Genetic competence in bacteria leads to horizontal gene transfer, which can ultimately affect antibiotic resistance, adaptation to stress conditions, and virulence. While the mechanisms of pneumococcal competence signaling cascades have been well characterized, the molecular mechanism behind competence regulation is not fully understood. The bacterial second messenger c-di-AMP has previously been shown to play a role in bacterial physiology and pathogenesis. In this study, we provide compelling evidence for the interplay between c-di-AMP and the pneumococcal competence state. These findings not only attribute a new biological function to this dinucleotide as a regulator of competence, transformation, and survival under stress conditions in pneumococci but also provide new insights into how pneumococcal competence is modulated.
KEYWORDS: Streptococcus pneumoniae, c-di-AMP, stress response, CdaA, competence, potassium, CSP
ABSTRACT
Streptococcus pneumoniae (the pneumococcus) is a naturally competent organism that causes diseases such as pneumonia, otitis media, and bacteremia. The essential bacterial second messenger cyclic di-AMP (c-di-AMP) is an emerging player in the stress responses of many pathogens. In S. pneumoniae, c-di-AMP is produced by a diadenylate cyclase, CdaA, and cleaved by phosphodiesterases Pde1 and Pde2. c-di-AMP binds a transporter of K+ (Trk) family protein, CabP, which subsequently halts K+ uptake via the transporter TrkH. Recently, it was reported that Pde1 and Pde2 are essential for pneumococcal virulence in mouse models of disease. To elucidate c-di-AMP-mediated transcription that may lead to changes in pathogenesis, we compared the transcriptomes of wild-type (WT) and Δpde1 Δpde2 strains by transcriptome sequencing (RNA-Seq) analysis. Notably, we found that many competence-associated genes are significantly upregulated in the Δpde1 Δpde2 strain compared to the WT. These genes play a role in DNA uptake, recombination, and autolysis. Competence is induced by a quorum-sensing mechanism initiated by the secreted factor competence-stimulating peptide (CSP). Surprisingly, the Δpde1 Δpde2 strain exhibited reduced transformation efficiency compared to WT bacteria, which was c-di-AMP dependent. Transformation efficiency was also directly related to the [K+] in the medium, suggesting a link between c-di-AMP function and the pneumococcal competence state. We found that a strain that possesses a V76G variation in CdaA produced less c-di-AMP and was highly susceptible to CSP. Deletion of cabP and trkH restored the growth of these bacteria in medium with CSP. Overall, our study demonstrates a novel role for c-di-AMP in the competence program of S. pneumoniae.
IMPORTANCE Genetic competence in bacteria leads to horizontal gene transfer, which can ultimately affect antibiotic resistance, adaptation to stress conditions, and virulence. While the mechanisms of pneumococcal competence signaling cascades have been well characterized, the molecular mechanism behind competence regulation is not fully understood. The bacterial second messenger c-di-AMP has previously been shown to play a role in bacterial physiology and pathogenesis. In this study, we provide compelling evidence for the interplay between c-di-AMP and the pneumococcal competence state. These findings not only attribute a new biological function to this dinucleotide as a regulator of competence, transformation, and survival under stress conditions in pneumococci but also provide new insights into how pneumococcal competence is modulated.
INTRODUCTION
Streptococcus pneumoniae (the pneumococcus) is an extracellular human pathogen that frequently causes pneumonia, septicemia, otitis media, sinusitis, and meningitis. The available pneumococcal vaccines protect against only a subset of over 90 capsular serotypes, and the threats from serotype switching and antibiotic resistance are mounting. One of the hallmarks of pneumococcal research is the discovery of natural transformation, which is the ability of cells to acquire extracellular DNA (1). Genetic competence is an important bioprocess that is critical for pneumococcal evolution and adaptation to diverse stress and environmental conditions. The mechanisms of competence initiation, DNA uptake and recombination, and competence regulation have been extensively studied in this pathogen; however, the regulatory cascades of competence development have not been completely elucidated.
The natural competence program of S. pneumoniae is initiated by the expression, processing, and secretion of a small pheromone, competence-stimulating peptide (CSP) (2). CSP is recognized by the histidine sensor kinase ComD of a two-component system, ComDE (3–5). Phosphorylated ComE upregulates the expression of the early competence genes, including the alternative sigma factor, ComX. ComX drives expression of late genes that govern transformation. Competent cells also induce cell lysis of neighboring noncompetent bacteria in a process termed fratricide (6). Genomic DNA is liberated from a subpopulation of cells that undergoes autolysis or is lysed through fratricide by competent pneumococci releasing the murein hydrolase, LytA, and bacteriocins (6–11). Expression of immunity factors within competent bacteria protects cells from lysis (9, 12). CSP-induced lysis is thought to be important for procurement of DNA from closely related Streptococcus spp. and therefore offers a mechanism that can promote horizontal gene transfer of compatible and beneficial genes (13, 14).
Environmental stochasticity is an element of competence induction that can affect the production of and the response to CSP (15–17). Microaerobic conditions suppress the ability to induce competence (18–21), and pH can affect competence induction and autolysis (17, 22, 23). In addition, DNA-damaging agents and hypermutation trigger competence (24, 25). Recently, it was suggested that initiation of the competence program via CSP is not strictly a consequence of quorum sensing but can be modulated by the input of multiple factors, including cell density and environmental stimuli (15).
Cyclic di-AMP (c-di-AMP) is a signaling dinucleotide that has recently been distinguished as a crucial player in bacterial physiology and stress adaptation. c-di-AMP is produced from ATP or ADP by diadenylate cyclases and cleaved by c-di-AMP phosphodiesterases into pApA or AMP (26–28). In many species, c-di-AMP is an essential second messenger. The physiological outcomes of c-di-AMP signaling are mediated by the functions of c-di-AMP-binding effector proteins and RNAs. However, c-di-AMP-producing bacteria express diverse c-di-AMP signaling networks, and not all effector molecules are conserved among species. Mutation of genes encoding c-di-AMP-synthesizing or -hydrolyzing enzymes disrupts c-di-AMP homeostasis, which alters cell wall homeostasis, osmolyte transport, central metabolism, and virulence (29–34).
S. pneumoniae encodes an essential diadenylate cyclase, CdaA, and two c-di-AMP phosphodiesterases, Pde1 and Pde2 (35). Previously, we identified a c-di-AMP-binding protein, CabP, as a mediator of potassium uptake and c-di-AMP homeostasis (36, 37). Recently, we found that phosphodiesterase-deficient pneumococcal strains, which possess elevated levels of c-di-AMP, grow poorly under low [K+] conditions and are highly susceptible to UV treatment and acidic, osmotic, and heat stress conditions (35–37). In addition, in a heat shock suppressor screen, we recovered single-nucleotide changes in cdaA that restored bacterial growth and stress resistance in these suppressor mutant strains by diminishing the enzymatic activity of the cyclase (37).
In an effort to determine how c-di-AMP affects the pneumococcal transcriptome, in this work we analyzed the gene expression profiles of wild-type (WT) and Δpde1 Δpde2 bacteria by transcriptome sequencing (RNA-Seq). We found that a large number of competence genes were upregulated in the Δpde1 Δpde2 strain. Thus, we further explored whether c-di-AMP affects pneumococcal competence and revealed that high levels of c-di-AMP were detrimental to transformation, which was partly restored by either reducing c-di-AMP levels or adding K+ in the transformation medium. By introducing a V76G mutation in CdaA to construct a cdaA* strain, which produces lower levels of c-di-AMP, we compared and contrasted c-di-AMP-dependent phenotypes in pneumococci. We found that c-di-AMP affects transformation, responsiveness to CSP, and growth under stress conditions. The inability of the cdaA* strain to grow in medium with CSP was restored by deletions of the genes needed for K+ uptake in S. pneumoniae. Overall, this study signifies that c-di-AMP influences the competence program of S. pneumoniae partly through modulating K+ uptake.
RESULTS
RNA-Seq analysis reveals induction of competence genes in a Δpde1 Δpde2 strain.
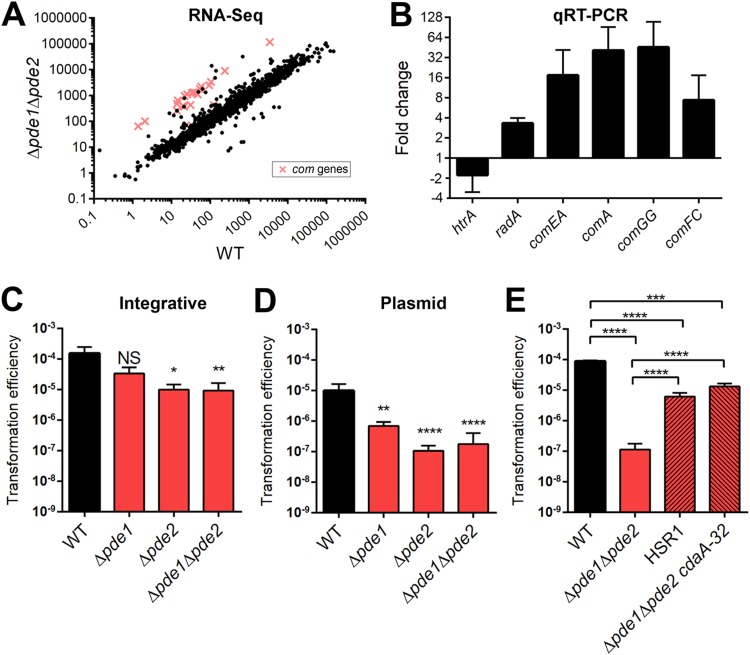
In a previous study, we demonstrated that Pde1 and Pde2 are phosphodiesterases that cleave c-di-AMP and that a Δpde1 Δpde2 strain has higher levels of c-di-AMP than the WT (35). Strains deficient for one or both Pde-encoding genes have been shown to have attenuated virulence in mouse models of otitis media, pneumonia, and bacteremia (35, 38). We hypothesized that maintaining c-di-AMP homeostasis is essential for pneumococcal virulence and the stress response since c-di-AMP, as a signaling nucleotide, modulates gene expression and protein activity. To address how c-di-AMP may augment the transcriptome, we compared the gene expression of the Δpde1 Δpde2 strain (with high c-di-AMP levels) to that of the WT (under homeostatic c-di-AMP conditions) by RNA-Seq analysis. We found that 126 genes were significantly differentially transcribed in the two strains using a fold change of ≥1.5 as a cutoff (see Table S1 in the supplemental material). Of these, 106 genes were upregulated and 20 genes were downregulated in the Δpde1Δpde2 strain. Strikingly, 59 of the differentially regulated genes have been reported to be CSP responsive, among which 20 of the upregulated genes in the Δpde1 Δpde2 strain were directly annotated as competence (com) genes (Fig. 1A) (differentially regulated com genes are shown in Table 1, and the full list of CSP-induced genes is shown in Table S2 in the supplemental material). To validate the RNA-Seq results, we performed quantitative reverse transcription-PCR (qRT-PCR) of a selection of differentially expressed genes. We found that the fold change between the WT and Δpde1 Δpde2 strains was comparable to that from the RNA-Seq data (Fig. 1B). These results suggest that c-di-AMP may play a role in the pneumococcal competence state.
FIG 1.
Differential gene expression between the WT and Δpde1 Δpde2 strains determined using RNA-Seq and the effect of mutations in cdaA on transformation of the Δpde1 Δpde2 strain. (A) Scatterplot of RNA reads of the WT and Δpde1 Δpde2 strains determined using RNA-Seq. The com genes upregulated in the Δpde1 Δpde2 strain are indicated in red. Each spot or cross represents the average reads from three independent biological replicates. (B) Validation of RNA-Seq results by qRT-PCR. Fold changes of selected genes were determined from cDNAs generated from the same RNA samples as used in RNA-Seq. (C, D, and E) The WT, the pde mutants, HSR1, and the Δpde1 Δpde2 cdaA-32 strain were given CSP to induce transformation. Strains were transformed with either an integrative fragment with a Janus cassette (C) or plasmid pVA838 (D and E). Transformation efficiencies were calculated as output/input as described in Materials and Methods. Data shown are the means from three independent biological replicates. Error bars indicate the standard errors of the means (SEMs). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant.
TABLE 1.
Differentially expressed com genes in the Δpde1 Δpde2 mutant compared to the WT
| Protein | Gene | Description | Fold change (log2)a | q value |
|---|---|---|---|---|
| ComM | SPD_1744 | Immunity factor | 3.8 | 0.0014 |
| ComEA | SPD_0843 | Late competence protein, DNA receptor | 4.1 | 0.0014 |
| ComEC | SPD_0844 | Late competence protein, DNA transport | 4.2 | 0.0014 |
| ComX1 | SPD_0014 | Competence-specific sigma factor | 4.3 | 0.0014 |
| ComX2 | SPD_1818 | Competence-specific sigma factor | 4.5 | 0.0014 |
| ComE | SPD_2063 | Response regulator | 4.7 | 0.0014 |
| ComA | SPD_0049 | CSP ABC transporter ATP-binding protein | 4.9 | 0.0014 |
| ComD | SPD_2064 | Histidine sensor kinase | 5.0 | 0.0014 |
| ComB | SPD_0050 | CSP ABC transporter permease | 5.0 | 0.0014 |
| ComC | SPD_2065 | CSP precursor | 5.1 | 0.0014 |
| ComGF | SPD_1858 | Late competence protein, DNA uptake | 5.1 | 0.0014 |
| ComGE | SPD_1859 | Late competence protein, DNA uptake | 5.2 | 0.0014 |
| ComW | SPD_0023 | Competence positive regulator | 5.2 | 0.0014 |
| ComGG | SPD_1857 | Late competence protein, DNA uptake | 5.2 | 0.0014 |
| ComGC | SPD_1861 | Late competence protein, DNA uptake | 5.2 | 0.0014 |
| ComGD | SPD_1860 | Late competence protein, DNA uptake | 5.4 | 0.0014 |
| ComGB | SPD_1862 | Late competence protein, DNA uptake | 5.4 | 0.0014 |
| ComGA | SPD_1863 | Late competence protein, DNA uptake | 5.5 | 0.0014 |
| ComFA | SPD_2035 | Late competence protein, DNA transport ATPase | 5.5 | 0.0014 |
| ComFC | SPD_2034 | Late competence protein, DNA uptake | 5.6 | 0.0014 |
Fold changes in RNA reads of upregulated com genes in the Δpde1 Δpde2 compared to those in the WT. Data listed are the average fold changes (Δpde1 Δpde2/WT) from three biological replicate experiments.
c-di-AMP phosphodiesterase mutants have a defect in transformation.
Due to the marked induction of competence genes in the Δpde1 Δpde2 strain, we hypothesized that c-di-AMP may affect the pneumococcal competence program. To test this hypothesis, we transformed the WT and the Δpde1, Δpde2, and Δpde1 Δpde2 mutants with linear DNA or shuttle plasmid pVA838. The linear DNA is comprised of the Janus cassette, which contains a kanamycin resistance marker (39), and homologous arms for replacing the arcA open reading frame (ORF). The plasmid harbors an erythromycin resistance marker. The results showed that all of the pde-deficient strains had significantly lower transformation efficiencies for both the integrative fragment and plasmid than the WT strain (Fig. 1C and D). There was no difference in the number of live cells at the time of plating. To investigate if this effect was due specifically to c-di-AMP dysregulation or another unknown function of the Pde proteins, we determined the transformation efficiency using two Δpde1 Δpde2-derived strains, HSR1 and a Δpde1 Δpde2 cdaA-32 strain. We previously showed that these strains lack pde1 and pde2 but that each harbors a point mutation in cdaA, which reduces intracellular c-di-AMP levels (37). HSR1 contains a single-nucleotide change in cdaA, which results in a V76G substitution. The Δpde1 Δpde2 cdaA-32 strain has an addition of an adenine immediately before the stop codon, which extends translation by 32 amino acids. The mutations in cdaA partially restored the transformation defect of pde-deficient pneumococci (Fig. 1E), signifying that high levels of c-di-AMP are detrimental for transformation of S. pneumoniae.
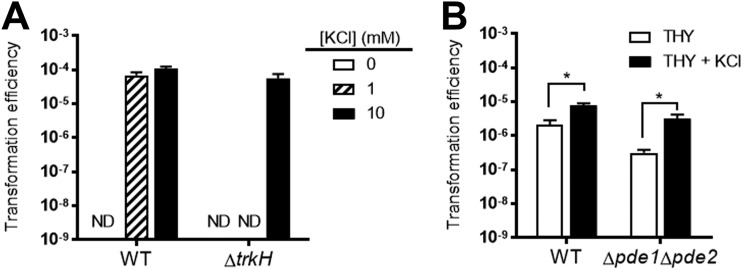
Controlling potassium transport is a known effector function of c-di-AMP in pneumococci (36). Previously, we demonstrated that a transporter of K+ (Trk) family protein, CabP, binds c-di-AMP. CabP facilitates K+ uptake via the transmembrane protein TrkH through a protein-protein interaction, but c-di-AMP-bound CabP dissociates from TrkH and impairs K+ uptake (36). In addition, Δpde1 Δpde2 bacteria, which possess high levels of c-di-AMP, have a greater growth defect in culture medium with a low [K+] (36). A study on competence in Streptococcus mutans reported that both a concentration of at least 25 mM environmental K+ and a potassium transporter, Trk2, were required for competence development and efficient transformation (40). Therefore, we hypothesized that high c-di-AMP levels diminishing K+ uptake may be impeding transformation. To explore the role of K+ transport in the transformation efficiency of S. pneumoniae, WT and ΔtrkH bacteria were transformed with plasmid in a modified protocol. Strains were grown in Todd-Hewitt broth containing 0.5% yeast extract (THY) before being thoroughly washed in a K+-free chemically defined medium (CDM) and resuspended in a CDM with 0, 1, or 10 mM K+, in which bacteria were incubated with CSP and the plasmid to initiate competence. The input, or number of live cells after incubation in the transformation medium, was similar between the two strains (see Table S3 in the supplemental material). The transformation efficiency was not significantly different between the WT and ΔtrkH strains after incubation in CDM with 10 mM K+ (Fig. 2A). However, ΔtrkH bacteria, which cannot grow but remain viable in medium containing 1 mM K+ (36) (Table S3), were unable to be transformed at this concentration of K+, while the WT still maintained a transformation efficiency similar to that in medium with 10 mM K+. Similarly, transformants were not recovered in either strain after incubation in K+-free medium (Fig. 2A; Table S3). These results demonstrate the essentiality of K+ in pneumococcal transformation. Next, the WT and Δpde1 Δpde2 strains were transformed in a modified protocol with or without the addition of excess K+ to THY medium. We found that addition of K+ significantly enhanced the transformation efficiencies of both the WT (2-fold) and Δpde1 Δpde2 (10-fold) strains (Fig. 2B). These findings highly suggest that the reduced transformation efficiency of the Δpde1 Δpde2 strain is partly due to impaired K+ uptake resulting from elevated c-di-AMP levels in this mutant.
FIG 2.
Effect of potassium on pneumococcal transformation efficiency. (A) WT and ΔtrkH bacteria were transformed in a modified protocol as described in Materials and Methods. Pneumococci were washed with potassium-free CDM and resuspended in CDM containing 0, 1, or 10 mM KCl. Transformation was initiated by addition of CSP and plasmid pVA838. (B) WT and Δpde1 Δpde2 bacteria were transformed with the plasmid in transformation medium either with or without addition of 20 mM KCl using a modified protocol as described in Materials and Methods. Transformation efficiencies were calculated as output/input as described in Materials and Methods. ND, under the level of detection (no colonies were recovered). Data shown are the means from three independent biological replicates. Error bars indicate the SEMs. *, P < 0.05.
Survival in media with different concentrations of glycine or pHs is dependent on c-di-AMP homeostasis.
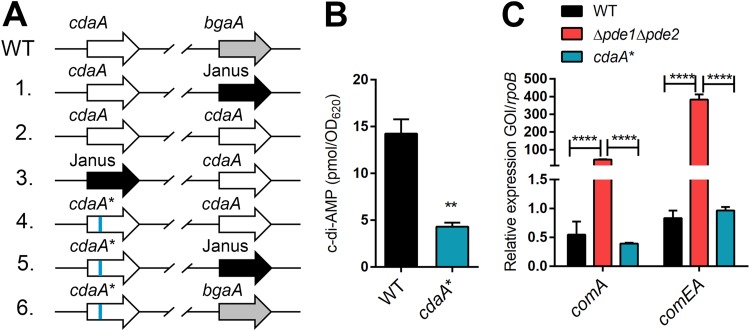
Based on the reduced transformation efficiency of the Δpde1 Δpde2 strain compared to that of the WT, we posited that low c-di-AMP levels may improve pneumococcal transformation efficiency. cdaA is an essential gene in S. pneumoniae, and we were unable to delete it directly (35). However, we were capable of deleting cdaA from the native locus after we inserted a second copy of cdaA at the bgaA locus. Ultimately, we introduced the V76G point mutation in S. pneumoniae through a six-step procedure (Fig. 3A). We designated this strain the cdaA* strain. As expected, the cdaA* strain produced lower levels of c-di-AMP than did WT bacteria (Fig. 3B). After construction of a low c-di-AMP-producing strain, we measured the expression of com genes in the cdaA* strain compared with the WT and Δpde1 Δpde2 strains. Analysis of relative quantification of a representative early gene, comA, and a late gene, comEA, showed that the Δpde1 Δpde2 strain displayed higher expression of both competence genes than WT and cdaA* bacteria (Fig. 3C). However, there was no significant difference in expression between the WT and cdaA* strains, demonstrating that lower c-di-AMP levels do not alter early and late com gene expression.
FIG 3.
Construction and characterization of cdaA* bacteria. (A) Construction of the cdaA* strain. The organizations of the cdaA locus and the bgaA locus are shown. Bacteria were transformed with the indicated ORFs flanked by the respective upstream and downstream homologous arms for integration and replacement of the previous gene as detailed in Materials and Methods. WT bacteria were transformed with the Janus cassette, which confers kanamycin resistance, to replace the native bgaA. The subsequent steps maintain the essential gene cdaA at the bgaA locus in order to introduce the cdaAV76G mutation at its native site. (B) Determination of c-di-AMP levels in the WT and cdaA* strains. (C) Expression of an early gene and a late gene in the WT, Δpde1 Δpde2, and cdaA* strains. Relative expression of comA and comEA was determined. Data shown in all graphs are the means from three independent biological replicates. The error bars indicate the SEMs. **, P < 0.01; ****, P < 0.0001.
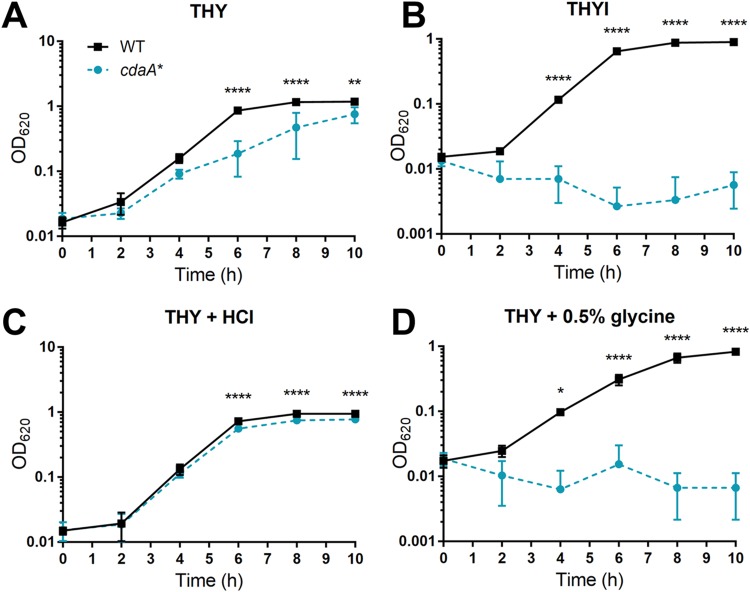
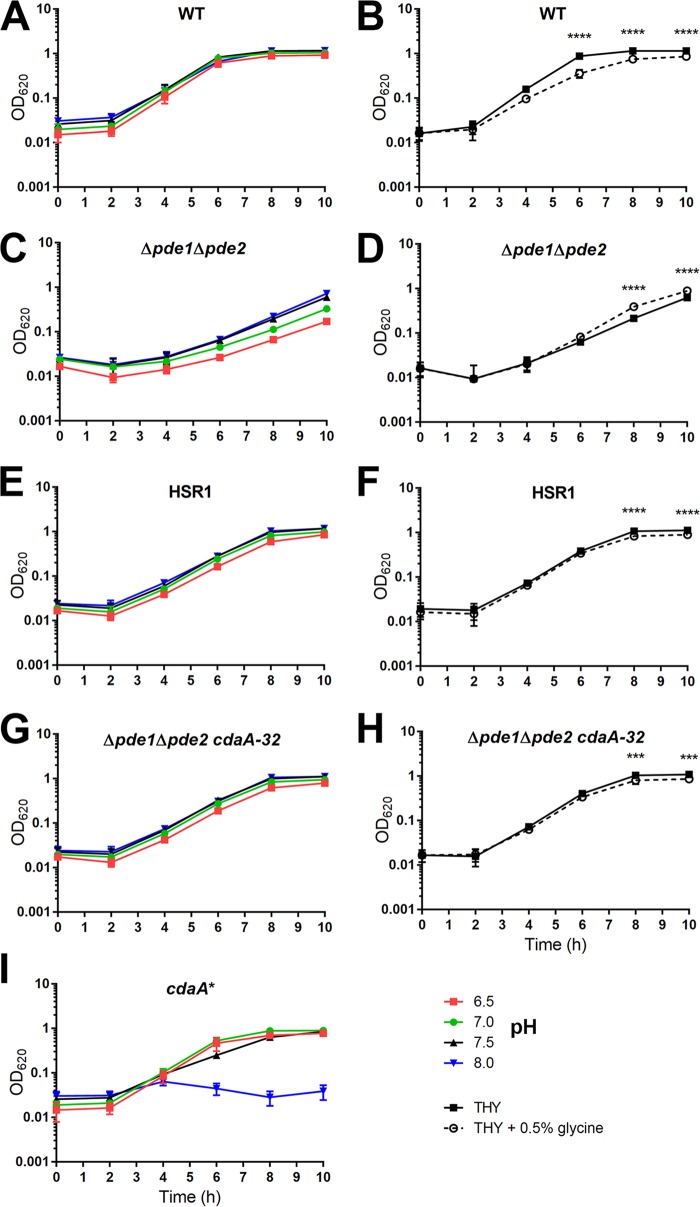
To explore the effect of low c-di-AMP on genetic competence, we sought to determine the transformation efficiency of the cdaA* strain. In the transformation protocol adapted from a report by van Opijnen et al. (41), the host bacteria are initially grown in THYI, which is Todd-Hewitt broth containing 0.5% yeast extract (THY) supplemented with 11 mM HCl and 0.5% glycine to reduce premature induction of competence and to increase permeability of the cell wall, respectively (42). Next, the bacteria are resuspended in competence-permissible medium THYII with addition of CSP and DNA. THYII is THY supplemented with components necessary for the induction: 2 mg · ml−1 bovine serum albumin (BSA), 1 mM CaCl2, and 10 mM NaOH (17, 42–44). Finally, the bacteria are recovered in THY before plating. Surprisingly, the cdaA* strain had a minor growth defect in THY medium but failed to grow in THYI (Fig. 4A and B). Upon division of the different additions in THYI, we found that the cdaA* strain exhibited a doubling time of 75.3 ± 4.3 min in THY, compared to the WT with a doubling time of 49.1 ± 1.9 min, which could be partially restored with the addition of HCl (doubling time, 51.9 ± 1.4 min) (Fig. 4A and C). Most noticeably, the cdaA* strain was unable to grow in THY with the addition of 0.5% glycine (Fig. 4D), which was comparable to the growth in THYI. To further examine the role of c-di-AMP in pneumococcal growth under these conditions, we grew the WT, Δpde1 Δpde2, HSR1, Δpde1 Δpde2 cdaA-32, and cdaA* strains in THY adjusted to pH 6.5, 7.0, 7.5, or 8.0 or in THY with the addition of 0.5% glycine and monitored the growth over time. The WT was able to grow under all pH conditions but had a slight growth defect from the addition of glycine (Fig. 5A and B). The Δpde1 Δpde2 mutant favored growth in more-basic media and had significantly better growth with glycine addition (Fig. 5C and D). In contrast, the cdaA* strain preferred media at pH 6.5 to 7.0 and had growth arrest at pH 8.0 (Fig. 5I). HSR1 and the Δpde1 Δpde2 cdaA-32 strain, two mutants in the Δpde1 Δpde2 background with lowered levels of c-di-AMP, corrected the growth phenotype of the Δpde1 Δpde2 strain in medium with either low pH or with the addition of glycine (Fig. 5E to G and H). Next, to verify that c-di-AMP levels control pneumococcal survival under basic conditions or in medium supplemented with glycine, we introduced native cdaA, or the Janus cassette as a control, at the bgaA locus in cdaA* and tested the growth of these strains. We found that complementation with native cdaA significantly increased c-di-AMP levels (see Fig. S1A in the supplemental material) and restored the bacterial growth in medium adjusted to pH 8.0 or supplemented with glycine (Fig. S1B and C). Taking these results together, the preferential growth of strains with dysregulated c-di-AMP suggests that c-di-AMP controls the stress response to pH and affects growth in media with additional glycine, which are components associated with pneumococcal competence.
FIG 4.
Effect of pH and glycine on growth of the WT and cdaA* strains. The WT and cdaA* strains were grown in THY (A), in THYI medium for transformation (B), in THY with the same HCl concentration as in THYI (pH 7.1) (C), or in THY with the same glycine concentration (0.5%) as in THYI (D). Bacterial growth was monitored by as OD620 every 2 h. Data shown are the means from three independent biological replicates. The error bars indicate the standard deviations (SD). *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
FIG 5.
Effect of pH and glycine on growth of the WT, Δpde1 Δpde2, HSR1, and Δpde1 Δpde2 strains with CdaA-32 addition. The WT and the indicated mutant strains were grown in THY (the pH of this broth is 7.5) (A, C, E, G, and I) or in THY with the pH adjusted to 8.0 with NaOH or adjusted to 7.0 or 6.5 with HCl or in THY or THY with the same glycine concentration (0.5%) as in THYI (B, D, F, and H). Bacterial growth was monitored as OD620 every 2 h. Data shown are the means from three independent biological replicates. The error bars indicate the SD. ***, P < 0.001; ****, P < 0.0001.
CSP is detrimental to low-c-di-AMP-producing bacteria through K+ uptake by CabP and TrkH.
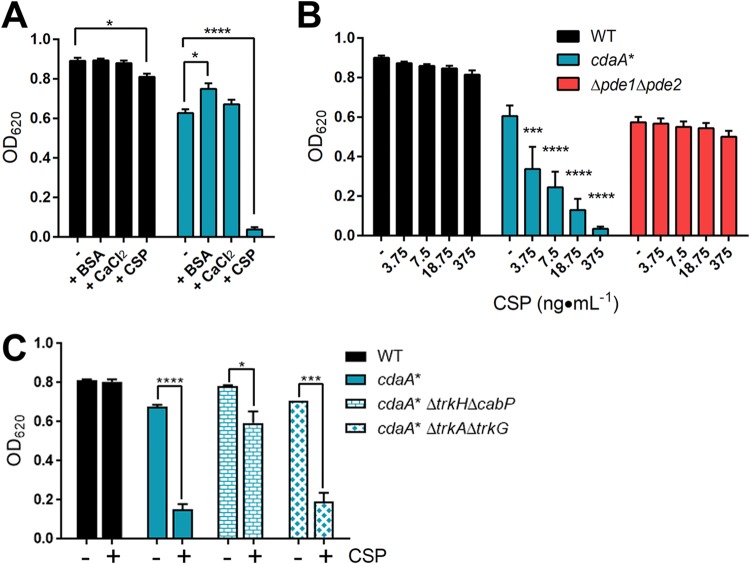
Since we found that cdaA* bacteria are highly susceptible to the addition of 0.5% glycine and basic conditions, we replaced the initial THYI medium for transformation with THY and removed the NaOH addition in THYII to allow growth of this strain in the transformation procedure. However, after induction with CSP and growth in a modified THYII without NaOH, there was still no growth of the cdaA* strain during transformation. We hypothesized that one of the components in THYII or CSP inhibits the growth of the cdaA* strain. We examined the growth of WT and cdaA* bacteria in THY or THY with the addition of BSA, CaCl2, or CSP individually at the same concentration as what is found in THYII broth. Addition of BSA or CaCl2 did not affect growth of either the WT or the cdaA* strain (Fig. 6A). However, CSP reduced the growth of the WT by 9% and the growth of the cdaA* strain by 94% compared to that in THY alone (Fig. 6A). The growth defect of the cdaA* strain could be restored by complementation (Fig. S1D). The reduction in growth of the WT is consistent with reports of CSP-dependent cell lysis termed fratricide (7–9, 12). The concentration of CSP in the transformation protocol is 375 ng · ml−1. In order to obtain a suitable concentration at which cdaA* may be able to grow, we diluted the concentration of CSP from 20-fold to 100-fold and measured the impact on growth of the WT, cdaA*, and Δpde1 Δpde2 strains. While the WT and Δpde1 Δpde2 strains were unaffected by the CSP additions, the cdaA* strain displayed CSP dose-dependent growth, as well as marked growth defects even with 3.75 ng · ml−1 of CSP added (Fig. 6B). Next, since the K+ content affects transformation efficiency, we explored the role of Trk family proteins in the susceptibility of cdaA* bacteria to CSP. S. pneumoniae expresses two sets of Trk family proteins, (i) CabP and TrkH and (ii) TrkA (SPD_0430) and TrkG (SPD_0429), which have high sequence similarity, respectively, but TrkA and TrkG are not involved in K+ uptake (36). The WT, cdaA*, cdaA* ΔtrkH ΔcabP, and cdaA* ΔtrkA ΔtrkG strains were grown in THY or THY with the addition of CSP. The deletion of the K+ uptake proteins CabP and TrkH restored growth of the cdaA* strain with CSP, while the strain with deletion of TrkA and TrkG was still susceptible to CSP addition (Fig. 6C). These results indicate that the toxicity of CSP in cdaA* bacteria was mediated by c-di-AMP effector proteins CabP and TrkH.
FIG 6.
Effect of CSP addition on growth of the WT, cdaA*, and Δpde1 Δpde2 strains. (A) The WT and cdaA* strains were grown in THY or in THY with 2 mg · ml−1 BSA, 1 mM CaCl2, or 375 ng · ml−1 CSP. Bacterial growth was measured as OD620 after 9 h of incubation. (B) The WT, cdaA*, and Δpde1 Δpde2 strains were grown in THY supplemented with the indicated concentrations of CSP. Bacterial growth was measured as OD620 after 10 h of incubation. (C) Growth of the indicated trk mutants in the presence and absence of 375 ng · ml−1 CSP. Bacterial growth was measured as OD620 after 8 h of incubation. Data shown for all panels are the means from three independent biological replicates. The error bars indicate the SEMs. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
Role of CSP signaling in susceptibility of the cdaA* strain to pH, glycine, and CSP.
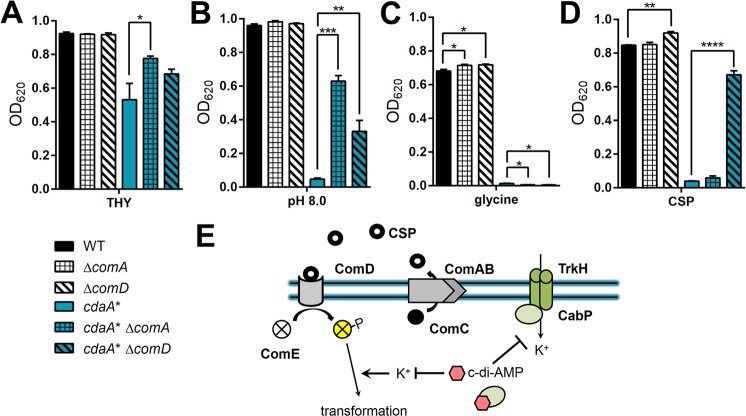
The two-component system ComDE recognizes CSP for competence induction and CSP-dependent cell lysis (3, 4). In addition, ComA is an ABC transporter that is required for releasing mature CSP (45, 46). We hypothesized that the susceptibility of the cdaA* strain to pH, glycine, and CSP is due to a response through this signaling pathway. We constructed ΔcomA and ΔcomD mutants in WT and cdaA* backgrounds to assess the role of bacteria secreting CSP or responding to CSP in survival within basic medium and medium with the addition of glycine or CSP. We found that deletions of comA and comD did not obviously affect the growth of WT bacteria in all of the tested media (Fig. 7A, B, and C). However, the cdaA* ΔcomA strain had significantly enhanced bacterial growth compared to the cdaA* strain in THY and THY at pH 8.0 but not in medium with additional glycine (Fig. 7A, B, and C). The deletion of comD in the cdaA* strain reestablished growth in THY at pH 8.0 (Fig. 7B). These results indicate that the susceptibility of the cdaA* strain to basic conditions is partially reliant on self-induction of CSP-dependent signaling. After subjecting the strains to CSP, the ΔcomD strain grew slightly better than the WT, as expected (Fig. 7D). Furthermore, the deletion of comD in cdaA* bacteria restored growth to levels comparable to that in THY alone, while the comA deletion had no appreciable effect (Fig. 7D). Taken together, these results indicate that the susceptibility of the cdaA* strain to CSP and basic pH is through a CSP-dependent signaling pathway, whereas the susceptibility to glycine is CSP independent.
FIG 7.
Impact of ComA and ComD on survival of the cdaA* strain in basic medium or with the addition of glycine or CSP. (A to D) The indicated strains were grown in THY (A), THY at pH 8.0 (B), THY with the addition of 0.5% glycine (C), or THY with the addition of 375 ng · ml−1 CSP (D). Bacterial growth was measured as OD620 after 10 h of incubation. Data shown are the means from three independent biological replicates. The error bars indicate the SEMs. (E) Hypothetical model. c-di-AMP blocks K+ transport by directly binding CabP, which inhibits TrkH transport. Regulation of K+ is required for effective transformation in the presence of CSP, and therefore, modulation of c-di-AMP levels can control pneumococcal competence. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
DISCUSSION
The competence state is an important process for bacterial horizontal gene transfer, stress adaptation, and virulence (14, 17, 47). Available pneumococcal vaccines and treatments for pneumococcal disease are being undermined by capsule switching and acquisition of antibiotic resistance due to effective genetic competence (48–51). S. pneumoniae utilizes a secreted small peptide, CSP, to induce natural competence. However, the regulation of the competence signaling cascade can be affected by environmental conditions and regulatory proteins, which have been extensively studied but still not fully deciphered. In this work, Δpde1 Δpde2 bacteria exhibited decreased transformation efficiency, which was validated as being c-di-AMP dependent through the employment of two mutants with distinct variations in CdaA that decrease c-di-AMP production. Increasing the K+ concentration in transformation media improved transformation efficiency, linking an effector function of c-di-AMP with genetic competence. Additionally, multiple components in transformation media, including CSP, are toxic to cdaA* bacteria, which produce lower levels of c-di-AMP than the WT. Toxicity of CSP in cdaA* bacteria was mediated by c-di-AMP effector proteins CabP and TrkH. Thus, we describe a novel regulation of the pneumococcal competence state by the bacterial second messenger c-di-AMP.
Unexpectedly, the induction of many competence-associated genes in the Δpde1 Δpde2 mutant did not confer enhanced transformation (Fig. 1). The reduced transformation efficiencies of the pneumococcal pde mutants could be caused by c-di-AMP-mediated dysregulation of (i) responses to environmental conditions, (ii) competence regulation, and/or (iii) cell wall homeostasis. During acclimatization to environmental conditions, pneumococci can engage the competence machinery as part of the stress response (15, 17, 22, 25, 52–54). The results of this work suggest that differential control of K+ uptake by c-di-AMP affects competence and transformation in Δpde1 Δpde2 bacteria. Control of K+ transport as an osmolyte by c-di-AMP has been shown to affect cell wall morphology (34), which may alter transformation. Previously, Binepal et al. reported that in Streptococcus mutans transformation efficiency is reduced in medium containing low K+ or in a strain lacking Trk2, the major potassium transporter (40). Similarly, our work demonstrates that ΔtrkH pneumococci had a defect in transformation after incubation in a medium with low K+. Therefore, despite S. mutans encoding a different competence regulon, which includes two secreted competence-inducing peptides instead of one as in S. pneumoniae, there is a shared influence of K+ concentration on competence regulation and transformation.
Competence development is an elaborate process that has a fine-tuned temporal pattern of gene expression (55–59). Dysregulation of competence kinetics can lead to alterations in transformation efficiencies. Imbalances in competence gene expression have been shown to negatively affect the competence state (5, 60–62). After the competence state is induced and subsequently shut off, there is a refractory period with a lack of responsiveness to CSP (63, 64). The mechanism of CSP tolerance could be affected by c-di-AMP. To observe the extent of competence gene induction after CSP addition, we utilized a late-gene ssbB promoter-reporter assay, which indicates entering the competence state (24, 25, 65). After addition of CSP, the WT and cdaA* strains displayed similar kinetics and amplitude in expression driven from the ssbB promoter (see Fig. S2 in the supplemental material). However, the Δpde1 Δpde2 mutant had a significantly reduced induction of this promoter (Fig. S2). These results suggest that low c-di-AMP does not affect CSP-dependent induction of the competence state but that high c-di-AMP impairs the response to CSP. Therefore, the Δpde1Δpde2 strain has diminished transformability and a delay in the CSP-dependent induction of the competence state, which may be caused by a dysregulation in the response to CSP and a prolonged refractory phase. The increased expression of com genes in the Δpde1 Δpde2 strain at a later stage in growth could be indicative of a misappropriation of competence induction or the inability to turn off competence. The increased com gene expression during mid-log phase did not confer transformability, as DNA added at this time did not result in transformants. In a previous report, we demonstrated that c-di-AMP signaling is involved in the adaptation to multiple stress conditions, including heat shock (35, 37). Heat shock proteins are expressed during competence and have implications in the stress response, competence, and virulence (54, 55). It is likely that c-di-AMP affects a shared mechanism that regulates these complex processes.
Our results obtained by constructing a low-c-di-AMP-producing strain, the cdaA* strain, suggest that c-di-AMP homeostasis must be maintained for competence program integrity. The cdaA* strain was unable to grow under multiple conditions commonly utilized to stimulate pneumococcal transformation, including the addition of glycine or CSP or the adjustment of medium to basic pH. In this study, we found that all of these growth phenotypes were mediated by c-di-AMP. Glycine is an essential amino acid in S. pneumoniae (66). The addition of 0.5% glycine is toxic for pneumococci with low levels of c-di-AMP, which is possibly due to increased glycine uptake and/or a buildup of glycine by enzyme inhibition/activity. c-di-AMP has been shown to regulate amino acid transport via OpuC proteins in Listeria monocytogenes and Staphylococcus aureus (67, 68). Previous studies have revealed that peptide and amino acid transport systems are detrimental in c-di-AMP-deficient strains (69, 70). There are no homologs for OpuC proteins in S. pneumoniae; however, these results suggest a conserved role for c-di-AMP. Also, we found that the sensitivity of the cdaA* strain to CSP addition could be abolished by deletion of comD, which encodes the known receptor for the initiation of competence (3, 4), or by the deletion of trkH-cabP, which encodes a K+ uptake system that is controlled by c-di-AMP. It is not clear whether CSP susceptibility due to excess K+ uptake is driven by a misappropriation of proteins induced by the competence cascade, including lysins and bacteriocins, by the inability to be protected by the immunity proteins, or by some other mechanism. CSP addition to the cdaA* strain did not affect cell survival and may inhibit growth of cdaA* pneumococci rather than cause cell lysis (see Fig. S3 in the supplemental material). Lastly, we noted that the cdaA* strain could not grow in basic medium at pH 8.0 and preferred more-acidic medium (pH 6.5 to 7.0). This is consistent with pde-deficient pneumococci, which contain high levels of c-di-AMP and grow poorly in medium with acidic pH (37). However, phosphodiesterase mutants of L. monocytogenes, Lactococcus lactis, and Bacillus subtilis have been shown to display acid resistance, and in S. aureus, bacteria elevate c-di-AMP levels in response to low-pH medium (32, 71–73). The differences in observations might reflect a divergence of c-di-AMP function with regard to pH adaptation. Overall, we report that cdaA* bacteria are hypersensitive to conditions utilized in transformation media.
In this study, we describe that c-di-AMP signaling can mediate pneumococcal survival under several environmental conditions, including differential pH and glycine concentrations. These results provide novel insight into the repercussions of c-di-AMP dysregulation and suggest that there may be additional, unidentified pneumococcal c-di-AMP effectors. In particular, competence induction is less permissible in acidic media than under alkaline conditions (15, 64). It has been reported that pneumococcal autolysis is driven by a CSP-dependent pathway during alkaline pH stress but is CSP independent and ComE dependent during acidic pH stress (22, 23, 74). We propose that c-di-AMP, through altering environmental stimuli such as K+ uptake, modulates competence initiation, thereby affecting transformation and bacterial growth (Fig. 7E). Future work will focus on defining the precise molecular mechanisms that are controlled by K+-mediated c-di-AMP signaling in S. pneumoniae.
In summary, we describe a modulation of pneumococcal competence development and transformation via the bacterial signaling molecule c-di-AMP. This work, as well as our previous reports (35–37), indicates that the newly discovered second messenger c-di-AMP controls pneumococcal physiology and stress response.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains utilized in this study are listed in Table S4 in the supplemental material. S. pneumoniae D39 (serotype 2; ATCC) and its derivatives were grown in Todd-Hewitt broth (BD Biosciences) containing 0.5% yeast extract (THY). Frozen bacterial stocks were prepared by growing the strains in THY to an optical density at 620 nm (OD620) of 0.4 before addition of glycerol to a final concentration of 10 to 15% and storing at –80°C. Pneumococcal strains were grown on tryptic soy agar (Fluka Analytical) plates supplemented with 5% sheep blood. For transformation or growth assays, THYI was prepared as 250 ml of THY with the addition of 185 mM HCl and 0.5% glycine. THYII was prepared as THY with the addition of 2 mg · ml−1 bovine serum albumin (BSA), 1 mM CaCl2, and 10 mM NaOH. Both THYI and THYII were sterile filtered. For pneumococcal growth in broth or on plates, bacteria were incubated at 37°C with 5% CO2. Antibiotics were supplemented with 4 μg · ml−1 erythromycin, 200 μg · ml−1 kanamycin, or 125 μg · ml−1 streptomycin as needed for selection. From frozen stocks prepared in 50% glycerol, Escherichia coli strains were streaked overnight on Luria-Bertani (LB) agar plates, and single colonies were inoculated in LB broth with shaking at 220 rpm at 37°C. Antibiotics were supplemented with 100 μg · ml−1 ampicillin, or 25 μg · ml−1 kanamycin as needed.
Transformation of pneumococci.
Pneumococcal stocks were inoculated in THYI medium at an OD620 of 0.01. To maintain the same density at the time of competence induction, cells were grown to an OD620 of 0.05 or 0.1, and 200 μl was harvested and resuspended in THYII. Integrative or plasmid DNA was added along with CSP1 at 375 ng · ml−1. After 2 h of incubation at 37°C, 800 μl THY was added and incubated for an additional 2 h. Transformed cells were selected on plates with the appropriate antibiotic as output or on plates without antibiotic selection as input. The transformation efficiency was calculated as the number of transformed cells divided by the total number of cells at the time of plating (output/input).
For transformation in CDM, WT and ΔtrkH bacteria were grown in 5 ml THY to an OD620 of 0.05. Bacteria were harvested, washed once with 200 ml K+-free CDM, and then incubated in 1 ml K+-free CDM twice for 30 min each at 37°C to deplete K+. Subsequently, bacteria in 200 μl K+-free CDM were harvested and resuspended in medium similar to THYII but replacing THY with CDM containing 0, 1, or 10 mM KCl. Plasmid pVA838 (100 ng) was added along with CSP1 at 375 ng · ml−1. After 2 h of incubation at 37°C, 800 μl CDM containing the same concentration of KCl as in transformation medium was added and incubated for an additional 1 h. Transformation efficiency was determined similarly as described above.
To test the effect of K+ on transformation of the Δpde1 Δpde2 strain, the WT and Δpde1 Δpde2 strains were grown in THY to an OD620 of 0.05, and 200 μl was harvested and resuspended in THYII in the presence or absence of 20 mM KCl. The other steps were as in the regular transformation protocol using 100 ng pVA838 plasmid.
Construction of pneumococcal mutants.
S. pneumoniae strains are listed in Table S4. Mutants were generated by homologous recombination as described previously (75) using primers listed in Table S5 in the supplemental material. All mutant strains originate from the ST581 strain, which is referred to as the WT. Generation of the Δpde1, Δpde2, Δpde1 Δpde2, HSR1, and Δpde1 Δpde2 cdaA-32 strains has been reported in our previous studies (35, 37). The cdaA* strain was generated in a six-step process utilizing the bgaA locus to express a second copy of the WT cdaA gene in order to replace the native locus with cdaAV76G. The plasmid pBluescript SK(−) (Stratagene) was used as the backbone to construct an integrative locus. The upstream and downstream homologous arms of the bgaA locus were modeled after the design for the pPP2 plasmid (76). The upstream and downstream arms were amplified from D39 genomic DNA with primers Pr3332/Pr3333 and Pr3336/Pr3337, respectively. The PCR product of the upstream arm was digested with KpnI and XhoI and ligated with pBluescript SK(−) digested with the same enzymes to construct pST3167. Next, the downstream-arm PCR product was digested with SacI and SacII and ligated with pST3167 to construct pST3172. The Janus cassette was amplified with primers Pr3334 and Pr3335, digested with EagI and SacII, and ligated to digested pST3172 to construct pST3222. The entire integrative fragment (in the order of upstream, multiple-cloning site [MCS], Janus cassette, and downstream) was amplified from pST3222 with primers Pr3332 and Pr3404. The PCR product was digested with KpnI and SphI and ligated into pUC19 digested with the same enzymes to form the integrative plasmid pST3277. The promoter and ORF of cdaA were amplified with primers Pr3342 and Pr2879, digested with BamHI and SalI, and inserted in the MCS to construct pST3286. All plasmids were confirmed by PCR, digestion, and sequencing.
For the six-step process to generate the cdaA* strain, the Janus cassette, which has kanamycin resistance and a dominant rpsL+ allele, was utilized for selection of kanamycin-resistant, streptomycin-sensitive colonies. In contrast, unmarked strains are kanamycin sensitive and streptomycin resistant. WT bacteria were transformed with pST3286, which contains a Janus cassette, to insert at the bgaA locus to make ST3293. Next, this locus was unmarked to make ST3313 by introduction of a cdaA knock-in, which was amplified from pST3286 with primers Pr3332/Pr3434 and Pr3336/3404, digested with SacII, and ligated. The native cdaA gene was replaced in ST3313 by overlapping PCR of the homologous arms and the Janus cassette to make ST3334. The upstream arm, Janus cassette, and downstream arm were amplified with primers Pr2874/Pr3470, Pr3471/Pr3472, and Pr3473/Pr2877, respectively. Fusion PCR was performed with the upstream arm and the Janus cassette and subsequently with this fragment and the downstream arm. Next, the Janus cassette at the cdaA locus was replaced with the cdaAV76G gene to make ST3405. This gene with the homologous arms was amplified from HSR1 genomic DNA with primers Pr2874 and Pr2877. The cdaA copy at the bgaA locus was removed by transformation with pST3277 to introduce the Janus cassette and make ST3406. At the last step, the Janus cassette was removed by transformation of the PCR product from the D39 bgaA locus with primers Pr3332 and Pr3404 to make ST3409. ST3409 is referred to as the cdaA* strain. The complemented strains ST3466 and ST3467 were made by transforming the cdaA* strain with integration of the Janus cassette from pST3277 as a negative control or with integration of cdaA and the Janus cassette from pST3286 at the bgaA locus, respectively.
The ΔcomA and ΔcomD mutants were generated as follows. For comA, the upstream and downstream arms were amplified with primers Pr2084/Pr2085 and Pr2086/Pr2087, respectively. The comD upstream and downstream arms were amplified with primers Pr3529/Pr3530 and Pr3531/Pr3532, respectively. The upstream and downstream arms were digested with XbaI and XhoI, respectively, before ligation with the Janus cassette amplified with Pr1097 and Pr1098 and digested with the same enzymes. These ligations were transformed into the WT and cdaA* strains. The ΔtrkH ΔcabP and ΔtrkA ΔtrkG mutants in the cdaA* background were constructed by amplifying the genetic loci of existing mutants from our previously study using primers Pr2976/Pr2973 and Pr2994/Pr3001, respectively, and transforming into the cdaA* strain (36). All strains were confirmed by PCR, and mutations with cdaA were confirmed by sequencing as well.
RNA extraction.
RNA was extracted using the RNeasy kit (Qiagen) according to the manufacturer’s instructions. Briefly, pneumococcal strains were grown in THY to an OD620 of 0.4 to 0.5. Five to 10 milliliters was harvested by centrifugation at 3,500 rpm for 10 min. The supernatant was removed, and each pellet was resuspended in 500 μl RLT buffer. Suspensions were transferred to 2-ml Safelock tubes with beads. Cells were disrupted in a mini-bead beater (Biospec Products) at 4,200 rpm for 50 s twice. After disruption, lysates were centrifuged for 30 s at 13,000 rpm. The supernatant above the beads was removed and transferred to a new tube. An equal volume of 70% ethanol was added before transferring the lysate to an RNeasy spin column (Qiagen). Lysates were centrifuged for 30 s at top speed. RW1 was added and spun through before an on-column digest with the RNase-free DNase I set (Qiagen) according to the manufacturer’s instructions. RW1 was added and spun through again before washing with RPE buffer twice. The spin column was placed in a fresh tube for collection of the RNA in water. The extracted RNA was subjected to two off-column DNase I digests in solution along with collections by sodium acetate and isopropanol precipitation and was checked by PCR for the absence of contaminating DNA. The RNA concentration was measured by A260, and RNA was assessed for purity by reading the A260/A280 and A260/A230 ratios. Lack of degradation of the RNA preparation was confirmed by running it on an agarose gel and observing intact rRNA bands.
RNA-Seq analysis.
RNA samples for RNA-Seq analysis were prepared and analyzed as described previously (77). Briefly, rRNA was removed from the samples using the Ribo-Zero magnetic kit (Epicentre), and strand-specific DNA libraries for Illumina sequencing were made with the ScriptSeq Complete kit (Epicentre). The sequencing was performed at the University at Buffalo Next Generation Sequencing and Expression Analysis Core Facility. The sequencing files were uploaded to RNA Rocket, trimmed, aligned, and assembled to the annotation of S. pneumoniae D39 in the PATRIC database (http://patricbrc.org). Assembled RNA-Seq data were also analyzed on the Rockhopper system (Wellesley College) (78, 79).
cDNA synthesis and qRT-PCR.
RNA was converted to cDNA as previously described (80). Quantitative reverse transcription-PCR (qRT-PCR) was run and analyzed on a Step One Plus real-time PCR System (Applied Biosystems, Life Technologies) using iQ SYBR green supermix (Bio-Rad) according to the manufacturer’s instructions. Primers for qRT-PCR were designed with annealing temperatures of 60°C and to amplify ∼200-bp regions internal to the ORF using Primer3Plus (http://bioinformatics.nl/primer3plus). Semiquantitative PCR amplifying rpoB was utilized to normalize the dilution of cDNA prior to qRT-PCR. The threshold cycle (CT) value of each gene was compared to that of rpoB as a constitutive control in the WT for relative quantification or ΔΔCT. The expression of rpoB was not altered in the RNA-Seq analysis between the WT and Δpde1 Δpde2 strains. Furthermore, rpoB has been previously shown not to be differentially expressed with competence induction and has been used as a control (54, 55, 81).
Measurement of c-di-AMP concentration by ELISA.
Determinations of c-di-AMP in S. pneumoniae were performed as described previously (37). Briefly, pneumococcal stocks were grown in THY to an OD620 of 0.4. Five milliliters was harvested by centrifugation at 3,500 rpm for 10 min, washed, and resuspended in 500 μl of 50 mM Tris-HCl (pH 8.0). Cells were sonicated by two 30-s pulses and boiled for 10 min in order to measure intracellular accumulation of c-di-AMP. Whole-cell lysates were assayed for c-di-AMP through a competitive enzyme-linked immunosorbent assay (ELISA) developed previously (82). The c-di-AMP concentration was normalized to the OD620. Three independent biological replicates were measured for each sample.
β-Galactosidase assays.
The promoter-reporter fusion plasmids of pPP2-erm (pST2868) and pPP2-erm:PʹgyrA (pST2885) were utilized as the positive and negative controls as described previously (37). The promoter region of PʹssbB was amplified using the D39 equivalent primers Pr3421 and Pr3422 described by Chastanet et al. (83). The promoter fragment was digested with EcoRI and BamHI and ligated with pST2868 to generate pST3302. Pneumococcal cultures were grown in THY to OD620 of 0.1. Next, 375 ng · ml−1 CSP1 was added, and samples were incubated for the time indicated. Samples were prepared and analyzed as described previously (37) on an iD3 SpectraMax (Molecular Devices). The initial slope of the first six readings (OD450) every 10 min were used to calculate the Miller units.
Statistical analysis.
Log-transformed data from the transformation efficiencies were analyzed by one-way analysis of variance (ANOVA) with Dunnett tests to correct for multiple comparisons. Growth curves were analyzed by two-way ANOVA with the Bonferroni correction. All other data were analyzed by one-way ANOVA with Dunnett tests to correct for multiple comparisons to the control or by two-tailed t tests for single comparisons, with at least three independent replicates for each sample.
Accession number(s).
The RNA-Seq data have been deposited at NCBI Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE113231.
Supplementary Material
ACKNOWLEDGMENTS
We thank Adam Underwood, Manuel A. Pazos II, and Jianle Gao for expert technical assistance, Joseph Wade and Jing Wang for help with RNA-Seq analysis, and the Metzger lab members and Yanina Tovpeko for helpful advice and discussions. We are indebted to the Next Generation Sequencing Core Facility of the University at Buffalo for RNA-Seq.
This work was partially supported by National Institutes of Health (NIH) grant R01DC006917 to D.W.M. G.B. is a subrecipient of NIH grants R56AI122763 and R35HL135756.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Griffith F. 1928. The significance of pneumococcal types. J Hyg (Lond) 27:113–159. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Havarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Havarstein LS, Gaustad P, Nes IF, Morrison DA. 1996. Identification of the streptococcal competence-pheromone receptor. Mol Microbiol 21:863–869. doi: 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- 4.Pestova EV, Havarstein LS, Morrison DA. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol 21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 5.Martin B, Soulet AL, Mirouze N, Prudhomme M, Mortier-Barriere I, Granadel C, Noirot-Gros MF, Noirot P, Polard P, Claverys JP. 2013. ComE/ComE∼P interplay dictates activation or extinction status of pneumococcal X-state (competence). Mol Microbiol 87:394–411. doi: 10.1111/mmi.12104. [DOI] [PubMed] [Google Scholar]
- 6.Claverys JP, Martin B, Havarstein LS. 2007. Competence-induced fratricide in streptococci. Mol Microbiol 64:1423–1433. doi: 10.1111/j.1365-2958.2007.05757.x. [DOI] [PubMed] [Google Scholar]
- 7.Steinmoen H, Knutsen E, Havarstein LS. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc Natl Acad Sci U S A 99:7681–7686. doi: 10.1073/pnas.112464599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinmoen H, Teigen A, Havarstein LS. 2003. Competence-induced cells of Streptococcus pneumoniae lyse competence-deficient cells of the same strain during cocultivation. J Bacteriol 185:7176–7183. doi: 10.1128/jb.185.24.7176-7183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guiral S, Mitchell TJ, Martin B, Claverys JP. 2005. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci U S A 102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wholey WY, Kochan TJ, Storck DN, Dawid S. 2016. Coordinated bacteriocin expression and competence in Streptococcus pneumoniae contributes to genetic adaptation through neighbor predation. PLoS Pathog 12:e1005413. doi: 10.1371/journal.ppat.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veening JW, Blokesch M. 2017. Interbacterial predation as a strategy for DNA acquisition in naturally competent bacteria. Nat Rev Microbiol 15:621–629. doi: 10.1038/nrmicro.2017.66. [DOI] [PubMed] [Google Scholar]
- 12.Havarstein LS, Martin B, Johnsborg O, Granadel C, Claverys JP. 2006. New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol Microbiol 59:1297–1307. doi: 10.1111/j.1365-2958.2005.05021.x. [DOI] [PubMed] [Google Scholar]
- 13.Johnsborg O, Eldholm V, Bjornstad ML, Havarstein LS. 2008. A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae and related commensal species. Mol Microbiol 69:245–253. doi: 10.1111/j.1365-2958.2008.06288.x. [DOI] [PubMed] [Google Scholar]
- 14.Johnsborg O, Havarstein LS. 2009. Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol Rev 33:627–642. doi: 10.1111/j.1574-6976.2009.00167.x. [DOI] [PubMed] [Google Scholar]
- 15.Moreno-Gamez S, Sorg RA, Domenech A, Kjos M, Weissing FJ, van Doorn GS, Veening JW. 2017. Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat Commun 8:854. doi: 10.1038/s41467-017-00903-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claverys JP, Havarstein LS. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front Biosci 7:d1798–d1814. doi: 10.2741/claverys. [DOI] [PubMed] [Google Scholar]
- 17.Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol 60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 18.Echenique JR, Chapuy-Regaud S, Trombe MC. 2000. Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol Microbiol 36:688–696. doi: 10.1046/j.1365-2958.2000.01891.x. [DOI] [PubMed] [Google Scholar]
- 19.Echenique JR, Trombe MC. 2001. Competence repression under oxygen limitation through the two-component MicAB signal-transducing system in Streptococcus pneumoniae and involvement of the PAS domain of MicB. J Bacteriol 183:4599–4608. doi: 10.1128/JB.183.15.4599-4608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Echenique JR, Trombe MC. 2001. Competence modulation by the NADH oxidase of Streptococcus pneumoniae involves signal transduction. J Bacteriol 183:768–772. doi: 10.1128/JB.183.2.768-772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapuy-Regaud S, Duthoit F, Malfroy-Mastrorillo L, Gourdon P, Lindley ND, Trombe MC. 2001. Competence regulation by oxygen availability and by Nox is not related to specific adjustment of central metabolism in Streptococcus pneumoniae. J Bacteriol 183:2957–2962. doi: 10.1128/JB.183.9.2957-2962.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinas GE, Cortes PR, Orio AG, Echenique J. 2008. Acidic stress induces autolysis by a CSP-independent ComE pathway in Streptococcus pneumoniae. Microbiology 154:1300–1308. doi: 10.1099/mic.0.2007/015925-0. [DOI] [PubMed] [Google Scholar]
- 23.Cortes PR, Pinas GE, Cian MB, Yandar N, Echenique J. 2015. Stress-triggered signaling affecting survival or suicide of Streptococcus pneumoniae. Int J Med Microbiol 305:157–169. doi: 10.1016/j.ijmm.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- 25.Gagne AL, Stevens KE, Cassone M, Pujari A, Abiola OE, Chang DJ, Sebert ME. 2013. Competence in Streptococcus pneumoniae is a response to an increasing mutational burden. PLoS One 8:e72613. doi: 10.1371/journal.pone.0072613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corrigan RM, Grundling A. 2013. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol 11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 27.Huynh TN, Woodward JJ. 2016. Too much of a good thing: regulated depletion of c-di-AMP in the bacterial cytoplasm. Curr Opin Microbiol 30:22–29. doi: 10.1016/j.mib.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pham TH, Liang ZX, Marcellin E, Turner MS. 2016. Replenishing the cyclic-di-AMP pool: regulation of diadenylate cyclase activity in bacteria. Curr Genet 62:731–738. doi: 10.1007/s00294-016-0600-8. [DOI] [PubMed] [Google Scholar]
- 29.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. 2011. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog 7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Y, Helmann JD. 2012. Analysis of the role of Bacillus subtilis sigma(M) in beta-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol 83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stulke J. 2013. Cyclic di-AMP homeostasis in Bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem 288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witte CE, Whiteley AT, Burke TP, Sauer JD, Portnoy DA, Woodward JJ. 2013. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. mBio 4:e00282-13. doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whiteley AT, Garelis NE, Peterson BN, Choi PH, Tong L, Woodward JJ, Portnoy DA. 2017. c-di-AMP modulates Listeria monocytogenes central metabolism to regulate growth, antibiotic resistance and osmoregulation. Mol Microbiol 104:212–233. doi: 10.1111/mmi.13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Commichau FM, Gibhardt J, Halbedel S, Gundlach J, Stulke J. 2018. A delicate connection: c-di-AMP affects cell integrity by controlling osmolyte transport. Trends Microbiol 26:175–185. doi: 10.1016/j.tim.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Bai Y, Yang J, Eisele LE, Underwood AJ, Koestler BJ, Waters CM, Metzger DW, Bai G. 2013. Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J Bacteriol 195:5123–5132. doi: 10.1128/JB.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bai Y, Yang J, Zarrella TM, Zhang Y, Metzger DW, Bai G. 2014. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J Bacteriol 196:614–623. doi: 10.1128/JB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zarrella TM, Metzger DW, Bai G. 2018. Stress suppressor screening leads to detection of regulation of cyclic di-AMP homeostasis by a Trk family effector protein in Streptococcus pneumoniae. J Bacteriol 200:e00045-18. doi: 10.1128/JB.00045-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cron LE, Stol K, Burghout P, van Selm S, Simonetti ER, Bootsma HJ, Hermans PW. 2011. Two DHH subfamily 1 proteins contribute to pneumococcal virulence and confer protection against pneumococcal disease. Infect Immun 79:3697–3710. doi: 10.1128/IAI.01383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Binepal G, Wenderska IB, Crowley P, Besingi RN, Senadheera DB, Jeannine Brady L, Cvitkovitch DG. 2017. K+ modulates genetic competence and the stress regulon of Streptococcus mutans. Microbiology 163:719–730. doi: 10.1099/mic.0.000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Opijnen T, Lazinski DW, Camilli A. 2015. Genome-wide fitness and genetic interactions determined by Tn-seq, a high-throughput massively parallel sequencing method for microorganisms. Curr Protoc Microbiol 36:1–24. doi: 10.1002/0471142727.mb0716s106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bricker AL, Camilli A. 1999. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol Lett 172:131–135. doi: 10.1111/j.1574-6968.1999.tb13460.x. [DOI] [PubMed] [Google Scholar]
- 43.Trombe MC, Rieux V, Baille F. 1994. Mutations which alter the kinetics of calcium transport alter the regulation of competence in Streptococcus pneumoniae. J Bacteriol 176:1992–1996. doi: 10.1128/jb.176.7.1992-1996.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morrison DA, Trombe MC, Hayden MK, Waszak GA, Chen JD. 1984. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAM beta 1. J Bacteriol 159:870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishii S, Yano T, Hayashi H. 2006. Expression and characterization of the peptidase domain of Streptococcus pneumoniae ComA, a bifunctional ATP-binding cassette transporter involved in quorum sensing pathway. J Biol Chem 281:4726–4731. doi: 10.1074/jbc.M512516200. [DOI] [PubMed] [Google Scholar]
- 46.Hui FM, Zhou L, Morrison DA. 1995. Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene 153:25–31. doi: 10.1016/0378-1119(94)00841-f. [DOI] [PubMed] [Google Scholar]
- 47.Lin J, Zhu L, Lau GW. 2016. Disentangling competence for genetic transformation and virulence in Streptococcus pneumoniae. Curr Genet 62:97–103. doi: 10.1007/s00294-015-0520-z. [DOI] [PubMed] [Google Scholar]
- 48.Johnston C, Campo N, Berge MJ, Polard P, Claverys JP. 2014. Streptococcus pneumoniae, le transformiste. Trends Microbiol 22:113–119. doi: 10.1016/j.tim.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, Konradsen HB, Nahm MH. 2015. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev 28:871–899. doi: 10.1128/CMR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chewapreecha C, Harris SR, Croucher NJ, Turner C, Marttinen P, Cheng L, Pessia A, Aanensen DM, Mather AE, Page AJ, Salter SJ, Harris D, Nosten F, Goldblatt D, Corander J, Parkhill J, Turner P, Bentley SD. 2014. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet 46:305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coffey TJ, Enright MC, Daniels M, Morona JK, Morona R, Hryniewicz W, Paton JC, Spratt BG. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol 27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 52.Slager J, Kjos M, Attaiech L, Veening JW. 2014. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell 157:395–406. doi: 10.1016/j.cell.2014.01.068. [DOI] [PubMed] [Google Scholar]
- 53.Engelmoer DJ, Rozen DE. 2011. Competence increases survival during stress in Streptococcus pneumoniae. Evolution 65:3475–3485. doi: 10.1111/j.1558-5646.2011.01402.x. [DOI] [PubMed] [Google Scholar]
- 54.Dagkessamanskaia A, Moscoso M, Henard V, Guiral S, Overweg K, Reuter M, Martin B, Wells J, Claverys JP. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol Microbiol 51:1071–1086. doi: 10.1111/j.1365-2958.2003.03892.x. [DOI] [PubMed] [Google Scholar]
- 55.Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol 51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 56.Peterson S, Cline RT, Tettelin H, Sharov V, Morrison DA. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J Bacteriol 182:6192–6202. doi: 10.1128/jb.182.21.6192-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alloing G, Martin B, Granadel C, Claverys JP. 1998. Development of competence in Streptococcus pneumonaie: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol Microbiol 29:75–83. doi: 10.1046/j.1365-2958.1998.00904.x. [DOI] [PubMed] [Google Scholar]
- 58.Rimini R, Jansson B, Feger G, Roberts TC, de Francesco M, Gozzi A, Faggioni F, Domenici E, Wallace DM, Frandsen N, Polissi A. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol Microbiol 36:1279–1292. doi: 10.1046/j.1365-2958.2000.01931.x. [DOI] [PubMed] [Google Scholar]
- 59.Weyder M, Prudhomme M, Berge M, Polard P, Fichant G. 2018. Dynamic modeling of Streptococcus pneumoniae competence provides regulatory mechanistic insights into its tight temporal regulation. Front Microbiol 9:1637. doi: 10.3389/fmicb.2018.01637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guiral S, Henard V, Granadel C, Martin B, Claverys JP. 2006. Inhibition of competence development in Streptococcus pneumoniae by increased basal-level expression of the ComDE two-component regulatory system. Microbiology 152:323–331. doi: 10.1099/mic.0.28425-0. [DOI] [PubMed] [Google Scholar]
- 61.Martin B, Prudhomme M, Alloing G, Granadel C, Claverys JP. 2000. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol Microbiol 38:867–878. doi: 10.1046/j.1365-2958.2000.02187.x. [DOI] [PubMed] [Google Scholar]
- 62.Johnston C, Mortier-Barriere I, Khemici V, Polard P. 2018. Fine-tuning cellular levels of DprA ensures transformant fitness in the human pathogen Streptococcus pneumoniae. Mol Microbiol 105:663–675. doi: 10.1111/mmi.14068. [DOI] [PubMed] [Google Scholar]
- 63.Morrison DA. 1997. Streptococcal competence for genetic transformation: regulation by peptide pheromones. Microb Drug Resist 3:27–37. doi: 10.1089/mdr.1997.3.27. [DOI] [PubMed] [Google Scholar]
- 64.Chen JD, Morrison DA. 1987. Modulation of competence for genetic transformation in Streptococcus pneumoniae. J Gen Microbiol 133:1959–1967. doi: 10.1099/00221287-133-7-1959. [DOI] [PubMed] [Google Scholar]
- 65.Luo P, Li H, Morrison DA. 2003. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol Microbiol 50:623–633. doi: 10.1046/j.1365-2958.2003.03714.x. [DOI] [PubMed] [Google Scholar]
- 66.Hartel T, Eylert E, Schulz C, Petruschka L, Gierok P, Grubmuller S, Lalk M, Eisenreich W, Hammerschmidt S. 2012. Characterization of central carbon metabolism of Streptococcus pneumoniae by isotopologue profiling. J Biol Chem 287:4260–4274. doi: 10.1074/jbc.M111.304311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huynh TN, Choi PH, Sureka K, Ledvina HE, Campillo J, Tong L, Woodward JJ. 2016. Cyclic di-AMP targets the cystathionine beta-synthase domain of the osmolyte transporter OpuC. Mol Microbiol 102:233–243. doi: 10.1111/mmi.13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schuster CF, Bellows LE, Tosi T, Campeotto I, Corrigan RM, Freemont P, Grundling A. 2016. The second messenger c-di-AMP inhibits the osmolyte uptake system OpuC in Staphylococcus aureus. Sci Signal 9:ra81. doi: 10.1126/scisignal.aaf7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeden MS, Schuster CF, Bowman L, Zhong Q, Williams HD, Grundling A. 2018. Cyclic di-adenosine monophosphate (c-di-AMP) is required for osmotic regulation in Staphylococcus aureus but dispensable for viability in anaerobic conditions. J Biol Chem 293:3180–3200. doi: 10.1074/jbc.M117.818716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whiteley AT, Pollock AJ, Portnoy DA. 2015. The PAMP c-di-AMP is essential for Listeria monocytogenes growth in rich but not minimal media due to a toxic increase in (p)ppGpp. Cell Host Microbe 17:788–798. doi: 10.1016/j.chom.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bowman L, Zeden MS, Schuster CF, Kaever V, Grundling A. 2016. New insights into the cyclic di-adenosine monophosphate (c-di-AMP) degradation pathway and the requirement of the cyclic dinucleotide for acid stress resistance in Staphylococcus aureus. J Biol Chem 291:26970–26986. doi: 10.1074/jbc.M116.747709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. 2010. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem 285:473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rallu F, Gruss A, Ehrlich SD, Maguin E. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol Microbiol 35:517–528. doi: 10.1046/j.1365-2958.2000.01711.x. [DOI] [PubMed] [Google Scholar]
- 74.Pinas GE, Reinoso-Vizcaino NM, Yandar Barahona NY, Cortes PR, Duran R, Badapanda C, Rathore A, Bichara DR, Cian MB, Olivero NB, Perez DR, Echenique J. 2018. Crosstalk between the serine/threonine kinase StkP and the response regulator ComE controls the stress response and intracellular survival of Streptococcus pneumoniae. PLoS Pathog 14:e1007118. doi: 10.1371/journal.ppat.1007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu L, Ma Y, Zhang JR. 2006. Streptococcus pneumoniae recruits complement factor H through the amino terminus of CbpA. J Biol Chem 281:15464–15474. doi: 10.1074/jbc.M602404200. [DOI] [PubMed] [Google Scholar]
- 76.Halfmann A, Hakenbeck R, Bruckner R. 2007. A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol Lett 268:217–224. doi: 10.1111/j.1574-6968.2006.00584.x. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y, Yang J, Bai G. 2018. Regulation of the CRISPR-associated genes by Rv2837c (CnpB) via an Orn-like activity in TB complex mycobacteria. J Bacteriol 200:e00743-17. doi: 10.1128/JB.00743-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. 2013. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tjaden B. 2015. De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biol 16:1. doi: 10.1186/s13059-014-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gazdik MA, McDonough KA. 2005. Identification of cyclic AMP-regulated genes in Mycobacterium tuberculosis complex bacteria under low-oxygen conditions. J Bacteriol 187:2681–2692. doi: 10.1128/JB.187.8.2681-2692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Berge MJ, Mercy C, Mortier-Barriere I, VanNieuwenhze MS, Brun YV, Grangeasse C, Polard P, Campo N. 2017. A programmed cell division delay preserves genome integrity during natural genetic transformation in Streptococcus pneumoniae. Nat Commun 8:1621. doi: 10.1038/s41467-017-01716-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Underwood AJ, Zhang Y, Metzger DW, Bai G. 2014. Detection of cyclic di-AMP using a competitive ELISA with a unique pneumococcal cyclic di-AMP binding protein. J Microbiol Methods 107:58–62. doi: 10.1016/j.mimet.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chastanet A, Prudhomme M, Claverys JP, Msadek T. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J Bacteriol 183:7295–7307. doi: 10.1128/JB.183.24.7295-7307.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.