Abstract
The development of dwarf wheat cultivars combined with high levels of agrochemical inputs during the green revolution resulted in high yielding cropping systems. However, changes in wheat cultivars were made without considering impacts on plant and soil microbe interactions. We studied the effect of these changes on root traits and on the assembly of rhizosphere bacterial communities by comparing eight wheat cultivars ranging from tall to semi-dwarf plants grown under field conditions. Wheat breeding influenced root diameter and specific root length (SRL). Rhizosphere bacterial communities from tall cultivars were distinct from those associated with semi-dwarf cultivars, with higher differential abundance of Actinobacteria, Bacteroidetes and Proteobacteria in tall cultivars, compared with a higher differential abundance of Verrucomicrobia, Planctomycetes and Acidobacteria in semi-dwarf cultivars. Predicted microbial functions were also impacted and network analysis revealed a greater level of connectedness between microbial communities in the tall cultivars relative to semi-dwarf cultivars. Taken together, results suggest that the development of semi-dwarf plants might have affected the ability of plants to recruit and sustain a complex bacterial community network in the rhizosphere.
Subject terms: Soil microbiology, Microbiome
Introduction
Wheat domestication originated in the Near East and the crop has undergone a series of crosses and modifications leading to the hexaploid bread species, Triticum aestivum L1. Wheat is a key crop for global food security, providing 20% of dietary requirements for calories and protein2. To achieve food demand of an increasing global population, projections forecast a need to increase wheat production by 11% by 2026 with just 1.8% increase in cultivation areas3. During the Green Revolution, Reduced height (Rht) dwarfing genes were introduced in modern wheat cultivars4, resulting in high-yielding wheat plants, which, when combined with agrochemical management and optimal conditions, increased yields, without productivity losses caused by lodging5. Plant height is a complex trait controlled by several Quantitative Trait Loci (QTLs) and more than 20 Rht genes have been described, however, most of them have low potential for successful breeding programs6. Most breeding programs take into consideration the improvement of above ground parts of plants, with little attention to below ground parts due to the inherent challenges of root analysis coupled with the influence of soil type on root traits7,8. As such, the effect of domestication and modern breeding on belowground traits in wheat largely remains unclear. However, Bai et al.9 assessed a diverse set of wheat germplasm [199 double-haploid progeny derived from a cross between Avalon and Cadenza (Triticum aestivum L.)] and found that plant height was positively correlated to some evaluated root traits such as: total root length, seminal laterals length, seminal axes length, seminal laterals surface area, seminal laterals volume and root dry weight. In addition, Figueroa-Bustos et al.10 reported a positive correlation between plant height and specific root length (SRL). The selection of desirable root traits through “root breeding”, would enable plants to explore soil more efficiently, thus improving water and nutrient acquisition11. It has also become clear that future breeding programs should consider plant-microbe interactions12 as plants are complex holobiont organisms influenced both positively and negatively by their microbial communities13.
It is now known that the wheat microbiome can be influenced by host genotype14,15, fertilization regime16, land management and seed load17, irrigation18, seed germination and sterilization19, tissue type and growth stage20, plant organ, host age and management strategy21. However, there are few studies correlating root traits with microbiome structure. Pérez-Jaramillo et al.22 found that 11.4% of bacterial community composition was explained by root traits, e.g. SRL, in common bean cultivars. Saleem et al.23, studying the impact of root system architecture on rhizosphere and root microbiome, found that fine roots harboured higher microbial richness than secondary and primary roots of nicotiana cultivars.
The selective breeding of wheat cultivars led to structural changes in wheat24,25, which could also have affected associated microbial communities. Similar to the gut microbiome, which is considered to play an important role in host health26, the microbiome of plants helps them tolerate biotic and abiotic stresses27. The hypothesis that plant breeding has influenced microbial communities of wheat was previously tested by Germida and Siciliano28, who addressed the diversity of culturable bacteria from ancient and modern wheat cultivars. Due to technological limitations, a deeper total community analysis was not possible in their study, however, with the development of high throughput next generation sequencing technologies the study of complex microbial communities can now be achieved in unprecedented detail.
We hypothesize that wheat breeding has affected root morphological traits, impacting bacterial community structure, diversity and 16S rRNA gene-predicted functions in the wheat rhizosphere. We tested this hypothesis by comparing the rhizosphere bacterial communities from a range of tall and semi-dwarf wheat plants grown under field conditions.
Material and Methods
Rhizosphere sampling of wheat accessions
A “heritage wheat” experiment at Rothamsted Research consisted of hexaploid wheat varieties planted in triplicate in 1 m2 plots randomly designed with approximately 350 seeds per plot. Nitrogen was applied as ammonium nitrate at 210 kg/ha with other inputs according to standard agronomic practice. Three plants at flowering stage were harvested from each plot in July (2014), and rhizosphere soil was collected by gently discarding loose soil and vigorously shaking the roots inside a polythene bag to release tightly attached soil, which was considered as rhizosphere16. Rhizosphere soil was homogenized, and a 5 g subsample was taken and stored at −80 °C prior to soil DNA extraction. Experimental design consisted of 8 cultivars × 3 replicates, resulting in 24 samples. Varieties range from Chidham White Chaff, a long straw variety from 1790 to the modern variety Crusoe from 2012. These varieties were grouped into two categories: “Tall” representing cultivars which were obtained before the Green Revolution and “Semi-dwarf” representing cultivars of which functional Reduced height (Rht) dwarfing genes have been incorporated by selective breeding (Table 1) (Supplementary Fig. S1).
Table 1.
Cultivars chosen for the current study and some characteristics, such as year of release, pedigree and height.
| Cultivar | Year | Pedigree | Heighta |
|---|---|---|---|
| Chidham White Chaff | 1790 | Not recorded | Tall |
| Red Lammas | 1850 | Not recorded | Tall |
| Victor | 1908 | (Squarehead*Red King)*Talavera | Tall |
| Avalon | 1980 | TJB 30/148* TL 365a/34/5 | Semi-dwarf |
| Hereward | 1989 | Norman’sib’*Disponent | Semi-dwarf |
| Malacca | 1997 | Riband*(Rendezvous)*Apostle | Semi-dwarf |
| Gallant | 2009 | (Malacca*Charger)*Xi-19 | Semi-dwarf |
| Crusoe | 2012 | Cordiale*Gulliver | Semi-dwarf |
Root morphological traits
Wheat seeds were surface sterilised following the protocol by Robinson et al.31 and pre-germinated in Petri dishes with filter paper soaked in autoclaved distilled water. Analysis of root morphology was performed according to Bai et al.9. Full details are given in Supplementary Methods. Specific root length (SRL) and statistical analyses were calculated in R (version 3.5.0) as described by Pérez-Jaramillo et al.22. Normality and homogeneity of variances were checked using Shapiro–Wilk test and Levene’s test, respectively, and One-way ANOVA and post-hoc test (Tukey HSD) were used to assess differences.
Soil DNA extraction and quantification
For each sample, DNA was extracted from 0.25 g of rhizosphere soil using the MoBio PowerSoil™ DNA Isolation Kit (Carlsbad, CA, USA). Extractions were performed according to the manufacturer’s instructions but with the use of the MP Biomedicals FastPrep-24 machine twice for 30 s at 5.5 m.s−1. DNA purity and concentration were determined by NanoDrop spectrophotometry (Thermo Scientific, Wilmington, DE, USA) as well as a Qubit 2.0 Fluorimeter using ds DNA HS assay kit (Thermo Fisher).
Illumina bacterial 16S rRNA gene sequencing
Briefly, PCR amplicon libraries targeting the 16S rRNA encoding gene present in metagenomic DNA were produced using a barcoded primer set adapted for the Illumina HiSeq. 2000 and MiSeq32. DNA sequence data was generated using Illumina paired-end sequencing at the Environmental Sample Preparation and Sequencing Facility (ESPSF) at Argonne National Laboratory. Full details are given in Supplementary Methods.
Metataxonomic sequence analysis pipeline
16S rRNA gene sequences were analysed using the pipeline proposed by the Brazilian Microbiome Project (BMP) available at http://brmicrobiome.org 33 with some modifications. Details are described in Supplementary Methods. It uses Quantitative Insights Into Microbial Ecology (QIIME) (version 1.8.0)34 and USEARCH 9.035. Operational taxonomic units (OTUs) were defined to 97% sequence identity against SILVA 128 database36. The generated OTU table was filtered using QIIME scripts to extract Bacterial domain and remove chloroplasts as suggested by Pérez-Jaramillo et al.22. Differences in bacterial community structure were investigated by Permutational Analysis of Variance (PERMANOVA)37 in Paleontological Statistics Software Package for Education and Data Analysis (PAST)38. PCoA plots were obtained using the same software. The number of observed OTUs and diversity based on Shannon index were calculated in QIIME. Statistical analyses of alpha diversity indexes were performed in R as described by Pérez-Jaramillo et al.22. Briefly, normality and homogeneity of variances were checked using Shapiro–Wilk test and Levene’s test, respectively, and One-way ANOVA and post-hoc test (Tukey HSD) were used to assess differences.
Analysis of differentially abundant OTUs
The online tool for comprehensive statistical, visual and meta-analysis of microbiome data called Microbiome Analyst39 was used for detecting OTUs which were differentially abundant between tall and semi-dwarf cultivars, using DESeq. 2, which has high sensitivity for small datasets (<20 samples per group). Besides, False Discovery Rate (FDR) values are relatively low for data sets with similar sizes when compared to other tools such as metagenomeSeq40. Further details are supplied in Supplementary Methods.
Network analysis
Network analyses were performed using the Molecular Ecological Network Analyses (MENA) pipeline41 available at http://ieg4.rccc.ou.edu/mena/. Network topological properties were calculated and evaluated41. Phylogenetic molecular ecological networks (pMENs) were visualised using Cytoscape (v.3.4.0)42. Full details are provided in Supplementary Methods. Additionally, co-occurrence analyses were performed using python module SparCC43. For this, the filtered OTU table previously obtained (with at least 0.01% of total abundance) was used. For each network, P-values were obtained by 100 permutations of random selections of the data table. Statistically significant (p < 0.01) SparCC correlations with a magnitude of > 0.7 or < −0.7 were included into the network analyses. Networks were visualized with Cytoscape (v.3.4.0)42.
Functional prediction from 16S rRNA gene data
In addition to the taxonomic analysis, 16S rRNA gene libraries were used to predict metagenome function using the PICRUSt software44 within the GALAXY server provided by the Langille Lab (v1.1.1) (http://galaxy.morganlangille.com/). Details on PICRUSt analyses are given in Supplementary Methods. The table with 16S rRNA gene-predicted functions was exported into Statistical Analysis of Metagenomic Profiles (STAMP) software (version 2.1.3)45 to test for statistical differences with ANOVA. Extended error plots were obtained, and they show the proportion of sequences (%) that were significantly different using a post-hoc test (Tukey-Kramer), Eta-squared, to measure effect size and Benjamini-Hochberg-FDR as a multiple test correction (p < 0.05). Relevant KEGG orthologs (KOs) were selected for further analyses.
Results and Discussion
Effect of wheat breeding on root morphological traits
Breeding process appears to have altered some root traits, especially root diameter and specific root length (SRL). Tall plants (old cultivars), when compared to semi-dwarf plants (recent cultivars), showed statistically higher mean SRL (tall = 113.63 m.g−1 and semi-dwarf = 91.62 m.g−1) (One-Way ANOVA, p < 0.05). The opposite general trend is observed for root diameter (tall = 0.44 mm and semi-dwarf = 0.49 mm) (One-Way ANOVA, p < 0.05) (Supplementary Fig. S2). Tall cultivars tend to have longer thinner roots, whereas shorter thicker roots are observed for semi-dwarf cultivars. Pérez-Jaramillo et al.22 also observed higher SRL values for wild common bean accessions, which could be useful for enhanced water and nutrient uptake in stressed soils, than the lower SRL of short varieties, suggesting semi-dwarf cultivars could be more dependent on fertile soils. Indeed, modern Italian bread wheat cultivars increased their N demand over time when compared to old tall cultivars46. Plant height reduction was achieved during the 20th century wheat breeding47, though little attention was given to assessing the effects of these programmes on wheat root traits. In an attempt to study whether Rht genes control both shoot height and seedling root growth, Bai et al.9 evaluated QTLs for plant height, root and seed traits from different wheat lines and identified some coincident QTLs for roots and height, concluding that the introduction of some known Rht dwarfing genes reduced both plant height and root proliferation. Breeding for higher yields in Australian wheat cultivars led to a reduced root length density (RLD) and total root length and increased efficiency of N uptake48. For other crops, such as barley, common bean, rice and maize, it has also been shown that both domestication and breeding caused changes in root architectural traits, leading to a differential spatial arrangement of roots49.
Effect of breeding process on bacterial structure, richness, diversity and differential recruitment of bacterial taxa
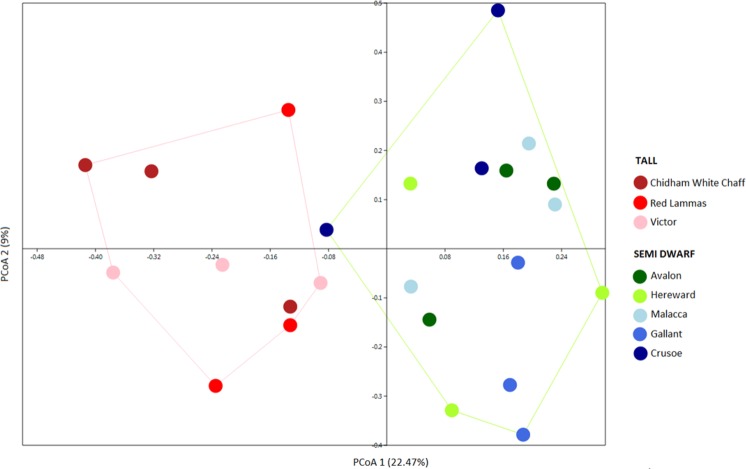
Bacterial communities from the wheat rhizosphere were mainly represented by Proteobacteria (36–41.7%), followed by Actinobacteria (12.1–16.1%), Bacteroidetes (11.1–16%) and Acidobacteria (8.7–13%) (Supplementary Fig. S3). The PCoA plot shows rhizosphere bacterial communities from tall cultivars differ from those associated with semi-dwarf cultivars (PERMANOVA, F = 0.827, p = 0.0001), with the first PCoA axis corresponding to 22.47% of the variation (Fig. 1). In general, tall plants when compared to semi-dwarf plants showed lower diversity (based on Shannon index) (tall = 9.068 and semi-dwarf = 9.423) and lower number of observed OTUs (tall = 1642.289 and semi-dwarf = 1810.667) (Supplementary Fig. S4).
Figure 1.
PCoA based on Bray-Curtis similarity distance matrix showing the structure of wheat rhizosphere bacterial communities associated with tall (dark red, red and pink) and semi-dwarf cultivars (green and blue).
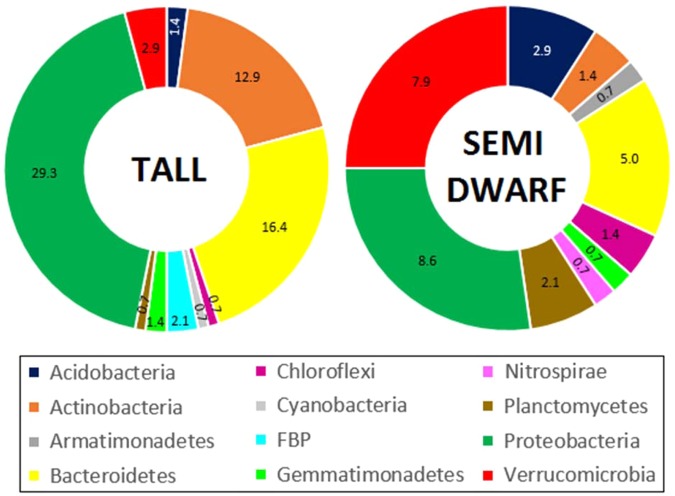
From a total of 140 significantly differentially abundant OTUs detected with DESeq. 2, 29.3% of OTUs belonging to the phylum Proteobacteria were significantly more abundant in the rhizosphere of tall cultivars, as opposed to 8.6% associated to semi-dwarf cultivars (Fig. 2).
Figure 2.
Doughnut charts showing the values (%) of differentially abundant OTUs grouped at phylum level, from a total of 140 detected among tall and semi-dwarf cultivars.
Tall cultivars also showed a higher percentage of differentially abundant OTUs belonging to the phyla Bacteroidetes (16.4%), Actinobacteria (12.9%) and Gemmatimonadetes (1.4%). On the other hand, semi-dwarf cultivars showed a larger proportion of OTUs belonging to the phyla Verrucomicrobia (7.9%), Acidobacteria (2.9%), Planctomycetes (2.1%) and Chloroflexi (1.4%). Additionally, OTUs differentially abundant and assigned to Armatimonadetes and Nitrospirae were only observed in semi-dwarf cultivars (0.7% in both cases) and significantly more OTUs assigned to Cyanobacteria and FBP were only detected in association with tall cultivars (0.7% and 2.1%, respectively).
The rhizospheres of semi-dwarf cultivars are enriched in members belonging to the phyla Acidobacteria and Verrucomicrobia, which are typically more associated with bulk soil rather than the wheat rhizosphere16. A closer analysis at genus level, indicates that tall cultivars are enriched with OTUs assigned to genera normally associated with plant growth promotion (PGP), especially members of the phylum Proteobacteria (Table 2) such as Brevundimonas (indole-3-acetic acid (IAA) production and P solubilisation)50; Devosia (N2 fixation)51; Rhizomicrobium (reduction of nitrate to nitrite)52; Sphingomonas (IAA and gibberellin production)53; Sphingopyxis (IAA production and N2 fixation)54; Massilia (IAA, siderophore and proteolytic enzyme production)55; Nitrosospira (ammonia-oxidizing bacterium (AOB))56 and Bradyrhizobium, Rhizobium, Methylobacterium, Variovorax, Klebsiella and Pseudomonas which typically possess genes contributing to plant-beneficial functions57. Although some differentially recruited OTUs within tall rhizospheres have been assigned to genera commonly related to plant growth promotion, these abilities are normally strain-specific, thus caution must be taken when analysing the results. Further work should evaluate these differences based on metagenomic datasets as well as functional characterisation of microbial isolates.
Table 2.
Proteobacteria genera assigned to OTUs which were found to be enriched in the rhizosphere of tall and semi-dwarf cultivars.
| Height | Class | Genus |
|---|---|---|
| Tall | Alphaproteobacteria | Bradyrhizobium, Brevundimonas, Devosia, |
| Methylobacterium, Rhizobium, Rhizomicrobium, | ||
| Skermanella, Sphingomonas, Sphingopyxis, | ||
| Variibacter, uncultured Caulobacteraceae, | ||
| unclassified Aurantimonadaceae, unclassified Rhizobiaceae, | ||
| unclassified Rickettsiales and unclassified Sphingomonadaceae | ||
| Betaproteobacteria | Massilia, Nitrosospira, Verticia, Variovorax, | |
| uncultured order B1-7BS, unclassified Oxalobacteraceae and | ||
| unclassified Comamonadaceae | ||
| Deltaproteobacteria | Bacteriovorax | |
| Gammaproteobacteria | Dyella, Klebsiella, Lysobacter, | |
| Pseudomonas, Rhodanobacter and Stenotrophomonas | ||
| Semi-dwarf | Betaproteobacteria | Uncultured Alcaligenaceae and uncultured Nitrosomonadaceae |
| Deltaproteobacteria | Haliangium, Phaselicystidaceae and uncultured Myxococcales | |
| Gammaproteobacteria | Polycyclovorans and uncultured Xanthomonadales |
The major metabolic consequence of plant dwarfing is a reduction in response to the plant phytohormone gibberellin58 and an increase of active endogenous gibberellin levels when compared to wild-type plants not carrying Rht dwarfing genes59,60. The correlation that the rhizosphere of short plants with decreased gibberellin sensitivity is associated with an increased colonisation of bacteria normally associated with bulk soil and a decrease of microbial phyla associated with plant growth promotion, suggests that gibberellin sensitivity influences the composition of the root microbiome. It is known that some microbes, such as PGP members of Rhizobiaceae family61 as well as some Sphingomonas spp53. produce this hormone, and we find these bacteria to be differentially more abundant in tall cultivar rhizospheres. It could follow that plant gibberellin insensitivity leads to a reduction in plant-microbe communication in the root environment and could reduce selection of gibberellin-producing microbes in the root zone. Apart from controlling the growth and development of plants, gibberellins also play a role in plant signalling60, affecting defence response mechanisms dependent on jasmonic acid (JA) or salicylic acid (SA)62, rhizobial infection of legumes63 mediated via DELLA proteins and can suppress arbuscular colonisation and development of mycorrhizal symbiosis64. Work should be conducted to determine if the root microbiome of these crop accessions diverges further when tall and shorts cultivars are cultured under low nutrient conditions. This would support the hypothesis that the tall cultivars are more capable of recruiting beneficial microbes, and when under stress they do this more readily than short cultivars. Furthermore, microbes have been shown to be capable of producing a range of phytohormones65. It will be interesting to determine if the plant breeding process has influenced the plant hormone status for other classes of phytohormones including abscisic acid, auxins, brassinosteroids, ethylene, jasmonates, salicyclic acid and strigolactones, as there is a strong interaction among them66 and whether their levels influence microbiome composition.
In addition to plant hormonal levels, the observed differences in microbiome structure between short and tall cultivars could be attributed to root exudation profiles23. Iannucci et al.67 showed that even though soil type dramatically changed the composition of root exudates, domestication and breeding also had major effects on root exudates in the rhizosphere of ten tetraploid wheat genotypes. The sensing and metabolism of root metabolites are likely important factors in determining microbiome assembly68.
Effect of wheat breeding on key rhizosphere taxa and network complexity
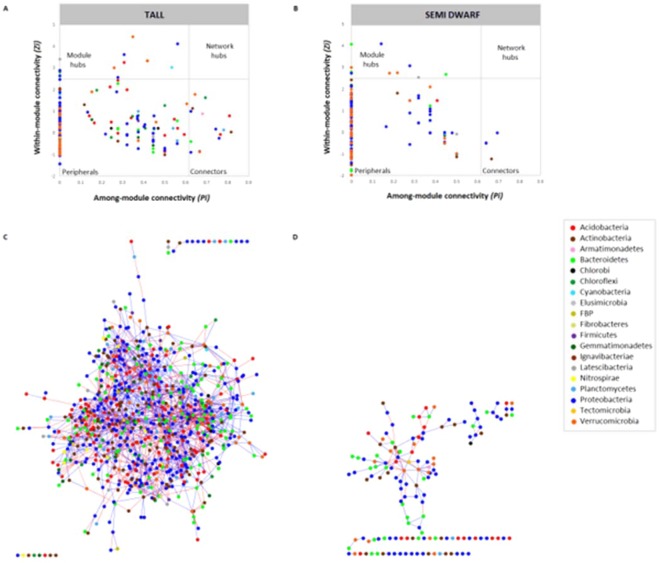
We found that for both tall and semi-dwarf networks, zero network hubs were identified and that most nodes were categorised as peripherals (Fig. 3A,B). However, fifteen nodes were classified as module hubs in tall cultivars and they represented a variety of taxonomic groups: five belonged to Proteobacteria (genera Pseudomonas, Luteibacter, Sorangium and orders Myxococcales and Xanthomonadales), three to Verrucomicrobia, two to Acidobacteria and one each from Bacteroidetes, Chloroflexi, Cyanobacteria, Gemmatimonadetes and Latescibacteria. Ten module hubs were identified in semi-dwarf cultivars and three belonged to Proteobacteria (one from ß-Proteobacteria, one from the family Rhodospirillaceae and one from the family Nitrosomonadaceae), three from Bacteroidetes (genera Parafilimonas and Fluviicola and family Saprospiraceae), three from Verrucomicrobia and one from Latescibacteria. Connectors were also detected in both cases, however, semi-dwarf cultivars displayed only four connectors, identified as belonging to genera Asticcacaulis and Pseudohongiella, both from phylum Proteobacteria, genus Luedemannella (Actinobacteria) and one Acidobacteria genus (RB41). On the other hand, tall cultivars showed fifteen connectors: two Acidobacteria (classes Subgroup 22 and Holophagae), one Actinobacteria (class MB-A2-108), one Armatimonadetes (family Fimbriimonadaceae), one Bacteroidetes (genus Pedobacter), one Chlorobi (order Ignavibacteriales), one Chloroflexi (class S085), one Planctomycetes (family Tepidisphaeraceae), two Verrucomicrobia (one identified as the genus Chthoniobacter and other belonging to the family Verrucomicrobiaceae) and five Proteobacteria (two belonging to the genus Haliangium, one betaproteobacterium from the family Rhodocyclaceae, one deltaproteobacterium from the family Sandaracinaceae and one α-Proteobacterium from the order Rhodospirillales (family DA111)). Members of the phylum Proteobacteria were found to be the most prominent putative keystone taxa, accounting for more than 30% of all module hubs and connectors in both tall and semi-dwarf cultivar networks. Members of Bacteroidetes and Verrucomicrobia were the second most prominent keystone taxa for semi-dwarf cultivars, each accounting for 21.43% of module hubs. On the other hand, members of Bacteroidetes accounted for less than 7% of module hubs and connectors and Verrucomicrobia members accounted for 16.67% in tall cultivars. Module hub and connector OTUs have been proposed as putative keystone taxa and ecological generalists, critical for maintaining community structure and function; peripherals on the other hand being considered isolated specialists41,69–74. As such, Proteobacteria seem to be critical in maintaining the structure and functionality of bacterial communities in wheat rhizosphere for both tall and semi dwarf cultivars and these results are in accordance with data provided by Oberholster et al.71, who also found Proteobacteria to be important keystone taxa for sorghum and sunflower rhizospheres.
Figure 3.
Classification of nodes (OTUs) to identify putative keystone OTUs within rhizosphere networks from tall (A) and semi-dwarf (B) cultivars. Node topologies are placed into four categories: module hubs (Zi > 2.5) are highly connected nodes within modules; connectors (Pi > 0.62) are nodes responsible for connecting modules; network hubs (Zi > 2.5 and Pi > 0.62) are highly connected nodes within the entire network and peripherals (Zi < 2.5 and Pi < 0.62) are nodes connected in modules with few outside connections. Bacterial co-occurrence networks from tall (C) and semi-dwarf (D) cultivars, calculated with sparCC, with correlations with a magnitude of > 0.7 or < −0.7. Each node (dot) represents an OTU and each colour indicates one respective phylum. The edges are represented by lines, with positive co-occurrence patterns between two nodes shown in blue and negative co-occurrence patterns shown in red.
Phylogenetic molecular ecological networks (pMENs) were obtained for both tall and semi-dwarf cultivars and the overall structure of each pMEN is presented in Supplementary Table S1. Topological properties indicated that R2 values for both networks (0.907 for tall cultivars and 0.899 for semi-dwarf cultivars) followed a power-law model, indicating that only a few nodes have many connections with other nodes41. Average path distance (GD) obtained for tall cultivars was lower than semi-dwarf cultivars, indicating that nodes in the network from tall cultivars are closer to each other. The average degree (avgK) obtained for tall cultivars indicated a more complex network structure, as a higher value was obtained. This higher complexity was also highlighted by networks calculated with sparCC represented in Fig. 3C,D. High values of modularity for both networks (tall = 0.749 and semi-dwarf = 0.890) indicate that they can be separated into modules using fast greedy modularity optimisation. Modularity values range from 0 to 1 and they measure the degree to which the network was organised into clearly delimited modules74 and modular structure of complex networks display critical roles in their functionality75. The number of modules obtained for tall cultivars was ninety-four, whereas semi-dwarf cultivars resulted in one-hundred and fifteen modules. A module is defined as a group of OTUs which are highly connected, however these are poorly connected with OTUs outside a given module41. We found that modules comprised of 4 nodes or more numbered 38 for dwarf varieties compared to 21 for tall varieties, indicating that the OTUs are better connected within the tall cultivar network (Supplementary Fig. S5).
The number of edges obtained in the pMEN from bacterial communities from tall cultivars was higher than those from semi-dwarf cultivars and this was also highlighted by the connectedness index which was 0.498 as opposed to 0.293 for semi-dwarf cultivars. This index ranges from 0 to 1, with values close to 1 indicating a highly-connected graph41. The ratio of positive to negative co-occurrence patterns for tall cultivars was 3.094, whereas the ratio of positive to negative interactions for semi-dwarf cultivars was 5.331, indicating that bacterial communities from semi-dwarf cultivars displayed more positive co-occurrence relationships than tall cultivars. The composition and number of nodes per modules varied and the composition of modules is shown in Supplementary Fig. S6. Similarly to nodes identified as keystone taxa, the majority of nodes were classified as Proteobacteria, followed by Acidobacteria, Bacteroidetes, Actinobacteria and Verrucomicrobia. For tall cultivars, the biggest module (#6) comprised of 70 nodes, with more than 32% of nodes belonging to Proteobacteria. As for semi-dwarf cultivars, the largest module (#1) was composed of 71 nodes, with more than 35% belonging to Proteobacteria.
In the present study, tall cultivars showed bacterial lower diversity than semi-dwarf cultivars as well as a more complex bacterial network structure in the rhizosphere. Shi et al.69 observed that microbial diversity decreased as network size and connectivity increased, which resulted in an increased community organisation. It has also been shown that more complex networks are able to cope with environmental changes76, increase crop productivity73 or suppress soil borne pathogen infection on plants77. Indeed, semi-dwarf wheat cultivars carrying Rht alleles were previously shown to be more susceptible than wild cultivars to initial infection by some pathogens78,79. This might be linked to the severity of DELLA effect on plant stature, however, differences in differential abundance of specific bacterial keystone taxa might point out some microbial influence on disease susceptibility. For instance, in the network from tall cultivars, OTUs 1135 and 427 in modules 2 and 3, respectively, were classified as belonging to the genus Haliangium, whose species have been described as putative biocontrol agents80 and another three-keystone species, from the same order, Myxococcales, were also observed in tall cultivars. Myxobacteria include Gram-negative gliding bacteria with predatory features, mostly due to the direct cell to cell contact, production of hydrolytic enzymes and secondary metabolites with antibiotic activity81. The third module in tall cultivars show one module hub (OTU 575 – classified as belonging to the genus Chryseobacterium) which has previously been described in the rhizosphere of wheat [14, 82;], and these bacteria have been shown to display plant growth-promotion82 and are enriched in the presence of 2,4-DAPG-producing species, such as Pseudomonas fluorescens83. Interestingly, the other module hub identified in the same module as Chryseobacterium was OTU 171 (assigned to the genus Pseudomonas) and they positively co-occur, meaning that their abundance changed along the same trend but not necessarily meaning they directly interact with each other74. OTU 280, assigned to the genus Pedobacter and OTU 563 (Luteibacter) have also been identified as keystone taxa in tall cultivars. They have been found to inhibit the growth of root pathogens, such as the fungus Rhizoctonia solani84,85. When comparing both networks, only two OTUs (601 and 118) assigned to the genus RB41 (Blastocatellaceae family from the phylum Acidobacteria) were commonly found and were identified as keystone species. The breeding process seems to have reduced the amount of keystone taxa known for their potential to antagonise pathogens, possibly affecting the degree of resistance of semi-dwarf cultivars to specific diseases. Future work should investigate whether tall and semi-dwarf cultivars select different beneficial bacteria with PGP abilities, as well as antagonistic activities against economically important soil-borne pathogens.
Effect of wheat breeding on 16S rRNA gene-predicted functions
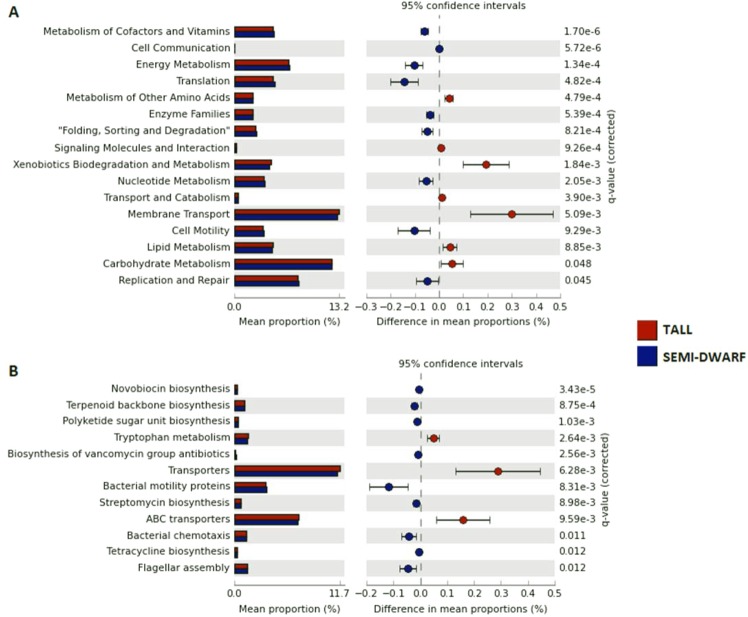
NSTI values obtained for 16S rRNA gene-predicted functions on rhizosphere soil samples are on average 0.1679 ± 0.013 for all samples and this value is in accordance to what is observed for soils44 and is lower when compared to other rhizosphere soils86. Rhizosphere samples collected from tall cultivars, on average have a NSTI value of 0.1534 ± 0.007 which is lower than NSTI value obtained from the rhizosphere of semi-dwarf cultivars (0.1766 ± 0.006). Functional prediction resulted in two hundred and twenty-one KEGG orthologs (KOs), of which one hundred and forty-one were differentially abundant (p < 0.05). Of these, 45.39% were significantly more abundant in tall cultivars and the remaining 54.61% were significantly more abundant in semi-dwarf cultivars. Seven 16S rRNA gene-predicted pathways were significantly enriched in rhizosphere bacterial communities of tall cultivars, as opposed to nine pathways enriched in the rhizosphere of semi-dwarf cultivars (Fig. 4A). Tall cultivars were predicted to be enriched in functions related to membrane transport, such as ABC transporters (Fig. 4B). Mahoney et al.14 suggested that differences in 16S rRNA gene-predicted functions observed between wheat cultivars could be driven by differential root exudate chemistry. The breeding process may have affected the quality and quantity of root exudates, impacting predicted functions in the rhizosphere. The presence of bespoke solute binding protein-dependent ABC transporters may be required for the use of root exudates by bacteria, aiding their colonisation of this niche87. When analysing specific KOs, tryptophan metabolism was also predicted to be enriched in bacterial communities from the rhizosphere of tall cultivars (Fig. 4B) and this suggests higher production of indole-3-acetic acid (IAA) hormone, as tryptophan is the main precursor for IAA biosynthesis, acting as a signalling molecule between plants and microbes as well as for plant growth promotion88. It could also mean that tall wheat plants are secreting more tryptophan upon colonization by specific bacterial groups89. On the other hand, bacterial communities from semi-dwarf cultivars were predicted to be enriched in predicted functions related to cell motility, such as bacterial motility proteins, bacterial chemotaxis and flagellar assembly (Fig. 4B). These features would facilitate bacterial migration towards roots, followed by attachment and establishment90. Some root exudates are known to induce motility in bacteria, which is an advantageous characteristic for root colonisation, when compared to non-motile bacteria91,92. 16S rRNA gene-predicted functions of streptomycin, novobiocin, tetracycline and vancomycin biosynthesis were significantly enriched in the rhizosphere of semi-dwarf cultivars (Fig. 4B) and this might have some relation to the decrease in keystone taxa which resulted in a more hostile, less coordinated environment compared with tall cultivars, where microbes compete with one another in a less structured manner to gain access to rich nutrients in the rhizosphere. It should be stated that PICRUSt is a tool which provides a functional prediction of microbiome based on marker genes, but it is not an actual measurement of such functions93. Future work on shotgun metagenomics would enable to assess these predicted observations.
Figure 4.
Statistical comparison using Welch’s t-test between the 16S rRNA-predicted functions of rhizosphere samples associated to tall and semi dwarf cultivars using Benjamini-Hochberg FRD correction (p < 0.05), showing the significant pathways (A) and specific selected KOs (B).
Conclusions, Limitations and Future Directions
In recent years, there has been a growing interest in modifying plant traits to change the ability of plants to interact with beneficial microbes13,94. Collectively, our results showed that wheat breeding from tall to semi-dwarf plants resulted in plants less able to select and sustain a complex rhizosphere. By evaluating differences in root traits of tall and semi-dwarf wheat cultivars and the structure, composition and differential recruitment of specific taxa and keystone species, we can start to understand the impacts of breeding process on root biology. Future work should assess the effect that the breeding process has had on plant hormonal status, exudation profile, plant performance and microbiome selection under low and high nutrient supplementation. This will allow the identification of keystone species and the development of synthetic communities of increased complexity, so the significance of their presence and absence in facilitating the development of a healthy microbiome can be tested.
Supplementary information
Acknowledgements
This research was funded by the Natural Environment Research Council (NERC) and the Biotechnology and Biological Sciences Research Council (BBSRC) under research program NE/N018125/1 LTS-M ASSIST – Achieving Sustainable Agricultural Systems www.assist.ceh.ac.uk. We also thank Bilateral BBSRC-Embrapa grant on “Exploitation of the rhizosphere microbiome for sustainable wheat production” (BB/N016246/1); “Optimization of nutrients in soil-plant systems: How can we control nitrogen cycling in soil?” (BBS/E/C/00005196) and “S2N – Soil to nutrition – Work package 1 – Optimizing nutrient flows and pools in the soil-plant-biota system” (BBS/E/C/000I0310). V.N.K. was also supported by São Paulo Research Foundation (FAPESP) (2013/08144-1 and 2014/16041-0). We thank Dr. Till Pellny and Dr. Alison Lovegrove from Plant Sciences department at Rothamsted Research for providing wheat seeds and for allowing rhizosphere collection from wheat plants in the field. We are grateful to Dr. Caihong Bai from Plant Sciences department at Rothamsted Research for kindly showing how to set up and analyse root morphological traits of wheat seedlings.
Author contributions
I.C., P.R.H. and T.H.M. designed the experiment. R.J.R. and T.H.M. collected the samples. V.N.K. processed, generated and analysed the data. D.H. developed an automated script for running QIIME analyses. V.N.K., I.C., I.S.M., P.R.H., R.M. and T.H.M. discussed the results. V.N.K and T.H.M. wrote the manuscript. V.N.K., I.C., M.R., P.R.H., R.M. and T.H.M. edited and commented on the manuscript. T.H.M. supervised the work. I.C., P.R.H., R.M. and T.H.M. were involved in resources and funding acquisition.
Data availability
16S rRNA gene amplicon data are available at the NCBI Sequence Read Archive (SRA) under accession number: PRJNA601112.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-58402-y.
References
- 1.Salamini F, Özkan H, Brandolini A, Schäfer-Pregl R, Martin W. Wild cereal domestication in the Near East. Nat. Rev. Genet. 2002;3:429–441. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- 2.Shiferaw B, et al. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Security. 2013;5(3):291–317. doi: 10.1007/s12571-013-0263-y. [DOI] [Google Scholar]
- 3.OECD/FAO. OECD-FAO Agricultural Outlook 2017–2026, OECD Publishing, Paris, 10.1787/agr_outlook-2017-en (2017).
- 4.Borojevic K, Borojevic K. The transfer and history of “Rht” genes in wheat from Japan to Europe. J. Hered. 2005;96(4):455–459. doi: 10.1093/jhered/esi060. [DOI] [PubMed] [Google Scholar]
- 5.Hedden P. The genes of the Green Revolution. Trends Genet. 2003;19(1):5–9. doi: 10.1016/S0168-9525(02)00009-4. [DOI] [PubMed] [Google Scholar]
- 6.Würschum T, Langer SM, Longin FH, Tucker MR, Leiser WL. A modern Green Revolution gene for reduced height in wheat. Plant J. 2017;92:892–903. doi: 10.1111/tpj.13726. [DOI] [PubMed] [Google Scholar]
- 7.Narayanan S, Mohan A, Gill KS, Prasad PVV. Variability of root traits in spring wheat germplasm. PLoS ONE. 2014;9(6):e100317. doi: 10.1371/journal.pone.0100317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wasson AP, et al. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J. Exp. Bot. 2012;63(9):3485–3498. doi: 10.1093/jxb/ers111. [DOI] [PubMed] [Google Scholar]
- 9.Bai C, Liang Y, Hawkesford MJ. Identification of QTLs associated with seedling root traits and their correlation with plant height. J. Exp. Bot. 2013;64(6):1745–1753. doi: 10.1093/jxb/ert041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueroa-Bustos V, Palta JA, Chen Y, Siddique KHM. Characterization of root and shoot traits in wheat cultivars with putative differences in root system size. Agronomy. 2018;8:109. doi: 10.3390/agronomy8070109. [DOI] [Google Scholar]
- 11.Paez-Garcia A, et al. Root traits and phenotyping strategies for plant improvement. Plants. 2015;4:334–355. doi: 10.3390/plants4020334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sessitsch A, Mitter B. 21st century agriculture: integration of plant microbiomes for improved crop production and food security. Microb. Biotechnol. 2015;8(1):32–33. doi: 10.1111/1751-7915.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Z, Jousset A. Plant breeding goes microbial. Trends Plant Sci. 2017;22(7):555–558. doi: 10.1016/j.tplants.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Mahoney AK, Yin C, Hulbert SH. Community structure, species variation, and potential functions of rhizosphere-associated bacteria of different winter wheat (Triticum aestivum) cultivars. Front. Plant Sci. 2017;8:132. doi: 10.3389/fpls.2017.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mauchline TH, et al. An analysis of Pseudomonas genomic diversity intake-all infected wheat fields reveals the lasting impact of wheat cultivars on the soil microbiota. Environ. Microbiol. 2015;17(11):4764–4778. doi: 10.1111/1462-2920.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kavamura VN, et al. Inorganic nitrogen application affects both taxonomical and predicted functional structure of wheat rhizosphere bacterial communities. Front. Microbiol. 2018;9:1074. doi: 10.3389/fmicb.2018.01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kavamura VN, et al. Land management and microbial seed load effect on rhizosphere and endosphere bacterial community assembly in wheat. Front. Microbiol. 2019;10:2625. doi: 10.3389/fmicb.2019.02625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mavrodi DV, et al. Long-term irrigation affects the dynamics and activity of the wheat rhizosphere microbiome. Front. Plant Sci. 2018;9:345. doi: 10.3389/fpls.2018.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Kuang Z, Wang W, Cao L. Exploring potential bacterial and fungal biocontrol agents transmitted from seeds to sprouts of wheat. Biol. Control. 2016;98:27–33. doi: 10.1016/j.biocontrol.2016.02.013. [DOI] [Google Scholar]
- 20.Robinson RJ, et al. Endophytic bacterial community composition in wheat (Triticum aestivum) is determined by plant tissue type, developmental stage and soil nutrient availability. Plant Soil. 2016;405:381–396. doi: 10.1007/s11104-015-2495-4. [DOI] [Google Scholar]
- 21.Gdanetz K, Trial F. The wheat microbiome under four management strategies, and potential for endophytes in disease protection. Phytobiomes J. 2017;1(3):158–168. doi: 10.1094/PBIOMES-05-17-0023-R. [DOI] [Google Scholar]
- 22.Pérez-Jaramillo JE, et al. Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits - scripts for statistical analysis and graphs. ISME J. 2017;11(10):2244–2257. doi: 10.1038/ismej.2017.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saleem M, et al. Impact of root system architecture on rhizosphere and root microbiome. Rhizosphere. 2018;6:47–51. doi: 10.1016/j.rhisph.2018.02.003. [DOI] [Google Scholar]
- 24.Bertin C, Yang XH, Weston LA. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil. 2003;256:67–83. doi: 10.1023/A:1026290508166. [DOI] [Google Scholar]
- 25.Graaff M-A, Six J, Jastrow JD, Schadt CW, Wullschleger SD. Variation in root architecture among switchgrass cultivars impacts root decomposition rates. Soil Biol. Biochem. 2013;58:198–206. doi: 10.1016/j.soilbio.2012.11.015. [DOI] [Google Scholar]
- 26.Lamoureux EV, Grandy SA, Langille MGI. Moderate exercise has limited but distinguishable effects on the mouse microbiome. mSystems. 2017;2:e00006–17. doi: 10.1128/mSystems.00006-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendes R, Raaijmakers JM. Cross-kingdom similarities in microbiome functions. ISME J. 2015;9:1905–1907. doi: 10.1038/ismej.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Germida J, Siciliano S. Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol. Fert. Soils. 2001;33:410–415. doi: 10.1007/s003740100343. [DOI] [Google Scholar]
- 29.Shewry PR, Pellny TK, Lovegrove A. Is modern wheat bad for health? Nat. Plants. 2016;2:1–3. doi: 10.1038/nplants.2016.97. [DOI] [PubMed] [Google Scholar]
- 30.Pask, A.J.D., Pietragalla, J., Mullan, D.M., Reynolds, M.P. (eds.). Physiological Breeding II: A Field Guide to Wheat Phenotyping. CIMMYT: Mexico, D.F, (2012).
- 31.Robinson RJ, Fraaije BA, Jackson RW, Hirsch PR, Mauchline TM. Wheat seed embryo excision enables the creation of axenic seedlings and Koch’s postulates testing of putative bacterial endophytes. Sci. Rep. 2016;6:25581. doi: 10.1038/srep25581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pylro VS, et al. Data analysis for 16S microbial profiling from different benchtop sequencing platforms. J. Microbiol. Methods. 2014;107:30–37. doi: 10.1016/j.mimet.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 34.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 36.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26(1):32–46. [Google Scholar]
- 38.Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001;4(1):9pp. [Google Scholar]
- 39.Dhariwal A, et al. MicrobiomeAnalyst - a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45(W1):W180–188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss S, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng Y, et al. Molecular ecological network analyses. BMC Bioinformatics. 2012;13:113. doi: 10.1186/1471-2105-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. Plos Comput. Biol. 2012;508:8–e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langille MGI, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31(9):814–823. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guarda G, Padovan S, Delogu G. Grain yield, nitrogen-use efficiency and baking quality of old and modern Italian bread-wheat cultivars grown at different nitrogen levels. Europ. J. Agronomy. 2004;21:181–192. doi: 10.1016/j.eja.2003.08.001. [DOI] [Google Scholar]
- 47.Ormoli L, Costa C, Negri S, Perenzin M, Vaccino P. Diversity trends in bread wheat in Italy during the 20th century assessed by traditional and multivariate approaches. Sci. Rep. 2015;5:17. doi: 10.1038/srep08574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aziz MM, Palta JA, Siddique KHM, Sadras VO. Five decades of selection for yield reduced root length density and increased nitrogen uptake per unit root length in Australian wheat varieties. Plant Soil. 2017;413:181–192. doi: 10.1007/s11104-016-3059-y. [DOI] [Google Scholar]
- 49.Dorlodot S, et al. Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 2007;12(10):474–481. doi: 10.1016/j.tplants.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 50.Rana A, et al. Identification of multi-trait PGPR isolates and evaluating their potential as inoculants for wheat. Ann. Microbiol. 2011;61:893–900. doi: 10.1007/s13213-011-0211-z. [DOI] [Google Scholar]
- 51.Schlatter DC, Yin C, Hulbert S, Burke I, Paulitz T. Impacts of repeated glyphosate use on wheat-associated bacteria are small and depend on glyphosate use history. Appl. Environ. Microbiol. 2017;83:e01354–17. doi: 10.1128/AEM.01354-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kodama Y, Watanabe K. Rhizomicrobium electricum sp. nov., a facultatively anaerobic, fermentative, prosthecate bacterium isolated from a cellulose-fed microbial fuel cell. Int. J. Syst. Evol. Microbiol. 2011;61:1781–1785. doi: 10.1099/ijs.0.023580-0. [DOI] [PubMed] [Google Scholar]
- 53.Khan AL, Waqas M, Kang SM. Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 2014;52(8):689–695. doi: 10.1007/s12275-014-4002-7. [DOI] [PubMed] [Google Scholar]
- 54.Cardinale M, et al. Paradox of plant growth promotion potential of rhizobacteria and their actual promotion effect on growth of barley (Hordeum vulgare L.) under salt stress. Microbiol. Res. 2015;181:22–32. doi: 10.1016/j.micres.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Chimwamurombe PM, Grönemeyer JL, Reinhold-Hurek B. Isolation and characterization of culturable seed-associated bacterial endophytes from gnotobiotically grown Marama bean seedlings. FEMS Microbiol. Ecol. 2016;92(6):fiw083. doi: 10.1093/femsec/fiw083. [DOI] [PubMed] [Google Scholar]
- 56.Hayatsu M, Tago K, Saito M. Various players in the nitrogen cycle: Diversity and functions of the microorganisms involved in nitrification and denitrification. J. Soil Sci. Plant Nutr. 2008;54:33–45. doi: 10.1111/j.1747-0765.2007.00195.x. [DOI] [Google Scholar]
- 57.Bruto M, Prigent-Combaret C, Muller D, Moënne-Loccoz Y. Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci. Rep. 2014;4:6261. doi: 10.1038/srep06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pearce S, et al. Molecular characterization of Rht-1 dwarfing genes in hexaploidy wheat. Plant Physiol. 2011;157:1820–1831. doi: 10.1104/pp.111.183657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng J, et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- 60.Richards DE, King KE, Ait-Ali T, Harberd NP. How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signalling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:67–88. doi: 10.1146/annurev.arplant.52.1.67. [DOI] [PubMed] [Google Scholar]
- 61.Nett R, et al. Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution. Nat. Chem. Biol. 2017;13:69–74. doi: 10.1038/nchembio.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 63.Fonouni-Farde C, et al. DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat. Commun. 2016;7:12636. doi: 10.1038/ncomms12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foo E, Ross JJ, Jones WT, Reid JB. Plant hormones in arbuscular mycorrhizal symbioses: an emerging role for gibberellins. Ann. Bot. 2013;111:769–779. doi: 10.1093/aob/mct041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Egamberdieva D, Wirth SJ, Alqarawi AA, Abd_Allah EF, Hashem A. Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front. Microbiol. 2017;8:2104. doi: 10.3389/fmicb.2017.02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang YH, Irving HR. Developing a model of plant hormone interactions. Plant Signal Behav. 2011;6(4):494–500. doi: 10.4161/psb.6.4.14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iannucci A, Fragasso M, Beleggia R, Nigro F, Papa R. Evolution of the crop rhizosphere: impact of domestication on root exudates in tetraploid wheat (Triticum turgidum L.) Front. Plant Sci. 2017;8:2124. doi: 10.3389/fpls.2017.02124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sasse J, Martinoia E, Northen T. Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 2018;23(1):25–41. doi: 10.1016/j.tplants.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 69.Shi S, et al. The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages. Ecol. Lett. 2016;19(8):926–936. doi: 10.1111/ele.12630. [DOI] [PubMed] [Google Scholar]
- 70.Jiang Y, et al. Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol. Biochem. 2017;109:145–155. doi: 10.1016/j.soilbio.2017.02.010. [DOI] [Google Scholar]
- 71.Oberholster T, Vikram S, Cowan D, Valverde A. Key microbial taxa in the rhizosphere of sorghum and sunflower grown in crop rotation. Sci. Total Environ. 2018;624:530–539. doi: 10.1016/j.scitotenv.2017.12.170. [DOI] [PubMed] [Google Scholar]
- 72.Olesen J, Bascompte J, Dupont Y, Jordano P. The modularity of pollination networks. PNAS. 2007;104:19891–19896. doi: 10.1073/pnas.0706375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tao J, et al. Integrated network analysis reveals the importance of microbial interactions for maize growth. Appl. Microbiol. Biotechnol. 2018;102:3805–3818. doi: 10.1007/s00253-018-8837-4. [DOI] [PubMed] [Google Scholar]
- 74.Zhou J, Deng Y, Luo F, He Z, Yang Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio. 2011;2(4):e00122–11. doi: 10.1128/mBio.00122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guimerà R, Amaral LAN. Cartography of complex networks: modules and universal roles. J. Stat. Mech. 2005;P02001:P02001-1-P02001-13. doi: 10.1088/1742-5468/2005/02/P02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berry D, Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014;5(219):219. doi: 10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang H, et al. An integrated insight into the relationship between soil microbial community and tobacco bacterial wilt disease. Front. Microbiol. 2017;8:2179. doi: 10.3389/fmicb.2017.02179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saville RJ, et al. The ‘Green Revolution’ dwarfing genes play a role in disease resistance in Triticum aestivum and Hordeum vulgare. J. Exp. Bot. 2012;63(3):1271–1283. doi: 10.1093/jxb/err350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Srinivasachary Gosman N, et al. Semi-dwarfing Rht-B1 and Rht-D1 loci of wheat differ significantly in their influence on resistance to Fusarium head blight. Theor. Appl. Genet. 2009;118:695–702. doi: 10.1007/s00122-008-0930-0. [DOI] [PubMed] [Google Scholar]
- 80.Fudou R, et al. Haliangicin, a novel antifungal metabolite produced by a marine myxobacterium. Isolation and structural elucidation. J. Antibiot. 2001;54(2):153–156. doi: 10.7164/antibiotics.54.153. [DOI] [PubMed] [Google Scholar]
- 81.Harwani D. Myxobacteria as a promising source of novel natural products. IJRASET. 2017;5:2654–2660. [Google Scholar]
- 82.Gontia-Mishra I, Sapre S, Kachare S, Tiwari S. Molecular diversity of 1-aminocyclopropane-1-carboxylate (ACC) deaminase producing PGPR from wheat (Triticum aestivum L.) rhizosphere. Plant Soil. 2017;414:213–227. doi: 10.1007/s11104-016-3119-3. [DOI] [Google Scholar]
- 83.Landa BB, Mavrodi DM, Thomashow LS, Weller DM. Interactions between strains of 2, 4-diacetylphloroglucinol-producing Pseudomonas fluorescens in the rhizosphere of wheat. Phytopathology. 2003;93:982–994. doi: 10.1094/PHYTO.2003.93.8.982. [DOI] [PubMed] [Google Scholar]
- 84.de Boer W, Wagenaar A-M, Klein, Gunnewiek PJA, van Veen JA. In vitro suppression of fungi caused by combinations of apparently non-antagonistic soil bacteria. FEMS Microbiol. Ecol. 2007;59:177–185. doi: 10.1111/j.1574-6941.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- 85.Yin C, et al. Role of bacterial communities in the natural suppression of Rhizoctonia solani bare patch disease of wheat (Triticum aestivum L.) Appl. Environ. Microbiol. 2013;79(23):7428–7438. doi: 10.1128/AEM.01610-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lopes LD, Silva MDCP, Andreote FD. Bacterial abilities and adaptation toward the rhizosphere colonization. Front. Microbiol. 2016;7:1341. doi: 10.3389/fmicb.2016.01341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mattarozzi M, et al. A metaproteomic approach dissecting major bacterial functions in the rhizosphere of plants living in serpentine soil. Anal. Bioanal. Chem. 2017;409:2327–2339. doi: 10.1007/s00216-016-0175-8. [DOI] [PubMed] [Google Scholar]
- 88.Li M, et al. Indole-3-acetic acid biosynthesis pathways in the plant-beneficial bacterium Arthrobacter pascens ZZ21. Int. J. Mol. Sci. 2018;19:443. doi: 10.3390/ijms19020443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu Y, et al. Plant-microbe communication enhances auxin biosynthesis by a root-associated bacterium, Bacillus amyloliquefaciens SQR9. MPMI. 2016;29(4):324–330. doi: 10.1094/MPMI-10-15-0239-R. [DOI] [PubMed] [Google Scholar]
- 90.Poole P. Shining a light on the dark world of plant root–microbe interactions. PNAS. 2017;114(17):4281–4283. doi: 10.1073/pnas.1703800114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barahona E, et al. Pseudomonas fluorescens F113 can produce a second flagellar apparatus, which is important for plant root colonization. Front. Microbiol. 2016;7:1471. doi: 10.3389/fmicb.2016.01471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turnbull GA, Morgan AW, Whipps JM, Saunders JR. The role of bacterial motility in the survival and spread of Pseudomonas fluorescens in soil and in the attachment and colonisation of wheat roots. FEMS Microbiol. Ecol. 2001;36:21–31. doi: 10.1111/j.1574-6941.2001.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 93.Voogd, N.J., Cleary, D.F.R., Polonia, A.R.M. & Gomes, N.C.M. Bacterial community composition and predicted functional ecology of sponges, sediment and seawater from the thousand islands reef complex, West Java, Indonesia. FEMS Microbiol. Ecol. 91 (2015). [DOI] [PubMed]
- 94.Gopal M, Gupta A. Microbiome selection could spur next-generation plant breeding strategies. Front. Microbiol. 2016;7:1971. doi: 10.3389/fmicb.2016.01971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA gene amplicon data are available at the NCBI Sequence Read Archive (SRA) under accession number: PRJNA601112.