Foliar plant pathogens, like Pseudomonas syringae, adjust their physiology and behavior to facilitate host colonization and disease, but the full extent of these adaptations is not known. Plant immune systems are triggered by bacterial molecules, such as the proteins that make up flagellar filaments. In this study, we found that during plant infection, AlgU, a gene expression regulator that is responsive to external stimuli, downregulates expression of fliC, which encodes the flagellin protein, a strong elicitor of plant immune systems. This change in gene expression and resultant change in behavior correlate with reduced plant immune activation and improved P. syringae plant colonization. The results of this study demonstrate the proximate and ultimate causes of flagellar regulation in a plant-pathogen interaction.
KEYWORDS: agricultural research, extracytoplasmic sigma factor, plant immunity, Pseudomonas syringae, flagellar gene regulation, plant-microbe interactions
ABSTRACT
Flagella power bacterial movement through liquids and over surfaces to access or avoid certain environmental conditions, ultimately increasing a cell’s probability of survival and reproduction. In some cases, flagella and chemotaxis are key virulence factors enabling pathogens to gain entry and attach to suitable host tissues. However, flagella are not always beneficial; both plant and animal immune systems have evolved receptors to sense the proteins that make up flagellar filaments as signatures of bacterial infection. Microbes poorly adapted to avoid or counteract these immune functions are unlikely to be successful in host environments, and this selective pressure has driven the evolution of diverse and often redundant pathogen compensatory mechanisms. We tested the role of AlgU, the Pseudomonas extracytoplasmic function sigma factor σE/σ22 ortholog, in regulating flagellar expression in the context of Pseudomonas syringae-plant interactions. We found that AlgU is necessary for downregulating bacterial flagellin expression in planta and that this results in a corresponding reduction in plant immune elicitation. This AlgU-dependent regulation of flagellin gene expression is beneficial to bacterial growth in the course of plant infection, and eliminating the plant’s ability to detect flagellin makes this AlgU-dependent function irrelevant for bacteria growing in the apoplast. Together, these results add support to an emerging model in which P. syringae AlgU functions at a key control point that serves to optimize the expression of bacterial functions during host interactions, including minimizing the expression of immune elicitors and concomitantly upregulating beneficial virulence functions.
IMPORTANCE Foliar plant pathogens, like Pseudomonas syringae, adjust their physiology and behavior to facilitate host colonization and disease, but the full extent of these adaptations is not known. Plant immune systems are triggered by bacterial molecules, such as the proteins that make up flagellar filaments. In this study, we found that during plant infection, AlgU, a gene expression regulator that is responsive to external stimuli, downregulates expression of fliC, which encodes the flagellin protein, a strong elicitor of plant immune systems. This change in gene expression and resultant change in behavior correlate with reduced plant immune activation and improved P. syringae plant colonization. The results of this study demonstrate the proximate and ultimate causes of flagellar regulation in a plant-pathogen interaction.
INTRODUCTION
Bacterial flagella are complex macromolecular machines, composed of more than 30 different proteins expressed from an intricately regulated gene expression network (1, 2). The most numerous flagellar protein is the flagellin subunit (encoded by the fliC gene in Pseudomonas and Enterobacteriaceae species) that makes up the flagellar filament, which can include up to 20,000 flagellin monomers and extend 10 to 15 μm from the cell surface (3). Flagellin proteins are composed of multiple domains, with the most conserved domains providing the structural elements for filament polymerization, but they also include epitopes that are recognized by plant and animal immune systems (4–7). The immune functions responding to these types of molecules are referred to as innate immunity in animals and pattern-triggered immunity (PTI) in plants. Both plants and animals have pattern recognition receptors (PRRs) on the surface of their cells to detect conserved microbe/pathogen-associated molecular patterns (M/PAMPs) as an indicator of microbial infection (5, 8, 9). Upon flagellin binding, the PRRs for flagellin recognition in plants (flagellin-sensitive 2 [FLS2] and FLS3) (10, 11) and in animals (Toll-like receptor 5 [TLR5]) (12) initiate a series of molecular events that make the conditions at the site of infection less hospitable for the invading microbe (13, 14). Plant PTI responses include bursts of reactive oxygen species (ROS) (15), changes in gene expression (16, 17), production of antimicrobials (18, 19), changes in cell membrane structure (20), and blockage of bacterial type three secretion system (T3SS)-dependent effector secretion (21, 22).
Bacterial pathogens have evolved multiple, redundant countermeasures to evade or suppress plant immune responses induced by flagellin and other PAMPs. Examples of these adaptations include T3SS-dependent delivery of effectors into host cell cytoplasm that interfere with the PRR signaling (23–26), production of toxins (e.g., coronatine) that can reverse stomata closure in response to flagellin (27), proteolytic degradation of free flagellin monomers (28, 29), flagellin sequence polymorphisms (7, 30, 31), posttranslational modifications of flagellin that alter recognition (32), and inhibition of plant enzymes that expose flagellin immunogenic epitopes (33). Downregulation of flagellin expression often occurs after pathogens reach suitable host tissues (34–38), and there is some evidence that this is an adaptive immune avoidance strategy in animal infection models. For example, downregulation of Salmonella flagella prevents host proinflammatory cell death response and enhances colonization of systemic sites in mice (39). Additionally, a greater proportion of gut microbiomes are flagellated in TLR5−/− mice (40), consistent with the idea that immune activation selects for or induces deflagellation in a wide range of host-adapted bacteria. Pseudomonas aeruginosa isolates from chronically infected cystic fibrosis patients are usually nonmotile and mucoid. This phenotypic conversion often results from mutations that abolish the anti-sigma factor function of MucA or MucB; these proteins normally act together to repress the activity of the extracytoplasmic function (ECF) sigma factor AlgU under noninducing conditions (41–44).
Here, we report the effects of conditional flagellar regulation in the context of a plant-microbe interaction. This study was prompted by several observations regarding flagellar regulation and AlgU function in Pseudomonas syringae. First, P. syringae flagellar genes are downregulated in the apoplast compared to when these bacteria are growing on the outer surfaces of leaves (45), suggesting that these bacteria have a mechanism for changing flagellar expression as they transition from epiphytic to pathogenic growth. Second, flagella are not necessary for P. syringae virulence if the normal infection route is bypassed by pressure infiltration into the apoplast (7, 46), and P. syringae typically does not migrate within plant tissues after entering the apoplast (47), supporting the idea that flagella do not contribute to disease processes after accessing the apoplast. Third, AlgU is active while P. syringae is in plants, where it functions as a major regulator of gene expression and benefits bacterial growth and disease in this context (48, 49). Finally, P. syringae AlgU downregulates fliC expression in vitro and coordinates this with upregulation of other genes that have well-established roles in virulence and plant disease (46, 48, 50). Based on these observations, we tested whether AlgU-dependent regulation of flagellin expression helps minimize plant immune elicitation and whether this regulation is an important component of the infection process. We found that in the absence of AlgU, flagella are a liability when P. syringae is in plant apoplasts and that this results from FLS2-dependent plant immune functions. Furthermore, AlgU is responsible for downregulating P. syringae flagellin gene expression in planta, helping to reduce plant immune activation and the negative effects this has on bacterial growth in planta. These findings demonstrate the beneficial role of AlgU-dependent regulation of flagellin expression in the context of a plant-pathogen interaction and help explain why AlgU is important for these bacteria to successfully colonize and cause disease in plants.
RESULTS
AlgU suppresses swimming motility.
Transcriptomic analysis revealed that P. syringae pv. tomato DC3000 algU downregulates flagellar and chemotaxis genes in vitro (48). We used a plate-based swimming assay to test whether these AlgU-dependent transcriptional changes correlate with differences in bacterial behavior (Fig. 1). We found that AlgU was capable of suppressing flagellum-mediated motility in the absence of mucAB anti-sigma factor genes. This result confirms that AlgU regulatory functions affect flagellar motility and that AlgU activity is repressed by MucAB in this growth medium. This is consistent with previous results in which other algU-dependent phenotypes were repressed by MucAB when the bacteria were grown in noninducing conditions (48).
FIG 1.
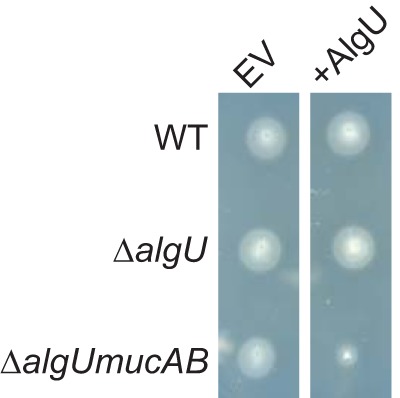
AlgU reduces swimming motility. Swimming motility of P. syringae pv. tomato DC3000 wild type (WT) and indicated mutants with empty vector (EV), pJN105 (79), or +AlgU expression vector pEM53 (80). Each strain was inoculated on Kings B (KB) medium (77) with 0.3% agar and incubated at 28°C for 24 h.
AlgU is necessary for downregulating P. syringae pv. tomato DC3000 fliC expression during infection.
We tested whether AlgU regulates DC3000 flagellar motility gene expression during plant infection by using reverse transcription-quantitative PCR (qRT-PCR) to compare the relative expression differences of these genes in DC3000 D36E (51) to that of the isogenic DC3000 D36E ΔalgU mucAB mutant 6 h after syringe infiltration in tomato leaves (see below for an explanation of why DC3000 D36E was used in these experiments). There are more than 40 regulatory and structural genes expressed in four hierarchical levels that provide the ordered assembly of Pseudomonas flagella (1). We examined the AlgU-dependent regulation of flagellar genes that were previously found to be downregulated by AlgU overexpression in vitro (48) and fliA and flgM because of their key function in regulating fliC and the class IV flagellar genes (1). The set of genes tested included representatives from all four transcriptional classes, but more class IV genes were included because this class was overrepresented, compared to the others, in our genome-wide AlgU regulon analysis (Table 1) (48). We found that for DC3000 D36E in tomato, the fliC gene was 2.7-fold more highly expressed in the strain lacking algU but found no evidence for differential regulation of any of the other genes tested. This result indicates that AlgU is necessary for downregulating flagellin expression while DC3000 is in plants and that AlgU directs downregulation of fliC without affecting the other genes we tested in the flagellar gene expression pathway.
TABLE 1.
Differences in flagellar and chemotaxis gene expression between P. syringae pv. tomato DC3000 D36E algU mucAB and DC3000 D36E in tomato infection
| Locus tag | Gene name | Regulatory class(es)a | Mean (SD) |
|---|---|---|---|
| PSPTO_1949 | fliC | IV | 2.70 (0.68) |
| PSPTO_0915 | cheY-1 | IV | 0.97 (0.21) |
| PSPTO_1925 | flgM | II and IV | 0.91 (0.37) |
| PSPTO_1933 | flgB | III | 0.99 (0.33) |
| PSPTO_1979 | fliA | I | 1.03 (0.27) |
| PSPTO_1980 | cheY-2 | IV | 0.90 (0.13) |
| PSPTO_1982 | cheA-2 | IV | 1.02 (0.21) |
| PSPTO_1987 | cheW | IV | 1.15 (0.04) |
| PSPTO_1988 | cheW-2 | IV | 0.97 (0.17) |
Regulatory class assignments were made based on those of the P. aeruginosa orthologs described by Dasgupta et al. (1).
AlgU-dependent downregulation of fliC expression reduces plant immune responses.
We tested whether AlgU-dependent downregulation of flagellin expression affected the degree to which PTI was induced in plants infected with P. syringae. For these experiments, we used P. syringae pv. tomato DC3000 D36E (52), a laboratory strain in which all known T3SS effectors have been deleted, some of which have well-established roles in suppressing PTI induced by flagellin (24–26). Using this strain and isogenic ΔalgU mucAB and ΔfliC mutants allowed us to focus the analysis on AlgU function in avoiding PTI without the redundant action of PTI-suppressing effectors.
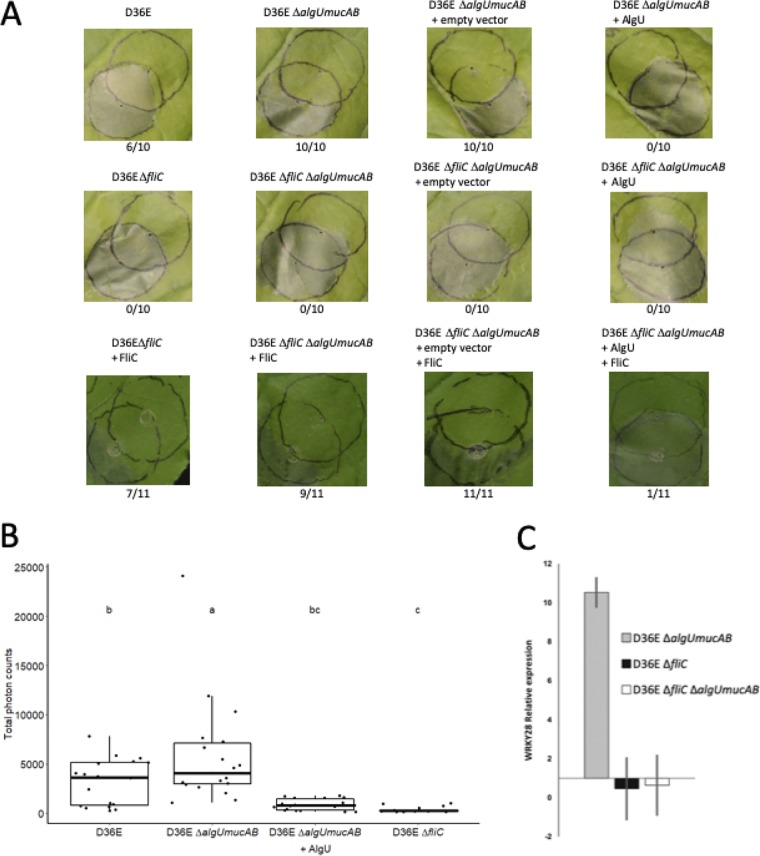
We first tested whether algU altered PTI by evaluating whether these strains could induce PTI-dependent suppression of effector translocation from a subsequent challenge inoculum of wild-type P. syringae pv. tomato DC3000. Wild-type DC3000 normally elicits an effector-dependent hypersensitive response (HR) when inoculated on Nicotiana benthamiana plants (21, 22). However, if PTI is induced in plant tissue before the wild-type strain is inoculated, then effector translocation is blocked and HR does not happen. We found that the challenge inoculum was capable of producing an HR in N. benthamiana plants infected with test strains expressing AlgU (Fig. 2A), suggesting that AlgU function in DC3000 D36E reduced PTI. Strains with wild-type levels of AlgU still show some PTI; we take this to indicate that AlgU reduces but does not eliminate this part of PTI completely. In contrast, the challenge inoculum was not able to produce an HR when algU was deleted from the test strain, indicating that in the absence of AlgU PTI was strongly induced. This strong PTI stimulated by DC3000 D36E ΔalgU mucAB was eliminated when fliC was also deleted, indicating that the effect of AlgU on flagellin expression is responsible for minimizing PTI. The challenge inoculum HR could be fully restored by transformation of the ΔalgU mucAB test strain with an AlgU-expressing plasmid, confirming that algU is responsible for minimizing PTI. The dependence on fliC was also tested by complementation, confirming that flagellin is the regulated elicitor of PTI in these interactions.
FIG 2.
AlgU-dependent downregulation of fliC expression reduces indicators of PTI in N. benthamiana and tomato plants. (A) PTI-dependent HR cell death suppression assay. N. benthamiana leaves were inoculated with a suspension containing 5 × 107 CFU/ml of test strains (top circles) and then incubated 6 hours before partially overlapping challenge inoculation with a suspension of 2 × 107 CFU/ml of the cell death-eliciting wild-type DC3000 strain (bottom circles) and incubated an additional 48 hours before evaluation. The genotype of each test strain and the number of times each test strain induced PTI as a fraction of number of times tested are shown above and below each photograph, respectively. +AlgU expression and empty vector control are the same as in Fig. 1. +FliC expression was provided by pME6010-fliC, which carries DC3000 fliC under the control of its native promoter (7). (B) L-012 chemiluminescence ROS assay for N. benthamiana leaves at 15 h after inoculation with the indicated strains at 5 × 108 CFU/ml. Individual points show total photon counts collected over 30 minutes for 18 replicates; box plots show the interquartile range (IQR), split by the median; and whiskers show the range of data that are within 1.5 × IQR above and below the first and third quartiles, respectively. Statistical significance was tested using linear mixed effects modeling. Letters indicate Tukey-Kramer honestly significant difference (HSD) mean comparison results; means not connected by letters are significantly different. (C) Mean relative expression level of WRKY28 (16) in tomato plants (Solanum lycopersicum) 6 h after inoculation with 5 × 105 CFU/ml of algU mucAB and/or fliC mutant derivatives relative to plants infected with DC3000 D36E. Error bars show standard deviation.
Second, we tested whether ROS production indicative of flagellin-induced PTI (51) was altered in the absence of AlgU. We found that flagellin-dependent ROS production at 15 h postinoculation (51) was significantly increased in N. benthamiana plants infected with the ΔalgU mucAB strain compared with DC3000 D36E (Fig. 2B). This phenotype was also complemented with AlgU expressed in trans, and ROS were eliminated by fliC deletion, confirming the requirement for flagellin in this aspect of PTI.
Finally, we tested the effect of algU and fliC on PTI-specific gene expression in tomato plants infected with DC3000 D36E and the isogenic ΔalgU mucAB and ΔfliC mutants. In this experiment, we measured gene expression of WRKY28, a tomato gene encoding a transcription regulator that is induced during PTI in tomato plants (16). We found that WRKY28 gene expression was increased more than 10-fold in tomato leaf tissue infected with the DC3000 D36E ΔalgU mucAB strain relative to DC3000 D36E (Fig. 2C) and that this induction was eliminated by deletion of fliC from the ΔalgU mucAB strain. Additionally, we found evidence that AlgU lowers flagellin expression to levels below the threshold for detection and activation of WRKY28. There was no difference in WRKY28 induction between DC3000 D36E ΔfliC and DC3000 D36E, suggesting that AlgU reduces flagellin expression to levels that are functionally equivalent to that in a fliC deletion strain.
Considering the combined results of the HR suppression assay in N. benthamiana (Fig. 2A), ROS generation in N. benthamiana (Fig. 2B), and WRK28 induction in tomato (Fig. 2C), the data indicate that AlgU-dependent regulation of fliC expression reduces PTI induced by DC3000 in tomato and N. benthamiana plants, two important plant hosts that are routinely used as models to study plant disease.
AlgU-dependent flagellar regulation is beneficial for P. syringae pv. tomato DC3000 growth in planta.
Evidence of increased PTI in plants infected with algU-deficient strains suggested that AlgU-dependent downregulation of fliC helps reduce the pathogen’s exposure to the antimicrobial functions of PTI. Furthermore, strains encoding a functional AlgU should be able to grow better in plants than strains without this function. To test this hypothesis, we determined whether AlgU-dependent regulation of fliC expression provided a growth benefit to bacteria during the course of a plant infection. In this experiment, we infected N. benthamiana plants with DC3000 D36E or the isogenic algU mucAB and fliC deletion mutants and then determined the number of bacteria present at the site of infection 3 days after infiltration. We found that there were 9.8-fold more DC3000 D36E cells than those of the ΔalgU mucAB mutant and that the growth defect of the ΔalgU mucAB cells could be fully suppressed by deletion of fliC (Fig. 3). Note that deleting fliC alone does not provide a growth benefit, as might be predicted if flagellin expression was not downregulated. However, the benefit of deleting fliC is apparent in the algU mucAB mutant. This suggests that AlgU is able to lower flagellin expression to levels that are functionally equivalent (in terms of cell growth) to that of the fliC deletion mutant, as was observed for WRKY28 induction (Fig. 2C).
FIG 3.
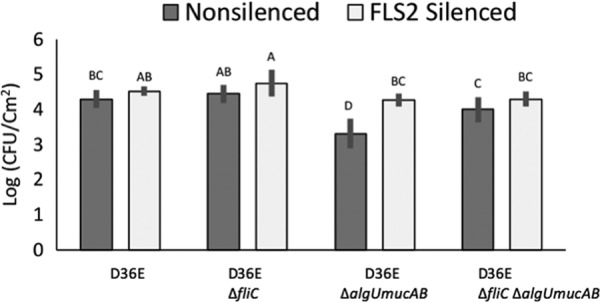
AlgU is required for growth during plant infection. Deleting algU mucAB reduces DC3000 D36E growth in N. benthamiana, but this phenotype can be suppressed by deleting fliC or eliminating FLS2 in host plants. The indicated strains were syringe infiltrated at 3 × 104 CFU/ml in N. benthamiana with (FLS2 silenced) or without (nonsilenced) FLS2 virus-induced gene silencing (VIGS). Bacteria were enumerated 3 days postinoculation. Error bars show standard deviation. Letters indicate Tukey-Kramer HSD mean comparison results; means not sharing letters are significantly different.
These data are consistent with the hypothesis that AlgU-dependent downregulation of fliC is an adaptive trait for bacteria growing in planta because of reduced immune elicitation. If this is true, then eliminating the plant’s ability to detect flagellin and trigger the PTI response should also negate the requirement for AlgU. We tested this by determining the growth of DC3000 D36E ΔalgU mucAB and ΔfliC mutants in N. benthamiana plants with the gene responsible for flagellin detection, the FLS2 gene (11), silenced by virus-induced gene silencing (VIGS) (53). As predicted, we found that in FLS2-silenced plants, bacteria lacking algU mucAB grew to levels comparable to strains with the intact algU mucAB locus (Fig. 3). Finding that AlgU is not needed if plants are unable to detect flagellin confirms that flagellin is the primary liability in the absence of AlgU. Together, these results indicate that DC3000 gains a fitness benefit from AlgU-dependent regulation of flagellin expression during plant infection and that this benefit is due to a lessened plant immune response.
DISCUSSION
Plant pathogens like P. syringae are equipped with motility systems that enable them to move from the surfaces of leaves into the apoplast, which gives them access to nutrients and shelter from desiccation and UV damage. P. syringae flagellar motility also provides a benefit to cells growing epiphytically on plant surfaces (54). After entering the apoplast, P. syringae transitions into a sessile lifestyle, giving rise to biofilm-like microcolonies that eventually stimulate disease lesions in the surrounding plant tissue (47, 55). These types of plant interactions do not result in systemic infections, and the characteristic speck and spot diseases that develop are the result of bacterial proliferation at or very near the site of primary infection and the associated disease lesion (47, 55). The bacteria undergo extensive transcriptional reprograming to express virulence genes and other functions needed for growth in plant tissue. These transcriptional changes occur in response to conditions in the plant host tissues (45, 49, 56–58) and are coordinated through a sophisticated network of sensors and regulators that are only partially characterized (45, 46, 48, 59–63). Studies focused on control of expression and production of the factors necessary for virulence have been particularly fruitful, such as those describing T3SS and the effectors translocated into plant cells. However, as we show here, there are additional adaptations that evolved to promote successful colonization, in particular, that downregulating flagellin expression lowers immune elicitation, helping maintain the apoplast as an environment favorable for bacterial growth.
There is a great deal of redundancy in bacterial systems that prevent plant immune response to flagellin. In our experiments testing the effect of AlgU-mediated regulation of fliC on plant immune responses, we used the P. syringae pv. tomato DC3000 D36E mutant, which carries a functional T3SS but lacks all known effector genes (52). We chose this strain because many of the T3SS-secreted effectors interfere with the plant’s ability to detect or mount an immune response to flagellin (24–26). This strain allowed us to observe AlgU’s role in the infection process without the redundant action of those effectors. In addition to the T3SS-dependent effectors, P. syringae also has other adaptations to help avoid flagellin-induced PTI, including flagellin glycosylation that blocks its detection (32), an inhibitor of a plant enzyme that exposes flagellin’s immunogenic epitopes (33), and a secreted protease that degrades free flagellin monomers (28, 29). The degree of redundancy that evolved to foil plant surveillance of flagellin certainly underscores the importance of this molecule in the evolutionary arms race for exploitation of valuable plant resources.
AlgU is also responsible for downregulating flagellin expression in P. aeruginosa strains. This is particularly apparent in P. aeruginosa isolates from chronically infected cystic fibrosis patients, which are usually mucoid and nonmotile (64, 65). Most of these strains have mutations in the mucA anti-sigma factor gene that inhibit their ability to regulate AlgU function (66). Additionally, P. aeruginosa flagellar expression is suppressed in response to mucopurulent respiratory fluid from cystic fibrosis (CF) patients (67), suggesting that flagella may also be downregulated in acute infections. These observations have been interpreted as evidence that flagellar regulation is an important immune avoidance mechanism (68).
P. aeruginosa and P. syringae use different genes to downregulate fliC expression in host tissues. P. aeruginosa AlgU downregulates flagellar production by stimulating the expression of AmrZ, which, in turn, represses the expression of fleQ, the gene encoding the master regulator of flagellar production (69). This regulatory circuit is not likely to function in P. syringae; expression of the fleQ gene is not affected by P. syringae AlgU (70), and AmrZ increases motility and flagellar expression in P. syringae (71). We should also add that AlgU does not downregulate flagellin expression in all P. syringae pathovars; for example, P. syringae pv. syringae B728a flagellar expression is downregulated in leaf apoplasts (45, 49), but this does not require algU (49).
AlgU has a central role in regulating genes over the course of P. syringae plant interactions (49). We found that AlgU-dependent regulation of fliC expression is apparent at 6 h after infection (Table 1), and such early changes in gene expression in response to the plant environment correlate closely with the overall success of colonization and growth in host tissues (56). AlgU is a member of the ECF class of sigma factors, which, as a group, provide a mechanism for adjusting bacterial physiology and behavior in response to external environmental conditions (72). The periplasmic protease AlgW is a crucial component of the signal transduction pathway that activates AlgU (73, 74) and is necessary for flagellar downregulation and virulence upregulation in P. syringae pv. maculicola ES4326 (46, 75). However, AlgW-dependent downregulation of flagellar expression does not affect ES4326 growth in Arabidopsis thaliana Columbia (Col-0) (46). This result conflicts with our observations of DC3000 interactions with tomato and N. benthamiana, but this discrepancy is likely due to diversification within the ES4326 flagellin flg-22 epitope that prevents its detection by Arabidopsis FLS2 (7), making downregulation to avoid immunity redundant.
Evidence suggests that AlgW-dependent virulence gene induction is likely the dominant sector of this regulatory pathway for the ES4326-Arabidopsis interactions (46). As stated above, AlgU is important for P. syringae pv. syringae B728a-plant interactions, but it does not regulate flagellin expression (49). These observations are consistent with the idea that AlgU’s principal role can vary depending on the details of each specific pathovar-host interactions (48, 49). However, the pleiotropic nature of AlgU-dependent regulation is likely maintained among the various pathovars because it provides for interactions when alternate sets of functions are necessary for colonization of different plants. From a more general perspective, at least part of AlgU’s function in these host-pathogen systems is to help avoid and suppress plant immunity through the combination of upregulating the T3SS, effector genes (46, 48), and alginate production (76) and downregulating flagellin expression to minimize the presentation of immune-eliciting PAMPs.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Except where noted, P. syringae pv. tomato DC3000 and derivative strains were grown at 28°C in Kings B (KB) medium (77) or on KB medium solidified with 1.5% (wt/vol) agar with 10 μg/ml gentamicin, 50 μg/ml kanamycin, or 10 μg/ml tetracycline added for plasmid maintenance when necessary. Escherichia coli DH5α was used as the host for molecular cloning and other plasmid manipulations used in this work. E. coli was grown at 37°C in LB medium or LB medium solidified with 1.5% (wt/vol) agar. All bacterial strains and plasmids used in these experiments are shown in Table S1 in the supplemental material. Bacterial swimming was tested on KB medium solidified with 0.3% agar and incubated for 24 hours at 28°C. DC3000 D36E ΔalgU mucAB and DC3000 D36E ΔalgU mucAB ΔfliC mutant strains were made by marker exchange mutagenesis using the deletion constructs to make the ΔalgU mucAB mutants described in reference 48 with DC3000 D36E and DC3000 D36E ΔfliC as the targeted recipient strains, respectively. Strains were confirmed by DNA sequencing of mutant loci.
RNA isolation and qRT-PCR.
Five-week-old tomato (Solanum lycopersicum cv. Moneymaker) plants were syringe inoculated with a bacterial suspension of 5 × 106 CFU/ml in 10 mM MgCl2. Leaf tissue was harvested 6 hours after infiltration and frozen in liquid nitrogen. Total (plant and bacterial) RNA was prepared with on-column DNase treatment using the RNeasy plant minikit (Qiagen, Valencia, CA). Reverse transcription was performed with 200 ng of total RNA for testing plant genes or 2 μg of total RNA for bacterial genes using a iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). The cDNA was diluted 10-fold (for testing plant genes) or 2.5-fold (for bacterial genes) with water, and 1 μl was used for quantitative PCR analysis. The qRT-PCR was performed on the Bio-Rad CFX Connect real-time PCR detection system using SsoAdvanced universal SYBR green (Bio-Rad) with primers at 2 μM. Cycling conditions during qRT-PCR were 95°C for 3 minutes, followed by 40 cycles of 95°C for 15 seconds, 52°C for 30 seconds, and 60°C for 30 seconds. Primer sequences of the DC3000 genes tested are listed in Table S2 in the supplemental material. The gap-1 expression was used for relative quantification of bacterial gene expression. Tomato Actin expression was used for relative quantification of WRKY28 gene expression (16).
Cell death suppression assay.
Primary streaks of P. syringae pv. tomato DC3000 D36E and its derivative strains were made from isolated colonies on KB agar with appropriate antibiotics and grown at 28°C overnight. The bacterial cells were scraped from plates with a sterile pipette tip and suspended in 10 mM MgCl2. To test the PTI-dependent inhibition of effector secretion, test strains were inoculated at 5 × 107 CFU/ml into 6-week-old N. benthamiana using a blunt syringe, and after 6 h, a suspension of DC3000 at 2 × 107 CFU/ml was challenge inoculated on the edge of the preinfiltrated area so as to partially overlap the test inoculum. Inoculated plants were incubated in a growth chamber with 16-hour light and 8-hour dark cycles at 20 to 25°C with 60% to 70% humidity for 48 hours. Three or four N. benthamiana plants were used in each replicate that was repeated three times.
Reactive oxygen species assay.
Primary streaks of P. syringae pv. tomato DC3000 D36E and its derivative strains were made from colonies isolated on KB agar with appropriate antibiotics and grown at 28°C overnight. Bacteria were scraped from the plates and suspended in 10 mM MgCl2 at 5 × 108 CFU/ml and infiltrated using a blunt syringe into 6-week-old N. benthamiana leaves. Inoculated plants were incubated in a growth chamber at 20 to 25°C with 60% to 70% humidity for 15 hours; 0.5-cm-diameter leaf disks were cut from leaves and placed into 96-well plates with 10 μl of sterile water and 100 μl of 0.5 mM L-012 (Wako, Japan) in 10 mM morpholinepropanesulfonic acid-KOH buffer (pH 7.4). The intensity of ROS generation was determined by monitoring the chemiluminescence using a Synergy 2 microplate reader (BioTek, USA). Three biological repeats were carried out.
Virus-induced gene silencing.
VIGS was performed as described in Velasquez et al. (53) and Rosli et al. (17). Briefly, fresh Agrobacterium tumefaciens GV2260 clones carrying either pTRV1, pQ11-EC1 (modified pTRV2 as control) or pTRV2-FLS2 were used to inoculate 2 ml of LB liquid medium containing 50 μg/ml rifampin and 50 μg/ml kanamycin and were grown at 28°C overnight. Cells from each culture were pelleted and resuspend in 1 ml of Agrobacterium induction medium (53) to an optical density at 600 nm (OD600) of 0.5 and then were incubated at 28°C for 3 hours prior to pelleting and resuspending cells in 1 ml infiltration buffer. TRV1 and silencing construct (e.g., TRV2::gene) cultures were mixed at a 1:1 ratio and infiltrated into 3-week-old N. benthamiana seedling leaves. Plants were incubated in a growth chamber with 16-hour light and 8-hour dark cycles at 20 to 22°C with 60% to 70% relative humidity for at least 3.5 weeks before they were used for bacterial growth assays. Silencing of phytoene desaturase (PDS) was used as a visual control for silencing efficiency based on its ability to cause plant photobleaching.
In planta bacterial growth.
Six- to 7-week-old N. benthamiana plant leaves were inoculated with a 3 × 104 CFU/ml bacterial suspension using a blunt syringe and were incubated in a growth chamber with 16-hour light and 8-hour dark cycles at 20 to 25°C with 60% to 70% humidity. Bacteria were recovered from plants by sampling leaf tissue at the site of infection using a number 2 disk punch (3 disks; total area, 0.589 cm2) at 3 days postinoculation. Leaf disks were homogenized by mechanical disruption in 300 μl of 10 mM MgCl2. Serial dilutions of the tissue homogenate were plated on LM (78) agar supplemented with 50 μg/ml rifampin, and the number of CFU per square centimeter leaf tissue was calculated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brian Kvitko and Alan Collmer for many helpful discussions and critical reading of the manuscript and Boris Vinatzer for pME6010-fliC.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol 50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 2.Soutourina OA, Bertin PN. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev 27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 3.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, Aderem A. 2005. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci U S A 102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai R, Lewis J, Yan S, Liu H, Clarke CR, Campanile F, Almeida NF, Studholme DJ, Lindeberg M, Schneider D, Zaccardelli M, Setubal JC, Morales-Lizcano NP, Bernal A, Coaker G, Baker C, Bender CL, Leman S, Vinatzer BA. 2011. The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog 7:e1002130. doi: 10.1371/journal.ppat.1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. 2006. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18:465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke CR, Chinchilla D, Hind SR, Taguchi F, Miki R, Ichinose Y, Martin GB, Leman S, Felix G, Vinatzer BA. 2013. Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytol 200:847–860. doi: 10.1111/nph.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boutrot F, Zipfel C. 2017. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol 55:257–286. doi: 10.1146/annurev-phyto-080614-120106. [DOI] [PubMed] [Google Scholar]
- 9.Janeway CA Jr., Medzhitov R. 2002. Innate immune recognition. Annu Rev Immunol 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 10.Hind SR, Strickler SR, Boyle PC, Dunham DM, Bao Z, O'Doherty IM, Baccile JA, Hoki JS, Viox EG, Clarke CR, Vinatzer BA, Schroeder FC, Martin GB. 2016. Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat Plants 2:16128. doi: 10.1038/nplants.2016.128. [DOI] [PubMed] [Google Scholar]
- 11.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. 2004. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 13.Couto D, Zipfel C. 2016. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16:537–552. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- 14.Jones JD, Vance RE, Dangl JL. 2016. Intracellular innate immune surveillance devices in plants and animals. Science 354:aaf6395. doi: 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- 15.Apel K, Hirt H. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 16.Kim J-G, Li X, Roden JA, Taylor KW, Aakre CD, Su B, Lalonde S, Kirik A, Chen Y, Baranage G, Mclane H, Martin GB, Mudgett MB. 2009. Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 21:1305–1323. doi: 10.1105/tpc.108.063123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosli HG, Zheng Y, Pombo MA, Zhong S, Bombarely A, Fei Z, Collmer A, Martin GB. 2013. Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biol 14:R139. doi: 10.1186/gb-2013-14-12-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahuja I, Kissen R, Bones AM. 2012. Phytoalexins in defense against pathogens. Trends Plant Sci 17:73–90. doi: 10.1016/j.tplants.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Bednarek P. 2012. Chemical warfare or modulators of defence responses—the function of secondary metabolites in plant immunity. Curr Opin Plant Biol 15:407–414. doi: 10.1016/j.pbi.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Gómez-Gómez L, Felix G, Boller T. 1999. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18:277–284. doi: 10.1046/j.1365-313X.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- 21.Crabill E, Joe A, Block A, van Rooyen JM, Alfano JR. 2010. Plant immunity directly or indirectly restricts the injection of type III effectors by the Pseudomonas syringae type III secretion system. Plant Physiol 154:233–244. doi: 10.1104/pp.110.159723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh H-S, Collmer A. 2005. Basal resistance against bacteria in Nicotiana benthamiana leaves is accompanied by reduced vascular staining and suppressed by multiple Pseudomonas syringae type III secretion system effector proteins. Plant J 44:348–359. doi: 10.1111/j.1365-313X.2005.02529.x. [DOI] [PubMed] [Google Scholar]
- 23.Göhre V, Spallek T, Häweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S. 2008. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol 18:1824–1832. doi: 10.1016/j.cub.2008.10.063. [DOI] [PubMed] [Google Scholar]
- 24.Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J. 2008. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, Zhou J-M. 2008. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18:74–80. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 26.Zhou J, Wu S, Chen X, Liu C, Sheen J, Shan L, He P. 2014. The Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant J 77:235–245. doi: 10.1111/tpj.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melotto M, Underwood W, Koczan J, Nomura K, He SY. 2006. Plant stomata function in innate immunity against bacterial invasion. Cell 126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 28.Bardoel BW, van der Ent S, Pel MJC, Tommassen J, Pieterse CMJ, van Kessel KPM, van Strijp J. 2011. Pseudomonas evades immune recognition of flagellin in both mammals and plants. PLoS Pathog 7:e1002206. doi: 10.1371/journal.ppat.1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pel MJC, van Dijken AJH, Bardoel BW, Seidl MF, van der Ent S, van Strijp JAG, Pieterse C. 2014. Pseudomonas syringae evades host immunity by degrading flagellin monomers with alkaline protease AprA. Mol Plant Microbe Interact 27:603–610. doi: 10.1094/MPMI-02-14-0032-R. [DOI] [PubMed] [Google Scholar]
- 30.Pfund C, Tans-Kersten J, Dunning FM, Alonso JM, Ecker JR, Allen C, Bent AF. 2004. Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol Plant Microbe Interact 17:696–706. doi: 10.1094/MPMI.2004.17.6.696. [DOI] [PubMed] [Google Scholar]
- 31.Sun W, Dunning FM, Pfund C, Weingarten R, Bent AF. 2006. Within-species flagellin polymorphism in Xanthomonas campestris pv. campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 18:764–779. doi: 10.1105/tpc.105.037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imanishi T, Itoh T, Suzuki Y, O'Donovan C, Fukuchi S, Koyanagi KO, Barrero RA, Tamura T, Yamaguchi-Kabata Y, Tanino M, Yura K, Miyazaki S, Ikeo K, Homma K, Kasprzyk A, Nishikawa T, Hirakawa M, Thierry-Mieg J, Thierry-Mieg D, Ashurst J, Jia L, Nakao M, Thomas MA, Mulder N, Karavidopoulou Y, Jin L, Kim S, Yasuda T, Lenhard B, Eveno E, Suzuki Y, Yamasaki C, Takeda J-i, Gough C, Hilton P, Fujii Y, Sakai H, Tanaka S, Amid C, Bellgard M, Bonaldo MdF, Bono H, Bromberg SK, Brookes AJ, Bruford E, Carninci P, Chelala C, Couillault C, de Souza SJ, Debily M-A, et al. 2004. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol 2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buscaill P, Chandrasekar B, Sanguankiattichai N, Kourelis J, Kaschani F, Thomas EL, Morimoto K, Kaiser M, Preston GM, Ichinose Y, van der Hoorn R. 2019. Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 364:1–10. doi: 10.1126/science.aav0748. [DOI] [PubMed] [Google Scholar]
- 34.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 35.Gauger EJ, Leatham MP, Mercado-Lubo R, Laux DC, Conway T, Cohen PS. 2007. Role of motility and the flhDC operon in Escherichia coli MG1655 colonization of the mouse intestine. Infect Immun 75:3315–3324. doi: 10.1128/IAI.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamp HD, Higgins DE. 2011. A protein thermometer controls temperature-dependent transcription of flagellar motility genes in Listeria monocytogenes. PLoS Pathog 7:e1002153. doi: 10.1371/journal.ppat.1002153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapatral V, Olson JW, Pepe JC, Miller VL, Minnich SA. 1996. Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol Microbiol 19:1061–1071. doi: 10.1046/j.1365-2958.1996.452978.x. [DOI] [PubMed] [Google Scholar]
- 38.Ott M, Messner P, Heesemann J, Marre R, Hacker J. 1991. Temperature-dependent expression of flagella in Legionella. J Gen Microbiol 137:1955–1961. doi: 10.1099/00221287-137-8-1955. [DOI] [PubMed] [Google Scholar]
- 39.Stewart MK, Cummings LA, Johnson ML, Berezow AB, Cookson BT. 2011. Regulation of phenotypic heterogeneity permits Salmonella evasion of the host caspase-1 inflammatory response. Proc Natl Acad Sci U S A 108:20742–20747. doi: 10.1073/pnas.1108963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA, Walter J, Vijay-Kumar M, Gewirtz AT, Ley RE. 2013. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie ZD, Hershberger CD, Shankar S, Ye RW, Chakrabarty AM. 1996. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol 178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathee K, McPherson CJ, Ohman DE. 1997. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J Bacteriol 179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schurr MJ, Yu H, Martinez-Salazar JM, Boucher JC, Deretic V. 1996. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol 178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin DW, Schurr MJ, Mudd MH, Deretic V. 1993. Differentiation of Pseudomonas aeruginosa into the alginate-producing form: inactivation of mucB causes conversion to mucoidy. Mol Microbiol 9:497–506. doi: 10.1111/j.1365-2958.1993.tb01711.x. [DOI] [PubMed] [Google Scholar]
- 45.Yu X, Lund SP, Scott RA, Greenwald JW, Records AH, Nettleton D, Lindow SE, Gross DC, Beattie GA. 2013. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc Natl Acad Sci U S A 110:E425–34. doi: 10.1073/pnas.1221892110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schreiber KJ, Desveaux D. 2011. AlgW regulates multiple Pseudomonas syringae virulence strategies. Mol Microbiol 80:364–377. doi: 10.1111/j.1365-2958.2011.07571.x. [DOI] [PubMed] [Google Scholar]
- 47.Chakravarthy S, Worley JN, Montes-Rodriguez A, Collmer A. 2018. Pseudomonas syringae pv. tomato DC3000 polymutants deploying coronatine and two type III effectors produce quantifiable chlorotic spots from individual bacterial colonies in Nicotiana benthamiana leaves. Mol Plant Pathol 19:935–947. doi: 10.1111/mpp.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Markel E, Stodghill P, Bao Z, Myers CR, Swingle B. 2016. AlgU controls expression of virulence genes in Pseudomonas syringae pv. tomato DC3000. J Bacteriol 198:2330–2344. doi: 10.1128/JB.00276-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu X, Lund SP, Greenwald JW, Records AH, Scott RA, Nettleton D, Lindow SE, Gross DC, Beattie GA, Beattie A, Beattie GA. 2014. Transcriptional analysis of the global regulatory networks active in Pseudomonas syringae during leaf colonization. mBio 5:e01683. doi: 10.1128/mBio.01683-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishiga T, Ishiga Y, Betsuyaku S, Nomura N. 2018. AlgU contributes to the virulence of Pseudomonas syringae pv. tomato DC3000 by regulating production of the phytotoxin coronatine. J Gen Plant Pathol 84:189–201. doi: 10.1007/s10327-018-0775-6. [DOI] [Google Scholar]
- 51.Wei H-L, Zhang W, Collmer A. 2018. Modular study of the type III effector repertoire in Pseudomonas syringae pv. tomato DC3000 reveals a matrix of effector interplay in pathogenesis. Cell Rep 23:1630–1638. doi: 10.1016/j.celrep.2018.04.037. [DOI] [PubMed] [Google Scholar]
- 52.Wei H-L, Chakravarthy S, Mathieu J, Helmann TC, Stodghill P, Swingle B, Martin GB, Collmer A. 2015. Pseudomonas syringae pv. tomato DC3000 type III secretion effector polymutants reveal an interplay between HopAD1 and AvrPtoB. Cell Host Microbe 17:752–762. doi: 10.1016/j.chom.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velasquez AC, Chakravarthy S, Martin GB. 2009. Virus-induced gene silencing (VIGS) in Nicotiana benthamiana and tomato. J Vis Exp doi: 10.3791/1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haefele DM, Lindow SE. 1987. Flagellar motility confers epiphytic fitness advantages upon Pseudomonas syringae. Appl Environ Microbiol 53:2528–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee J, Teitzel GM, Munkvold K, del Pozo O, Martin GB, Michelmore RW, Greenberg JT. 2012. Type III secretion and effectors shape the survival and growth pattern of Pseudomonas syringae on leaf surfaces. Plant Physiol 158:1803–1818. doi: 10.1104/pp.111.190686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nobori T, Velásquez AC, Wu J, Kvitko BH, Kremer JM, Wang Y, He SY, Tsuda K. 2018. Transcriptome landscape of a bacterial pathogen under plant immunity. Proc Natl Acad Sci U S A 115:E3055. doi: 10.1073/pnas.1800529115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson JC, Wan Y, Kim YM, Pasa-Tolic L, Metz TO, Peck SC. 2014. Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proc Natl Acad Sci U S A 111:6846–6851. doi: 10.1073/pnas.1403248111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lovelace AH, Smith A, Kvitko BH. 2018. Pattern-triggered immunity alters the transcriptional regulation of virulence-associated genes and induces the sulfur starvation response in Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 31:750–765. doi: 10.1094/MPMI-01-18-0008-R. [DOI] [PubMed] [Google Scholar]
- 59.Lan L, Deng X, Zhou J, Tang X. 2006. Genome-wide gene expression analysis of Pseudomonas syringae pv. tomato DC3000 reveals overlapping and distinct pathways regulated by hrpL and hrpRS. Mol Plant Microbe Interact 19:976–987. doi: 10.1094/MPMI-19-0976. [DOI] [PubMed] [Google Scholar]
- 60.Lam HN, Chakravarthy S, Wei H-L, BuiNguyen H, Stodghill PV, Collmer A, Swingle BM, Cartinhour SW. 2014. Global analysis of the HrpL regulon in the plant pathogen Pseudomonas syringae pv. tomato DC3000 reveals new regulon members with diverse functions. PLoS One 9:e106115. doi: 10.1371/journal.pone.0106115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jovanovic M, James EH, Burrows PC, Rego FG, Buck M, Schumacher J. 2011. Regulation of the co-evolved HrpR and HrpS AAA+ proteins required for Pseudomonas syringae pathogenicity. Nat Commun 2:177. doi: 10.1038/ncomms1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jovanovic M, Lawton E, Schumacher J, Buck M. 2014. Interplay among Pseudomonas syringae HrpR, HrpS and HrpV proteins for regulation of the type III secretion system. FEMS Microbiol Lett 356:201–211. doi: 10.1111/1574-6968.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Records AR, Gross DC. 2010. Sensor kinases RetS and LadS regulate Pseudomonas syringae type VI secretion and virulence factors. J Bacteriol 192:3584–3596. doi: 10.1128/JB.00114-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garrett ES, Perlegas D, Wozniak DJ. 1999. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J Bacteriol 181:7401–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Firoved AM, Boucher JC, Deretic V. 2002. Global genomic analysis of AlgU (sigma(E))-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J Bacteriol 184:1057–1064. doi: 10.1128/jb.184.4.1057-1064.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tart AH, Wolfgang MC, Wozniak DJ. 2005. The alternative sigma factor AlgT represses. J Bacteriol 187:7955–7962. doi: 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolfgang MC, Jyot J, Goodman AL, Ramphal R, Lory S. 2004. Pseudomonas aeruginosa regulates flagellin expression as part of a global response to airway fluid from cystic fibrosis patients. Proc Natl Acad Sci U S A 101:6664–6668. doi: 10.1073/pnas.0307553101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cobb LM, Mychaleckyj JC, Wozniak DJ, López-Boado YS. 2004. Pseudomonas aeruginosa flagellin and alginate elicit very distinct gene expression patterns in airway epithelial cells: implications for cystic fibrosis disease. J Immunol 173:5659–5670. doi: 10.4049/jimmunol.173.9.5659. [DOI] [PubMed] [Google Scholar]
- 69.Tart AH, Blanks MJ, Wozniak DJ. 2006. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J Bacteriol 188:6483–6489. doi: 10.1128/JB.00636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Markel E, Dalenberg H, Monteil CL, Vinatzer BA, Swingle B. 2018. An AlgU-regulated antisense transcript encoded within the Pseudomonas syringae fleQ gene has a positive effect on motility. J Bacteriol 200:e00576-17. doi: 10.1128/JB.00576-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prada-Ramírez HA, Pérez-Mendoza D, Felipe A, Martínez-Granero F, Rivilla R, Sanjuán J, Gallegos M-T. 2016. AmrZ regulates cellulose production in Pseudomonas syringae pv. tomato DC3000. Mol Microbiol 99:960–977. doi: 10.1111/mmi.13278. [DOI] [PubMed] [Google Scholar]
- 72.Mascher T. 2013. Signaling diversity and evolution of extracytoplasmic function (ECF) sigma factors. Curr Opin Microbiol 16:148–155. doi: 10.1016/j.mib.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 73.Qiu D, Eisinger VM, Rowen DW, Yu HD. 2007. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wood LF, Ohman DE. 2009. Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol 72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 75.Yorgey P, Rahme LG, Tan M-W, Ausubel FM. 2001. The roles of mucD and alginate in the virulence of Pseudomonas aeruginosa in plants, nematodes and mice. Mol Microbiol 41:1063–1076. doi: 10.1046/j.1365-2958.2001.02580.x. [DOI] [PubMed] [Google Scholar]
- 76.Aslam SN, Newman MA, Erbs G, Morrissey KL, Chinchilla D, Boller T, Jensen TT, De Castro C, Ierano T, Molinaro A, Jackson RW, Knight MR, Cooper RM. 2008. Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr Biol 18:1078–1083. doi: 10.1016/j.cub.2008.06.061. [DOI] [PubMed] [Google Scholar]
- 77.King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44:301–307. [PubMed] [Google Scholar]
- 78.Butcher BG, Bao Z, Wilson J, Stodghill P, Swingle B, Filiatrault M, Schneider D, Cartinhour S. 2017. The ECF sigma factor, PSPTO_1043, in Pseudomonas syringae pv. tomato DC3000 is induced by oxidative stress and regulates genes involved in oxidative stress response. PLoS One 12:e0180340. doi: 10.1371/journal.pone.0180340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. doi: 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 80.Clarke CR, Hayes BW, Runde BJ, Markel E, Swingle BM, Vinatzer BA. 2016. Comparative genomics of Pseudomonas syringae pathovar tomato reveals novel chemotaxis pathways associated with motility and plant pathogenicity. PeerJ 4:e2570. doi: 10.7717/peerj.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.