The most common intracellular symbiont on the planet—Wolbachia pipientis—is infamous largely for the reproductive manipulations induced in its host. However, more recent evidence suggests that this bacterium may also serve as a nutritional mutualist in certain host backgrounds and for certain metabolites. We performed a large-scale analysis of conserved gene content across all sequenced Wolbachia genomes to infer potential nutrients made by these symbionts.
KEYWORDS: Insects, metabolites, mutualism, symbiosis
ABSTRACT
The most common intracellular symbiont on the planet—Wolbachia pipientis—is infamous largely for the reproductive manipulations induced in its host. However, more recent evidence suggests that this bacterium may also serve as a nutritional mutualist in certain host backgrounds and for certain metabolites. We performed a large-scale analysis of conserved gene content across all sequenced Wolbachia genomes to infer potential nutrients made by these symbionts. We review and critically evaluate the prior research supporting a beneficial role for Wolbachia and suggest future experiments to test hypotheses of metabolic provisioning.
INTRODUCTION
Wolbachia is a ubiquitous bacterial symbiont of nematodes, insects, and other arthropods. It is infamous for its prevalence across insects, where 40 to 60% of species harbor the infection (1, 2). One way that Wolbachia can spread through insect populations is by reproductive manipulations, which include sperm-egg incompatibility (so-called cytoplasmic incompatibility [CI]), parthenogenesis induction, male killing, and feminization (3). However, reproductive manipulation alone cannot explain Wolbachia’s prevalence and frequency within insects (4). For example, Wolbachia bacteria that do not cause any reproductive manipulations, such as strain wAu, can spread quite efficiently (5), suggesting that Wolbachia provides some other benefit to its hosts that allows them to increase in frequency. Additionally, Wolbachia infections entering new populations will find it quite difficult to establish themselves using reproductive manipulations alone; assuming a net fitness cost to the host, the Wolbachia infection frequency in a population must be above some threshold for reproductive manipulations to be effective at maintaining the infection (6–8). This begs the question: how is Wolbachia maintained without reproductive manipulation?
One way that Wolbachia may be maintained in insect populations is by providing mutualistic benefits. Although Wolbachia is occasionally horizontally transmitted (9) or introgressed into new host backgrounds (10), it is primarily maternally transmitted (3, 11). Therefore, Wolbachia’s fitness is dependent on that of the host. Any mutualistic benefit Wolbachia can provide to the host, assuming that the immediate cost to itself is not too great, would increase its own fitness (12). Indeed, many maternally transmitted bacterial symbionts provide nutrients to their insect hosts, and by examining the genomic content of a symbiont, one can hypothesize as to potential nutrients supplied (13). But could Wolbachia be both Jekyll and Hyde (14)? Here, we review what is known and unknown about Wolbachia with regard to nutritional mutualism. Based on a large-scale genomic comparison (natural experiments of gene gain and loss), we suggest methods to directly test hypotheses of nutrient supplementation by Wolbachia.
WOLBACHIA IS NOT MONOLITHIC: STRAIN AND HOST DIVERSITY
Wolbachia strains infect a diversity of hosts, from spiders to isopods and from nematodes to insects (1, 2). We have the most genomic sampling from filarial nematode and insect-infecting strains (clades A, B, C, and D). In addition, Wolbachia-induced effects have been best explored in these two broad host types. The symbionts, however, will differ in regard to the phenotype induced in the host and also their tissue localization. For example, in leaf-cutting ants, Wolbachia is found in the foregut, both intra- and extracellularly, and therefore seems well poised to provide nutritional supplementation (15). Importantly, to understand the data on Wolbachia nutritional mutualisms, we must highlight that Wolbachia strains infecting the filarial nematodes are quite different from the insect-associated strains. Filarial nematode-infecting Wolbachia strains (clades C and D) have smaller genomes than their arthropod-associated counterparts (clades A and B) (16–18), and their genomes harbor fewer predicted type IV secretion system effectors (16, 19). These facts, coupled to prior observations that nematode-associated Wolbachia strains exhibit a pattern of cocladogenesis with their hosts (20, 21) and that clearing of the symbiont is detrimental to the nematode (22), have led researchers to suggest that the C and D clade Wolbachia may be mutualistic (18). Wolbachia is believed to supplement a large array of nutrients in the nematodes, ranging from heme to riboflavin/flavin adenine dinucleotide (FAD) (16, 23), although few have been experimentally tested. Therefore, because their hosts and symbiotic contexts are so different, and because the Wolbachia clades have been diverging for millions of years (20, 24), there are likely significant differences in the mechanisms by which the different clades of symbionts interact with their hosts. Evidence taken from experiments based on one specific host and symbiont may not apply to other Wolbachia strains.
MAKING A CASE FOR NUTRITIONAL MUTUALISM
When the genome of the Wolbachia pipientis strain wMel was first sequenced, it was hypothesized that Wolbachia strains are auxotrophs, consuming host amino acids, as they cannot synthesize their own (25). More recently, comparative genomics and metabolic modeling support the idea of Wolbachia strains as metabolic parasites (26). However, although the microbes may acquire amino acids from the host, many new studies have suggested that Wolbachia strains are nutritional mutualists (a comprehensive review of older literature can be found in reference 12). The use of comparative genomics to infer Wolbachia's nutritional contributions to the host are fraught with caveats (Fig. 1). If a Wolbachia genome is truly missing the genes which encode enzymes in a specific pathway, then that Wolbachia strain probably does not make that product. However, the absence of a gene is often not confirmed, as Wolbachia assemblies are often incomplete and highly fragmented due to repetitive regions in the genome. On top of that, if a Wolbachia genome includes the genes for enzymes to synthesize a specific product, this does not necessarily imply that it shares that product with the host, and even if Wolbachia could share the product, the host may not need it. Indeed, all Wolbachia genomes encode the ability to convert fructose to phosphoenolpyruvate via the glycolytic pathway, but this pathway is largely redundant with the host metabolic capabilities and perhaps is not useful under most ecological contexts. So, in order for Wolbachia’s nutritional supplementation to be selected for, (i) Wolbachia has to be capable of making the nutrient, (ii) must supply it to the host somehow (either Wolbachia is consumed intracellularly, the nutrient leaks through the membrane, or there is a specialized transporter), and (iii) the nutrient should be relevant to host fitness (Fig. 1). Most of the evidence in support of Wolbachia as a nutritional mutualist has come from genomic studies, and in few cases have authors cleared hosts of Wolbachia and supplemented them with nutrients assumed to be provisioned.
FIG 1.
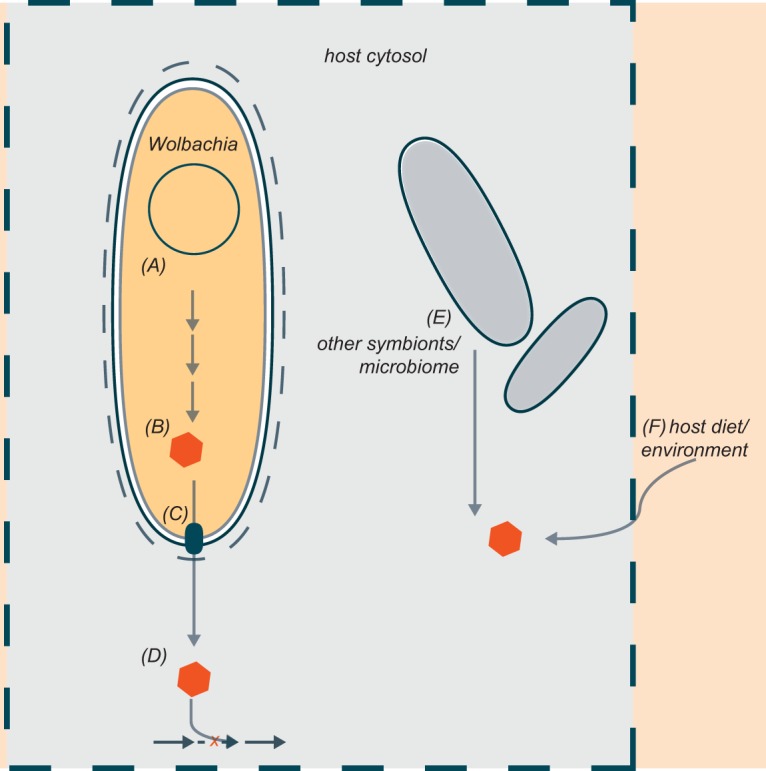
Schematic representation of Wolbachia supplementation of host nutrients. Wolbachia must (A) encode the enzymes for synthesis of said nutrient in its genome, (B) express the enzymes and generate the nutrient, and (C) export the nutrient somehow to the host, where it (D) complements a missing pathway important for host physiology. Ecological context impinges upon any nutrient supplementation if (E) other symbionts and the microbiome can supply the nutrient or (F) the host can acquire it from the surrounding environment or its diet.
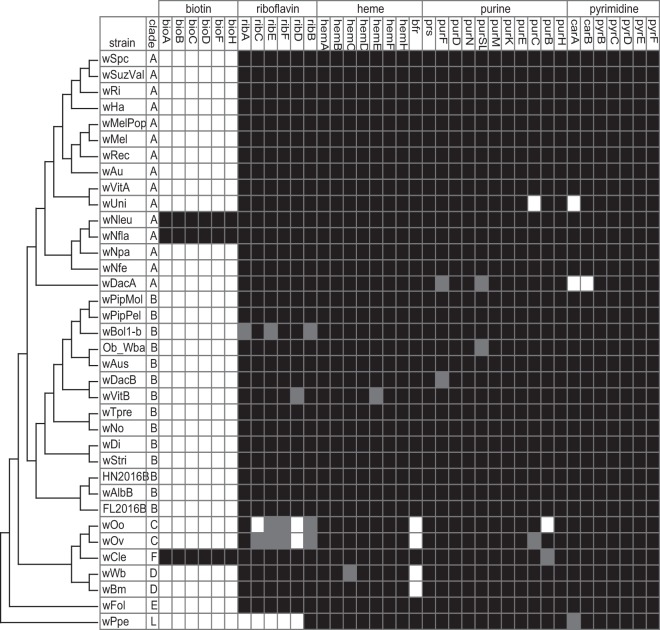
In the literature, several metabolic pathways in the host have been speculated to be supplemented or modulated by Wolbachia (15, 23, 27–32), from B vitamins to nucleotides. Comparative genomics is a first step to identifying metabolic capabilities across Wolbachia strains that could be involved in nutritional mutualism. Additionally, identifying Wolbachia strains that differ in their abilities to make specific nutrients leads to straightforward laboratory assays to confirm the nutritional supplementation suggested by genomic content. Below, we analyze the 36 publicly available Wolbachia genomes spanning clades A, B, C, D, E, F, and L for the presence/absence of genes encoding enzymes involved in the production of these nutrients (Fig. 2). We discuss previous experiments performed to support the provisioning of specific nutrients and suggest future areas of research to test Wolbachia’s role as a “Jekyll and Hyde” symbiont.
FIG 2.
The presence of enzyme-encoding loci involved in biotin, riboflavin, heme, purine, and pyrimidine biosynthesis is shown across 36 publicly available Wolbachia genomes. Existing functional annotation data (NCBI’s Prokaryotic Genome Annotation Pipeline [PGAP]) were used to identify enzymes across draft genomes and orthologs defined by complete linkage clustering of reciprocal blast hits. Black, present in the assembly; gray, present but pseudogenized (as defined by NCBI); white, absent from the assembly.
EVIDENCE FOR WOLBACHIA AS A NUTRITIONAL MUTUALIST
B vitamins.
The most obvious case where we find clear evidence for Wolbachia’s role as a nutritional mutualist in insects is in the bedbug, where Wolbachia pipientis strain wCle, naturally infecting the bedbug Cimex lectularius, provisions B vitamins to its host. In this system, we see clear cocladogenesis between the bedbugs and the Wolbachia infecting them (33). If cleared of the infection with antibiotics, bedbugs survive only if supplemented with B vitamins (34). It was subsequently suggested that biotin (B7) might be the vitamin in question, as genomic analyses found a biotin operon in the genome of strain wCle (35). Importantly, Wolbachia in the bedbug is found in bacteriocytes (34), specialized cells that house nutritional symbionts in various insects. Wolbachia is also bacteriocyte associated in the termite, infected by the related strain wCtub; this localization suggests that these specific symbionts may be nutritional mutualists. Interestingly, the bioA gene was amplified from the wCtub symbionts, suggesting that they may also harbor the biotin biosynthesis operon (30). This operon was unique to the F clade symbionts until the Wolbachia strains that infect Nomada bees (wNfla/wNleu) were sequenced (28). The biotin operon in the Nomada bee Wolbachia is flanked by insertion sequence (IS) elements, suggesting that it may have been horizontally acquired. So far, the biotin biosynthesis loci (bioABCDFH) have been identified only in these two disparate clades of Wolbachia, the F and A clades (Fig. 2).
Based on the genomics of conserved B-vitamin pathways in Wolbachia genomes, riboflavin (vitamin B2) has also been suggested as something the symbiont could contribute. The enzymes encoding the synthesis of riboflavin are conserved across many Wolbachia genomes (32) (although, interestingly, not in the clade C or L symbionts) (Fig. 2) and experiments in the bedbug system suggest that this nutrient also benefits the host (32). One might assume, then, that if the host were unable to take up riboflavin from its environment, it could rely on Wolbachia to generate that vitamin. In contrast, when mosquito cells harboring Wolbachia strain wStri (from the leafhopper Laodelphax striatellus) are inhibited from taking up riboflavin in their media, Wolbachia loads are reduced intracellularly (27); this is perhaps not what you would expect if the symbiont is specifically provisioning riboflavin. It could be that the host consumes Wolbachia intracellularly and therefore avails itself of all Wolbachia-derived nutrients, leading to the loss of the infection. However, the experiments are confounded by the fact that wStri is not the native Wolbachia strain for Aedes albopictus. Further research is needed to understand the mechanism by which host and Wolbachia B vitamin metabolism are related, the conditions under which this vitamin is provisioned, and how the vitamin is provided to the host.
Heme and iron homeostasis.
Does Wolbachia provide the host with heme or modulate host iron metabolism (29)? Cells use iron for a variety of purposes, including energy conservation, oxygen transport, and production of heme. Heme itself is an important cofactor in many metalloproteins, the most famous of which is hemoglobin, which coordinates the transfer of oxygen throughout the bloodstream (36, 37). Heme is also found in a variety of other metalloproteins, where it is involved in electron transfer and catalysis. The loci encoding the enzymatic components of this pathway are hemABCDEFH and are found complete across all sequenced Wolbachia genomes; therefore, all sequenced Wolbachia strains seem metabolically capable of synthesizing heme (Fig. 2). Importantly, since many sequenced nematodes do not encode enzymes to synthesize heme and other vitamins (16, 38–40), the suggestion that Wolbachia might provide them may be true. In contrast, Caenorhabditis elegans does not encode heme biosynthesis and is thought to acquire it from its environment; therefore, just because the animal cannot make a nutrient does not necessarily mean that it requires a symbiont to provide it (Fig. 1).
Evidence for Wolbachia’s supplementation of host heme in filarial nematodes comes by way of both comparative genomics and chemical inhibition (23, 41). Because the filarial nematode genome of Brugia malayi does not encode enzymes for heme biosynthesis and the associated Wolbachia strain wBm does, it is hypothesized that Brugia might get heme from Wolbachia (23). In a subsequent study, the authors identified a chemical inhibitor of the wBm δ-aminolevulinic acid dehydratase (encoded by the hemB gene in the Wolbachia species strain wBm genome) (41). Treatment of a different Wolbachia-infected filarial nematode (Litomosoides sigmodontis) with this inhibitor resulted in death of the nematode (41), suggesting that Wolbachia’s supplementation of heme to the host is a critical component of the mutualism. However, we cannot rule out that hemB is critical to Wolbachia’s own ability to function and that disruption of Wolbachia cell biology results in filaricide. Nonetheless, targeting heme biosynthesis is a promising avenue for pursuit of antifilarial therapies (23).
How would Wolbachia-synthesized heme be transported to the host cell? Many other pathogenic, invading microbes perform the opposite task, scavenging iron from the host for survival, and many do this via heme transporters (42). For example, the outer membrane heme transporter HutA in Vibrio cholerae binds to host heme and is upregulated under low iron conditions (43, 44). Wolbachia, if providing heme to the host, would have to have a transporter function in the opposite direction (Fig. 1). All sequenced Wolbachia genomes encode an inner membrane heme exporter (ccmA and ccmC, based on existing NCBI annotations), and in Escherichia coli, these proteins are predicted to be involved in the microbe’s cytochrome maturation (45). However, although uncharacterized outer membrane proteins are encoded by Wolbachia genomes (46, 47), no evidence exists that these proteins participate in heme transport.
Regardless of whether Wolbachia supplements host heme, as an intracellular bacterial symbiont, Wolbachia likely acquires iron from the host. Therefore, Wolbachia infection may alter host iron metabolism in ways that are phenotypically perceptible. Indeed, Wolbachia pipientis strain wMel seems to increase fecundity in fruit flies (Drosophila melanogaster) under high-iron diets; rearing flies on diets rich in FeCl3 reduces fecundity, but a Wolbachia infection seems to modulate that reduction (48). Under a low-iron diet, the results were more mixed, with a modest and less consistent effect of Wolbachia infection (48). In a related study, Wolbachia bacterioferritin gene expression increased in the presence of high iron in three different host backgrounds, while host ferritin was downregulated (31), suggesting that Wolbachia may be modulating iron homeostasis; Wolbachia bacterioferritin may be binding available iron, leaving the host with a reduced need for cytosolic ferritin (31). Interestingly, the Wolbachia infecting filarial nematodes (clades C and D) lack bacterioferritin in their genomes (Fig. 2). As Wolbachia completely depends on the host for iron, it and the host must compete or share the iron in the host’s diet. As yet, it is unclear what the ecological relevance of Wolbachia’s modulation of high iron toxicity would be for infected insects; certainly, the importance of the phenotype depends on the host’s diet. If hosts encounter high-iron environments with some frequency, then Wolbachia could provide an iron homeostasis benefit.
Nucleotides.
Many rickettsia species are well-known nucleotide parasites, encoding their own ATP/ADP translocase to siphon off this important currency from host cells (49). Metabolic modeling suggests that many rickettsia species may also consume host GMP and UMP (50). Wolbachia genomes, however, do not encode a translocase. Instead, it was recently suggested that Wolbachia might actually provision ATP (17). In a nematode symbiont (wOo), researchers found that Wolbachia highly expresses nucleotide and nucleoside metabolism proteins (17). In contrast, the authors suggested that vitamins and cofactors are not exchanged because enzymes for these pathways were poorly represented in their proteomics analysis (17). Based on genomic comparisons across Wolbachia, a connection between iron metabolism and nucleotide provisioning was suggested; perhaps Wolbachia take up host iron to synthesize components of their electron transport chain, by which they can synthesize ATP for the host (29). The nematode-associated strain wBm, for example, retains the de novo nucleotide biosynthesis pathways, and perhaps, when demand is high, wBm could supply these to the host (16). Indeed, de novo nucleotide biosynthesis genes of both purines and pyrimidines are conserved across most Wolbachia strains sequenced (Fig. 2). Therefore, many Wolbachia strains could potentially supplement host nucleotide metabolism.
Future directions.
Wolbachia is a primarily maternally transmitted symbiont that is most well known for its ability to manipulate host reproduction (3). Because Wolbachia is primarily maternally transmitted, we expect that benefits provided to the host would be selected for, as they would increase host fitness and, by proxy, Wolbachia fitness (48). Additionally, because many Wolbachia strains do not manipulate host reproduction (4), the maintenance of Wolbachia within host populations must rely on other phenotypes, possibly nutritional mutualism. From B vitamins to iron to nucleotides, all of the hypotheses outlined above for Wolbachia’s nutritional mutualism require testing. The only strong physiological evidence for direct metabolic provisioning comes from the bedbug-wCle model, where B-vitamin supplementation rescues Wolbachia clearing. However, the comparative genomics analyses performed here suggest natural comparisons that can be performed across host-symbiont combinations (Fig. 2). For example, nutritional provisioning in the case of the Nomada bees and their biotin operon-containing Wolbachia strains is questionable, given the vitamin content of pollen (28). However, this hypothesis can be tested, as two Wolbachia strains infecting the Nomada bees (wNfla and wNleu) would be expected to express the biotin biosynthesis operon, and this expression should be absent from the related strains wNfe and wNpa (Fig. 2), which lack the operon. One might also expect that bees with strain wNfla or wNfleu would be able to subsist on diets low in biotin, while the opposite would be true of bees with strains wNfe or wNpa.
We propose the Drosophila system as a powerful and straightforward genetic model system in which to identify metabolites supplemented by Wolbachia. Drosophila melanogaster is an excellent model host that has been used successfully to mechanistically identify Wolbachia-host interactions (51–57). Additionally, the flies can be transinfected with other Wolbachia strains, both in cell lines and in whole animals, and the microbiome easily controlled. Based on the genomic analyses, for example, strain wMel could supplement host heme, purines, pyrimidines, and riboflavin but not biotin (Fig. 2). Hypotheses based on genomic content would be straightforward to test using defined diets coupled to fly genetics and labeled precursors. For example, one could test whether Wolbachia infection increases fecundity using a holidic diet without riboflavin. A negative control could be a diet excluding biotin, which Wolbachia species strain wMel is not expected to synthesize and therefore would not rescue. Similarly, one could track the production of nucleotides by Wolbachia by providing labeled amino acid precursors to a Drosophila mutant unable to generate its own nucleotides (mutations in enzymes such as rudimentary or rudimentary-like [58]). We would then expect that any mRNAs produced by the host would include the label from Wolbachia-derived nucleotides.
In summary, comparative genomics is a powerful starting point in understanding host-microbe symbioses, and increasing evidence suggests that Wolbachia can supplement host nutrition in certain contexts. This nutritional mutualism could explain some of Wolbachia’s prevalence across insect populations and fitness effects for which we do not understand the underlying mechanism (8). For the filarial-nematode infecting strains, nutritional mutualisms could be targets for drug development, as knocking out Wolbachia results in death of the nematode or reduced fecundity. In the future, we expect that specific metabolic contributions from Wolbachia strains to host metabolism and fitness will be identified. The evolution of these biosynthetic pathways across both the Wolbachia and host phylogeny could unveil selective pressures on the symbiosis.
Biography

Irene L. G. Newton is an associate professor in the Department of Biology at Indiana University, Bloomington. She received her Ph.D. from Harvard in organismic and evolutionary biology focused on functional genomics of deep-sea hydrothermal vent symbioses. She switched gears to the more tractable Wolbachia-Drosophila model symbiosis during her postdoctoral fellowship with Ralph Isberg at Tufts University. She has been working in the symbiosis field for over 15 years.
REFERENCES
- 1.Zug R, Hammerstein P. 2012. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7:e38544. doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinert LA, Araujo-Jnr EV, Ahmed MZ, Welch JJ. 2015. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc Biol Sci 282:20150249. doi: 10.1098/rspb.2015.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 4.Hamm CA, Begun DJ, Vo A, Smith CCR, Saelao P, Shaver AO, Jaenike J, Turelli M. 2014. Wolbachia do not live by reproductive manipulation alone: infection polymorphism in Drosophila suzukii and D. subpulchrella. Mol Ecol 23:4871–4885. doi: 10.1111/mec.12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriesner P, Hoffmann AA. 2018. Rapid spread of a Wolbachia infection that does not affect host reproduction in Drosophila simulans cage populations. Evolution 72:1475. doi: 10.1111/evo.13506. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann AA, Turelli M, Harshman LG. 1990. Factors affecting the distribution of cytoplasmic incompatibility in Drosophila simulans. Genetics 126:933–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turelli M. 2010. Cytoplasmic incompatibility in populations with overlapping generations. Evolution 64:232–241. doi: 10.1111/j.1558-5646.2009.00822.x. [DOI] [PubMed] [Google Scholar]
- 8.Zug R, Hammerstein P. 2018. Evolution of reproductive parasites with direct fitness benefits. Heredity (Edinb) 120:266–281. doi: 10.1038/s41437-017-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown AN, Lloyd VK. 2015. Evidence for horizontal transfer of Wolbachia by a Drosophila mite. Exp Appl Acarol 66:301–311. doi: 10.1007/s10493-015-9918-z. [DOI] [PubMed] [Google Scholar]
- 10.Cooper BS, Vanderpool D, Conner WR, Matute DR, Turelli M. 2019. Wolbachia acquisition by Drosophila yakuba: clade hosts and transfer of incompatibility loci between distantly related Wolbachia. Genetics 212:1399–1419. doi: 10.1534/genetics.119.302349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raychoudhury R, Baldo L, Oliveira DC, Werren JH. 2009. Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution 63:165–183. doi: 10.1111/j.1558-5646.2008.00533.x. [DOI] [PubMed] [Google Scholar]
- 12.Zug R, Hammerstein P. 2015. Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biol Rev Camb Philos Soc 90:89–111. doi: 10.1111/brv.12098. [DOI] [PubMed] [Google Scholar]
- 13.Moran NA, Plague GR, Sandstrom JP, Wilcox JL. 2003. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc Natl Acad Sci U S A 100(Suppl 2):14543–14548. doi: 10.1073/pnas.2135345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiggins FM, Hurst GD. 2011. Microbiology. Rapid insect evolution by symbiont transfer. Science 332:185–186. doi: 10.1126/science.1205386. [DOI] [PubMed] [Google Scholar]
- 15.Andersen SB, Boye M, Nash DR, Boomsma JJ. 2012. Dynamic Wolbachia prevalence in Acromyrmex leaf-cutting ants: potential for a nutritional symbiosis. J Evol Biol 25:1340–1350. doi: 10.1111/j.1420-9101.2012.02521.x. [DOI] [PubMed] [Google Scholar]
- 16.Foster J, Ganatra M, Kamal I, Ware J, Makarova K, Ivanova N, Bhattacharyya A, Kapatral V, Kumar S, Posfai J, Vincze T, Ingram J, Moran L, Lapidus A, Omelchenko M, Kyrpides N, Ghedin E, Wang S, Goltsman E, Joukov V, Ostrovskaya O, Tsukerman K, Mazur M, Comb D, Koonin E, Slatko B. 2005. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol 3:e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darby AC, Armstrong SD, Bah GS, Kaur G, Hughes MA, Kay SM, Koldkjær P, Rainbow L, Radford AD, Blaxter ML, Tanya VN, Trees AJ, Cordaux R, Wastling JM, Makepeace BL. 2012. Analysis of gene expression from the Wolbachia genome of a filarial nematode supports both metabolic and defensive roles within the symbiosis. Genome Res 22:2467–2477. doi: 10.1101/gr.138420.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comandatore F, Cordaux R, Bandi C, Blaxter M, Darby A, Makepeace BL, Montagna M, Sassera D. 2015. Supergroup C Wolbachia, mutualist symbionts of filarial nematodes, have a distinct genome structure. Open Biol 5:150099. doi: 10.1098/rsob.150099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rice DW, Sheehan KB, Newton ILG. 2017. Large-scale identification of Wolbachia pipientis effectors. Genome Biol Evol 9:1925–1937. doi: 10.1093/gbe/evx139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandi C, Anderson TJ, Genchi C, Blaxter ML. 1998. Phylogeny of Wolbachia in filarial nematodes. Proc Biol Sci 265:2407–2413. doi: 10.1098/rspb.1998.0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefoulon E, Bain O, Makepeace BL, d’Haese C, Uni S, Martin C, Gavotte L. 2016. Breakdown of coevolution between symbiotic bacteria Wolbachia and their filarial hosts. PeerJ 4:e1840. doi: 10.7717/peerj.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genchi C, Sacchi L, Bandi C, Venco L. 1998. Preliminary results on the effect of tetracycline on the embryogenesis and symbiotic bacteria (Wolbachia) of Dirofilaria immitis. An update and discussion. Parassitologia 40:247–249. [PubMed] [Google Scholar]
- 23.Wu B, Novelli J, Foster J, Vaisvila R, Conway L, Ingram J, Ganatra M, Rao AU, Hamza I, Slatko B. 2009. The heme biosynthetic pathway of the obligate Wolbachia endosymbiont of Brugia malayi as a potential anti-filarial drug target. PLoS Negl Trop Dis 3:e475. doi: 10.1371/journal.pntd.0000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werren JH, Zhang W, Guo LR. 1995. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc Biol Sci 261:55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- 25.Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, McGraw EA, Martin W, Esser C, Ahmadinejad N, Wiegand C, Madupu R, Beanan MJ, Brinkac LM, Daugherty SC, Durkin AS, Kolonay JF, Nelson WC, Mohamoud Y, Lee P, Berry K, Young MB, Utterback T, Weidman J, Nierman WC, Paulsen IT, Nelson KE, Tettelin H, O’Neill SL, Eisen JA. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol 2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jimenez NE, Gerdtzen ZP, Olivera-Nappa A, Salgado JC, Conca C. 2019. A systems biology approach for studying Wolbachia metabolism reveals points of interaction with its host in the context of arboviral infection. PLoS Negl Trop Dis 13:e0007678. doi: 10.1371/journal.pntd.0007678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fallon AM, Baldridge GD, Carroll EM, Kurtz CM. 2014. Depletion of host cell riboflavin reduces Wolbachia levels in cultured mosquito cells. In Vitro Cell Dev Biol Anim 50:707–713. doi: 10.1007/s11626-014-9758-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerth M, Bleidorn C. 2016. Comparative genomics provides a timeframe for Wolbachia evolution and exposes a recent biotin synthesis operon transfer. Nat Microbiol 2:16241. doi: 10.1038/nmicrobiol.2016.241. [DOI] [PubMed] [Google Scholar]
- 29.Gill AC, Darby AC, Makepeace BL. 2014. Iron necessity: the secret of Wolbachia’s success? PLoS Negl Trop Dis 8:e3224. doi: 10.1371/journal.pntd.0003224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hellemans S, Kaczmarek N, Marynowska M, Calusinska M, Roisin Y, Fournier D. 2019. Bacteriome-associated Wolbachia of the parthenogenetic termite Cavitermes tuberosus. FEMS Microbiol Ecol 95. doi: 10.1093/femsec/fiy235. [DOI] [PubMed] [Google Scholar]
- 31.Kremer N, Voronin D, Charif D, Mavingui P, Mollereau B, Vavre F. 2009. Wolbachia interferes with ferritin expression and iron metabolism in insects. PLoS Pathog 5:e1000630. doi: 10.1371/journal.ppat.1000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriyama M, Nikoh N, Hosokawa T, Fukatsu T. 2015. Riboflavin provisioning underlies Wolbachia’s fitness contribution to its insect host. mBio 6:e01732-15. doi: 10.1128/mBio.01732-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balvin O, Roth S, Talbot B, Reinhardt K. 2018. Co-speciation in bedbug Wolbachia parallel the pattern in nematode hosts. Sci Rep 8:8797. doi: 10.1038/s41598-018-25545-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hosokawa T, Koga R, Kikuchi Y, Meng XY, Fukatsu T. 2010. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci U S A 107:769–774. doi: 10.1073/pnas.0911476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikoh N, Hosokawa T, Moriyama M, Oshima K, Hattori M, Fukatsu T. 2014. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc Natl Acad Sci U S A 111:10257–10262. doi: 10.1073/pnas.1409284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y, Berry SM, Pfister TD. 2001. Engineering novel metalloproteins: design of metal-binding sites into native protein scaffolds. Chem Rev 101:3047–3080. doi: 10.1021/cr0000574. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y, Yeung N, Sieracki N, Marshall NM. 2009. Design of functional metalloproteins. Nature 460:855–862. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, Angiuoli SV, Creasy T, Amedeo P, Haas B, El-Sayed NM, Wortman JR, Feldblyum T, Tallon L, Schatz M, Shumway M, Koo H, Salzberg SL, Schobel S, Pertea M, Pop M, White O, Barton GJ, Carlow CKS, Crawford MJ, Daub J, Dimmic MW, Estes CF, Foster JM, Ganatra M, Gregory WF, Johnson NM, Jin J, Komuniecki R, Korf I, Kumar S, Laney S, Li B-W, Li W, Lindblom TH, Lustigman S, Ma D, Maina CV, Martin DM, McCarter JP, et al. 2007. Draft genome of the filarial nematode parasite Brugia malayi. Science 317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godel C, Kumar S, Koutsovoulos G, Ludin P, Nilsson D, Comandatore F, Wrobel N, Thompson M, Schmid CD, Goto S, Bringaud F, Wolstenholme A, Bandi C, Epe C, Kaminsky R, Blaxter M, Maser P. 2012. The genome of the heartworm, Dirofilaria immitis, reveals drug and vaccine targets. FASEB J 26:4650–4661. doi: 10.1096/fj.12-205096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C, Samuel TK, Krause M, Dailey HA, Hamza I. 2012. Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by heme-responsive gene-2. J Biol Chem 287:9601–9612. doi: 10.1074/jbc.M111.307694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lentz CS, Halls V, Hannam JS, Niebel B, Strubing U, Mayer G, Hoerauf A, Famulok M, Pfarr KM. 2013. A selective inhibitor of heme biosynthesis in endosymbiotic bacteria elicits antifilarial activity in vitro. Chem Biol 20:177–187. doi: 10.1016/j.chembiol.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 42.Tong Y, Guo M. 2009. Bacterial heme-transport proteins and their heme-coordination modes. Arch Biochem Biophys 481:1–15. doi: 10.1016/j.abb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henderson DP, Payne SM. 1994. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J Bacteriol 176:3269–3277. doi: 10.1128/jb.176.11.3269-3277.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. 2005. Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect Immun 73:8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren Q, Thony-Meyer L. 2001. Physical interaction of CcmC with heme and the heme chaperone CcmE during cytochrome c maturation. J Biol Chem 276:32591–32596. doi: 10.1074/jbc.M103058200. [DOI] [PubMed] [Google Scholar]
- 46.Baldo L, Desjardins CA, Russell JA, Stahlhut JK, Werren JH. 2010. Accelerated microevolution in an outer membrane protein (OMP) of the intracellular bacteria Wolbachia. BMC Evol Biol 10:48. doi: 10.1186/1471-2148-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiggins FM, Hurst GD, Yang Z. 2002. Host-symbiont conflicts: positive selection on an outer membrane protein of parasitic but not mutualistic Rickettsiaceae. Mol Biol Evol 19:1341–1349. doi: 10.1093/oxfordjournals.molbev.a004195. [DOI] [PubMed] [Google Scholar]
- 48.Brownlie JC, Cass BN, Riegler M, Witsenburg JJ, Iturbe-Ormaetxe I, McGraw EA, O’Neill SL. 2009. Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog 5:e1000368. doi: 10.1371/journal.ppat.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitz-Esser S, Linka N, Collingro A, Beier CL, Neuhaus HE, Wagner M, Horn M. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J Bacteriol 186:683–691. doi: 10.1128/jb.186.3.683-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Driscoll TP, Verhoeve VI, Guillotte ML, Lehman SS, Rennoll SA, Beier-Sexton M, Rahman MS, Azad AF, Gillespie JJ. 2017. Wholly rickettsia! Reconstructed metabolic profile of the quintessential bacterial parasite of eukaryotic cells. mBio 8:e00859-17. doi: 10.1128/mBio.00859-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teixeira L, Ferreira A, Ashburner M. 2008. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol 6:e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serbus LR, White PM, Silva JP, Rabe A, Teixeira L, Albertson R, Sullivan W. 2015. The impact of host diet on Wolbachia titer in Drosophila. PLoS Pathog 11:e1004777. doi: 10.1371/journal.ppat.1004777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schultz MJ, Tan AL, Gray CN, Isern S, Michael SF, Frydman HM, Connor JH. 2018. Wolbachia wStri blocks Zika virus growth at two independent stages of viral replication. mBio 9:e00738-18. doi: 10.1128/mBio.00738-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frydman HM, Li JM, Robson DN, Wieschaus E. 2006. Somatic stem cell niche tropism in Wolbachia. Nature 441:509–512. doi: 10.1038/nature04756. [DOI] [PubMed] [Google Scholar]
- 55.Ferree PM, Frydman HM, Li JM, Cao J, Wieschaus E, Sullivan W. 2005. Wolbachia utilizes host microtubules and Dynein for anterior localization in the Drosophila oocyte. PLoS Pathog 1:e14. doi: 10.1371/journal.ppat.0010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fast EM, Toomey ME, Panaram K, Desjardins D, Kolaczyk ED, Frydman HM. 2011. Wolbachia enhance Drosophila stem cell proliferation and target the germline stem cell niche. Science 334:990–992. doi: 10.1126/science.1209609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beckmann JF, Ronau JA, Hochstrasser M. 2017. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol 2:17007. doi: 10.1038/nmicrobiol.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Freund JN, Jarry BP. 1987. The rudimentary gene of Drosophila melanogaster encodes four enzymic functions. J Mol Biol 193:1–13. doi: 10.1016/0022-2836(87)90621-8. [DOI] [PubMed] [Google Scholar]