The clinically relevant pathogen Pseudomonas aeruginosa uses diverse organic compounds as electron donors and possesses multiple enzymes that transfer electrons from central metabolism to O2. These pathways support a balanced intracellular redox state and produce cellular energy. P. aeruginosa also reduces secondary metabolites called phenazines to promote redox homeostasis and virulence. In this study, we examined the reciprocal relationship between these primary and secondary routes of electron flow. We found that phenazines affect respiratory function and that the complement of phenazines produced is strongly affected by growth in assemblages called biofilms. These results provide a more nuanced understanding of P. aeruginosa redox metabolism and may inform strategies for treating persistent infections caused by this bacterium.
KEYWORDS: Pseudomonas aeruginosa, biofilms, electron transport chain, extracellular electron transfer, phenazines
ABSTRACT
Extracellular electron transfer (EET), the reduction of compounds that shuttle electrons to distal oxidants, can support bacterial survival when preferred oxidants are not directly accessible. EET has been shown to contribute to virulence in some pathogenic organisms and is required for current generation in mediator-based fuel cells. In several species, components of the electron transport chain (ETC) have been implicated in electron shuttle reduction, raising the question of how shuttling-based metabolism is integrated with primary routes of metabolic electron flow. The clinically relevant bacterium Pseudomonas aeruginosa can utilize carbon sources (i.e., electron donors) covering a broad range of reducing potentials and possesses a branched ETC that can be modulated to optimize respiratory efficiency. It also produces electron shuttles called phenazines that facilitate intracellular redox balancing, increasing the complexity of its metabolic potential. In this study, we investigated the reciprocal influence of respiratory metabolism and phenazine-associated physiology in P. aeruginosa PA14. We found that phenazine production affects respiratory activity and terminal oxidase gene expression and that carbon source identity influences the mechanisms enabling phenazine reduction. Furthermore, we found that growth in biofilms, a condition for which phenazine metabolism is critical to normal development and redox balancing, affects the composition of the P. aeruginosa phenazine pool. Together, these findings can aid interpretation of P. aeruginosa behavior during host infection and provide inroads to understanding the cross talk between primary metabolism and shuttling-based physiology in the diverse bacteria that carry out EET.
IMPORTANCE The clinically relevant pathogen Pseudomonas aeruginosa uses diverse organic compounds as electron donors and possesses multiple enzymes that transfer electrons from central metabolism to O2. These pathways support a balanced intracellular redox state and produce cellular energy. P. aeruginosa also reduces secondary metabolites called phenazines to promote redox homeostasis and virulence. In this study, we examined the reciprocal relationship between these primary and secondary routes of electron flow. We found that phenazines affect respiratory function and that the complement of phenazines produced is strongly affected by growth in assemblages called biofilms. These results provide a more nuanced understanding of P. aeruginosa redox metabolism and may inform strategies for treating persistent infections caused by this bacterium.
INTRODUCTION
Members of the domain Bacteria exhibit a vast variety of pathways and substrates that can be used to generate energy (1). This metabolic diversity allows bacteria to persist in conditions that do not support eukaryotes, which typically produce energy by redox transformations of oxygen (O2) and water (2). An additional advantage for many bacterial species is metabolic versatility, the ability to use multiple substrates and/or pathways for energy generation. Metabolic versatility provides resilience in heterogeneous or changing conditions and is an important feature of bacteria that form multicellular assemblages called biofilms.
Biofilms are clusters of microbes that adhere to each other by producing complex, polysaccharide-based matrices. In these structures, the combined effects of diffusion and metabolism generate resource gradients and unique conditions that differ from those found in historically popular, well-mixed liquid cultures. Importantly, biofilm-specific conditions can induce distinct physiologies that contribute to stress and antibiotic resistance, which in turn have important implications for virulence (3, 4). We seek to understand how redox metabolisms are integrated to support multicellular growth in the bacterium Pseudomonas aeruginosa, a common cause of chronic, biofilm-based infections.
P. aeruginosa uses organic compounds as carbon and energy sources. It can generate ATP by fermenting these compounds or by coupling their oxidation to generation of a proton motive force that powers ATP synthase (i.e., respiration). Results from our work, using a colony morphology model to study P. aeruginosa biofilm metabolism and development, support the model that electron flow to two major oxidants supports energy generation and redox homeostasis in biofilms: (i) O2 and (ii) phenazines, redox-active metabolites that can mediate extracellular electron transfer (EET) to oxidants available outside the cell and at a distance (Fig. 1A) (5, 6). Genetic analyses and microelectrode profiling studies indicate that P. aeruginosa’s ability to reduce O2 and phenazines depends on the composition of its electron transport (i.e., respiratory) chain (ETC) (7).
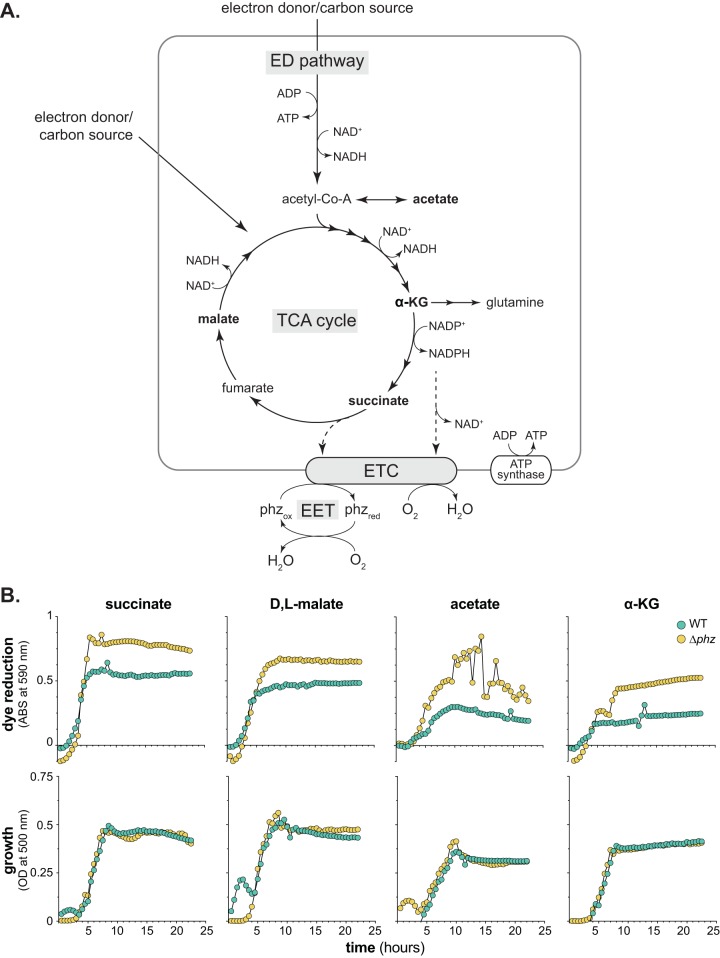
FIG 1.
Phenazines influence tetrazolium dye reduction. (A) Simplified representation of major redox pathways operating in P. aeruginosa. ED, Entner-Doudoroff; TCA, tricarboxylic acid; ETC, electron transport chain; α-KG, α-ketoglutarate. Electron transfer through the ETC can be coupled to the generation of a proton motive force that powers the ATP synthase. ATP is also generated by the ED pathway. Electrons can originate from diverse carbon sources that can enter the ED pathway or TCA cycle. Phenazine-mediated extracellular electron transfer (EET) is depicted as phenazines receiving electrons from the ETC and transferring them to an extracellular electron acceptor such as oxygen (O2) (phzred and phzox denote reduced and oxidized phenazines, respectively). Carbon sources identified in panel B are in bold. (B) Dye reduction (top) and growth (bottom) of WT PA14 and the phenazine-null mutant (Δphz) on succinate, d,l-malate, acetate, and α-KG. Curves show data from one replicate, which is representative of data from three biological replicates; see Fig. S3 in the supplemental material for the full data set.
In contrast to the mitochondrial ETC, which has one route for electrons to be delivered to O2, bacterial respiratory chains often have multiple routes (8). Electrons fed into P. aeruginosa’s ETC are ultimately transferred to one of five canonical terminal oxidases, the enzymes that catalyze the final electron transfer step to reduce O2 to water (9, 10). Each of these terminal oxidases possesses unique characteristics, including different expression patterns and affinities for O2 (11–14). Four terminal oxidases, Cox, Cyo, Cco1, and Cco2 (or, alternatively, aa3, bo3, cbb3-1, and cbb3-2, respectively) are members of the heme-copper oxidase superfamily; Cio is the only copperless terminal oxidase, rendering it resistant to cyanide, which is endogenously produced by P. aeruginosa at high cell densities (15, 16). Cox, Cco1, and Cco2 are cytochrome c oxidases, while Cyo and Cio are quinol oxidases. The Ccos have high affinities for O2 and have been shown to be the main terminal oxidases contributing to P. aeruginosa growth under laboratory conditions (14). They are sufficient for wild-type (WT) colony biofilm morphogenesis and phenazine reduction in biofilms grown on a complex medium (7). Furthermore, the Ccos, together with Cio, support microaerobic growth of this bacterium (10, 12). The loci encoding Cox and Cyo, on the other hand, are upregulated under conditions of nutrient starvation (13). The stationary-phase sigma factor RpoS induces expression of cox, while the transcription factor Anr, which regulates gene expression during the transition from aerobic to anaerobic growth (17), induces cco2 expression (11, 13). The two-component response regulator RoxSR also modulates the expression of all five P. aeruginosa terminal oxidases, although the exact mechanisms through which this system regulates their expression remain largely unelucidated (13, 18).
Though metabolism is often conceptualized as a modular process in which the oxidation of individual carbon sources can be paired with the respiration of electron acceptors via common, central pathways, this is an oversimplification. Different carbon sources (i) offer various degrees of reducing power, (ii) are routed through specific pathways that will differ in their yields of direct electron donors to the ETC, and (iii) can directly or indirectly act as regulatory cues affecting the expression or activity of ETC components (19, 20). Furthermore, changes in electron flow through the ETC can affect the expression of terminal oxidase genes (21). Here, we examined the physiological impacts of phenazine production and ETC composition by surveying their relevance for respiratory activity during the use of diverse carbon sources in P. aeruginosa. We tested whether carbon source identity affects the ability to grow with specific respiratory chain components and the ability to produce and reduce specific phenazines during P. aeruginosa biofilm growth. Our results indicate that the identity of the provided carbon source has profound effects on PA14 growth and the mechanisms enabling phenazine production and utilization. Furthermore, our data suggest that phenazines themselves can influence the expression of terminal oxidases within a biofilm.
RESULTS AND DISCUSSION
Phenazine production and ETC composition influence tetrazolium dye reduction in P. aeruginosa.
To assess how phenazine production affects P. aeruginosa PA14 respiration, we measured respiratory activity using commercially available “phenotype microarray” plates, which contain a different carbon source in each well. The manufacturer-provided growth medium for these plates contains a tetrazolium dye that is reduced by the electron transport chain (ETC) and thereby undergoes an irreversible color change (22, 23). First, we used phenotype microarray plates to compare tetrazolium dye reduction by wild-type (i.e., phenazine-producing) PA14 (WT) and the phenazine-null Δphz mutant. We found that on four carbon sources (succinate, d,l-malate, acetate, and α-ketoglutarate [α-KG]), phenazine production inhibited tetrazolium dye reduction (Fig. 1B). On all other carbon sources, respiratory activity was similar between the WT and Δphz strains (Fig. 2B). During incubation in defined, morpholinepropanesulfonic acid (MOPS)-buffered media containing each of the 95 “phenotype microarray” carbon sources, 32 supported growth (see File S1 in the supplemental material), and the WT and Δphz strains grew similarly on all of these carbon sources (see Fig. S1 in the supplemental material). These results suggest that during growth on succinate, d,l-malate, acetate, and α-KG, electrons that would have been transferred to tetrazolium dye are instead transferred to phenazines. (Although some synthetic phenazines can act as mediators to tetrazolium dyes in vitro [24], we did not observe phenazine-enhanced dye reduction in this experiment.) All four of the carbon sources that gave rise to phenazine-dependent inhibition of dye reduction are intermediates in or branchpoints of the tricarboxylic acid (TCA) cycle (Fig. 1A), a major source of reducing power for respiration. α-KG in particular is notable for its role as a precursor for glutamine and phenazine syntheses (25, 26).
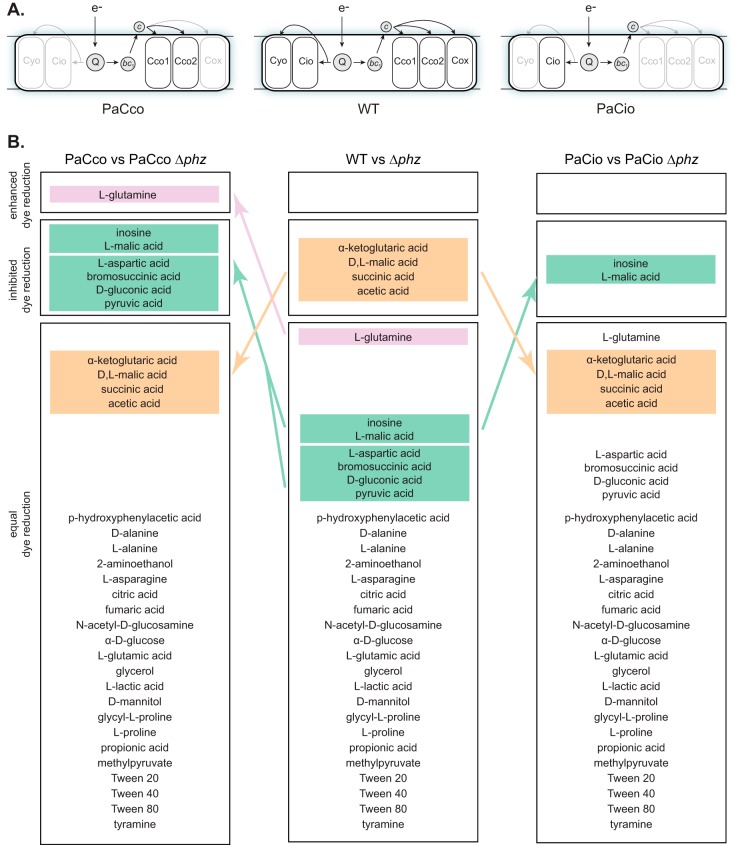
FIG 2.
ETC composition influences tetrazolium dye reduction. (A) Depiction of the terminal oxidase complexes present in the WT (center) and the strains PaCco (left) and PaCio (right). Electrons enter the ETC via primary dehydrogenases (not shown) and are transferred to the quinone pool (Q). Reduced Q can act as a substrate for the quinol oxidases (Cyo and Cio) or the cytochrome bc1 complex (bc1). A cytochrome c protein mediates electron transfer between bc1 and the terminal oxidases Cco1, Cco2, and Cox. (B) The 32 carbon sources that support PA14 growth, grouped based on tetrazolium dye reduction patterns of each phenazine-producing and phenazine-null strain. Groups were designated based on data from three experiments. Carbon sources highlighted in colored boxes are those on which dye reduction patterns changed relative to the strains with the full complement of terminal oxidases (i.e., WT and Δphz, the center column). Carbon source descriptors are as provided by the manufacturer.
In a previous study, we found that electron transfer to phenazines depended on the composition of the ETC during growth in biofilms (7). We therefore suspected that the effects of phenazines on respiratory activity would also depend on the composition of the ETC. We generated strains that contained only the two Ccos (“PaCco,” Δcox Δcyo Δcio) or Cio (“PaCio,” Δcox Δcyo Δcco1 cco2) (Fig. 2A), because prior work has suggested that these terminal oxidases make the greatest contributions to aerobic and microaerobic growth in P. aeruginosa under laboratory conditions (10, 12). We then measured the respiratory activity and growth of these strains on each of the “phenotype microarray” carbon sources. While strains with all terminal oxidases (WT and Δphz) and those with just the Ccos (PaCco and PaCco Δphz) exhibited similar growth kinetics, the growth of PaCio strains was generally abrogated (File S1; representative growth curves are shown in Fig. S2 in the supplemental material), in line with previous findings that mutants lacking functional Cco terminal oxidases have growth defects (12, 14).
To test whether ETC composition influences the effects of phenazines on respiratory activity, we conducted phenotype microarray experiments for the PaCco and PaCio strains in phenazine-producing and phenazine-null backgrounds. We compared each phenazine-null mutant to its parent strain and categorized each carbon source depending on whether it enhanced tetrazolium dye reduction, inhibited dye reduction, or had no effect (Fig. 2B; see File S2 in the supplemental material for raw data). During growth on most carbon sources, phenazine production did not affect respiratory activity, regardless of ETC composition (Fig. 2B). However, we were intrigued to find that the phenazine-dependent inhibition of dye reduction, seen during growth on α-KG, d,l-malate, succinate, and acetate (Fig. 1B), was not observed in PaCco and PaCio, indicating that the full complement of terminal oxidases is required for this effect. Interestingly, deletion of all non-Cco terminal oxidases revealed just one condition, growth on l-glutamine, in which phenazine production enhanced dye reduction. Characterization of the PaCco and PaCio strains also revealed several new carbon sources for which phenazines inhibited dye reduction. Together, these results suggest that phenazine production differentially affects tetrazolium dye reduction and, by extension, electron flow through the ETC, depending on which terminal oxidases are present.
Phenazine production affects terminal oxidase gene expression.
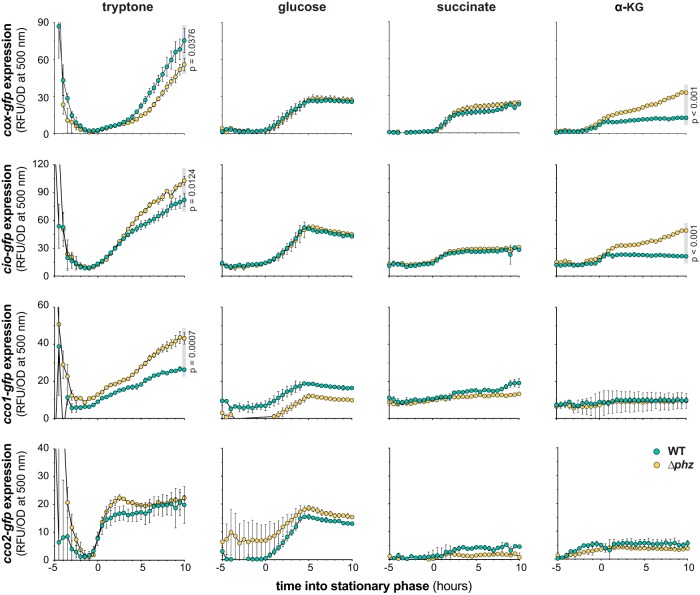
Previous studies have shown that phenazines oxidize the cellular redox state (5, 27) and that this property affects the activities of regulators that control terminal oxidase expression (11, 13, 18, 28, 29). To further characterize the effects of phenazines on the ETC, we created reporter strains corresponding to each of the loci encoding the five major P. aeruginosa terminal oxidases. We measured relative expression levels in liquid cultures and biofilms grown on the carbon sources succinate, α-KG, glucose, and tryptone. Succinate and α-KG constitute two of the four carbon sources that showed phenazine-dependent inhibition of tetrazolium dye reduction in the presence of all terminal oxidases (Fig. 1B). Glucose and succinate are popular carbon sources for laboratory media and represent P. aeruginosa’s two main pathways for carbon source oxidation, the Entner-Doudoroff pathway and the TCA cycle, respectively (Fig. 1A). Tryptone, an undefined protein digest, is also a standard carbon source used to grow P. aeruginosa in the laboratory and is of interest because it represents a complex condition that may be relevant for P. aeruginosa growth in natural settings (30). We found that the WT and Δphz strains showed similar growth kinetics and yields on all of these carbon sources (see Fig. S4 in the supplemental material). We observed a longer lag phase on glucose than on tryptone, succinate, and α-KG, reflecting P. aeruginosa’s preference for TCA cycle intermediates over sugars (31).
Expression of the P. aeruginosa terminal oxidases Cox, Cio, Cco1, and Cco2 during growth in liquid cultures and biofilms is shown in Fig. 3 and 4, respectively. (We detected expression of P. aeruginosa’s fifth major terminal oxidase, Cyo, only in late stationary phase during planktonic growth on tryptone and at low or negligible levels under all other conditions examined.) For reporter strains grown in liquid cultures, we identified four combinations of reporters and conditions that showed higher expression in late stationary phase in the Δphz mutant than in the phenazine-producing background: the Cio and Cco1 reporter strains grown on tryptone and the Cox and Cio reporter strains grown on α-KG. We have previously reported that respiratory chain components involved in nitrate respiration are induced by defects in phenazine production (32), and our observation that the Cox, Cio, and Cco1 terminal oxidases are induced in the Δphz background under some conditions is consistent with this theme. Enhancement of terminal oxidase expression by phenazine deficiency is supported by the notion that terminal oxidases are regulated in response to cellular redox conditions and that phenazines constitute an alternate method of redox balancing when aerobic respiratory activity is limited (5, 33, 34). The two-component response regulator RoxSR, which is believed to sense the redox state of the cell by detecting electron flow through the ETC, induces expression of all terminal oxidase gene clusters except that of cox, which it represses (13, 18, 28). The observation that cox-gfp was the only reporter for which higher expression was seen in the phenazine-producing background during growth in tryptone may be a consequence of its unique regulation by RoxSR and the stationary-phase sigma factor RpoS (13, 28).
FIG 3.
Electron donor and phenazines affect terminal oxidase expression in liquid culture. Mean fluorescence (RFU) of reporter strains engineered to express GFP under the control of the cox, cio, cco1, or cco2 promoter during liquid culture growth on the indicated carbon sources is shown. Mean fluorescence values were corrected for cell density (OD at 500 nm), and the fluorescence values of a strain expressing GFP without a promoter (the MCS control) were subtracted from each data point. Onset of stationary phase (time = 0 h) was determined for each strain from the respective growth curves (see Fig. S4 in the supplemental material), and fluorescent reporter expression data are plotted from 5 h prior to onset of stationary phase to 10 h into stationary phase. Gray boxes indicate the time points for which P values were determined using unpaired, two-tailed t tests. Data represent the mean from three biological replicates; error bars denote standard deviation and are not drawn in instances where they would be obscured by point markers.
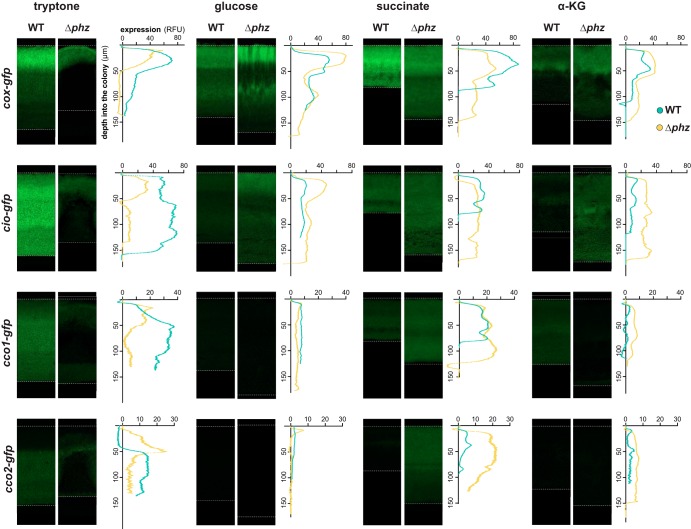
FIG 4.
Electron donor and phenazines affect terminal oxidase expression in colony biofilms. Representative images show expression of GFP under the control of the cox, cio, cco1, or cco2 promoter in thin sections prepared from biofilms grown for 3 days on the indicated carbon sources. Reporter fluorescence is shown in green and is overlain on the DIC image of each colony biofilm. Dotted lines indicate the air-biofilm (top) or agar-biofilm (bottom) interfaces. Graphs show fluorescence values (RFU) relative to biofilm depth of the corresponding images. Fluorescence values of the MCS control (a strain expressing GFP without a promoter) in either the WT or Δphz background have been subtracted from each plot. The y axis for each graph provides scale for the respective image, which is representative of at least six biological replicates.
Biofilm growth promotes the formation of resource gradients, which in turn leads to a spatial heterogeneity that is not observed in liquid culture. To elucidate how this gradient-induced heterogeneity might influence electron flow toward a particular respiratory branch and/or terminal oxidase, we quantified spatially resolved gene expression patterns of terminal oxidases in biofilms grown on tryptone, glucose, succinate, and α-KG (Fig. 4). Vertical sectioning of biofilms enabled visualization of changes in gene expression across depth. We identified several loci and conditions for which expression was generally enhanced in the absence of phenazines (Fig. 4), again consistent with complementary roles of the terminal oxidases and phenazines in redox balancing. However, this effect was limited to biofilms grown on defined media containing distinct carbon sources. For example, cio-gfp was more expressed in the phenazine-null mutant when biofilms were grown on α-KG (Fig. 4). This is similar to what we observed for this reporter during the stationary phase of planktonic growth (Fig. 3), a growth phase that shares some characteristics, such as high cell density and nutrient limitation, with the biofilm mode of growth. In contrast, biofilms grown on tryptone consistently showed higher levels of terminal oxidase gene expression in the phenazine-producing background than in the phenazine-null background. This may be because of enhanced matrix production in tryptone-grown Δphz biofilms that leads to the formation of thinner biofilms with vertical structures, called “wrinkles,” which increase the overall surface area and therefore cellular access to O2 (5, 35, 36). We previously attributed the thickness of WT biofilms to the physiological benefit of phenazine production (5, 7). Interestingly, in this study we found that WT biofilms grown on a defined medium containing glucose, succinate, or α-KG were thinner than Δphz biofilms and that biofilm thickness showed overall variation between conditions (see Fig. S5 in the supplemental material). Furthermore, only growth on tryptone led to the formation of significant wrinkles in the Δphz mutant by day three of biofilm development, and Δphz biofilms grown on glucose, succinate, or α-KG remained relatively featureless. The metabolic pathways that function during growth on distinct carbon sources may differentially affect redox homeostasis and the response to O2 limitation. Glucose, for example, can be oxidized through respiration but can also be fermented to lactate, whereas succinate and α-KG are not fermentable carbon sources. Finally, we note that the phenazine pyocyanin has been shown to have both beneficial and toxic effects that can influence biofilm development in a condition-dependent manner (37).
Phenazine derivatization is highly dependent on carbon source identity.
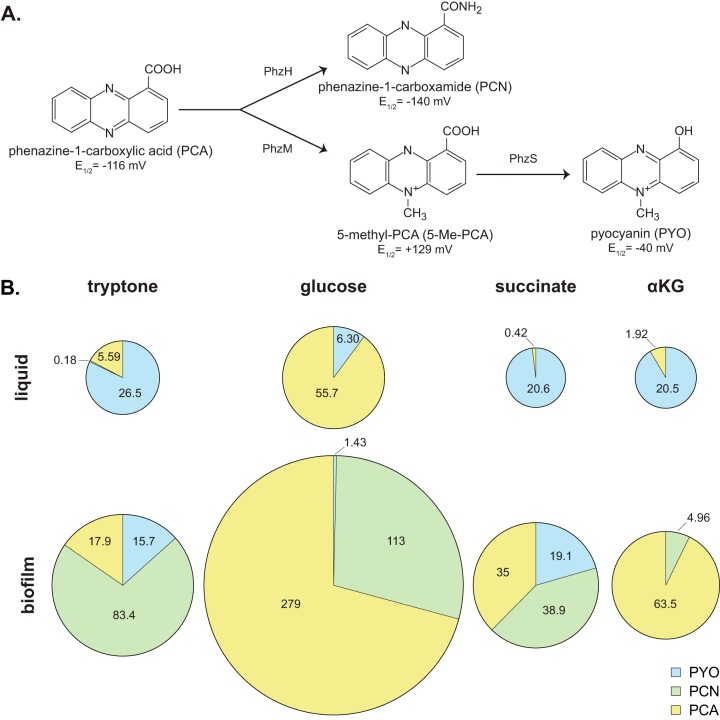
The data presented thus far suggest that PA14’s branched ETC is modulated in response to electron donor identity and the production of phenazines. PA14 makes at least four phenazines (Fig. 5A), each having a unique redox potential (38, 39). We therefore next asked if the composition of the phenazine pool changes depending on the carbon source. We grew PA14 in liquid cultures and as biofilms on tryptone, glucose, succinate, and α-KG and quantified phenazine production using high-performance liquid chromatography (HPLC).
FIG 5.
Phenazine production is affected by carbon source. (A) The precursor phenazine phenazine-1-carboxylic acid (PCA) can be modified by PhzH to produce phenazine-1-carboxamide (PCN) and/or by PhzM to produce 5-methyl-PCA (5-Me-PCA). 5-Me-PCA can be further modified by PhzS to yield pyocyanin (PYO). The redox potential of each phenazine is indicated (E1/2 versus a normal hydrogen electrode at pH 7) (39, 44). (B) Phenazines produced by WT PA14 in liquid cultures (top) and biofilms (bottom) grown on the indicated carbon sources. The area of each pie represents the combined concentration of all measured phenazines, and each “slice” represents the mean concentration in micromolar (listed for each phenazine produced alongside the respective “slice”) of PCA, PCN, or PYO. Mean values are representative of data from at least six biological replicates. For numerical data, including error and total n, see Fig. S6 in the supplemental material.
The precursor phenazine, phenazine-1-carboxylic acid (PCA), can be derivatized to phenazine-1-carboxamide (PCN), 5-methyl-PCA (5-Me-PCA), or pyocyanin (PYO) (Fig. 5A). Production and derivatization of phenazines are known to be regulated by quorum sensing and fine-tuned by diverse environmental factors (40), but the precise mechanisms of regulation remain to be elucidated. We focused our analysis on the production of PCA, PCN, and PYO because 5-Me-PCA is highly reactive and unstable (41) and therefore difficult to detect by HPLC. Notably, while we found that the biofilm growth conditions favored the production of PCN, this phenazine was not detectable in samples from liquid cultures (Fig. 5B, bottom panel versus top panel; see Fig. S6 in the supplemental material). In contrast, while PYO generally contributed the most to the phenazine complement in liquid culture, it was the least-produced phenazine in biofilms. The extent of PCA derivatization, primarily to PYO (liquid culture) or PCN (biofilm), was dependent on the type of carbon source (Fig. 5B). These dynamics in phenazine production indicate that the carbon source influences the kinds and amount of phenazines produced by PA14 and highlight the plasticity of phenazine derivatization in response to environmental conditions.
Mechanisms of phenazine reduction in biofilms differ depending on carbon source.
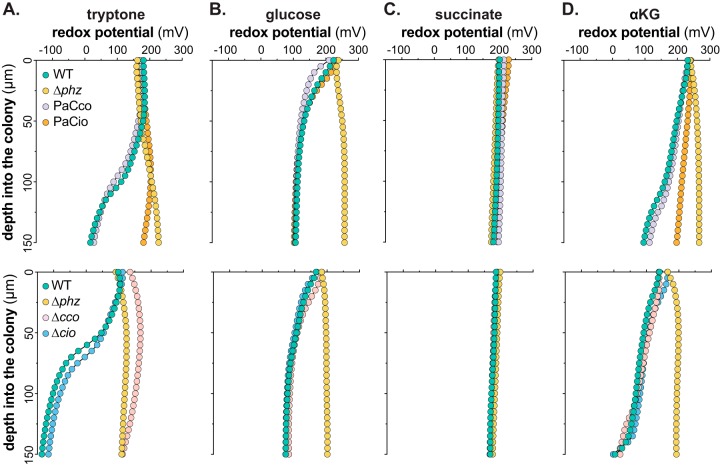
Our observations regarding the condition-dependent effects of phenazines on respiratory activity and terminal oxidase gene expression (Fig. 1B to 4) and the high condition dependence of phenazine derivatization (Fig. 5) raise the question of how carbon source identity influences phenazine utilization. We have previously shown that phenazine reduction in tryptone-grown biofilms is detectable as a gradient of an increasingly reduced extracellular redox state approaching the biofilm base and that it requires a Cco terminal oxidase (7). For this study, we used redox microelectrodes to measure phenazine reduction in biofilms grown on defined media containing our carbon sources of interest (Fig. 6). We note that the redox profiles do not indicate where the microelectrode transitions from the biofilm to the underlying growth medium. We therefore measured biofilm thicknesses based on the thin sections presented in Fig. 4 (Fig. S5) and focused our analysis on the part of the redox profiles that correspond to the biofilms rather than the growth media. Phenazine reduction was detectable during growth on glucose (Fig. 6B) and α-KG (Fig. 6D) but with reduction profiles different from those observed on tryptone (Fig. 6A): on glucose, phenazines were reduced in the topmost, aerobic zone of the biofilm, while on α-KG, reduction was gradual across biofilm depth. Therefore, carbon source identity also affects where and to what degree a particular phenazine complement is utilized within WT biofilms (Fig. 6, green traces).
FIG 6.
Carbon source alters reduction of endogenously produced phenazines. Changes in redox potential across depth for WT, Δphz, and terminal oxidase mutant colony biofilms grown for 3 days on tryptone (A), glucose (B), succinate (C), and α-KG (D) are shown. The top panels show redox profiles for the WT, Δphz, PaCco, and PaCio strains, while the bottom panels show redox profiles for the WT, Δphz, Δcco1 cco2 (Δcco), and Δcio strains. Data are representative of at least six biological replicates.
In contrast to those grown on tryptone, glucose, or α-KG, we did not detect any phenazine reduction in biofilms grown on succinate (Fig. 6C), which may be attributed to potential phenazine toxicity on this carbon source. Biofilms grown on succinate are noticeably thin, and we have previously correlated this phenotype with decreased fitness in biofilms (7). This phenotype is rescued upon deletion of phenazines (Fig. 4), indicating that phenazines may have adverse effects on biofilms grown on succinate. Thus, we were surprised to find that WT biofilms grown on succinate produced appreciable amounts of all three detectable phenazines (Fig. 5B) but that none of these seemed to be reduced. This suggests that in succinate-grown biofilms, phenazines take on functions outside redox balancing (38). The carbon-nitrogen (C/N) ratio is an important intracellular cue for metabolism, with nitrogen limitation leading to blockage of glucose uptake (42). Low glutamine levels indicate nitrogen limitation, which manifests as a buildup of α-KG, the precursor for glutamine synthesis (Fig. 1A). Because the syntheses of PCA and PCN require an investment of two and three glutamine molecules, respectively, it is possible that these phenazines serve as nitrogen sinks, functioning to balance the intracellular C/N ratio during limited access to nitrogen and thereby to foster metabolic activity.
To test the contributions of specific terminal oxidases to phenazine utilization on different carbon sources, we next measured redox profiles for strains that can express only Ccos (PaCco) or Cio (PaCio) and strains with deletions of the respective genes, i.e., Δcco and Δcio strains. Because redox measurements taken at different times can exhibit some variation due to fluctuations in the ambient environment, on each graph we have shown WT and Δphz controls that were measured at the same time as the presented terminal oxidase mutant strains. On tryptone, Cco terminal oxidases are both sufficient (Fig. 6A, top panel) and necessary (Fig. 6A, bottom panel) for phenazine reduction, which is consistent with our observation that Cio is neither sufficient nor necessary for phenazine reduction on tryptone (Fig. 6A). While we cannot directly equate expression with activity, we note that the expression patterns of the Cco terminal oxidases on tryptone (Fig. 4) coincide with the portion of the biofilm with the most phenazine reduction (Fig. 6A).
Phenazine reduction on glucose was comparable in PaCco and WT biofilms (Fig. 6B, top panel). However, unlike in biofilms grown on tryptone, Ccos were not necessary for phenazine reduction on glucose, a result confirmed by WT-like phenazine reduction in PaCio (Fig. 6B, top panel) as well as in the Δcco strain (Fig. 6B, bottom panel). Because of the interconnectivity and built-in redundancy of the P. aeruginosa ETC, we cannot rule out the possibility that deletion of specific terminal oxidases alters expression patterns of respiratory chain components. However, these redox profiles are consistent with our finding that cco1-gfp and cco2-gfp expression levels were lower in WT biofilms grown on glucose than in those grown on tryptone (Fig. 4). Similarly, Cio was sufficient (Fig. 6B, top panel) but not necessary (Fig. 6B, bottom panel) for phenazine reduction on glucose. Because electron flow to the quinol and cytochrome c oxidases branches off at the quinone pool, these results suggest that phenazine reduction occurs at or upstream of the quinone pool on glucose.
On α-KG, we also observed that the Ccos were sufficient, but not necessary, for phenazine reduction (Fig. 6D). In contrast to the case with glucose, however, PaCio exhibited a phenazine reduction defect relative to the WT and PaCco (Fig. 6D, top panel). This defect cannot be fully attributed to the lack of Ccos, as the Δcco strain shows WT-like phenazine reduction on this carbon source (Fig. 6D, bottom panel). It is therefore possible that in biofilms grown on α-KG, phenazine reduction relies upon electron flow through the bc1 complex, which is dependent upon the presence of a cytochrome c oxidase or upon an ETC-independent mechanism(s) (34).
Because PA14 synthesizes multiple phenazines that have distinct chemical properties (e.g., redox potentials) and may be reduced by distinct mechanisms, we created a “clean” strain background that would allow us to measure the reduction profiles of each phenazine in isolation for a more in-depth characterization of phenazine utilization. To this end, we generated a mutant, referred to as “ΔHMS Δphz,” that is unable to produce or modify any of PA14’s known phenazines. We grew this strain on agar plates each containing one pure phenazine compound and measured its reduction (see Fig. S7 in the supplemental material). Due to the aforementioned instability of 5-Me-PCA, we used its synthetic analog phenazine methosulfate (PMS) as a proxy to study 5-Me-PCA reduction (43). Like 5-Me-PCA, PMS is hydrophilic and has a significantly higher redox potential than PCA, PCN, and PYO (Fig. 5A) (39, 44). Previous work has shown that 5-Me-PCA is required for WT colony biofilm morphogenesis (43) and that the addition of PMS restores the Δphz redox profile to that of the WT on tryptone (7).
We observed that on tryptone, only the methylated phenazines PMS and PYO were reduced within the biofilm (Fig. S7A). Although on tryptone, biofilms predominantly produced PCN (Fig. 5B), we did not detect significant PCN reduction when it was added exogenously to tryptone-grown ΔHMS Δphz biofilms (Fig. S7A). In contrast, on glucose, PCA and PCN showed the most substantial redox profiles (Fig. S7B), which was consistent with them being the prevalent phenazines made under those conditions (Fig. 5B). These findings support the notion that a phenazine like PCN may contribute differently to biofilm physiology, depending on growth conditions: on glucose, PCN may mediate extracellular electron shuttling (EET), while on tryptone, it may take on a different function.
Although we did not observe phenazine reduction in biofilms grown on succinate (Fig. 6C), we detected reduction of exogenously provided PCN and PYO on this carbon source (Fig. S7C). This discrepancy could be attributed to an effect of phenazine biosynthesis itself on preferred pathways of carbon source utilization and/or cellular redox state and to the fact that PA14 responds differently to phenazines provided exogenously, which come without the “cost” of production. Alternatively, different mechanisms of phenazine reduction may operate more readily when phenazines are produced in the cytoplasm.
Together, the results of this study show the adaptability and interdependence of electron transfer schemes that can operate in P. aeruginosa. Our surveys of respiratory activity, phenazine production, and phenazine reduction under distinct conditions of electron donor (carbon source) and electron acceptor availability reveal unexpected influences of these parameters on the details of electron flow. While we focused our analysis on the reciprocal relationships between carbon source identity and utilization of phenazines or specific terminal oxidases, the data produced by this study enable other comparisons of fundamental physiological features of P. aeruginosa grown under distinct conditions. We therefore hope that the observations presented here will prove informative for the interpretation of P. aeruginosa behavior in diverse contexts.
Concluding remarks.
The opportunistic pathogen P. aeruginosa is prevalent in health care settings, immunocompromised individuals, and people with the inherited disease cystic fibrosis (CF). P. aeruginosa is the leading cause of chronic pulmonary infections and morbidity and mortality in patients with CF (3, 45). It exhibits metabolic versatility in its use of diverse carbon sources (i.e., electron donors), its branched aerobic respiratory chain, its capacity for nitrate respiration, and its ability to ferment arginine and pyruvate (9). Studies examining P. aeruginosa evolution during CF lung infection have identified mutations associated with changes in nutrient uptake and metabolism (46, 47). Furthermore, ETC genes, including those encoding dehydrogenases and the Cco terminal oxidases, are upregulated under conditions that mimic those found in the CF lung (48). Though P. aeruginosa’s metabolic capacities are thought to contribute to its success as a pathogen, an understanding of the condition dependence and integration of its primary redox pathways is key to interpreting its behavior in hosts.
Beyond its canonical metabolic pathways, a further distinctive feature of P. aeruginosa is its production of phenazines, small molecules that can carry out EET. This property further enhances P. aeruginosa’s metabolic versatility because it provides a means to balance the intracellular redox state and generate some ATP when resources such as O2 and nitrate are not accessible (34). P. aeruginosa in O2-limited environments, such as hypoxic biofilm subzones, can reduce phenazines that then diffuse and transfer electrons to O2 or other oxidants available at a distance. Mechanisms supporting EET have been described for diverse bacterial species, including Shewanella oneidensis, Listeria monocytogenes, and Enterococcus faecalis, and it has been suggested that many more organisms have the potential to carry out this type of metabolism (49–52). EET therefore constitutes a mechanism whereby excreted metabolites can “fine-tune” primary pathways of electron flow and energy generation and thereby promote survival.
Several of our previous studies have highlighted the interdependence of primary metabolism and phenazine-associated physiology (5, 7, 32, 53). In this work, we assessed the effects of phenazine production on respiratory activity and terminal oxidase expression, the effect of growth conditions on phenazine production, and the roles of the Cco terminal oxidases in endogenous and exogenous phenazine utilization. We found that the “realms” of P. aeruginosa physiology concerning O2 and phenazine metabolism strongly influence each other and that these effects can differ dramatically depending on carbon source (i.e., electron donor) identity. These findings underscore the considerable effect of the electron donor(s) on downstream redox balancing mechanisms of P. aeruginosa. This work adds to our growing understanding of the different mechanisms that govern P. aeruginosa metabolism, which can be modulated precisely to allow for maximal growth and survival under a variety of conditions, including those found within a host.
MATERIALS AND METHODS
Strains and growth conditions.
Pseudomonas aeruginosa strain UCBPP-PA14 (54, 55) precultures were routinely grown for 12 to 16 h in 2 ml of lysogeny broth (LB) (56) in 13- by 100-mm borosilicate glass tubes at 37°C with shaking at 250 rpm. Precultures serving as biological replicates were inoculated from separate clonal source colonies grown on streaked LB–1.5% agar plates. Liquid subcultures were made from diluting precultures 1:100 (1:50 for PaCio, PaCio Δphz , Δcco, and ΔHMS Δphz Δcco) in 5 ml of LB and growing in 18- by 150-mm borosilicate glass tubes at 37°C with shaking at 250 rpm until mid-exponential phase (optical density [OD] at 500 nm of ∼0.5). Liquid subcultures were used for experiments unless otherwise noted. The strains used in this study are listed in Table 1.
TABLE 1.
Strains used in this study
| Species and strain | Strain no. | Description | Reference or source |
|---|---|---|---|
| Pseudomonas aeruginosa | |||
| UCBPP-PA14 | Clinical isolate | 54 | |
| PA14 Δphz | LD24 | PA14 with deletions in phzA1-G1 and phzA2-G2 operons | 60 |
| PA14 Δcco1 cco2 (∆cco) | LD1933 | PA14 with both cco operons cco1 (cbb3-1) and cco2 (cbb3-2) (PA14_44340-PA14_44400) deleted simultaneously | 7 |
| PA14 ∆cio | LD2076 | PA14 with the cioAB (PA14_13030-PA14_13040) operon deleted | This study |
| PA14 PaCco | LD2587 | PA14 with deletions in PA14_01290-01320 (cox/aa3 operon), PA14_47150-47210 (cyo/bo3 operon), and PA14_13030-13040 (cio operon) | 7 |
| PA14 PaCco Δphz | LD2588 | PA14 with only the Cco terminal oxidases remaining in the phenazine-null background; deletions in the phzA1-G1, phzA2-G2, cox/aa3, cyo/bo3, and cio operons | This study |
| PA14 PaCio | LD1989 | PA14 with only the Cio terminal oxidase remaining; deletions in the cox/aa3, cyo/bo3, cco1, and cco2 operons | This study |
| PA14 PaCio Δphz | LD1990 | PA14 with only the Cio terminal oxidase remaining in the phenazine-null background; deletions in the phzA1-G1, phzA2-G2, cox/aa3, cyo/bo3, cco1, and cco2 operons | This study |
| PA14 ΔphzH ΔphzM ΔphzS | LD3122 | PA14 with deletions in the phenazine biosynthetic genes phzH (PA14_00640), phzM (PA14_09490), phzS (PA14_09400), phzA1-G1, and phzA2-G2; also referred to as ΔHMS Δphz | This study |
| ΔphzA1-G1 | |||
| ΔphzA2-G2 | |||
| PA14 ΔphzH ΔphzM ΔphzS | LD3140 | PA14 with deletions in the phenazine biosynthetic genes phzH, phzM, phzS, phzA1-G1, and phzA2-G2 and the terminal oxidase operons of cco1 and cco2; made by mating LD1933 to LD3122; also referred to as ΔHMS Δphz Δcco | This study |
| ΔphzA1-G1 | |||
| ΔphzA2-G2 Δcco | |||
| PA14 attB::MCS-gfp | LD2820 | PA14 with MCS-gfp inserted at the attB site using pLD2722 | 29 |
| PA14 Δphz attB::MCS-gfp | LD2821 | PA14 Δphz with MCS-gfp inserted at the attB site using pLD2722 | 29 |
| PA14 attB::Pcox-gfp | LD2962 | PA14 with promoter of the cox/aa3 operon driving gfp expression inserted at the attB site using pLD2958 | This study |
| PA14 attB::Pcio-gfp | LD2969 | PA14 with promoter of the cio operon driving gfp expression inserted at the attB site using pLD2960 | This study |
| PA14 attB::Pcco-1-gfp | LD2784 | PA14 with promoter of the cco1 operon driving gfp expression inserted at the attB site using pLD2777 | 7 |
| PA14 attB::Pcco-2-gfp | LD2786 | PA14 with promoter of the cco2 operon driving gfp expression inserted at the attB site using pLD2778 | 7 |
| PA14 Δphz attB::Pcox-gfp | LD2963 | PA14 Δphz with promoter of the cox/aa3 operon driving gfp expression inserted at the attB site using pLD2958 | This study |
| PA14 Δphz attB::Pcio-gfp | LD3317 | PA14 Δphz with promoter of the cio operon driving gfp expression inserted at the attB site using pLD2960 | This study |
| PA14 Δphz attB::Pcco-1-gfp | LD2785 | PA14 Δphz with promoter of the cco1 operon driving gfp expression inserted at the attB site using pLD2777 | This study |
| PA14 Δphz attB::Pcco-2-gfp | LD2787 | PA14 Δphz with promoter of the cco2 operon driving gfp expression inserted at the attB site using pLD2778 | This study |
| E. coli | |||
| UQ950 | LD44 | DH5 λpir strain for cloning; F− Δ(argF-lac)169 ϕ80 dlacZ58(ΔM15) glnV44(AS) rfbD1 gyrA96(NaIr) recA1 endA1 spoT thi-1 hsdR17 deoR λpir+ | D. Lies |
| BW29427 | LD661 | Donor strain for conjugation; thrB1004 pro thi rpsL hsdS lacZΔM15RP4-1360 Δ(araBAD)567ΔdapA1314::[erm pir(wt)] | B. Wanner |
| S17-1 | LD2901 | Strr Tpr; F− RP4-2-Tc::Mu aphA::Tn7 recA λpir lysogen | 61 |
| S. cerevisiae InvSc1 | LD676 | MATa/MATα leu2/leu2 trp1-289/trp1-289 ura3-52/ura3-52 his3-Δ1/his3-Δ1 | Invitrogen |
The growth media used in this study were 1% tryptone and MOPS minimal medium (50 mM 4-morpholinepropanesulfonic acid [pH 7.2], 43 mM NaCl, 93 mM NH4Cl, 2.2 mM KH2PO4, 1 mM MgSO4·7H2O, 1 μg/ml FeSO4·7H2O, and 20 mM carbon source [d-glucose, sodium succinate hexahydrate, or α-ketoglutaric acid sodium salt]). Stock solutions of MgSO4, FeSO4, and carbon sources were filter sterilized. Agar-containing versions of all growth media contained 1% agar (Teknova A7777).
Construction of P. aeruginosa deletion mutant strains.
Markerless deletions mutants were created by amplifying ∼1 kb of flanking DNA sequence from each side of the gene(s) to be deleted using the primers listed in Table 2. These flanking sequences were inserted into pMQ30 via gap repair cloning in Saccharomyces cerevisiae strain InvSc1 (57). The resulting plasmids, listed in Table 3, were then transformed into Escherichia coli strain UQ950, verified by restriction digests and/or sequencing, and moved into PA14 using biparental conjugation using E. coli strain BW29427. Single recombinants in PA14 were selected using LB agar plates containing 100 μg/ml gentamicin. Markerless deletions (double recombinants) were selected on LB without NaCl containing 10% sucrose and confirmed by PCR. Combinatorial mutants were constructed by using single mutants as hosts for biparental conjugation as described in Table 1.
TABLE 2.
Primers used in this study
| Primer | Sequencea | Plasmid made |
|---|---|---|
| LD725 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatCCCCTCAGAGAAGTCAGTCG | pLD1929 (Δcco1 cco2 in pMQ30) |
| LD1063 | gttgcccaggtgttcctgtGGCGGACCACCTTGTAGTTA | |
| LD949 | ggaattgtgagcggataacaatttcacacaggaaacagctTGTAGTCGAGGGACTTCTTGC | |
| LD1064 | taactacaaggtggtccgccACAGGAACACCTGGGCAAC | |
| LD1118 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatTCTTCAGGTTCTCGCGGTAG | pLD1966 (Δcox in pMQ30) |
| LD1119 | aagtgccagtaccaactggcGCAGATCCAGAAGATGGTCA | |
| LD1120 | tgaccatcttctggatctgcGCCAGTTGGTACTGGCACTT | |
| LD1121 | ggaattgtgagcggataacaatttcacacaggaaacagctATCGCGAGACTCATGGTTTT | |
| LD1134 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatCGCTGCTTGTCGATCTGTT | pLD1967 (Δcyo [bo3] in pMQ30) |
| LD1135 | gcgacatgaccctgttcaacCTGACCGGCTACTGGACC | |
| LD1136 | ggtccagtagccggtcagGTTGAACAGGGTCATGTCGC | |
| LD1137 | ggaattgtgagcggataacaatttcacacaggaaacagctCCTCGGCGACCATGAATAC | |
| LD1126 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatTTCAGGTTCTTCGGGTTCTC | pLD2044 (Δcio in pMQ30) |
| LD1187 | aacagcgcgccgaccagcatCTCTTCGTTCGTTTTCAGCC | |
| LD1188 | ggctgaaaacgaacgaagagATGCTGGTCGGCGCGCTGTT | |
| LD1189 | ggaattgtgagcggataacaatttcacacaggaaacagctGCGTTGATGAAGCGGATAAC | |
| LD2247 | gattcgactgcactagtCTCGCCTTGCGGCTGGATGG | pLD2958 |
| LD2248 | gattcgactgcctcgagTCGGGTTCCCCTTATCGTTG | |
| LD2251 | gattcgactgcactagtCGCGGCGCTGGCGGAAACG | pLD2960 |
| LD2252 | gattcgactgcctcgaGGCAACTCCTCTTCAGGTTC | |
| LD2120 | gattcgacatcactagtACGCCCAGCTCCAACAAA | pLD2777 |
| LD2121 | gattcgatgccctcgaGCTAGGGGTTCCACGGTTAAT | |
| LD2122 | gattcgactgcactagtCATCGACTTGCCGCCCAG | pLD2778 |
| LD2123 | gattcgatgccctcgaGCTATGGGCTTCCATCCAC |
Underlining indicates introduction of a restriction digest site used for cloning; lowercase letters indicate homologous sequences used for recombination.
TABLE 3.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pMQ30 | 7.5-kb mobilizable vector; oriT sacB Gmr | 57 |
| pLD2722 | Gmr Tetr; flanked by Flp recombinase target (FRT) sites to resolve out resistance cassettes | 7 |
| pFLP2 | Flp recombinase-producing plasmid | 62 |
| pLD1929 | Δcco1 cco2 PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain InvSc1 | 7 |
| pLD1966 | Δcox (aa3) PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain InvSc1 | This study |
| pLD1967 | Δcyo (bo3) PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain InvSc1 | This study |
| pLD2044 | Δcio PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain InvSc1 | This study |
| pLD2958 | PCR-amplified cox (aa3) promoter ligated into pLD2722 using SpeI and XhoI | This study |
| pLD2960 | PCR-amplified cio promoter ligated into pLD2722 using SpeI and XhoI | This study |
| pLD2777 | PCR-amplified cco1 promoter ligated into pLD2722 using SpeI and XhoI | 7 |
| pLD2778 | PCR-amplified cco2 promoter ligated into pLD2722 using SpeI and XhoI | 7 |
Construction of reporter strains.
Reporter constructs for P. aeruginosa’s terminal oxidases (Cox, Cio, Cco1, and Cco2) were constructed using the primers listed in Table 2 to amplify the respective promoter regions (500 bp upstream of the respective terminal oxidase operon), adding an SpeI and an XhoI digest site to the 5′ and 3′ ends of the promoter, respectively. Purified PCR products were digested and ligated into the pLD2722 vector at the multiple-cloning site (MCS), upstream of the green fluorescent protein (GFP) gene sequence. The resulting plasmids (listed in Table 3) were transformed into E. coli strain UQ950, verified by sequencing, and moved into WT PA14 or the Δphz mutant using biparental conjugation with E. coli strain S17-1. Recombinants were selected for as described previously (7).
Growth assays and determination of carbon sources that support PA14 growth.
One milliliter of liquid subculture was washed two times in phosphate-buffered saline (PBS) (Cold Spring Harbor Laboratories), resuspended in 1 ml MOPS minimal medium without a carbon source, and diluted 1:100 in MOPS minimal medium without a carbon source. One hundred microliters of this cell suspension were dispensed into each well of a PM1 phenotype microarray carbon microplate (Biolog 12111) and incubated at 37°C with continuous shaking on the medium setting in a Synergy 4 plate reader (BioTek). Growth was measured by taking OD readings at 500 nm every 30 min for 20 to 24 h. Each strain was assayed in biological triplicate.
Carbon sources that support growth of PA14 were determined by monitoring growth of the WT over 22.5 h for each biological replicate. After subtracting the cell density (OD at 500 nm) values for the negative control (well A1 on Biolog plate PM1, in which there is no carbon source added), a cutoff of 0.15 was applied to the other 95 cell density values; anything falling below this cutoff was designated background. In the rare instances where there was a discrepancy between the biological replicates, the growth curve of each replicate was manually inspected to determine whether there was growth or not. We found that 32 of 95 carbon sources in Biolog plate PM1 allowed for growth of WT PA14 (see File S1 in the supplemental material). We ensured that only those carbon sources that support growth of WT also allow the terminal oxidase and/or phenazine mutants to grow by visually inspecting their growth.
Determination of carbon sources that support reduction of tetrazolium violet.
For dye reduction assays, PA14 cells were prepared and assayed according to the manufacturer’s instructions. Briefly, PBS-washed cells were resuspended in 1× IF-0 (IF-0a GN/GP Base Inoculating Fluid; Biolog 72268) to a percent transmittance of 42. The 42% transmittance cell suspension was diluted 1:5 in 1× IF-0 plus 1× Redox Dye A (tetrazolium violet; Biolog 74221). One hundred microliters of this cell-dye suspension was dispensed into each well of a PM1 microplate and incubated at 37°C with continuous shaking on the medium setting in a Synergy 4 plate reader (BioTek). Dye reduction was measured by taking absorbance readings at 590 nm, in accordance with the manufacturer’s instructions, every 30 min for 20 to 24 h. Absorbance values for a well containing no carbon source (A1) were subtracted from each data point. Tetrazolium dye reduction was determined only for carbon sources that supported PA14 growth (see File S1 and Fig. S1 in the supplemental material). Because we encountered variability in dye reduction between runs, we performed each experiment in biological triplicate and assigned a carbon source to a particular category if dye reduction trends were reproducible in at least two out of the three replicates (see File S2 in the supplemental material). The dye reduction patterns of WT, PaCco, and PaCio were compared to those of their respective phenazine-null counterparts, and the effect of phenazines on dye reduction in each carbon source was determined (Fig. 2B).
Liquid culture growth and terminal oxidase reporter expression assays.
One milliliter of liquid subcultures was washed two times in PBS and resuspended in 1 ml PBS. These cells were then diluted 1:100 in the respective growth medium in a clear, flat-bottom polystyrene black 96-well plate (VWR 82050-756) and incubated at 37°C with continuous shaking on the medium setting in a Synergy 4 plate reader (BioTek). Growth was monitored by taking OD readings at 500 nm and gfp expression was assessed by taking fluorescence readings at excitation and emission wavelengths of 480 nm and 510 nm, respectively, every 30 min for 20 to 24 h. Fluorescence and absorbance readings were taken simultaneously. For growth curves (see Fig. S4 in the supplemental material), values of OD at 500 nm of a growth medium blank were subtracted from the growth values of each strain. For the expression data shown in Fig. 3, relative fluorescence unit (RFU) values for each reporter were adjusted for cell density by dividing the fluorescence value at each time point by the corresponding cell density value. RFU values, also corrected for cell density, for a strain lacking a promoter upstream of the gfp (MCS-gfp) were subtracted from the fluorescence values for each reporter.
Thin-sectioning assays.
Thin-sectioning assays were performed as described previously (58). Briefly, 5 μl of subcultures was spotted onto 1% agar plates containing two-layer growth medium (1% tryptone, MOPS-glucose, MOPS-succinate, or MOPS–α-KG) and grown in the dark at 25°C with > 90% humidity (Percival CU-22L). After 3 days, colonies were covered with an agar layer and sandwiched colonies were lifted from the bottom layer, washed for 10 min in PBS (pH 7.4) at room temperature in the dark, and fixed in 4% paraformaldehyde in PBS overnight at room temperature in the dark. Fixed colonies were washed twice in PBS and dehydrated through a series of ethanol washes. Colonies were cleared via three 60-min incubations in Histoclear-II (National Diagnostics HS-202) and infiltrated with wax via two separate washes with 100% Paraplast Xtra paraffin wax (Thermo Fisher Scientific 50-276-89) for 2 h each at 55°C, and then colonies were allowed to polymerize overnight at 4°C. Tissue processing was performed using an STP120 tissue processor (Thermo Fisher Scientific 813150). Trimmed blocks were sectioned in 10-μm-thick sections perpendicular to the plane of the colony using an automatic microtome (Thermo Fisher Scientific 905200ER) and collected onto slides. Slides were air dried overnight, heat fixed on a hot plate for 1 h at 45°C, and rehydrated. Rehydrated colonies were immediately mounted in Tris-buffered 4′,6′-diamidino-2-phenylindole (DAPI)-Fluorogel (Thermo Fisher Scientific 50-246-93) and overlaid with a coverslip. Differential interference contrast (DIC) and fluorescent confocal images were captured from using an LSM800 confocal microscope (Zeiss, Germany). Images were taken from the center of the biofilm and, where applicable, in a region devoid of wrinkles (n ≥ 6 for each strain on each carbon source).
Thin-sectioning data analysis.
Fluorescence intensities of terminal-oxidase GFP reporters expressed in biofilm thin sections were determined using Fiji image processing software (59) and plotted after subtracting values for the respective strains containing a promoterless GFP reporter. Cropped versions of the analyzed images are shown. Background fluorescence was removed by aligning images for terminal-oxidase GFP reporters with those for the promoterless GFP reporter and subtracting their fluorescence values using Adobe Photoshop CC.
Measurement of biofilm thickness.
Images taken for thin-sectioning data analyses were measured to quantify thicknesses of WT and Δphz colony biofilms grown on tryptone, glucose, succinate, and α-KG. The data plotted represent mean values from six biological replicates; three technical measurements from the center of one biofilm were averaged to yield each biological replicate.
Phenazine detection and quantification.
(i) For phenazine extraction from liquid cultures, 1 ml of liquid subcultures was washed three times in 1× PBS and resuspended in 1 ml 1× PBS. Washed cells were diluted 1:100 in 200 μl of the respective growth medium in a clear, flat-bottom polystyrene 96-well plate (VWR 82050-716) and incubated at 37°C with continuous shaking on the medium setting in a Synergy 4 plate reader (BioTek). Growth was measured by taking OD readings at 500 nm every 30 min, and cultures were monitored to determine onset of stationary phase. Five hours after each culture reached stationary phase, samples were collected into a microcentrifuge tube and centrifuged for 2 min at 16,873 relative centrifugal force (rcf) to pellet cells. The supernatant of each sample was applied to a 0.22-μm Spin-X column (VWR 29442-754) and centrifuged for 2 min at 16,873 rcf, and 200 μl of the resulting cell-free flowthrough was loaded into an HPLC vial for analysis. For each growth condition, n = 10.
(ii) For phenazine extraction from biofilms, 1 ml of liquid subcultures was washed twice in 1× PBS and resuspended in 1 ml of 1× PBS. A 25-mm filter disk with a pore size of 0.2 μm (GE Healthcare 110606) was placed into the center of one 35- by 10-mm round petri dish (VWR 25373-041) filled with 4 ml of 1% tryptone agar, MOPS-glucose agar, MOPS-succinate agar, or MOPS–α-KG agar, and 10 μl of the washed cells was spotted onto the filter disk. Colony biofilms were grown for 3 days in the dark at 25°C with > 90% humidity, after which point each colony and filter were lifted off their plate. The agar upon which the biofilm had developed was placed into a 5-ml aliquot of 100% methanol in a polypropylene conical tube, and phenazines were extracted from the agar overnight at room temperature in the dark. Three hundred microliters of the phenazine extraction was filtered through a 0.22-μm Spin-X column as described above, and 200 μl of the cell-free flowthrough were loaded into an HPLC vial for analysis. For each growth condition, n ≥ 6.
(iii) Phenazines were identified using high-performance liquid chromatography (Agilent 1100 HPLC system) as described previously (43, 60) and by comparing sample peaks to peaks of pure phenazine standards run as controls. The height of each peak (arbitrary units [AU]) was used to determine the concentration of each phenazine. For phenazine quantification from biofilms, the calculated phenazine concentration was multiplied by 1.25 to account for dilution during the phenazine extraction step. Dilution standards of purified PYO, PCA, and PCN were prepared at known concentrations, and extinction coefficients (ε) were generated for each: εPYO = 1.34 μM/AU, εPCA = 7.686 μM/AU, and εPCN = 7.518 μM/AU.
Redox microprofiling.
(i) Extracellular redox states of day three (∼72-h) phenazine-producing colony biofilms were measured using a 25-μm-tip redox microelectrode and external reference (Unisense RD25 and REF-RM). Biofilms were grown by spotting 10 μl of liquid subcultures onto 60 ml of growth medium (1% tryptone, MOPS-glucose, MOPS-succinate, or MOPS–α-KG agar) in a 10- by 10- by 1.5-cm square petri dish (LDP D210-16). Calibration and redox measurements were performed using the SensorTrace Profiling software (Unisense) as described previously (7).
(ii) For measuring extracellular redox states of biofilms grown on exogenously added phenazines, purified phenazine compounds were added to the respective growth media (described above) to final concentrations of 200 μM after cooling to ∼55°C in a water bath. The phenazines tested were PCA (Apexmol), PCN (Apexmol), PMS (Thermo Fisher Scientific AC130160010), and PYO (Cayman Chemicals). PCA and PCN stock solutions were 25 mM in dimethyl sulfoxide (DMSO), while PYO was 200 mM in DMSO. The PMS stock solution was 200 mM in water. Extracellular redox states of biofilms were measured on DMSO-only plates to ensure that the solvent did not alter the extracellular redox state. Extracellular redox states of day three (∼72-h) biofilms grown on exogenously added phenazines were measured as described above. Redox potential values at the surface of the biofilm (0 μm) were set to 0 mV, and the remaining values for each plot were adjusted accordingly to facilitate comparisons of phenazine reduction patterns between conditions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH/NIAID grant R01AI103369, an NSF Career Award to L.E.P.D., and NIH training grant 5T32GM008798 to J.J.
We thank Hannah Dayton for proofreading the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Madigan MT, Martinko JM, Bender KS, Buckley DH, Stahl DA. 2015. Brock biology of microorganisms. Pearson, London, United Kingdom. [Google Scholar]
- 2.Poole RK, Cook GM. 2000. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv Microb Physiol 43:165–224. doi: 10.1016/s0065-2911(00)43005-5. [DOI] [PubMed] [Google Scholar]
- 3.Høiby N, Ciofu O, Johansen HK, Song Z-J, Moser C, Jensen PØ, Molin S, Givskov M, Tolker-Nielsen T, Bjarnsholt T. 2011. The clinical impact of bacterial biofilms. Int J Oral Sci 3:55–65. doi: 10.4248/IJOS11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elias S, Banin E. 2012. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev 36:990–1004. doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- 5.Dietrich LEP, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. 2013. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol 195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kempes CP, Okegbe C, Mears-Clarke Z, Follows MJ, Dietrich L. 2014. Morphological optimization for access to dual oxidants in biofilms. Proc Natl Acad Sci U S A 111:208–213. doi: 10.1073/pnas.1315521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jo J, Cortez KL, Cornell WC, Price-Whelan A, Dietrich LE. 2017. An orphan cbb3-type cytochrome oxidase subunit supports Pseudomonas aeruginosa biofilm growth and virulence. Elife 6:e30205. doi: 10.7554/eLife.30205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White D, Drummond JT, Fuqua C. 2012. The physiology and biochemistry of prokaryotes. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 9.Williams HD, Zlosnik JEA, Ryall B. 2007. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv Microb Physiol 52:1–71. doi: 10.1016/S0065-2911(06)52001-6. [DOI] [PubMed] [Google Scholar]
- 10.Arai H. 2011. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front Microbiol 2:103. doi: 10.3389/fmicb.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comolli JC, Donohue TJ. 2004. Differences in two Pseudomonas aeruginosa cbb3 cytochrome oxidases. Mol Microbiol 51:1193–1203. doi: 10.1046/j.1365-2958.2003.03904.x. [DOI] [PubMed] [Google Scholar]
- 12.Alvarez-Ortega C, Harwood CS. 2007. Responses of Pseudomonas aeruginosato low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol 65:153–165. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawakami T, Kuroki M, Ishii M, Igarashi Y, Arai H. 2010. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ Microbiol 12:1399–1412. doi: 10.1111/j.1462-2920.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- 14.Arai H, Kawakami T, Osamura T, Hirai T, Sakai Y, Ishii M. 2014. Enzymatic characterization and in vivo function of five terminal oxidases in Pseudomonas aeruginosa. J Bacteriol 196:4206–4215. doi: 10.1128/JB.02176-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham L, Williams HD. 1995. Isolation and characterization of mutants defective in the cyanide-insensitive respiratory pathway of Pseudomonas aeruginosa. J Bacteriol 177:432–438. doi: 10.1128/jb.177.2.432-438.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham L, Pitt M, Williams HD. 1997. The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol Microbiol 24:579–591. doi: 10.1046/j.1365-2958.1997.3561728.x. [DOI] [PubMed] [Google Scholar]
- 17.Ray A, Williams HD. 1997. The effects of mutation of the anr gene on the aerobic respiratory chain of Pseudomonas aeruginosa. FEMS Microbiol Lett 156:227–232. doi: 10.1111/j.1574-6968.1997.tb12732.x. [DOI] [PubMed] [Google Scholar]
- 18.Comolli JC, Donohue TJ. 2002. Pseudomonas aeruginosa RoxR, a response regulator related to Rhodobacter sphaeroides PrrA, activates expression of the cyanide-insensitive terminal oxidase. Mol Microbiol 45:755–768. doi: 10.1046/j.1365-2958.2002.03046.x. [DOI] [PubMed] [Google Scholar]
- 19.Saint-Amans S, Girbal L, Andrade J, Ahrens K, Soucaille P. 2001. Regulation of carbon and electron flow in Clostridium butyricum VPI 3266 grown on glucose-glycerol mixtures. J Bacteriol 183:1748–1754. doi: 10.1128/JB.183.5.1748-1754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aussel L, Pierrel F, Loiseau L, Lombard M, Fontecave M, Barras F. 2014. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys Acta 1837:1004–1011. doi: 10.1016/j.bbabio.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Otten MF, Stork DM, Reijnders WN, Westerhoff HV, Van Spanning RJ. 2001. Regulation of expression of terminal oxidases in Paracoccus denitrificans. Eur J Biochem 268:2486–2497. doi: 10.1046/j.1432-1327.2001.02131.x. [DOI] [PubMed] [Google Scholar]
- 22.Berridge MV, Herst PM, Tan AS. 2005. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol Annu Rev 11:127–152. doi: 10.1016/S1387-2656(05)11004-7. [DOI] [PubMed] [Google Scholar]
- 23.Tachon S, Michelon D, Chambellon E, Cantonnet M, Mezange C, Henno L, Cachon R, Yvon M. 2009. Experimental conditions affect the site of tetrazolium violet reduction in the electron transport chain of Lactococcus lactis. Microbiology 155:2941–2948. doi: 10.1099/mic.0.029678-0. [DOI] [PubMed] [Google Scholar]
- 24.Bernofsky C, Swan M. 1973. An improved cycling assay for nicotinamide adenine dinucleotide. Anal Biochem 53:452–458. doi: 10.1016/0003-2697(73)90094-8. [DOI] [PubMed] [Google Scholar]
- 25.Pierson LS III, Thomashow LS. 1992. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30-84. Mol Plant Microbe Interact 5:330–339. doi: 10.1094/MPMI-5-330. [DOI] [PubMed] [Google Scholar]
- 26.Culbertson JE, Toney MD. 2013. Expression and characterization of PhzE from P aeruginosa PAO1: aminodeoxyisochorismate synthase involved in pyocyanin and phenazine-1-carboxylate production. Biochim Biophys Acta 1834:240–246. doi: 10.1016/j.bbapap.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Price-Whelan A, Dietrich LEP, Newman DK. 2007. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J Bacteriol 189:6372–6381. doi: 10.1128/JB.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osamura T, Kawakami T, Kido R, Ishii M, Arai H. 2017. Specific expression and function of the A-type cytochrome c oxidase under starvation conditions in Pseudomonas aeruginosa. PLoS One 12:e0177957. doi: 10.1371/journal.pone.0177957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y-C, Cornell WC, Jo J, Price-Whelan A, Dietrich L. 2018. The Pseudomonas aeruginosa complement of lactate dehydrogenases enables use of d- and l-lactate and metabolic cross-feeding. mBio 9:e00961-18. doi: 10.1128/mBio.00961-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frimmersdorf E, Horatzek S, Pelnikevich A, Wiehlmann L, Schomburg D. 2010. How Pseudomonas aeruginosa adapts to various environments: a metabolomic approach. Environ Microbiol 12:1734–1747. doi: 10.1111/j.1462-2920.2010.02253.x. [DOI] [PubMed] [Google Scholar]
- 31.Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev 34:658–684. doi: 10.1111/j.1574-6976.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 32.Lin Y-C, Sekedat MD, Cornell WC, Silva GM, Okegbe C, Price-Whelan A, Vogel C, Dietrich L. 2018. Phenazines regulate Nap-dependent denitrification in Pseudomonas aeruginosa biofilms. J Bacteriol 200:e00031-18. doi: 10.1128/JB.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Kern SE, Newman DK. 2010. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J Bacteriol 192:365–369. doi: 10.1128/JB.01188-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glasser NR, Kern SE, Newman DK. 2014. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol Microbiol 92:399–412. doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okegbe C, Price-Whelan A, Dietrich L. 2014. Redox-driven regulation of microbial community morphogenesis. Curr Opin Microbiol 18:39–45. doi: 10.1016/j.mib.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okegbe C, Fields BL, Cole SJ, Beierschmitt C, Morgan CJ, Price-Whelan A, Stewart RC, Lee VT, Dietrich L. 2017. Electron-shuttling antibiotics structure bacterial communities by modulating cellular levels of c-di-GMP. Proc Natl Acad Sci U S A 114:E5236–E5245. doi: 10.1073/pnas.1700264114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meirelles LA, Newman DK. 2018. Both toxic and beneficial effects of pyocyanin contribute to the lifecycle of Pseudomonas aeruginosa. Mol Microbiol 110:995. doi: 10.1111/mmi.14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price-Whelan A, Dietrich LEP, Newman DK. 2006. Rethinking “secondary” metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol 2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Newman DK. 2008. Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ Sci Technol 42:2380–2386. doi: 10.1021/es702290a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakhtah H, Price-Whelan A, Dietrich L. 2013. Regulation of phenazine biosynthesis, p 19–42. In Chincholkar S, Thomashow L (ed), Microbial phenazines: biosynthesis, agriculture and health. Springer, Berlin, Germany. [Google Scholar]
- 41.Hansford GS, Holliman FG, Herbert RB. 1972. Pigments of Pseudomonas species. IV. In vitro and in vivo conversion of 5-methylphenazinium-1-carboxylate into aeruginosin A. J Chem Soc Perkin 1 1:103–105. doi: 10.1039/p19720000103. [DOI] [PubMed] [Google Scholar]
- 42.Doucette CD, Schwab DJ, Wingreen NS, Rabinowitz JD. 2011. α-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat Chem Biol 7:894–901. doi: 10.1038/nchembio.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakhtah H, Koyama L, Zhang Y, Morales DK, Fields BL, Price-Whelan A, Hogan DA, Shepard K, Dietrich L. 2016. The Pseudomonas aeruginosa efflux pump MexGHI-OpmD transports a natural phenazine that controls gene expression and biofilm development. Proc Natl Acad Sci U S A 113:E3538–47. doi: 10.1073/pnas.1600424113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng H, Kim J, Liew M, Yan JK, Herrera O, Bok JW, Kelleher NL, Keller NP, Wang Y. 2015. Redox metabolites signal polymicrobial biofilm development via the NapA oxidative stress cascade in Aspergillus. Curr Biol 25:29–37. doi: 10.1016/j.cub.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams HD, Davies JC. 2012. Basic science for the chest physician: Pseudomonas aeruginosa and the cystic fibrosis airway. Thorax 67:465–467. doi: 10.1136/thoraxjnl-2011-201498. [DOI] [PubMed] [Google Scholar]
- 46.Son MS, Matthews WJ Jr, Kang Y, Nguyen DT, Hoang TT. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun 75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoboth C, Hoffmann R, Eichner A, Henke C, Schmoldt S, Imhof A, Heesemann J, Hogardt M. 2009. Dynamics of adaptive microevolution of hypermutable Pseudomonas aeruginosa during chronic pulmonary infection in patients with cystic fibrosis. J Infect Dis 200:118–130. doi: 10.1086/599360. [DOI] [PubMed] [Google Scholar]
- 48.Eichner A, Günther N, Arnold M, Schobert M, Heesemann J, Hogardt M. 2014. Marker genes for the metabolic adaptation of Pseudomonas aeruginosa to the hypoxic cystic fibrosis lung environment. Int J Med Microbiol 304:1050–1061. doi: 10.1016/j.ijmm.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 49.Kotloski NJ, Gralnick JA. 2013. Flavin electron shuttles dominate extracellular electron transfer by Shewanella oneidensis. mBio 4:e00553-12. doi: 10.1128/mBio.00553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mevers E, Su L, Pishchany G, Baruch M, Cornejo J, Hobert E, Dimise E, Ajo-Franklin CM, Clardy J. 2019. An elusive electron shuttle from a facultative anaerobe. Elife 8:e48054. doi: 10.7554/eLife.48054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Light SH, Su L, Rivera-Lugo R, Cornejo JA, Louie A, Iavarone AT, Ajo-Franklin CM, Portnoy DA. 2018. A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature 562:140–144. doi: 10.1038/s41586-018-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keogh D, Lam LN, Doyle LE, Matysik A, Pavagadhi S, Umashankar S, Low PM, Dale JL, Song Y, Ng SP, Boothroyd CB, Dunny GM, Swarup S, Williams RBH, Marsili E, Kline KA. 2018. Extracellular electron transfer powers Enterococcus faecalis biofilm metabolism. mBio 9:e00626-17. doi: 10.1128/mBio.00626-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schiessl KT, Hu F, Jo J, Nazia SZ, Wang B, Price-Whelan A, Min W, Dietrich L. 2019. Phenazine production promotes antibiotic tolerance and metabolic heterogeneity in Pseudomonas aeruginosa biofilms. Nat Commun 10:762. doi: 10.1038/s41467-019-08733-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 55.Mathee K. 2018. Forensic investigation into the origin of Pseudomonas aeruginosa PA14—old but not lost. J Med Microbiol 67:1019–1021. doi: 10.1099/jmm.0.000778. [DOI] [PubMed] [Google Scholar]
- 56.Bertani G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol 186:595–600. doi: 10.1128/jb.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cornell WC, Morgan CJ, Koyama L, Sakhtah H, Mansfield JH, Dietrich L. 2018. Paraffin embedding and thin sectioning of microbial colony biofilms for microscopic analysis. J Vis Exp doi: 10.3791/57196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dietrich LEP, Price-Whelan A, Petersen A, Whiteley M, Newman DK. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol 61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 61.Teng F, Murray BE, Weinstock GM. 1998. Conjugal transfer of plasmid DNA from Escherichia coli to Enterococci: a method to make insertion mutations. Plasmid 39:182–186. [DOI] [PubMed] [Google Scholar]
- 62.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.