c-di-GMP is a ubiquitous bacterial second messenger that regulates motility, biofilm formation, and virulence of many bacterial pathogens. The PilZ domain is a widespread c-di-GMP receptor that binds c-di-GMP through its RXXXR and [D/N]hSXXG motifs; some PilZ domains lack these motifs and are unable to bind c-di-GMP. We used structural and sequence analysis to assess the diversity of PilZ-related domains and define their common features. We show that the hydrophobic residue h in the second position of the second motif is highly conserved; it may serve as a readout for c-di-GMP binding. We describe three principal classes of PilZ-related domains, canonical, tetramer-forming, and divergent PilZ domains, and propose the evolutionary pathways that led to the emergence of these PilZ types.
KEYWORDS: bacterial signaling, biofilm formation, ligand binding, cyclic nucleotide, PilZ, crystal structure, biofilms, c-di-GMP, cyclic nucleotides, evolution, protein domain, protein structure-function, receptor-ligand interaction
ABSTRACT
The widespread bacterial second messenger cyclic diguanylate (c-di-GMP) regulates a variety of processes, including protein secretion, motility, cell development, and biofilm formation. c-di-GMP-dependent responses are often mediated by its binding to the cytoplasmic receptors that contain the PilZ domain. Here, we present comparative structural and sequence analysis of various PilZ-related domains and describe three principal types of them: (i) the canonical PilZ domain, whose structure includes a six-stranded beta-barrel and a C-terminal alpha helix, (ii) an atypical PilZ domain that contains two extra alpha helices and forms stable tetramers, and (iii) divergent PilZ-related domains, which include the eponymous PilZ protein and PilZN (YcgR_N) and PilZNR (YcgR_2) domains. We refine the second c-di-GMP binding motif of PilZ as [D/N]hSXXG and show that the hydrophobic residue h of this motif interacts with a cluster of conserved hydrophobic residues, helping maintain the PilZ domain fold. We describe several novel PilZN-type domains that are fused to the canonical PilZ domains in specific taxa, such as spirochetes, actinobacteria, aquificae, cellulose-degrading clostridia, and deltaproteobacteria. We propose that the evolution of the three major groups of PilZ domains included (i) fusion of pilZ with other genes, which produced Alg44, cellulose synthase, and other multidomain proteins; (ii) insertion of an ∼200-bp fragment, which resulted in the formation of tetramer-forming PilZ proteins; and (iii) tandem duplication of pilZ genes, which led to the formation of PilZ dimers and YcgR-like proteins.
IMPORTANCE c-di-GMP is a ubiquitous bacterial second messenger that regulates motility, biofilm formation, and virulence of many bacterial pathogens. The PilZ domain is a widespread c-di-GMP receptor that binds c-di-GMP through its RXXXR and [D/N]hSXXG motifs; some PilZ domains lack these motifs and are unable to bind c-di-GMP. We used structural and sequence analysis to assess the diversity of PilZ-related domains and define their common features. We show that the hydrophobic residue h in the second position of the second motif is highly conserved; it may serve as a readout for c-di-GMP binding. We describe three principal classes of PilZ-related domains, canonical, tetramer-forming, and divergent PilZ domains, and propose the evolutionary pathways that led to the emergence of these PilZ types.
INTRODUCTION
Cyclic bis(3′→5′)-dimeric GMP (c-di-GMP) is a widespread bacterial second messenger that regulates a variety of processes, including DNA repair, transcription, mRNA decay, motility, protein and polysaccharide secretion, cell development, and biofilm formation (1–4). Cyclic di-GMP has a well-documented contribution to virulence of such important human pathogens as Vibrio cholerae, Pseudomonas aeruginosa, Salmonella enterica, Borrelia (Borreliella) burgdorferi, and Yersinia pestis and plant pathogens such as Xanthomonas campestris and Xanthomonas citri (1–7). Cellular responses to c-di-GMP-dependent signaling are mediated by specific receptors, which include two types of riboswitches and a wide variety of c-di-GMP-binding proteins. The structures and ligand-binding properties of experimentally characterized c-di-GMP receptor proteins have been the subject of several reviews (8–10). In the past several years, novel c-di-GMP-binding proteins have been discovered, with some being widespread in bacteria, such as MshEN (11, 12), and some showing relatively narrow phylogenetic distribution (13, 14). There are reasons to believe that the current list is still incomplete (9).
The ∼110-amino-acid (aa) PilZ domain (PF07238 in the Pfam database [15]) was the first c-di-GMP receptor to be identified (16), owing to its presence at the C terminus of the cellulose synthase catalytic subunit BcsA from Acetobacter xylinum (current name, Komagataeibacter xylinus), the protein whose studies led to the original discovery of c-di-GMP (17, 18). That work also described a number of other PilZ-containing domain architectures, including PilZ dimers, fusions of PilZ with the N-terminal PilZN (referred to as the YcgR domain PF07317 in Pfam) and PilZNR (YcgR_2, PF12945 in Pfam) domains, a fusion with HlyD family secretion protein in the alginate biosynthesis protein Alg44, as well as fusions of PilZ with REC, GGDEF, EAL, HD-GYP, chemoreceptor (MCPsignal), and CheC signaling domains (16).
Subsequent studies of various PilZ-containing proteins confirmed the ability of this domain to bind c-di-GMP and control a variety of c-di-GMP-regulated functions (19–30). It has become clear that different PilZ-containing proteins bind c-di-GMP with a wide range of affinities, which contributes to the diversity of c-di-GMP signaling mechanisms (31). Curiously, the eponymous PilZ protein of P. aeruginosa (genomic locus tag PA2960) does not bind c-di-GMP (21, 32). Instead, it appears to regulate formation of type IV pili through protein-protein interactions. In Xanthomonas spp., PilZ protein interacts with FimX (33, 34), while in P. aeruginosa, FimX does not do so and the PilZ-interacting partner remains to be characterized (35).
Over the years, structures of several PilZ domains and PilZ-containing proteins have been solved (22, 24, 25, 36–42), providing valuable insight into the mechanisms of c-di-GMP binding and the roles of the conserved RXXXR and [D/N]XSXXG c-di-GMP-binding motifs (reviewed in references 9 and 10). Here, we present a comparative analysis of PilZ sequences and structures, which allowed identification of additional conserved elements in PilZ domains and delineation of three major classes of PilZ-related domains. Based on this analysis, we propose a general scheme of PilZ evolution that involves three major evolutionary routes leading to (i) multidomain fusion proteins with canonical PilZ, (ii) tetrameric PilZ proteins, and (iii) two-domain proteins consisting of diverse PilZ-related N-terminal domains fused to the C-terminal canonical PilZ. Evolution along these routes is sometimes accompanied by the loss of c-di-GMP-binding motifs.
RESULTS
Structural conservation of canonical PilZ domains.
The typical PilZ domain core consists of six β-strands followed by a single long α-helix. These β-strands are arranged in two antiparallel β-sheets that form a β-barrel. In the past 12 years, more than 30 structures of various PilZ domains have been solved and deposited in the Protein Data Bank (PDB) (see Table S1 in the supplemental material). These include high-resolution crystal structures of the stand-alone PilZ domains as well as PilZ domains from the bacterial cellulose synthase (BcsA and AcsAB), alginate production protein Alg44, flagellar brake proteins YcgR, VCA0042, and MotI, and the transcriptional regulator MrkH (22, 25–27, 36, 37, 40, 41, 43–45). As noted in the respective reports, most of these domains are very similar in sequence and structure. Indeed, comparison of representative PilZ structures using the Dali server (46) revealed that, despite their relatively low sequence identity (≤20%), these domains readily align over their entire lengths, with root mean square deviations (RMSD) of the Cα traces within 3.2 Å (Table 1). Comparison of the same structures using the VAST tool, which computes an alignment of secondary structure elements, α-helices, and β-strands (47), brought similar results, with slightly shorter aligned regions but a bit higher sequence identity and even lower RMSD values (≤2.5 Å) (Table S2).
TABLE 1.
Structural similarity of canonical, tetrameric, and divergent PilZ domains
| Organism and protein name(s) (genomic locus taga) | PDB entry(ies), resolution (Å) | Alignment length (aa), % identity, RMSDb (Å) | Reference or sourcec |
|---|---|---|---|
| Canonical | |||
| Pseudomonas aeruginosa PilZ or MapZ (PA4608) | 5XLY_B, 1.76 | 109, 100, 0 | 42 |
| 2L74_A, NMRd | 121, 100, 2.1 | 38 | |
| Legionella pneumophila PilZ (Lpg0364) | 4Q63_A, 1.95 | 88, 11, 2.7 | NA, MCSG |
| Pseudomonas aeruginosa PilZ from Alg44 (PA3542) | 4RT1_B, 1.7 | 101, 17, 2.6 | 39 |
| Komagataeibacter xylinus PilZ from AcsAB | 4I86_B, 2.10 | 92, 20, 2.6 | 45 |
| Rhodobacter sphaeroides BcsA (RSP_0333, aa 578–691) | 5EJZ_A, 2.95 | 98, 12, 2.6 | 25 |
| Escherichia coli YcgR (b1194, aa 111–244) | 5Y6G_A, 2.30 | 101, 17, 2.7 | 51 |
| Pseudomonas putida YcgR or FlgZ (PP_4397, aa 121–238) | 3KYF_A, 2.10 | 101, 12, 2.5 | 43 |
| Vibrio cholerae PlzD (VCA0042, aa 134–247) | 1YLN_A, 2.20 | 99, 14, 3.7 | NA, MCSG |
| 2RDE_A, 1.92 | 99, 14, 3.1 | 22 | |
| Bacillus subtilis MotI or DgrA (BSU22910, aa 97–208) | 5VX6_A, 3.2 | 104, 16, 3.2 | 41 |
| Klebsiella pneumoniae MrkH (KP13_02245, aa 107–234) | 5KEC_D, 1.95 | 104, 11, 3.1 | 40 |
| Tetrameric (tPilZ): Xanthomonas campestris Xcc0612 (XccB100_2234) | 3RQA_A, 2.1 | 93, 12, 2.9 | 48 |
| Divergent (xPilZ) | |||
| Xanthomonas campestris or X. citri PilZ (Xcc1024, Xac1133) | 3CNR_B, 1.90; 3DSG_B, 2.09 | 72, 13, 2.9 | 36, 37 |
| Geobacter sulfurreducens GSU3033 (aa 1–102) | 2L1T_A, NMR | 85, 5, 3.1 | NA, JCSG |
| Escherichia coli YcgR_N (b1194, aa 1–110) | 5Y6H_A, 1.77 | 80, 4, 2.9 | 51 |
| Vibrio cholerae PlzD (VCA0042, aa 24–130) | 2RDE_A, 1.92 | 75, 9, 2.2 | 22 |
| Klebsiella pneumoniae MrkH (KP13_02245, aa 1–106) | 5KEC_D, 1.95 | 77, 10, 2.9 | 40 |
For multidomain structures, the amino acid boundaries of the respective PilZ domains are also shown in the parentheses.
The overlap length, percent identity, and RMSD values were taken from Dali and/or VAST (46, 47) alignments of the respective structures against the stand-alone PilZ domain in the Pseudomonas aeruginosa protein MapZ (PDB entry 5XLY_B).
NA, unpublished; MCSG, Midwest Center for Structural Genomics; JCSG, Joint Center for Structural Genomics.
NMR-derived structures are averaged from several traces and do not come with a resolution figure. Resolution information is available at Protein Data Bank.
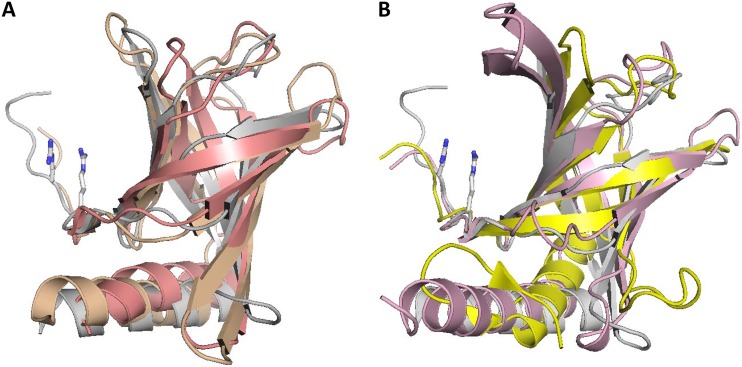
Structural superposition of PilZ domains from K. xylinus AcsAB and P. aeruginosa Alg44 and C-terminal domains of E. coli YcgR, V. cholerae PlzD, and K. pneumoniae MrkH against the high-resolution structure of the PilZ domain protein MapZ (PA4608) of P. aeruginosa (24, 26, 42) is shown in Fig. 1, while pairwise superpositions of the domains listed in Table 1 are shown in Fig. S1. A structure-guided sequence alignment of these domains is presented in Fig. S2 and, in an abbreviated form, in Fig. 2. These comparisons demonstrate that, despite some differences in the lengths of β-strands and an occasional split of a long β-strand into two, PilZ domains of the BcsA/AcsAB, YcgR, PlzD, MrkH, and Alg44 proteins adopt essentially the same core structure as stand-alone PilZ domains. Accordingly, they can all be considered canonical PilZ domains.
FIG 1.
Structural superposition of canonical PilZ domains. (A) Stand-alone PilZ domain protein MapZ (PA4608) from P. aeruginosa (PDB entry 5XLY_B, residues 4 to 107), shown in gray, superposed against PilZ domains from K. xylinus cellulose synthase (PDB entry 4I86, shown in salmon) and from P. aeruginosa Alg44 (PDB entry 4RT1, shown in wheat color). (B) MapZ structure (in gray) superposed against C-terminal PilZ domains of E. coli YcgR (PDB entry 5Y6G, shown in pink) and V. cholerae PlzD (PDB entry 2RDE, residues 134 to 237, shown in yellow). The Arg residues of the MapZ RXXXR motif are in stick representation.
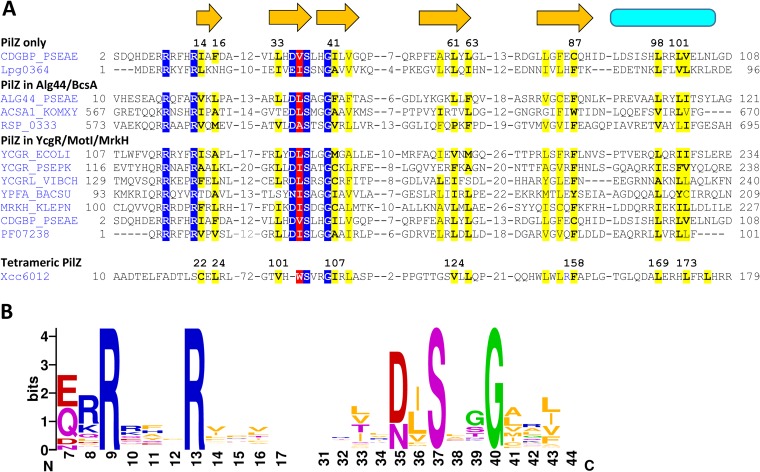
FIG 2.
Sequence conservation of canonical PilZ domains. (A) Structure-based sequence alignment of canonical PilZ domains. The residues of the c-di-GMP-binding motifs RXXXR and [D/N]XSXXG are shaded blue. The proteins are listed under their UniProt names or genomic locus tags. The secondary structure elements (β-strands [arrows] and the α-helix [cylinder]) are from the P. aeruginosa MapZ structure (PDB entry 5XLY_B). Conserved hydrophobic residues are shaded yellow, and those forming the hydrophobic pocket are shown in boldface and labeled by their numbers in the MapZ structure. The newly identified conserved h residue of the [D/N]hSXXG motif is shaded red. For comparison, the sequences of the PilZ domain entry in Pfam (PF07238) and the tetramer-forming tPilZ domain (PDB entry 3RQA) are shown at the bottom. The complete alignment is presented in Fig. S2. (B) Sequence logos of the c-di-GMP-binding motifs.
Another variation on the theme is the PilZ-PilZ domain architecture that represents tandem duplication of the canonical PilZ domain. In these proteins, exemplified by the previously described (16) Bll4394 and Blr5568 proteins from Bradyrhizobium japonicum (current name, B. diazoefficiens), both PilZ domains keep their C-terminal α-helices and often retain intact c-di-GMP-binding motifs. The current version of Pfam (15) lists more than 240 such PilZ-PilZ proteins, primarily from alphaproteobacteria but also some from gamma- and deltaproteobacteria, planctomycetes, and candidate division NC10. While analyzing PilZ fusion proteins (see below), we were able to substantially expand this list, with additional members again found primarily in alphaproteobacteria. The largest such family includes Sinorhizobium meliloti protein SMc00999 and more than a thousand closely related proteins. As discussed below, these proteins are important from an evolutionary point of view, as tandem duplications of the canonical PilZ domain, followed by diversification of the N-terminal domain, give rise to a variety of YcgR-like two-domain proteins.
Conservation of hydrophobic residues, the [D/N]hSXXG motif.
In addition to the well-known RXXXR and [D/N]XSXXG motifs, the original description of the PilZ domain (16) showed conservation of two hydrophobic residues. One was located at the end of the second β-strand between the D/N and S residues of the second c-di-GMP-binding motif, and the other (mostly Phe) was near the end of the last β-strand (Leu/Ile/Val/Phe-32 and Phe/Trp/Tyr-90, respectively [see Fig. 1 of reference 16]). However, the roles of these hydrophobic residues have not been investigated. Now, using structure-based sequence alignment of these domains, we were able to confirm strict conservation of these and several other hydrophobic residues (Fig. 2A). Therefore, the second c-di-GMP-binding motif of PilZ could be redefined as [D/N]hSXXG, where h stands for a hydrophobic residue, which is primarily Ile, Leu, or Val at this position (Fig. 2B). This observation prompted us to check the positions of this h residue in various PilZ structures.
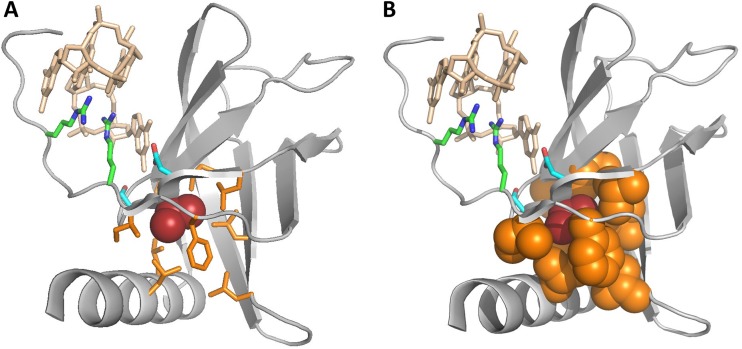
As shown in Fig. 3, the [I/L/V] hydrophobic residue, while located within the c-di-GMP-binding motif, does not interact with c-di-GMP. Instead, its side chain is oriented toward the other side of the β-strand and is located within a hydrophobic pocket formed by at least 7 other hydrophobic residues. Sequence alignment (Fig. 2A) showed that the positions of these residues were well conserved among various PilZ domains, although the residues themselves varied. The most conserved of them was the previously noted Phe residue (replaced by Cys-87 in P. aeruginosa MapZ), located near the end of the last β-strand of PilZ (Fig. 2A). Comparison of the PilZ structures showed that this Phe residue directly interacts with the h residue of the [D/N]hSXXG motif (Val-36 in P. aeruginosa MapZ) and apparently limits the mobility of the Gly residue of this motif. Another pocket-forming hydrophobic residue is located immediately after the Gly of the [D/N]hSXXG motif (Fig. 2). However, at that position, aliphatic residues (Ile, Leu, Val, and Ala) were found along with Met and Cys (Fig. 2), which is why we refrained from including this second h in the definition of the c-di-GMP binding motif as [D/N]hSXXGh.
FIG 3.
Hydrophobic pocket surrounding the h residue of the [D/N]hSXXG motif. The structure of the P. aeruginosa MapZ (PDB entry 5XLY_B) is in gray, the c-di-GMP dimer is in wheat color, and the c-di-GMP-binding Arg9, Arg13, Asp35, and Ser37 residues are shown in stick representation, with C atoms in green and blue. The h residue of the [D/N]hSXXG motif, Val36, is red. The Ile14, Phe16, Leu33, Ile41, Leu61, Leu63, Cys87, Leu98, and Leu101 residues (highlighted in Fig. 2) are in orange and shown in stick (A) or filled space (B) representation.
As seen in Fig. 2, two more residues that form the hydrophobic pocket are located after the RXXXR motif in the RXXXRhXh pattern. However, these two positions can be occupied by various uncharged (or even charged) residues, so we have chosen to leave the RXXXR motif as it is.
Tetramer-forming PilZ domains.
The Xcc0612 protein from X. campestris (GenBank accession no. ALE68703; PDB entry 3RQA) contains an atypical PilZ domain (PilZ_2, Pfam domain PF16823, referred to here as tPilZ) that displays essentially the same structure as canonical PilZ (RSMD, 2.2 to 2.9 Å), with the exception of a short α-helix at the N terminus and an ∼60-aa insert that forms two additional α-helices, α2 and α3 (Fig. 4A) (48). In the Xcc6012 structure, α3 helices from four PilZ protomers interact with each other via strong hydrophobic heptarepeats, leading to the formation of a very stable tetramer, which is strengthened further by intermolecular bridges between Lys-77 in the middle of the 75DAKLD79 motif with Asp75 and Asp79 of the adjacent monomer (48). Previously, tPilZ domains were thought to be limited to xanthomonads. We have now detected the respective genes in the genomes of a wide variety of gammaproteobacteria, as well as in certain members of other subdivisions of Proteobacteria and of the phyla Nitrospirae, Nitrospinae/Tectomicrobia, Deferribacteres, and Thermodesulfobacteria (Fig. S3 and Table S3), with more than 3,000 proteins in the current protein databases. All these proteins have similarly sized inserts that form two predicted α-helices but display certain variation of the domain-interlocking motif, which was generalized to [DENH]X[KR]h[DEN], where h again indicates a hydrophobic amino acid residue. Further, the tPilZ domain of Xcc0612 has disrupted RXXXR and [D/N]hSXXG motifs and is unable to bind c-di-GMP (48). In contrast, tPilZ domains from many other bacteria have these motifs intact, suggesting that they can still bind c-di-GMP. Indeed, P. aeruginosa protein PA2989 (which has a typical RXXXR motif but the [D/N] residue of the second motif is replaced by Ser) (Fig. S3) has been experimentally shown to bind c-di-GMP (49, 50). In contrast, in PA2989 homologs from some other Pseudomonas species, these motifs are disrupted (Fig. S3), indicating the loss of c-di-GMP binding within a single bacterial genus. Intact motifs are also present in the tPilZ protein Lpg1926 from the important pathogen Legionella pneumophila (Fig. S3), suggesting that it, too, has retained the ability to bind c-di-GMP.
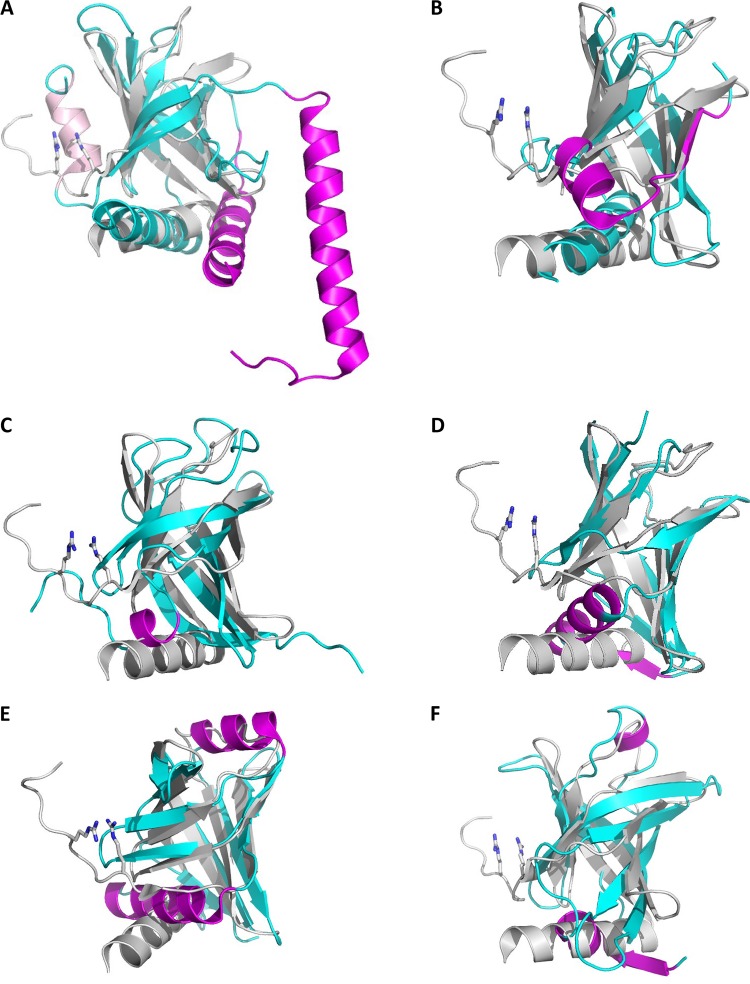
FIG 4.
Structural similarity of PilZ-related domains. Canonical PilZ domain protein MapZ (PA4608) from P. aeruginosa (PDB entry 5XLY_B, residues 4 to 107) superposed against the tetramer-forming PilZ domain from X. campestris (PDB entry 3RQA) (A), the stand-alone c-di-GMP-nonbinding PilZ protein from X. citri (Xac1133, PDB entry 3CNR) (B), the DUF5634_N domain from G. sulfurreducens protein GSU3033 (PDB entry 2L1T) (C), the N-terminal domains from E. coli YcgR (PDB entry 5Y6H) (D), V. cholerae PlzD (YcgR_2, PDB entry 2RDE, residues 24 to 130) (E), and K. pneumoniae MrkH (PDB entry 5KEC, residues 1 to 106) (F). The reference PilZ structure 5XLY is in gray, aligned structural elements from other proteins are in cyan, and structural elements that deviate from the canonical PilZ structure are in magenta. The Arg residues of the MapZ RXXXR motif are in stick representation.
Even when the [D/N]hSXXG motif is not conserved, tPilZ domains retain a hydrophobic residue (typically I/L/V but occasionally W or F) in the position of h and the constellation of hydrophobic residues to form a pocket around it (Fig. 2 and Fig. S3). One of these residues immediately precedes the (former) [D/N]hSXXG motif, whereas in canonical PilZ domains it is separated from this motif by an additional residue (Fig. S2 and S3). tPilZ domains also retain the same amphipathic C-terminal α-helix that provides two hydrophobic residues to this pocket. This α-helix typically contains a patch of Arg, Lys, and His residues and carries a strong positive charge (Fig. S3).
In contrast to the canonical PilZ, tPilZ domains are almost never seen forming fusions to other domains. In addition, these domains always appear to be encoded by a single copy per genome, irrespective of the total number of other PilZ domain proteins encoded by that genome (Table S3). The reasons for this behavior remain to be investigated.
Divergent PilZ-related domains.
While the canonical and tetramer-forming PilZ domains share the same core structure of a six-stranded β-barrel followed by an α-helix, members of the third group, referred to here as divergent PilZ-related domains, exhibit various deviations from that pattern.
One such deviation is seen in the stand-alone PilZ domain from two nearly identical proteins, Xc1028 from X. campestris and Xac1133 from X. citri (36, 37). These proteins lack the c-di-GMP-binding motifs and participate in the regulation of type IV pilus formation through their interaction with the EAL domain of the FimX protein and the pilus motor protein PilB (33, 34, 36). The loss of the N-terminal loop with its RXXXR motif is accompanied by the inversion of the direction of the N-terminal β-strand and conversion of the second β-strand into an α-helix (36, 37). Accordingly, the five remaining β-strands do not form a complete barrel, and the alignment of this structure against canonical PilZ is much shorter (72 aa residues), with an RMSD of 2.9 Å (Table 1 and Fig. 4B). The sequence of the eponymous PilZ protein (PA2960) from P. aeruginosa (32, 37) is very similar (67% identity) to those of Xc1028 and Xac1133, indicating that it, too, belongs to the group of divergent PilZ domains (abbreviated here as xPilZ).
The greatest variety of PilZ-related domains is found in a vast superfamily of two-domain proteins that combine various xPilZ domains at their N termini with canonical PilZ domains at their C termini. These proteins are generally similar to the previously described (16) alphaproteobacterial proteins with tandemly duplicated PilZ domains, such as B. diazoefficiens Bll4394 and Blr5568, but show various structural changes in their N-terminal PilZ-like domains. Based on the length and sequence similarity to the experimentally characterized flagellar brake proteins YcgR and MotI (22, 41, 43, 51), these proteins are often referred to as YcgR or YcgR-like. However, sequence similarity between these proteins is mostly limited to their common C-terminal PilZ domains. In contrast, their N-terminal domains exhibit various deviations from the PilZ core structure, as seen, for example, in the four structurally characterized domains: PilZN (YcgR, PF07317), PilZNR (YcgR_2, PF12945), DUF5634_N (PF18672), and the N-terminal domain of the transcriptional regulator MrkH (Fig. 4C to F). While all these domains have a β-barrel fold like the one in canonical PilZ, they all lack the C-terminal α-helix. They also show additional deviations from the PilZ-PilZ consensus of Bll4394 and Blr5568 proteins.
The fewest changes are seen in the GSU3033 protein of Geobacter sulfurreducens (GenBank accession no. AAR36425), which contains a canonical PilZ domain at its C terminus. The N-terminal domain of GSU3033 has been characterized by nuclear magnetic resonance (NMR) (PDB entry 2L1T) and, along with related deltaproteobacterial domains, placed in a separate Pfam family, DUF5634_N (PF18672). This domain, referred to as PilZN1 in Table 2, has the same N-terminal long loop followed by a six-stranded β-barrel as the canonical PilZ domain but lacks the C-terminal α-helix and both c-d-GMP-binding motifs; in addition, the N-terminal loop forms a short (3-aa) α-helix (Fig. 4C).
TABLE 2.
Noncanonical PilZ-related domains
| Domain name, Pfam entrya | Phylogenetic distribution | Representative example |
||
|---|---|---|---|---|
| Organism locus tagb | GenBank entry | Boundaries | ||
| tPilZ (PilZ_2), PF16823 | Proteobacteria, Nitrospirae, Thermodesulfobacteria | V. cholerae VC1885 | AAF95033 | 1–190 |
| P. aeruginosa PA2989 | AAG06377 | 1–230 | ||
| T. indicus Thein_0310 | AEH44194 | 1–190 | ||
| PilZN (YcgR), PF07317 | Betaproteobacteria, Gammaproteobacteria | E. coli YcgR | AAC74278 | 1–110 |
| P. putida PP_4397 | AAN69975 | 1–115 | ||
| PilZNR (YcgR_2), PF12945 | Proteobacteria, Firmicutes, others | V. cholerae VCA0042 | AAF95956 | 1–130 |
| B. subtilis BSU22910 | CAB14207 | 1–90 | ||
| PilZN1, PF18672 (DUF5634_N) | Desulfuromonadales | G. sulfurreducens GSU3033 | AAR36425 | 1–105 |
| PilZN2 | Enterobacteriales | K. pneumoniae MrkH (KP13_02245) | AEO27488 | 1–105 |
| PilZN3 | Spirochaetes | B. burgdorferi BB0733 | AAC67086 | 1–140 |
| T. denticola TDE0214 | AAS10711 | 1–140 | ||
| PilZN4 | Spirochaetes, Aquificae, Fibrobacteres | T. denticola TDE1318 | AAS11835 | 1–210 |
| A. aeolicus Aq_1917 | AAC07718 | 1–220 | ||
| PilZN5 | Actinobacteria | G. obscurus Gobs_0077 Gobs_0078 | ADB72887 | 1–95 |
| PilZN6 | Deltaproteobacteria | D. baarsii Deba_0512 | ADK83884 | 1–140 |
| D. baarsii Deba_3043 | ADK86396 | 1–140 | ||
| PilZN7 | Hungateiclostridiaceae | H. thermocellum Cthe_0108 | ABN51349 | 1–90 |
| H. thermocellum Cthe_0868 | ABN52102 | 1–100 | ||
PilZN1 to PilZN7 are tentative domain names, expected to be replaced as soon as these domains are experimentally characterized.
For complete names of the organisms, see the text or the respective GenBank entries.
The structure of MrkH, a transcriptional regulator of type 3 fimbria production in Klebsiella pneumoniae (52), reveals yet another N-terminal six-stranded β-barrel domain (PilZN2 in Table 2) fused to a canonical PilZ domain (27, 40). In PilZN2, the C-terminal α-helix is lost and the N-terminal loop is replaced by a short β-strand, followed by a 13-aa α-helix (Fig. 4D).
Like PilZN2, the PilZN (YcgR) domain structures from E. coli and Pseudomonas putida deviate from the canonical PilZ structure in the absence of the C-terminal α-helix and presence of a 13-aa α-helix after the β1 strand. In addition, they contain an 8-aa α-helix sandwiched between β5 and β6 (Fig. 4E). Finally, the PilZNR (YcgR_2) domain of the V. cholerae VCA0042 (PlzD) and B. subtilis MotI (DgrA) proteins contains mostly the same structural elements as the PilZN domain, but its first α-helix is much shorter, only 5 amino acid residues (Fig. 4F). In summary, all these domains show clear similarity to the canonical PilZ and can be considered divergent PilZ-related domains.
PilZ-related domains of unknown structure.
The 32nd release of the Pfam database (last accessed 31 August 2019) listed 8,676 sequences that had PilZ as their only recognized domain (15). More than 1,800 of them consisted of 180 to 350 amino acid residues, suggesting that, in addition to PilZ, they contained an unrecognized protein domain of a similar length. We have extracted such protein sequences from UniProt (53), clustered them by sequence similarity, and performed sequence analysis and secondary structure predictions for representatives of the largest clusters and diverse taxonomic lineages. Some of the largest clusters of proteins retrieved this way represented tPilZ domains and PilZ dimers, whose lengths also fall within the 180- to 350-aa range. However, this analysis also revealed several previously unrecognized domains, some of which, while not fitting the Pfam models for YcgR, YcgR_2, or DUF5634_N domains, were similar to the canonical PilZ in having the same (predicted) six-stranded β-barrel fold and likely represented additional xPilZ domains.
The list of previously and newly described noncanonical PilZ domains is presented in Table 2. Many of these domains show limited phylogenetic distribution, being found only in closely related members of a specific phylum, class, family, or just a single genus. For lack of a better description, we have tentatively named these novel variants PilZN3 and PilZN4 (found in spirochetes and members of Aquificae and Fibrobacteres), PilZN5 (found in actinomycetes), PilZN6 (found in Deltaproteobacteria), and PliZN7 (found in cellulose-degrading clostridia).
To our knowledge, B. burgdorferi PlzA (BB0733) and Treponema denticola TDE0214, which both have the PilZN3-PilZ domain architecture, are the only proteins with novel xPilZ domains that have been experimentally characterized; they both affect motility and virulence of the respective spirochetes (54–56). Secondary structure predictions indicate that the PilZN3 domain retains the C-terminal α-helix of PilZ and has three additional α-helices (Fig. S4A). While some spirochetes encode multiple PilZ-containing proteins, those with the PilZN3-PilZ architecture are the only ones found in Borrelia/Borreliella spp. and in Treponema pallidum, the causative agent of syphilis.
Treponema denticola also encodes another PilZ domain protein, TDE1318, which combines a canonical PilZ domain with a 210-aa region that consists of a six-stranded xPilZ domain without the C-terminal α-helix and a unique pentahelical subdomain at the N terminus (Fig. S4B). The first α-helix of this subdomain consists exclusively of uncharged (mostly hydrophobic) amino acid residues, indicating that it could localize to the membrane and serve as a membrane anchor for the entire protein. In addition to spirochetes, such proteins have been found in members of Aquificae and Fibrobacteres.
Finally, some new domain combinations can be found in the same genome in two or more paralogous forms. Thus, Geodermatophilus obscurus and several other actinobacteria encode two copies of the proteins with PilZN5-PilZ domain combination, Desulfarculus baarsii and other deltaproteobacteria encode two PilZN6-PilZ proteins, and such cellulose-degrading clostridia as Hungateiclostridium thermocellum and Pseudobacteroides cellulosolvens encode 9 and 11 proteins, respectively, with the PilZN7-PilZ domain architecture (Fig. S4C to E).
DISCUSSION
On the constantly growing list of c-di-GMP-binding proteins, the PilZ domain occupies a unique position, not only as the first c-di-GMP receptor to be identified and experimentally characterized (16, 19, 22–24) but also, along with the recently described MshEN (11, 12) and the inactivated GGDEF and EAL domains, as the only dedicated c-di-GMP-binding domains that are truly widespread in bacteria. Other c-di-GMP receptors have a relatively narrow phylogenetic distribution; many of them likely evolved as specific add-on adaptations to c-di-GMP binding within common protein superfamilies (13, 14, 57–59).
Full-length PilZ domains consisting of a six-stranded β-barrel and a C-terminal α-helix are encoded by the genomes from numerous bacterial phyla (see Pfam entry PF07238 or the web site https://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html). This widespread distribution, including their presence in such early-diverging phyla as Aquificae and Thermotogae, strongly suggests that this form of PilZ is the ancestral one, which is another reason why we refer to it as the canonical version. Structural comparisons show the presence of essentially the same canonical variant of PilZ in BcsA, YcgR, MotI, and MrkH proteins, where PilZ is the C-terminal domain, and also in Alg44, where PilZ is at the N terminus (Fig. 1). While certain stand-alone PilZ domains show disruption of the c-di-GMP-binding motifs and the resulting loss of c-di-GMP binding (Fig. 4B), PilZ domains in fusion proteins exhibit no such change and invariably retain the ability to bind c-di-GMP. It appears that the PilZ domains in BcsA, Alg44, and YcgR- and MotI-related proteins have evolved under strict selection that ensured preservation of c-di-GMP binding and, accordingly, their participation in the c-di-GMP signaling circuits.
In addition to these protein types, the PilZ domain can be found in other domain architectures (see the Pfam database [15] entry http://pfam.xfam.org/family/PilZ or Fig. 1 in references 10 and 22). Although PilZ domains in those proteins have not been structurally characterized, the high degree of sequence conservation suggests that they all retain the same PilZ structure, further expanding the list of canonical PilZ domains.
Except for the two-helical insert, tPilZ, the atypical tetramer-forming version of the PilZ domain, is very similar to the canonical version (Fig. 2 and 4A). The tPilZ domain is found in the Proteobacteria and in members of several other phyla (see Fig. S3 and Table S3 in the supplemental material). Because of the loss of c-di-GMP-binding motifs, members of the tPilZ family have often been overlooked. Thus, a comprehensive study of c-di-GMP regulation in L. pneumophila (60) did not even list the Lpg1926 protein as PilZ, even though this protein retains the c-di-GMP-binding motifs (Fig. S3). On the other hand, tPilZ domains of V. cholerae protein PlzB (VC1885) and P. aeruginosa PA2989 were just assumed to be the canonical PilZ domains (23, 49).
The tPilZ domain could have evolved fairly early in bacterial evolution through an insertion into the canonical pilZ gene of an ∼200-bp fragment encoding helices α2 and α3. However, outside gamma and delta subdivisions of Proteobacteria and the phylum Nitrospirae, the tPilZ domain is found in a relatively small number of organisms with a patchy phylogenetic distribution (Table S3). Thus, this version of PilZ likely spreads by horizontal gene transfer, which implies that it carries out some beneficial function(s). As is the case with canonical PilZ, certain members of the tetrameric PilZ family retain intact c-di-GMP-binding motifs, whereas others have totally lost them (Fig. S3) and appear to participate in regulation solely through protein-protein interactions. The recognition of the tPilZ domains as a specific subfamily of PilZ should stimulate investigation of their roles in bacterial regulation. Two tPilZ proteins, V. cholerae PlzB and X. campestris Xcc6021, have been reported to contribute to virulence despite their inability to bind c-di-GMP (23, 48), making a detailed characterization of this family an interesting and promising task.
Conservation of the C-terminal α-helix.
As noted above, canonical PilZ domains differ from xPilZ ones by the presence of a long C-terminal α-helix. This helix is preserved in the PilZ fusion proteins BcsA and Alg44 and PilZ dimers, as well as in the tetrameric PilZ domain (Fig. 2). Based on sequence comparisons, it has also been predicted in PilZ-containing chemotaxis receptors Tlp1 and Aer from Azospirillum brasilense (29, 30), Ser/Thr-type protein kinase Pkn1 (MXAN_1467) from Myxococcus xanthus (61), and many other PilZ fusion proteins. This helix is characterized by a large number of charged amino acid residues, particularly Arg and Lys, which makes it a prime candidate for protein-protein interactions. Indeed, in Caulobacter crescentus, overexpression of the PilZ protein DgrACC (GenBank accession no. AAK23578) caused motility block that could be alleviated by frameshift mutations in the C-terminal region of the protein, indicating the importance of this region for motility control (20). A detailed study of the YcgR protein from E. coli demonstrated the involvement of this α-helix (particularly residues Gln223 and lle227) in the interaction with the motor protein FliM (62). The role of the C-terminal α-helix has also been seen in other contexts. In P. aeruginosa MapZ, this helix mediates the interaction of PilZ with the methyltransferase CheR (63). In K. pneumoniae MrkH, it has been implicated in binding DNA (40). In the cellulose synthase structure, the C-terminal α-helix of PilZ is split into two, and both pack against the glycosyltransferase domain (BcsA has an additional long C-terminal α-helix after the PilZ domain) (25, 64). The C-terminal α-helix also plays a role in the interaction of the PilZ domain of X. citri with the type IV pilus biogenesis regulator FimX (34). Finally, in the PilZ domain of Alg44, C-terminal α-helices of two monomers interact, contributing to the formation of the PilZ dimer (39). These data clearly indicate that the C-terminal α-helix of PilZ, with its clustering of charged residues, is a key element mediating protein-protein interactions by the PilZ domain.
Role of the conserved hydrophobic residues.
While the presence of conserved hydrophobic residues in the PilZ domain could be seen even in its original description (16) and is clearly visible in the HMM logo of PilZ on the Pfam database (15) web site http://pfam.xfam.org/family/PF07238#logoBlock, the role of these residues has long remained overlooked. However, strict conservation of these residues in all canonical PilZ and tPilZ domains (Fig. 2), as well as limited conservation in xPilZ domains (Fig. S2 and data not shown), are hardly accidental. These residues appear to play several important roles in maintaining the structure and activity of various PilZ-related domains. First, by forming a hydrophobic core in the middle of the protein, these residues could play a key role in the folding of the PilZ domain. Given that these residues come from five different β-strands (Fig. 2), this hydrophobic core appears to hold the entire structure together, ensuring the rigidity of the entire β-barrel. This should be important irrespective of the ability of a particular PilZ domain to bind c-di-GMP and participate in protein-protein interactions. Second, in those PilZ domains that bind c-di-GMP, the fixed position of the central h residue of the c-di-GMP-interacting [D/N]hSXXG motif is involved in stabilizing the SXXG β-turn hairpin, which is important for c-di-GMP binding. Conversely, c-di-GMP binding can be expected to affect the organization of the hydrophobic pocket and the position of the C-terminal α-helix, which (i) provides two (sometimes three) hydrophobic residues to that pocket and (ii) plays a key role in protein-protein and protein-DNA interactions mediated by the PilZ domain (see above). This way, the h residue of the [D/N]hSXXG motif could serve as a molecular readout, connecting c-di-GMP binding to changes in the domain interactions in at least some PilZ domains (in cellulose synthase the read-out is movement of the gating loop, while in YcgR it is likely the distance between the two PilZ domains in a dimer [22, 25, 49]). However, verification of this proposed mechanism would require a detailed analysis of the subtle changes in high-resolution structures of PilZ with and without bound c-di-GMP.
Pathways of PilZ evolution.
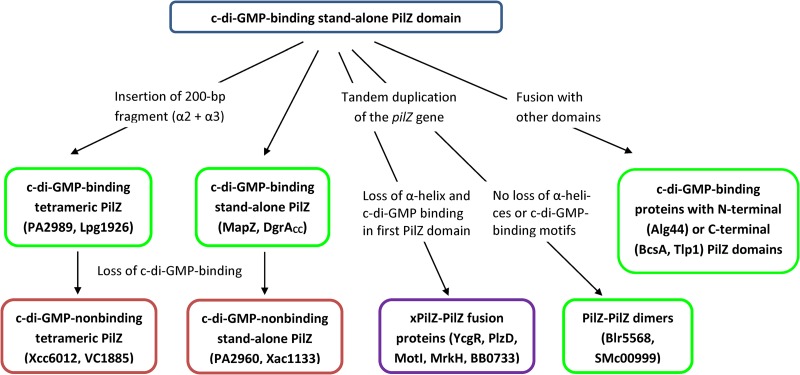
The widespread distribution of the canonical c-di-GMP-binding form of PilZ and its conservation in a variety of domain contexts suggests that the stand-alone form of PilZ was the ancestral version of this domain. From that, one of the pathways of PilZ evolution likely involved the insertion of a two-helical fragment that led to the formation of the tetramer-forming tPilZ domains (Fig. 5). For some reason, tetramer formation precluded domain fusions, such that tPilZ domains are almost always found in a stand-alone form. c-di-GMP binding motifs are seen in tPilZ proteins from several diverse phyla (Fig. S3 and Table S3), indicating that the c-di-GMP binding form was the ancestral one. However, the loss of the c-di-GMP binding motifs is a constant feature in the tPilZ family and can be traced even within a single genus, Pseudomonas (Fig. S3).
FIG 5.
Proposed pathways of PilZ evolution. c-di-GMP-binding versions are in green frames, nonbinding versions are in red frames, and the purple frame indicates that only the second (canonical) of the two PilZ-related domains is c-di-GMP binding. The proteins used as examples are described in the text or listed in Tables 1 and 2. See the text for details.
Another major pathway of PilZ evolution involved a variety of domain fusions. Some of them, such as those in BcsA, Alg44, Tlp1, and many other proteins, preserved the canonical PilZ structure and the ability to bind c-di-GMP. A special widespread variant of such fusions was the tandem duplication of the PilZ domain, which occasionally led to the formation of fusion proteins consisting of two canonical PilZ domains, such as, for example, B. diazoefficiens proteins Bll4394 and Blr5568 (16) and S. meliloti protein SMc00999. Alternatively, tandem duplication led to the loss of the C-terminal α-helix in the N-terminal PilZ domain, often accompanied by a variety of other structural changes, and ended up in xPilZ-PilZ domain architectures. This diversification of the N-terminal xPilZ domains gave rise to PilZN, PilZNR, PilZN1, PilZN2, and other domains, some of which are listed in Table 2. All these domains have lost the c-di-GMP binding motifs, relegating c-di-GMP binding to the interdomain linker at the beginning of the second (canonical) PilZ domain. The proposed pathways of PilZ evolution are schematically depicted in Fig. 5. We hope that this scheme will clarify the relationships between known PilZ, tPilZ, and xPilZ domains and will serve as a guide to discovering and characterizing new members of the PilZ superfamily.
MATERIALS AND METHODS
Structural alignment of PilZ-related domains.
The available three-dimensional structures of PilZ-related domains (see Table S1 in the supplemental material) were extracted from the PDB and MMDB databases and compared using Dali, VAST, and PDBeFold tools (46, 47, 65, 66). The lengths of aligned Cα traces, sequence identity in the aligned segments, and their root mean square deviation (RMSD) values were as reported by the Dali (Table 1) and VAST (Table S2) servers. Structural superpositions were generated with PyMOL (67) using the high-resolution structure of the stand-alone PilZ domain in the P. aeruginosa protein MapZ (PDB entry 5XLY_B, residues 4 to 107) as the reference and aligned β-strand segments from the Dali and VAST outputs as a guide. For clarity, the second C-terminal α-helix of MapZ (residues 109 to 121), which is not part of the PilZ domain core, was removed from Fig. 1 and 4 and Fig. S1. Structure-based sequence alignments of various PilZ domains were taken from Dali and VAST outputs and supplemented with the alignments generated by the HHpred tool (68) of the MPI Bioinformatics tool kit (69). The sequence logos of c-di-GMP-binding motifs were generated using the WebLogo program (70) from an alignment of 9,224 UniProt (53) sequences that was retrieved from Pfam (15) and manually edited to remove gaps and leave only those sequences where both motifs were intact.
Sequence analysis of tetramer-forming PilZ domains.
The initial lists of tPilZ domain-containing proteins were obtained using PSI-BLAST (71) searches against the NCBI protein database and HMMer (72) searches against UniProt (53) with the X. campestris protein XccB100_2234 (GenBank accession no. CAP51589, UniProt accession no. B0RT03, PDB entry 3RQA) as the query. We further carried out database searches with PSI-BLAST and HMMer, starting from the proteins in that initial list, and additionally employed PHI-BLAST (71) with specified KLD and, subsequently, expanded [KR]h[DEN] motifs. The taxonomic representation of tPilZ domains was evaluated through BLAST searches against phylum-specific protein sets (excluding Proteobacteria).
Identification of novel PilZ-related domains.
The list of UniProt entries containing PilZ as their only recognized domain was taken from Pfam (15) and sorted by length to extract those sequences whose lengths fell within the 180- to 350-aa range. These sequences were clustered with the MMseqs2 tool (73) of the MPI Bioinformatics tool kit (69), and representatives from the top 20 clusters were studied one by one. Additional clusters were included in the analyzed set based on the phylogenetic representation of their members. Selected representatives of each cluster were checked for the presence of known protein domains in their N-terminal regions using CD-Search (74) with relaxed parameters (expect value cutoff, 10), and the predicted structural similarity to PilZ was checked using HHPred (68, 69). Protein secondary structures were taken from the HHpred outputs and/or predicted using JPred (75).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NIH Intramural Research Program at the National Library of Medicine (M.Y.G.), by the Ministry of Education, Taiwan, Republic of China, under the ATU plan, and by the National Science Council, Taiwan, Republic of China (grant 102-2113-M005-006-MY3 to S.-H.C.).
We thank Rasika Harshey (University of Texas at Austin), Daria Shalaeva (University of Osnabrück), and Yuri Wolf (NCBI) for helpful suggestions.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teschler JK, Zamorano-Sanchez D, Utada AS, Warner CJ, Wong GC, Linington RG, Yildiz FH. 2015. Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol 13:255–268. doi: 10.1038/nrmicro3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conner JG, Zamorano-Sanchez D, Park JH, Sondermann H, Yildiz FH. 2017. The ins and outs of cyclic di-GMP signaling in Vibrio cholerae. Curr Opin Microbiol 36:20–29. doi: 10.1016/j.mib.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenal U, Reinders A, Lori C. 2017. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 5.Su J, Zou X, Huang L, Bai T, Liu S, Yuan M, Chou SH, He YW, Wang H, He J. 2016. DgcA, a diguanylate cyclase from Xanthomonas oryzae pv. oryzae regulates bacterial pathogenicity on rice. Sci Rep 6:25978. doi: 10.1038/srep25978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall CL, Lee VT. 2018. Cyclic-di-GMP regulation of virulence in bacterial pathogens. Wiley Interdiscip Rev RNA 9:10.1002/wrna.1454. doi: 10.1002/wrna.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valentini M, Filloux A. 2019. Multiple roles of c-di-GMP signaling in bacterial pathogenesis. Annu Rev Microbiol 73:387–406. doi: 10.1146/annurev-micro-020518-115555. [DOI] [PubMed] [Google Scholar]
- 8.Mills E, Pultz IS, Kulasekara HD, Miller SI. 2011. The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell Microbiol 13:1122–1129. doi: 10.1111/j.1462-5822.2011.01619.x. [DOI] [PubMed] [Google Scholar]
- 9.Chou SH, Galperin MY. 2016. Diversity of c-di-GMP-binding proteins and mechanisms. J Bacteriol 198:32–46. doi: 10.1128/JB.00333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheang QW, Xin L, Chea RYF, Liang ZX. 2019. Emerging paradigms for PilZ domain-mediated c-di-GMP signaling. Biochem Soc Trans 47:381–388. doi: 10.1042/BST20180543. [DOI] [PubMed] [Google Scholar]
- 11.Roelofs KG, Jones CJ, Helman SR, Shang X, Orr MW, Goodson JR, Galperin MY, Yildiz FH, Lee VT. 2015. Systematic identification of cyclic-di-GMP binding proteins in Vibrio cholerae reveals a novel class of cyclic-di-GMP-binding ATPases associated with type II secretion systems. PLoS Pathog 11:e1005232. doi: 10.1371/journal.ppat.1005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YC, Chin KH, Tu ZL, He J, Jones CJ, Sanchez DZ, Yildiz FH, Galperin MY, Chou SH. 2016. Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain. Nat Commun 7:12481. doi: 10.1038/ncomms12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubey BN, Lori C, Ozaki S, Fucile G, Plaza-Menacho I, Jenal U, Schirmer T. 2016. Cyclic di-GMP mediates a histidine kinase/phosphatase switch by noncovalent domain cross-linking. Sci Adv 2:e1600823. doi: 10.1126/sciadv.1600823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nesper J, Hug I, Kato S, Hee CS, Habazettl JM, Manfredi P, Grzesiek S, Schirmer T, Emonet T, Jenal U. 2017. Cyclic di-GMP differentially tunes a bacterial flagellar motor through a novel class of CheY-like regulators. Elife 6:e28842. doi: 10.7554/eLife.28842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 17.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 18.Römling U, Galperin MY. 2017. Discovery of the second messenger cyclic di-GMP. Methods Mol Biol 1657:1–8. doi: 10.1007/978-1-4939-7240-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryjenkov DA, Simm R, Römling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP. The PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 20.Christen M, Christen B, Allan MG, Folcher M, Jeno P, Grzesiek S, Jenal U. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci U S A 104:4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. 2007. The second messenger bis-(3’-5’)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol 65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 22.Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. 2007. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J 26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pratt JT, Tamayo R, Tischler AD, Camilli A. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem 282:12860–12870. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramelot TA, Yee A, Cort JR, Semesi A, Arrowsmith CH, Kennedy MA. 2007. NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins 66:266–271. doi: 10.1002/prot.21199. [DOI] [PubMed] [Google Scholar]
- 25.Morgan JL, McNamara JT, Zimmer J. 2014. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21:489–496. doi: 10.1038/nsmb.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan XF, Xin L, Yen JT, Zeng Y, Jin S, Cheang QW, Fong R, Chiam KH, Liang ZX, Gao YG. 2018. Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein. J Biol Chem 293:100–111. doi: 10.1074/jbc.M117.815704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, He Q, Su K, Gao F, Huang Y, Lin Z, Zhu D, Gu L. 2016. The PilZ domain of MrkH represents a novel DNA binding motif. Protein Cell 7:766–772. doi: 10.1007/s13238-016-0317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu L, Venkataramani P, Ding Y, Liu Y, Deng Y, Yong GL, Xin L, Ye R, Zhang L, Yang L, Liang ZX. 2016. A cyclic di-GMP-binding adaptor protein interacts with histidine kinase to regulate two-component signaling. J Biol Chem 291:16112–16123. doi: 10.1074/jbc.M116.730887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell MH, Bible AN, Fang X, Gooding J, Campagna S, Gomelsky M, Alexandre G. 2013. Integration of the second messenger c-di-GMP into the chemotactic signaling pathway promotes sensory adaptation. mBio 4:e00001-13. doi: 10.1128/mBio.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Neal L, Akhter S, Alexandre G. 2019. A PilZ-containing chemotaxis receptor mediates oxygen and wheat root sensing in Azospirillum brasilense. Front Microbiol 10:312. doi: 10.3389/fmicb.2019.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pultz IS, Christen M, Kulasekara HD, Kennard A, Kulasekara B, Miller SI. 2012. The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol Microbiol 86:1424–1440. doi: 10.1111/mmi.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alm RA, Bodero AJ, Free PD, Mattick JS. 1996. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol 178:46–53. doi: 10.1128/jb.178.1.46-53.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chin KH, Kuo WT, Yu YJ, Liao YT, Yang MT, Chou SH. 2012. Structural polymorphism of c-di-GMP bound to an EAL domain and in complex with a type II PilZ-domain protein. Acta Crystallogr D Biol Crystallogr 68:1380–1392. doi: 10.1107/S0907444912030594. [DOI] [PubMed] [Google Scholar]
- 34.Guzzo CR, Dunger G, Salinas RK, Farah CS. 2013. Structure of the PilZ-FimXEAL-c-di-GMP complex responsible for the regulation of bacterial type IV pilus biogenesis. J Mol Biol 425:2174–2197. doi: 10.1016/j.jmb.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 35.Qi Y, Xu L, Dong X, Yau YH, Ho CL, Koh SL, Shochat SG, Chou SH, Tang K, Liang ZX. 2012. Functional divergence of FimX in PilZ binding and type IV pilus regulation. J Bacteriol 194:5922–5931. doi: 10.1128/JB.00767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guzzo CR, Salinas RK, Andrade MO, Farah CS. 2009. PILZ protein structure and interactions with PILB and the FIMX EAL domain: implications for control of type IV pilus biogenesis. J Mol Biol 393:848–866. doi: 10.1016/j.jmb.2009.07.065. [DOI] [PubMed] [Google Scholar]
- 37.Li TN, Chin KH, Liu JH, Wang AH, Chou SH. 2009. XC1028 from Xanthomonas campestris adopts a PilZ domain-like structure without a c-di-GMP switch. Proteins 75:282–288. doi: 10.1002/prot.22330. [DOI] [PubMed] [Google Scholar]
- 38.Habazettl J, Allan MG, Jenal U, Grzesiek S. 2011. Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout. J Biol Chem 286:14304–14314. doi: 10.1074/jbc.M110.209007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitney JC, Whitfield GB, Marmont LS, Yip P, Neculai AM, Lobsanov YD, Robinson H, Ohman DE, Howell PL. 2015. Dimeric c-di-GMP is required for post-translational regulation of alginate production in Pseudomonas aeruginosa. J Biol Chem 290:12451–12462. doi: 10.1074/jbc.M115.645051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schumacher MA, Zeng W. 2016. Structures of the activator of K. pneumonia biofilm formation, MrkH, indicates PilZ domains involved in c-di-GMP and DNA binding. Proc Natl Acad Sci U S A 113:10067–10072. doi: 10.1073/pnas.1607503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian S, Gao X, Dann CE III, Kearns DB. 2017. MotI (DgrA) acts as a molecular clutch on the flagellar stator protein MotA in Bacillus subtilis. Proc Natl Acad Sci U S A 114:13537–13542. doi: 10.1073/pnas.1716231114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Y, Yuan Z, Gu L. 2017. Structural basis for the regulation of chemotaxis by MapZ in the presence of c-di-GMP. Acta Crystallogr D Struct Biol 73:683–691. doi: 10.1107/S2059798317009998. [DOI] [PubMed] [Google Scholar]
- 43.Ko J, Ryu KS, Kim H, Shin JS, Lee JO, Cheong C, Choi BS. 2010. Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by Pilz domain proteins. J Mol Biol 398:97–110. doi: 10.1016/j.jmb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Whitney JC, Hay ID, Li C, Eckford PD, Robinson H, Amaya MF, Wood LF, Ohman DE, Bear CE, Rehm BH, Howell PL. 2011. Structural basis for alginate secretion across the bacterial outer membrane. Proc Natl Acad Sci U S A 108:13083–13088. doi: 10.1073/pnas.1104984108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujiwara T, Komoda K, Sakurai N, Tajima K, Tanaka I, Yao M. 2013. The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A. Biochem Biophys Res Commun 431:802–807. doi: 10.1016/j.bbrc.2012.12.103. [DOI] [PubMed] [Google Scholar]
- 46.Holm L, Laakso LM. 2016. Dali server update. Nucleic Acids Res 44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madej T, Lanczycki CJ, Zhang D, Thiessen PA, Geer RC, Marchler-Bauer A, Bryant SH. 2014. MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res 42:D297–D303. doi: 10.1093/nar/gkt1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li TN, Chin KH, Fung KM, Yang MT, Wang AH, Chou SH. 2011. A novel tetrameric PilZ domain structure from xanthomonads. PLoS One 6:e22036. doi: 10.1371/journal.pone.0022036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. 2010. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laventie BJ, Nesper J, Ahrne E, Glatter T, Schmidt A, Jenal U. 2015. Capture compound mass spectrometry–a powerful tool to identify novel c-di-GMP effector proteins. J Vis Exp 97:e51404. doi: 10.3791/51404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hou Y, Yang W-S, Hong Y, Wang D-C, Li D-F. Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility in Escherichia coli. J Biol Chem, in press. doi: 10.1074/jbc.RA119.009739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilksch JJ, Yang J, Clements A, Gabbe JL, Short KR, Cao H, Cavaliere R, James CE, Whitchurch CB, Schembri MA, Chuah ML, Liang ZX, Wijburg OL, Jenney AW, Lithgow T, Strugnell RA. 2011. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog 7:e1002204. doi: 10.1371/journal.ppat.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.The UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res 43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freedman JC, Rogers EA, Kostick JL, Zhang H, Iyer R, Schwartz I, Marconi RT. 2010. Identification and molecular characterization of a cyclic-di-GMP effector protein, PlzA (BB0733): additional evidence for the existence of a functional cyclic-di-GMP regulatory network in the Lyme disease spirochete, Borrelia burgdorferi. FEMS Immunol Med Microbiol 58:285–294. doi: 10.1111/j.1574-695X.2009.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pitzer JE, Sultan SZ, Hayakawa Y, Hobbs G, Miller MR, Motaleb MA. 2011. Analysis of the Borrelia burgdorferi cyclic-di-GMP-binding protein PlzA reveals a role in motility and virulence. Infect Immun 79:1815–1825. doi: 10.1128/IAI.00075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bian J, Liu X, Cheng YQ, Li C. 2013. Inactivation of cyclic di-GMP binding protein TDE0214 affects the motility, biofilm formation, and virulence of Treponema denticola. J Bacteriol 195:3897–3905. doi: 10.1128/JB.00610-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsuyama BY, Krasteva PV, Baraquet C, Harwood CS, Sondermann H, Navarro MV. 2016. Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 113:E209–E218. doi: 10.1073/pnas.1523148113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Z, Wu Z, Zhang J. 2016. Crystal structure of the YajQ-family protein XC_3703 from Xanthomonas campestris pv. campestris. Acta Crystallogr F Struct Biol Commun 72:720–725. doi: 10.1107/S2053230X16013017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levi A, Folcher M, Jenal U, Shuman HA. 2011. Cyclic diguanylate signaling proteins control intracellular growth of Legionella pneumophila. mBio 2:e00316. doi: 10.1128/mBio.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Munoz-Dorado J, Inouye S, Inouye M. 1991. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell 67:995–1006. doi: 10.1016/0092-8674(91)90372-6. [DOI] [PubMed] [Google Scholar]
- 62.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu L, Xin L, Zeng Y, Yam JK, Ding Y, Venkataramani P, Cheang QW, Yang X, Tang X, Zhang LH, Chiam KH, Yang L, Liang ZX. 2016. A cyclic di-GMP-binding adaptor protein interacts with a chemotaxis methyltransferase to control flagellar motor switching. Sci Signal 9:ra102. doi: 10.1126/scisignal.aaf7584. [DOI] [PubMed] [Google Scholar]
- 64.Morgan JL, Strumillo J, Zimmer J. 2013. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493:181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holm L. 2019. Benchmarking fold detection by DaliLite v.5. Bioinformatics 2019:btz536. doi: 10.1093/bioinformatics/btz536. [DOI] [PubMed] [Google Scholar]
- 66.Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 67.DeLano WL. 2010. The PyMOL molecular graphics system. Schrödinger, LLC, New York, NY. [Google Scholar]
- 68.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. 2018. A completely reimplemented MPI Bioinformatics Toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 70.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST–a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. 2018. HMMER web server: 2018 update. Nucleic Acids Res 46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mirdita M, Steinegger M, Söding J. 2019. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics 35:2856–2858. doi: 10.1093/bioinformatics/bty1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Derbyshire MK, Gonzales NR, Lu S, He J, Marchler GH, Wang Z, Marchler-Bauer A. 2015. Improving the consistency of domain annotation within the Conserved Domain Database. Database 2015:bav012. doi: 10.1093/database/bav012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.