydrofolate and dihydrofolate signatures. Additional information is included in Table 1.
Lipid analysis with SERS is known to be challenging due to the aqueous environment of the colloidal particles [38]. Unsurprisingly, we only observe one metabolite in this category (lipoamide), which is shown in Figure 2. We expect that some metabolites could yield positive results in different solvent media or pH conditions, which can be addressed in future studies.
SERS generated highly selective spectral signatures for similar structures. Figure 3, for example, presents the SERS spectra of dihydrofolate and tetrahydrofolate, which structurally indistinguishable except for are one alkene and two hydrogens. While the spectra have a similar fingerprint, there are distinct differences. For example, they both contain a peak at 580 cm−1, but dihydrofolate’s spectrum exhibits an intense band at 529 cm−1 that is absent from tetrahydrofolate’s spectrum. In addition, tetrahydrofolate has a peak at 1269 cm−1 that is not present in the dihydrofolate spectrum.
The data presented in our database should be useful for characterization of = samples through reference spectral libraries. For example, Kauffman et al. developed a standardization method for Raman spectra across laboratory set-ups and handheld spectrometers through a resolution matching algorithm enabling accurate library verification of common pharmaceuticals [39]. In addition, principle component analysis has emerged as a powerful identification algorithm for the rapid analysis of SERS spectra [40, 41]. Recent studies indicate the promise of quantitative multiplexing of metabolites using SERS without the need for prior chromatographic separation [42, 43]. Coupled with our targeted approach towards experimentally analyzing chemical libraries via SERS, we envision that these computational tools will enable future high-throughput, reliable, user-friendly methods for large-scale studies. In addition, recent advances in interfacing capillary zone electrophoresis with SERS detection would allow generation of spectra from separated components [44].
4. Conclusion
We report the first SERS metabolite reference library. The spectra represent a wide array of biological/chemical classifications, demonstrating the technique’s viability over a large range of molecules. The widely used Lee and Meisel silver colloids were used for this work due to their facile synthesis and extensively characterized enhancement properties, boding well for future high-throughput studies. In addition, we have presented an optimized approach towards handling large data sets of metabolites using SERS. In addition, by inputting our library of metabolite spectra into a database, we envision that principle component analysis may be successfully employed to rapidly analyze unknown samples.
A companion paper presents electrophoretic mobility data for many of the same metabolites [26]. These two databases should be of value in a range of metabolome studies.
Supplementary Material
Highlights.
We randomly selected a set of metabolites from a large commercial library using citrate-capped silver nanoparticles as the substrate for SERS spectroscopy. 20% of these compounds successfully generated spectra. A reference database of 63 metabolite spectra is presented as a resource to the community.
Acknowledgments
This material is based on work supported by the National Science Foundation under grant number NSF/CHE-1709881 (L. M. S., L. F. P. K., M. G. T., and J.P.C.) and by the National Institutes of Health (R01GM096767; N.J.D.). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. The DLS analyses were conducted at the Center for Environmental Science and Technology (CEST) at the University of Notre Dame. L. M. S. acknowledges support from the Arthur J. Schmitt Foundation. L.F.P.K. acknowledges support from the German Academic Exchange Service.
Footnotes
Declaration of interest:
The authors declare no conflict of interest in this work
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A, HMDB 4.0: the human metabolome database for 2018, Nucleic Acids Research 46(D1) (2017) D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics. 3 (2007) 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lubes G, Goodarzi M, GC–MS based metabolomics used for the identification of cancer volatile organic compounds as biomarkers, Journal of Pharmaceutical and Biomedical Analysis 147 (2018) 313–322. [DOI] [PubMed] [Google Scholar]
- [4].Pontes JGM, Brasil AJM, Cruz GCF, de Souza RN, Tasic L, NMR-based metabolomics strategies: plants, animals and humans, Analytical Methods 9(7) (2017) 1078–1096. [Google Scholar]
- [5].Scalbert A, Brennan L, Fiehn O, Hankemeier T, Kristal BS, van Ommen B, Pujos-Guillot E, Verheij E, Wishart D, Wopereis S, Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research, Metabolomics 5(4) (2009) 435–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Faris GW, Copeland RA, Wavelength dependence of the Raman cross section for liquid water, Appl. Opt 36(12) (1997) 2686–2688. [DOI] [PubMed] [Google Scholar]
- [7].Prince RC, Frontiera RR, Potma EO, Stimulated Raman Scattering: From Bulk to Nano, Chemical Reviews 117(7) (2017) 5070–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Smith E, Dent G, Modern Raman Spectroscopy: A Practical Approach, John Wiley & Sons Ltd; 2005. [Google Scholar]
- [9].Albrecht MG, Creighton JA, Anomalously intense Raman spectra of pyridine at a silver electrode, Journal of the American Chemical Society 99(15) (1977) 5215–5217. [Google Scholar]
- [10].Gu X, Trujillo MJ, Olson JE, Camden JP, SERS Sensors: Recent Developments and a Generalized Classification Scheme Based on the Signal Origin, Annual Review of Analytical Chemistry 11(1) (2018) 147–169. [DOI] [PubMed] [Google Scholar]
- [11].Jeanmaire DL, Van Duyne RP, Surface raman spectroelectrochemistry: Part I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode, Journal of Electroanalytical Chemistry and Interfacial Electrochemistry 84(1) (1977) 1–20. [Google Scholar]
- [12].Cialla-May D, Zheng XS, Weber K, Popp J, Recent progress in surface-enhanced Raman spectroscopy for biological and biomedical applications: from cells to clinics, Chemical Society Reviews 46(13) (2017) 3945–3961. [DOI] [PubMed] [Google Scholar]
- [13].Stiles PL, Dieringer JA, Shah NC, Duyne RPV, Surface-Enhanced Raman Spectroscopy, Annual Review of Analytical Chemistry 1(1) (2008) 601–626. [DOI] [PubMed] [Google Scholar]
- [14].Alharbi O, Xu Y, Goodacre R, Simultaneous multiplexed quantification of caffeine and its major metabolites theobromine and paraxanthine using surface-enhanced Raman scattering, Analytical and Bioanalytical Chemistry 407(27) (2015) 8253–8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Guicheteau J, Argue L, Hyre A, Jacobson M, Christesen SD, Raman and surface-enhanced Raman spectroscopy of amino acids and nucleotide bases for target bacterial vibrational mode identification, SPIE Chemical and Biological Sensing VII 6218 (2006). [Google Scholar]
- [16].Radu AI, Kuellmer M, Giese B, Huebner U, Weber K, Cialla-May D, Popp J, Surface-enhanced Raman spectroscopy (SERS) in food analytics: Detection of vitamins B2 and B12 in cereals, Talanta 160 (2016) 289–297. [DOI] [PubMed] [Google Scholar]
- [17].Sui H, Wang Y, Zhang X, Wang X, Cheng W, Su H, Wang X, Sun X, Han XX, Zhao B, Ozaki Y, Ultrasensitive detection of thyrotropin-releasing hormone based on azo coupling and surface-enhanced resonance Raman spectroscopy, Analyst 141(17) (2016) 5181–5188. [DOI] [PubMed] [Google Scholar]
- [18].Farcau C, Astilean S, Mapping the SERS Efficiency and Hot-Spots Localization on Gold Film over Nanospheres Substrates, The Journal of Physical Chemistry C 114(27) (2010) 11717–11722. [Google Scholar]
- [19].Lee PC, Meisel D, Adsorption and surface-enhanced Raman of dyes on silver and gold sols, The Journal of Physical Chemistry 86(17) (1982) 3391–3395. [Google Scholar]
- [20].Dick LA, McFarland AD, Haynes CL, Van Duyne RP, Metal Film over Nanosphere (MFON) Electrodes for Surface-Enhanced Raman Spectroscopy (SERS): Improvements in Surface Nanostructure Stability and Suppression of Irreversible Loss, The Journal of Physical Chemistry B 106(4) (2002) 853–860. [Google Scholar]
- [21].Chien F-C, Huang WY, Shiu J-Y, Kuo CW, Chen P, Revealing the spatial distribution of the site enhancement for the surface enhanced Raman scattering on the regular nanoparticle arrays, Opt. Express 17(16) (2009) 13974–13981. [DOI] [PubMed] [Google Scholar]
- [22].Fang Y, Seong N-H, Dlott DD, Measurement of the Distribution of Site Enhancements in Surface-Enhanced Raman Scattering, Science 321(5887) (2008) 388. [DOI] [PubMed] [Google Scholar]
- [23].Kleinman SL, Ringe E, Valley N, Wustholz KL, Phillips E, Scheidt KA, Schatz GC, Van Duyne RP, Single-Molecule Surface-Enhanced Raman Spectroscopy of Crystal Violet Isotopologues: Theory and Experiment, Journal of the American Chemical Society 133(11) (2011) 4115–4122. [DOI] [PubMed] [Google Scholar]
- [24].Laibinis PE, Whitesides GM, Allara DL, Tao YT, Parikh AN, Nuzzo RG, Comparison of the structures and wetting properties of self-assembled monolayers of n-alkanethiols on the coinage metal surfaces, copper, silver, and gold, Journal of the American Chemical Society 113(19) (1991) 7152–7167. [Google Scholar]
- [25].Ulman A, Formation and Structure of Self-Assembled Monolayers, Chemical Reviews 96(4) (1996) 1533–1554. [DOI] [PubMed] [Google Scholar]
- [26].Petrov AP, Sherman LM, Camden JP, Dovichi NJ, Database of free solution mobilities for 276 metabolites, Talanta (2019) 10.1016/j.talanta.2019.120545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Agresti A, Franklin C, Statistics - The Art and Science of Learning from Data, 3rd ed., Pearson Education, United States, 2013. [Google Scholar]
- [28].Kennedy BJ, Spaeth S, Dickey M, Carron KT, Determination of the Distance Dependence and Experimental Effects for Modified SERS Substrates Based on Self-Assembled Monolayers Formed Using Alkanethiols, The Journal of Physical Chemistry B 103(18) (1999) 3640–3646. [Google Scholar]
- [29].Tripathi A, Emmons ED, Fountain AW, Guicheteau JA, Moskovits M, Christesen SD, Critical Role of Adsorption Equilibria on the Determination of Surface-Enhanced Raman Enhancement, ACS Nano 9(1) (2015) 584–593. [DOI] [PubMed] [Google Scholar]
- [30].Diaz Fleming G, Finnerty JJ, Campos-Vallette M, Célis F, Aliaga AE, Fredes C, Koch R, Experimental and theoretical Raman and surface-enhanced Raman scattering study of cysteine, Journal of Raman Spectroscopy 40(6) (2009) 632–638. [Google Scholar]
- [31].Jing C, Fang Y, Experimental (SERS) and theoretical (DFT) studies on the adsorption behaviors of l-cysteine on gold/silver nanoparticles, Chemical Physics 332(1) (2007) 27–32. [Google Scholar]
- [32].Nie S, Castillo CG, Bergbauer KL, Kuck JFR, Nabiev IR, Yu N-T, Surface-Enhanced Raman Spectra of Eye Lens Pigments, Applied Spectroscopy 44(4) (1990) 571–575. [Google Scholar]
- [33].Goto T, Watarai H, SERS Study of Rotational Isomerization of Cysteamine Induced by Magnetic Pulling Force, Langmuir 26(7) (2010) 4848–4853. [DOI] [PubMed] [Google Scholar]
- [34].Jiang X, Yang M, Meng Y, Jiang W, Zhan J, Cysteamine-Modified Silver Nanoparticle Aggregates for Quantitative SERS Sensing of Pentachlorophenol with a Portable Raman Spectrometer, ACS Applied Materials & Interfaces 5(15) (2013) 6902–6908. [DOI] [PubMed] [Google Scholar]
- [35].Michota A, Kudelski A, Bukowska J, Chemisorption of Cysteamine on Silver Studied by Surface-Enhanced Raman Scattering, Langmuir 16(26) (2000) 10236–10242. [Google Scholar]
- [36].Michota A, Kudelski A, Bukowska J, Influence of electrolytes on the structure of cysteamine monolayer on silver studied by surface-enhanced Raman scattering, Journal of Raman Spectroscopy 32(5) (2001) 345–350. [Google Scholar]
- [37].Michota A, Kudelski A, Bukowska J, Molecular structure of cysteamine monolayers on silver and gold substrates: Comparative studies by surface-enhanced Raman scattering, Surface Science 502-503 (2002) 214–218. [Google Scholar]
- [38].Li Y, Driver M, Decker E, He L, Lipid and lipid oxidation analysis using surface enhanced Raman spectroscopy (SERS) coupled with silver dendrites, Food Research International 58 (2014) 1–6. [Google Scholar]
- [39].Rodriguez JD, Westenberger BJ, Buhse LF, Kauffman JF, Standardization of Raman spectra for transfer of spectral libraries across different instruments, Analyst 136(20) (2011) 4232–4240. [DOI] [PubMed] [Google Scholar]
- [40].Patel IS, Premasiri WR, Moir DT, Ziegler LD, Barcoding bacterial cells: a SERS-based methodology for pathogen identification, Journal of Raman Spectroscopy 39(11) (2008) 1660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pozzi F, Porcinai S, Lombardi JR, Leona M, Statistical methods and library search approaches for fast and reliable identification of dyes using surface-enhanced Raman spectroscopy (SERS), Analytical Methods 5(16) (2013) 4205–4212. [Google Scholar]
- [42].Alharbi O, Xu Y, Goodacre R, Simultaneous multiplexed quantification of nicotine and its metabolites using surface enhanced Raman scattering, Analyst 139(19) (2014) 4820–4827. [DOI] [PubMed] [Google Scholar]
- [43].Muhamadali H, Watt A, Xu Y, Chisanga M, Subaihi A, Jones C, Ellis DI, Sutcliffe OB, Goodacre R, Rapid Detection and Quantification of Novel Psychoactive Substances (NPS) Using Raman Spectroscopy and Surface-Enhanced Raman Scattering, Frontiers in Chemistry 7(412) (2019) 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Negri P, Schultz ZD. Online SERS detection of the 20 proteinogenic L-amino acids separated by capillary zone electrophoresis. Analyst 139(22), (2014) 5989–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.