Abstract
Background
This study aimed to assess gene expression alterations related to T lymphocytes function and explore their potential association with hypoxemia among septic patients.
Methods
This is a retrospective cohort clinical study with laboratory investigations. We studied patients enrolled in sepsis biological specimen bank from Department of Critical Care Medicine, Zhongda Hospital, fulfilling consensus criteria for sepsis without any documented immune comorbidity admitted in ICU within 48 h after onset with whole blood samples drawn within 24 h of admission. Whole genome expression by microarray assay (Human LncRNA Microarray V4.0) was compared in hypoxemia cohort versus without. Differentially expressed (DE) genes with >1 log2[fold change (FC)] and false discovery rate (FDR) <0.20 that enriched in T cell related biological process entered the adjusted analysis to identify the candidate genes. The correlation analysis within candidate genes or with clinical parameters were performed. We assessed candidate expression ex vivo in co-culture system with RAW246.7 cells and validated genes identified in prior studies of sepsis-ARDS/hypoxemia within our present study.
Results
Septic patients (n=9) with hypoxemic phenotype held higher illness severity, serum lactate and creatine, and incidence of lymphopenia compared with non-hypoxemic group (n=6). Several gene signatures related to apoptosis, inhibitory receptors, T cell immunoreceptor, transcriptions factors, toll-like receptors and cytokine and effector molecules were upregulated in hypoxemic group. Candidate genes were identified after adjustment for age, sex and presence of lymphopenia with significantly negative correlations with partial pressure of O2 in an arterial blood (PaO2) and fraction of inspiration O2 (FiO2) ratio, among which NLRP3, SOS1, ELF1 and STAT3 held an increasing expression in ex vivo validation while the others, PSMA5, CLEC4D, CD300A, PRKD2 and PSMA2 showed the opposite alteration from those in vivo.
Conclusions
Higher illness severity and incidence of lymphopenia was observed following hypoxemia in sepsis and T cell-related gene signatures were associated with hypoxemia during sepsis.
Keywords: Hypoxemia; sepsis; T cell, differential expressed
Introduction
Sepsis is a public health problem worldwide and characterized by a dysregulated host response to infection, which includes T lymphocytes dysfunction (1-3). Damage associated molecular patterns following the hypoxemia or hypoperfusion in context of sepsis play a key role on sepsis-induced T cell dysfunction (4). The clinical founding of lymphopenia (including diminished T cell numbers) and induction of immunoparalysis (to varying degrees) (3), suggested the necessity to clarify the mechanism that how sepsis induces the T cell function alterations, including proliferation, apoptosis and cellular activation and exhaustion as well as hypo-responsiveness. Sepsis is a heterogeneous syndrome with variations of clinical and biological features in different subsets or phenotypes. Identification of T lymphocytes involvement in septic patients with distinct phenotypes may allow further understanding the mechanism and benefit precise therapy.
Respiratory organ failure is most prevalent, which could present as hypoxemia or (and) acute respiratory distress syndrome (ARDS), affecting as many as 87% organ failure patients in ICU stay (5). Critically ill patients with hypoxemia hold both higher mortality and severity of illness than those without (6). Hypoxemia is often an early complication of sepsis, which increases the death risk and could be a trigger to the dysregulated immune response. Evidence shows that the impact of hypoxia on immunity and inflammation can vary depending on the microenvironment and immune processes occurring in a given niche and on the other hand pathological hypoxia can drive tissue dysfunction and disease development through immune cell dysregulation (7). In this way, whether T lymphocytes function involved gene expression differs in septic patients with distinct hypoxemic phenotype is worth further investigation.
Based on the foundational concept that changes in gene transcription precede and guide the majority of important biological processes (8), we sought to understand alterations of T lymphocytes function involved genes, in our case of community-acquire sepsis within hours upon presentation to medical care, in whole blood gene transcription that distinguish septic patients that are complicated with hypoxemia from those who are not. The primary hypothesis was that T lymphocytes function involved genes in whole blood would reveal differential expression in septic patients between hypoxemic versus non-hypoxemic phenotypes.
Methods
This is a retrospective cohort clinical study with laboratory investigations.
Participants
This study was conducted in accordance with the amended Declaration of Helsinki. All the included subjects were from sepsis biological specimen bank from Department of Critical Care Medicine, Zhongda Hospital. The sepsis biological specimen bank was registered in 2017 by approval and supervise of Institutional Ethics Committee of Zhongda Hospital (Registration number: 2017ZDSYLL105). Written informed consent was obtained from all patients or their surrogate when possible. The Animal Care and Use Committee of Southeast University approved all experiments involving the use of animals.
To reduce sample heterogeneity, we took the time course and origin of sepsis into consideration thus only community-acquired cases were included. In addition, our primary interest was identifying differences in T cells-related gene expression in septic subjects with hypoxemia compared to those without. To avoid the mixed factors from the baseline of the subjects on immune function, cases with any past history of tumors, hematological or immunological disease, or treatment with chemotherapy agents or corticosteroids within 6 months prior to hospitalization were excluded. Fifteen patients were enrolled in general intensive care unit of a tertiary teaching hospital in Nanjing, China, of which all were between 18 and 90 years old with a diagnosis of community-acquired sepsis according to criteria of the Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016 (1). Since most of the critically ill patients admitted were in need of respiratory support, the fraction of inspiration O2 (FiO2) used should be taken into consideration. In this way, patients were divided into 2 groups according to pressure of O2 in an arterial blood (PaO2) or/and PaO2/FiO2: hypoxemic phenotype (PaO2 <80 mmHg or P/F <200 mmHg, n=9) and non-hypoxemic phenotype (PaO2 ≥80 mmHg and P/F ≥200 mmHg, n=6).
Biological sample collection, processing, and measurements
Whole blood was collected within 24 h of ICU admission for the isolation of RNA. Total RNA was extracted using the TRIZOL reagent (Invitrogen, US) according to the manufacturer’s instructions. mRNA was purified from total RNA after removal of rRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation Kit, Epicentre). Each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3’ bias utilizing a random priming method (Arraystar Flash RNA Labeling Kit, Arraystar) and the labeled cRNAs were purified by RNeasy Mini Kit (Qiagen). Then, the labeled cRNAs were hybridized onto a Human LncRNA Microarray V4.0 (Arraystar) chip, which is designed for 20,730 coding genes. The arrays were scanned using an Agilent G2505C scanner and the density of fluorescence was analyzed using Agilent Feature Extraction software.
Microarray analysis
Array images were analysed by Agilent Feature Extraction software (version 11.0.1.1). GeneSpring GX v12.1 software package (Agilent Technologies) were used to acquire the quantile normalization and subsequent data process. We compared T cell related gene expression in septic patients with hypoxemia and those without. T cell related genes in our study were identified according to the search strategy “(T cell) AND” Homo sapiens “[porgn: __txid9606] in PubMed. Differentially expressed (DE) mRNAs with statistical significance between the two groups were defined by P value/fold change (FC) (P<0.5, │log2FC│>0.25) along with the false discovery rate (FDR) <0.20, which calculated by the Benjamini-Hochberg procedure, used to indicate the expected fraction of falsely declared DE genes among the total set of DE genes (i.e., FDR <0.20 would indicate that the result is likely to be valid 4 of 5 times). Gene ontology (GO) analysis was applied to determine the roles of these DE mRNAs enriched in T cell related biological pathways or GO terms. Comparisons of gene signatures were illustrated by HemI (Heatmap Illustrator, version 1.0). The genes enriched in T cell related biological process were selected to enter the adjusted analysis. We included relevant clinical factors in the adjusted analysis such as age, gender, and lymphopenia, performed by SPSS Version 23 (IBM, Chicago, Ill, USA). The rationale for adjusting for the presence of lymphopenia was that decreased expression of genes in lymphocyte that was less than 1,000 cells/µL in peripheral blood might be associated with a decrease in its number rather than its functional regulation.
Ex vivo experiments
Mice
C57BL/6 male mice, 4–6 weeks of age, were purchased from the Comparative Medicine Centre, Yangzhou University (Yangzhou, China). The animals were housed 5 mice per cage in a laminar air flow room maintained at 22±2 °C with relative humidity of 55%±5%. Mice were cared and treated in accordance with the guidelines established by the Committee of Animal Care and Use of Southeast University.
Isolation, purification and identification of mouse spleen derived CD4+ T cells
CD4+ T cells derived from mouse spleen were isolated through positive CD4 selection, using CD4 (L3T4) MicroBeads, MS columns and MiniMACS™ Separator, according to manufacturer’s instructions (Miltenyi Biotech, Bergisch Gladbach, Germany). The identification was evaluated by positive expression of CD4 measured by flow cytometry (FCM).
CD4+ T Cell culture
5×106 CD4+ T cells in 1 mL RPMI 1640 culture medium (10 mM HEPES/2 mM l-glutamine/10% 0.22 µm filtered FBS/50 uM β-mercaptoethanol) were seeded into a well of the 24-well plate pre-coated with anti-CD3 (5 µg/mL) and anti-CD28 (2 µg/mL) in an atmosphere of 5% CO2 at 37 °C. Cells were treated overnight. Then, 106 RAW246.7 as antigen presenting cells were directly added. LPS (1,000 ng/mL) (mix of gel filtration chromatography purified LPS from Escherichia coli O111:B4, Sigma Aldrich, Saint Louis, MO, USA) was added as stimuli in an atmosphere of 5% CO2 and another group of naïve T cells with RAW246.7 were cultured with in an atmosphere of 5% CO2 and 5% O2 at 37 °C as hypoxia condition, set as non-hypoxic sepsis and hypoxic sepsis groups.
Quantitative reverse transcription real-time polymerase chain reaction
DE genes were selected for quantitative polymerase chain reaction (qPCR) confirmation. TaqMan-specific inventoried primers for NLR family, pyrin domain containing 3 (NLRP3), proteasome subunit alpha 5 (PSMA5), C-type lectin domain family 4 member D (CLEC4D), SOS Ras/Rac guanine nucleotide exchange factor 1 (SOS1), CD300a molecule (CD300A), E74-like factor 1 (ELF1), protein kinase D2 (PRKD2), signal transducer and activator of transcription 3 (STAT3), proteasome subunit alpha 2 (PSMA2). Housekeeping genes β-catenin were also measured. Real-time qPCR was performed with the ABI Prism 7000 Sequence Detection System (Life Technologies, Carlsbad, CA, USA). As run method, PCR activation at 95 °C for 20 s was followed by 40 cycles of 1 s at 95 °C and 20 s at 60 °C. The average threshold count (Ct) value of 2–3 technical replicates were used in all calculations. The average Ct values of the internal controls (housekeeping genes) was used to calculate Ct values for the samples. Data analysis was performed using the 2-ΔΔCt method.
Statistical analysis
Comparisons of characteristics of clinical variables between septic patients with hypoxemia and those without, were performed using unpaired t-tests, Mann-Whitney U tests, or Chi-squared tests, as appropriate by SPSS Version 23 (IBM, Chicago, IL, USA).
The candidate genes were identified with P<0.05 after adjusted analysis. A power calculation for the detected normalized intensity difference of candidate genes between groups with the present sample size was performed by Power Analysis and Sample Size (PASS) software (PASS 2008. NCSS, LLC. Kaysville, Utah, USA). The Spearman correlation analysis was performed between any two of the candidate genes or between the candidate gene and P/F ratio upon enrolment by GraphPad PRISM Version 5.3 (San Diego, CA, USA).
Comparisons of variables between groups in ex vivo experiments, were performed using unpaired t tests, Mann-Whitney U tests, or Chi-squared tests, as appropriate by GraphPad PRISM Version 5.3 (San Diego, CA, USA).
Results
Clinical presentation, inflammatory, immune indicators and organ function indicators
Septic patients with hypoxemia held a lower lymphocyte count [0.4 (0.3, 0.6) vs. 0.8 (0.5, 1.2), P=0.020] and higher N/L ratio than those without hypoxemia [17.9 (7.1, 23.4) vs. 6.8 (2.9, 12.3), P=0.050]. There was no statistical difference between two groups in terms with systemic inflammatory response presentations (heart rate, respiratory rate, white blood cell count or systolic blood pressure) as well as neutrophil count, hs-CRP, PCR and IL-6.
As summarized in Table 1, septic patients with hypoxemia presented higher SOFA scores than those without at admission (11±4 vs. 7±1, P=0.046), holding a higher level of illness severity. As for respective systemic organ function, septic patients with hypoxia exhibited a significantly higher level of serum creatine compared with those without [164 (79, 332) vs. 60 (55, 105), P=0.017], indicating higher risk of renal dysfunction. In addition, patients with hypoxemia held higher levels of serum lactate [2.2 (1.5, 3.4) vs. 1.2 (0.78, 2), P=0.039] compared with patients without hypoxemia. No statistical difference was observed in terms of other inflammatory and biochemical parameters.
Table 1. Clinical variables of septic patients: hypoxemic vs. non-hypoxemic.
| Clinical variable | Non-hypoxemic, N=6 | Hypoxemic, N=9 | P |
|---|---|---|---|
| Age, years | 58±7 | 46±14 | NS |
| Female gender | 2(33.3) | 2(9) | NS |
| Presumed infection site | |||
| Lung | 1 (17.7) | 6 (33.3) | |
| Abdomen | 5 (82.3) | 3 (66.7) | |
| PaO2/FiO2 ratio | 346±55 | 129±62 | <0.001 |
| APACHE II score | 18±5 | 24±9 | NS |
| SOFA score | 7±1 | 11±4 | 0.046 |
| SIRS indicators | |||
| Maximum temperature, °C | 37±0.8 | 38±0.6 | NS |
| Lowest systolic blood pressure, mmHg | 70±6 | 71±9 | NS |
| Maximum heart rate, beats per min | 110 (85,123) | 115 (105,146) | NS |
| Maximum respiratory rate, per min | 21 (18,22) | 27 (19,36) | NS |
| Maximal dose of vasopressors during 24 h upon enrollment, median IQR (25%, 75%), n (%) | 8 (5,12.5), 100 (100) | 20 (0,80), 6 (66.7) | NS |
| Organ function indicator | |||
| Platelets, thousands | 80 (47, 158) | 107 (66, 157) | NS |
| Urine output, prior 8 h, mL/h | 122 (79, 142) | 62 (35, 166) | NS |
| Serum creatinine, mg/dL | 60 (55, 105) | 164 (79, 332) | 0.017 |
| Total bilirubin | 17.2 (8.4, 42.4) | 20 (8.1, 54.4) | NS |
| Complicated with CNS dysfunction | 0 (0.0) | 2 (22.2) | NS |
| Peripheral blood count | |||
| White blood count (per μL) | 7.0 (4.0, 9.0) | 10.0 (4.5, 16.5) | NS |
| Neutrophil count (per μL) | 5.5 (2.9, 7.8) | 8.0 (4.0, 16.9) | NS |
| Lymphocyte count (per μL) | 0.8 (0.5, 1.2) | 0.4 (0.3, 0.6) | 0.020 |
| Neutrophil and Lymphocyte ratio (N/L) | 6.8 (2.9, 12.3) | 17.9 (7.1, 23.4) | 0.050 |
| Inflammatory indicator (serum) | |||
| hs-CRP, mg/L | 98 (78, 138) | 138 (50, 203) | NS |
| PCT, ug/L | 3.3 (8.4, 9.7) | 4.9 (0.8, 15.8) | NS |
| IL-6, pg/L | 538 (65, 2214) | 117 (19, 2691) | NS |
| Biochemical parameter | |||
| Albumin, g/L | 27.3±5.0 | 26.4±4.8 | NS |
| Lactate, mmol/L | 1.2 (0.78, 2) | 2.2 (1.5, 3.4) | 0.039 |
| pH | 7.43±0.04 | 7.34±0.06 | 0.019 |
| SB, mmol/L | 20.4±2.8 | 18.1±3.6 | NS |
| Base excess, mmol/L | −3 (−6.3, −0.9) | −8.4 (−13.6, −3.6) | NS |
Data are presented as percentage, mean ± SD, IQR (25%, 75%). FiO2, fraction of inspiration O2; PaO2, partial pressure of O2 in an arterial blood; APACHE, acute physiology and chronic health evaluation; SOFA, sequential organ failure assessment; SIRS, systemic inflammatory response syndrome; CNS, central nervous system; hs-CRP, hypersensitive C-reactive protein; PCT, procalcitonin; IL, interleukin; SB, standard bicarbonate. NS, not significant.
Upregulation of hypoxia related genes from whole blood in septic patients complicated with hypoxemia
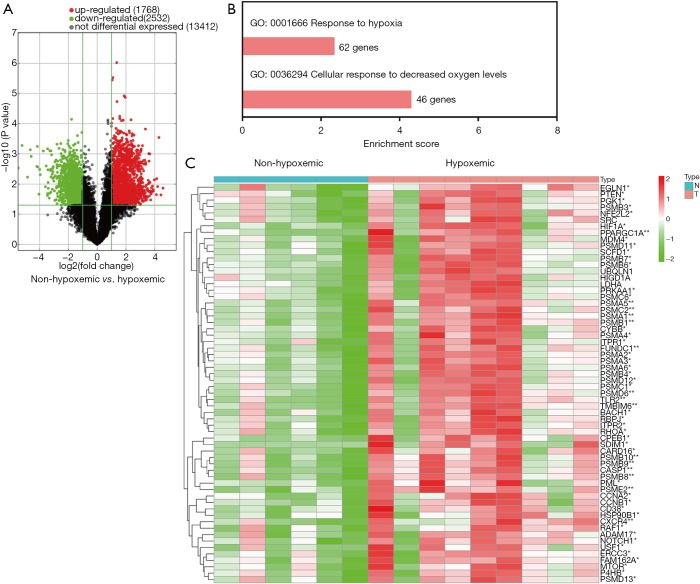
The annotated genes expression in two groups were shown in Figure 1A. According to GO analysis, 62 of the DE genes were enriched in hypoxia related biological process (Figure 1B). Go functions related to cell response to hypoxia were found to be significantly up-regulated in septic patients with hypoxemia, compared to those without (Figure 1C).
Figure 1.
Volcano plot of significantly differential experiment genes and significant GO analyses related to cell response to hypoxia. (A) Analyzing the differentially expressed genes by t test, log2FC was taken as abscissa and negative logarithm-log10 (P value) of P value was taken as ordinate; (B) the significant GO of differentially expressed genes related to cell response to hypoxia; (C) the details of differentially expressed genes in GO function related to cell response to hypoxia. *, P<0.05; **, P<0.01. GO, gene ontology.
DE T cell related genes and biological process involvement
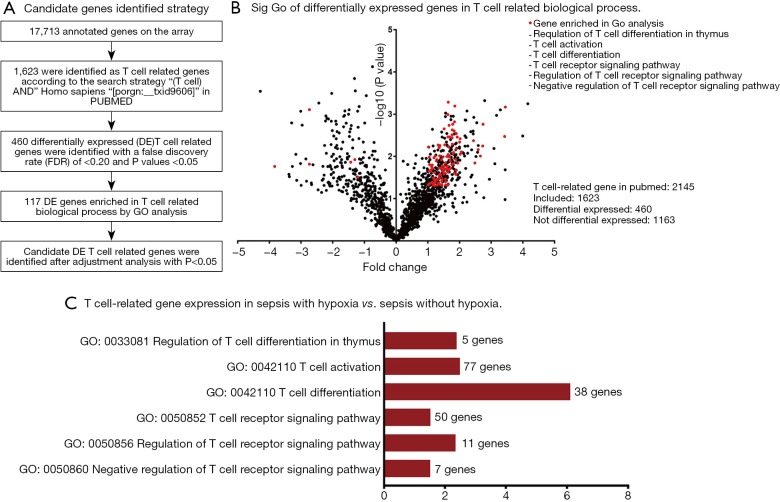
As shown in Figure 2, of the 17,713 annotated genes on the array, 1,623 were identified as T cell related genes and were analyzed for the differential expression. Among them, 460 were detected as DE genes with P value <0.05 (Figure 2A, http://cdn.amegroups.cn/static/application/4cb5beed9a1cc22e907db2464e50f777/10.21037atm.2019.12.63-1.pdf).
Figure 2.
Differentially expressed T cell related genes and biological process involvement. (A) Candidate genes identified strategy; (B) T cell-related gene expression in sepsis with hypoxia vs. sepsis without hypoxia; (C) Sig Go of DE genes in T cell related biological process.
According to GO analysis, 117 of the DE genes were enriched in several key biological process related to T cells, including regulation of T cell differentiation in thymus, T cell activation, T cell differentiation, T cell receptor signaling pathway, regulation of T cell receptor signaling pathway (Figure 2B, Table 2).
Table 2. Significant GO term of DE genes in T lymphocytes related biological process.
| GO:ID | Term, regulation in sepsis with hypoxic vs. non-hypoxic | Count | Fold. Enrichment | P value | Genes (117 genes in total) |
|---|---|---|---|---|---|
| GO:0033081 | Regulation of T cell differentiation in thymus, down | 5 | 2.76 | 0.030 | FOXJ1//SHH//TESPA1//FOXN1//CLPTM1 |
| GO:0042110 | T cell activation, up | 77 | 1.34 | 0.004 | BAX//IL15//LCP1//PSEN1//CLEC4E//CLEC4D//RHOA//PTPN22//RHOH//PTPN2//SP3//VAV1//RUNX2//TNFSF13B//CTLA4//GRB2//CD274//LYN//PAK2//PDPK1//PIK3CA//PTPN6//RAC1//SRC//CDC42//RIPK3//CTNNB1//JMJD6//GLI3//LIG4//B2M//ADAM17//SOS1//PRDM1//BMP4//RIPK2//HLA-E//CTPS1//PIK3CG//CORO1A//ANXA1//CD46//NCK1//HAVCR2//PYCARD//IL18//HLA-G//PELI1//RAB27A//FCGR2B//CEACAM1//CYLD//IRF1//TNFSF8//ATG5//BATF//PRELID1//CBFB//HLX//NLRP3//PRKAR1A//MICB//FAM49B//BCL10//CD47//CD300A//ZBTB16//AP3B1//STAT3//GSN//DDOST//PPP3CA//AZI2//CLEC7A//TREML2//CASP8//HSH2D |
| GO:0030217 | T cell differentiation, up | 38 | 1.36 | 0.030 | CLEC4E//CLEC4D//RHOA//RIPK3//CTNNB1//JMJD6//GLI3//LIG4//B2M//ADAM17//SOS1//PRDM1//BMP4//CYLD//IRF1//TNFSF8//CD46//ATG5//IL15//BATF//PRELID1//CTLA4//CBFB//HLA-G//ANXA1//HLX//RIPK2//NLRP3//IL18//ZBTB16//AP3B1//STAT3//PTPN2//PTPN22//RHOH//SP3//VAV1//RUNX2 |
| GO:0050852 | T cell receptor signaling pathway, up | 50 | 2.01 | 7.9E-07 | CD300A//BCL10//CYLD//PRKD2//ELF1//PTPN22//GBP1//PTPN2//PTPN6//CEACAM1//EZR//CHUK//DENND1B//LCP2//NCK1//NFKBIA//PAK2//PDE4B//PDPK1//PIK3CA//PIK3CB//PLCG2//PSEN1//PSMA1//PSMA2//PSMA3//PSMA4//PSMA5//PSMA6//PSMB1//PSMB3//PSMB4//PSMB6//PSMB7//PSMB8//PSMB9//PSMB10//PSMC1//PSMC2//PSMC6//PSMD11//PSMD12//PSMD13//PSME2//BTK//TEC//CUL1//RIPK2//THEMIS2//PSMD6 |
| GO:0050856 | Regulation of T cell receptor signaling pathway, up | 11 | 2.46 | 0.003 | CYLD//PRKD2//ELF1//PTPN22//GBP1//PTPN2//PTPN6//CEACAM1//EZR//CD300A//BCL10 |
| GO:0050860 | Negative regulation of T cell receptor signaling pathway, up | 7 | 3.13 | 0.004 | ELF1//PTPN22//GBP1//PTPN2//PTPN6//CEACAM1//EZR |
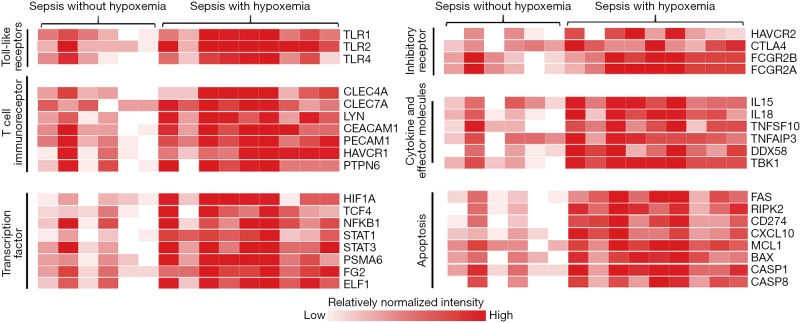
Heat map revealed difference detected that related to inhibitory receptor, cytokine and effector molecules, apoptosis, T cell immunoreceptor, toll-like receptors, and transcription factor between sepsis with hypoxemia and those without. As shown in Figure 3, compared with septic patients without hypoxemia, toll-like receptors of TLR1, TLR2 and TLR9 were more activated in those with hypoxemia. Genes of some interesting inhibitory receptors like Tim-3(HAVCR2), CTLA4, FcγRIIb (FCGR2B) as well as cell death genes of FAS, RIPK2, PD-1(CD274), BAX, CASP1 and CASP8 were up-regulated following hypoxemia. In addition, transcription factor and cytokine and effectors molecules that were responsive to hypoxia and inflammatory environment presented differently.
Figure 3.
Comparison of gene signature related to T cell function.
Candidate genes identified and adjusted for age, sex, and lymphopenia
Candidate genes identified strategy was illustrated in Figure 2A. Of 117 genes enriched in T cell biological process, candidate genes meeting a cut point of >1 log2 fold difference and P <0.05 in adjusted analysis for age, sex, and lymphopenia are presented in Table 2. These nine genes of NLRP3, PSMA2, PSMA5, CLEC4D, SOS1, CD300A, ELF1, PRKD2, STAT3 were upregulated in septic patients with hypoxemia compared with those without hypoxemia. There was no downregulated gene identified after adjustment for age, sex, and lymphopenia.
Associations of candidate genes identified with organ injury in early sepsis
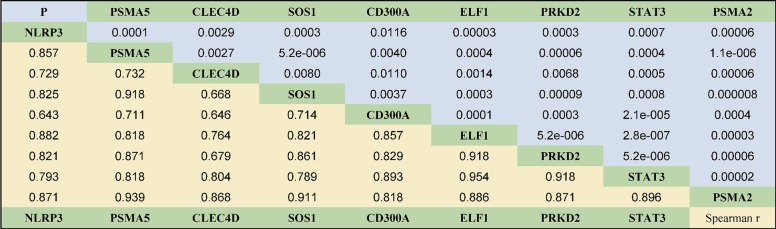
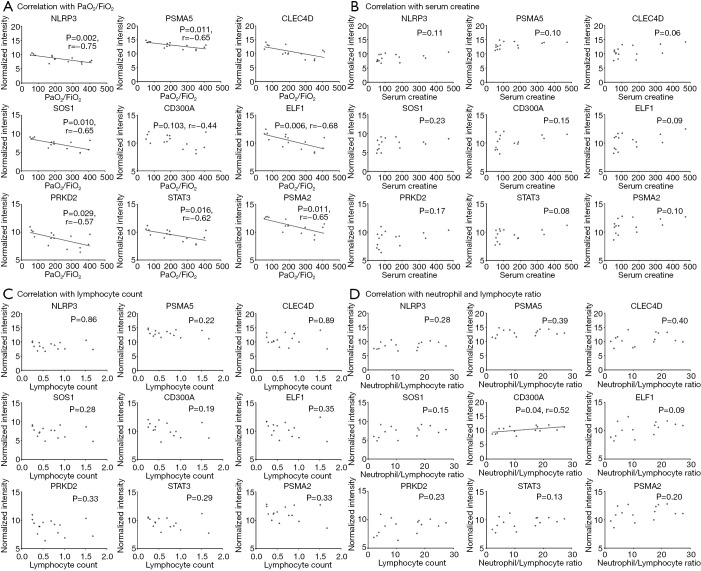
We further verified the correlation of the candidate genes with P/F ratio. There was a positive correlation between any two of these genes (all P values <0.05, Figure 4). As shown in Figure 5, except for CD300A, the candidate genes demonstrated a negative association with P/F ratios while CD300A gene expression was positively correlated to neutrophil and lymphocyte ratio (P=0.04). No significant correlation of candidate genes was detected with serum creatine or peripheral lymphocyte count. Two of the candidate genes in our study, NLRP3 and STAT3, have been previously described in studies of the sepsis or (and) hypoxia pathogenesis according to the literature search of the PubMed database while the other candidate genes identified in our study were found to have several or few relevant citations to sepsis or hypoxia process in the literature (Table 3).
Figure 4.
Spearman correlations between candidate genes identified in septic patients with hypoxemia.
Figure 5.
Correlation analysis of candidate genes with clinical indicators. Correlation analysis of candidate genes with PaO2/FiO2 (A), serum creatine (B), lymphocyte count (C) and neutrophil and lymphocyte ratio (D), respectively.
Table 3. Differentially expressed genes in sepsis with hypoxemic vs. non-hypoxemic phenotypes in model adjusted for age, sex, and lymphopenia.
| Gene symbol | ENTREZ ID | −Log2 fold change | Raw P value | FDR | No. PubMed references | ||
|---|---|---|---|---|---|---|---|
| Gene symbol | AND sepsis | AND hypoxia | |||||
| NLRP3 | 114548 | 3.03 | 0.010 | 0.11 | 4984 | 136 | 68 |
| PSMA5 | 5686 | 2.90 | 0.005 | 0.10 | 37 | 0 | 0 |
| CLEC4D | 338339 | 5.59 | 0.014 | 0.17 | 53 | 1 | 0 |
| SOS1 | 6654 | 3.39 | 0.016 | 0.12 | 746 | 0 | 2 |
| CD300A | 11314 | 2.74 | 0.017 | 0.12 | 69 | 1 | 3 |
| ELF1 | 1997 | 3.42 | 0.005 | 0.10 | 136 | 0 | 0 |
| PRKD2 | 25865 | 3.07 | 0.017 | 0.12 | 20 | 0 | 2 |
| STAT3 | 6774 | 2.57 | 0.006 | 0.10 | 20754 | 158 | 68 |
| PSMA2 | 5683 | 3.11 | 0.009 | 0.11 | 24 | 0 | 0 |
Values are adjusted for age, gender, and lymphopenia. PubMed Search completed December 1, 2018. FDR, false discovery rate.
Assessment of DE genes identified in LPS-stimulating T cells in ex vivo condition with hypoxia or normoxia
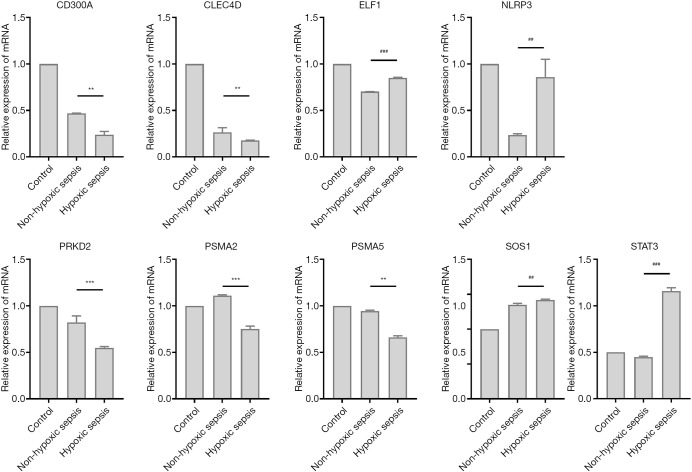
CD4+ T cells originated from mice spleen were sorted and then cultured with LPS stimulation in normoxia or hypoxia condition to confirm the differential candidate genes identified after adjustment as above. All the candidate genes of LPS-stimulated cells in hypoxia condition showed significantly different expression when compared with normoxia, of which the gene of NLRP3, SOS1, ELF1, STAT3 held an increasing expression. Among them, NLRP3 and STAT3 in hypoxia group with LPS stimulation showed more than twice times higher expression compared with that in normoxia group with LPS stimulation, which turned out to show similar with in vivo alteration, while the other gene of PSMA5, CLEC4D, CD300A, PRKD2, PSMA2 showed the opposite alteration of decreased expression from those in vivo (Figure 6).
Figure 6.
Assessment of candidate expression alterations ex vivo. **, P<0.01 with the opposite change direction as observed in vivo; ##, P<0.01 with the same change direction as observed in vivo; ***, P<0.001 with the opposite change direction as observed in vivo; ###, P<0.001 with the same change direction as observed in vivo.
Validation of DE genes in prior gene expression studies of sepsis-related ARDS
Using the literature search strategy as described in Kangelaris study (8), we compared our results to those of the four previous whole blood gene expression analysis in ARDS/acute lung injury (ALI) (8-11) (Table 4). Five of eight genes identified in Howrylak study (9) presented significantly differential expression with P<0.05 and FDR <0.20 in unadjusted model, including upregulated cyclin-dependent kinase inhibitor 1A (CDKN1A) and CREB/ATF bZIP transcription factor (CREBZF), and down-regulated ADP-ribosylation factor 3 (ARF3), patatin-like phospholipase domain containing 2 (PNPLA2) and aminopeptidase-like 1 (NPEPL1) in septic patients with hypoxemia. Two of the three genes identified in sepsis/ARDS patients compared with those of non-septic SIRS in Dolinay study (10), caspase 1, apoptosis-related cysteine peptidase (CASP1) and interleukin 18 (IL18) were found to be upregulated significantly in sepsis with hypoxemia, while only expression of CASP1 held an FDR<0.20. Chen et al. re-analyzed the public data from the Howrylak study and selected 12 DE genes related to acute lung injury (11). We found that two of the twelve genes were significantly upregulated with FDRs <0.20 in our study among septic patients with hypoxemia compared to those without: CDKN1A and histone cluster 2 H4 family member b (HIST2H4B). In a more recent study of 2015, Kangelaris et al. (8). reported some significant differentially expressed genes, including the downregulation of HCAR3, RBP7 and MME in early septic patients with ARDS compared to patients only with sepsis, which has been detected in our study with upregulation despite of no statistical difference in septic patients with hypoxemia compared to those without. All the genes above showed no associations with hypoxemia in septic patients compared to those without after the adjustment for age, gender and existence of lymphopenia.
Table 4. Association with candidate genes differentially expressed in prior gene expression studies of sepsis-related hypoxemia/ARDS.
| Gene | Hypoxemic vs. non-hypoxemic, regulation | Unadjusted | Adjusted in Model 1 | |||
|---|---|---|---|---|---|---|
| P | −Log2 fold change | FDR | P | OR | ||
| 2009, Howrylak et al. | ||||||
| CDKN1A† | Up | 0.036 | 1.71 | 0.14 | NS | – |
| FTH1 | Up | NS | 1.69 | 0.29 | – | – |
| ARF3* | Down | 0.021 | 2.42 | 0.12 | NS | – |
| BTG2 | Up | NS | 1.51 | 0.45 | – | – |
| NQO2 | Up | NS | 2.53 | 0.19 | – | – |
| PNPLA2† | Down | 0.010 | 1.52 | 0.11 | NS | – |
| NPEPL1 | Down | 0.022 | 4.59 | 0.12 | NS | – |
| CREBZF | Up | 0.016 | 2.12 | 0.12 | 0.050 | 7.253 |
| 2012, Dolinay et al. | ||||||
| CASP1† | Up | 0.001 | 4.59 | 0.09 | NS | – |
| IL18 | Up | 0.003 | 3.80 | 0.92 | NS | – |
| IL1B | Down | NS | 1.48 | 0.51 | – | – |
| 2013, Chen et al. | ||||||
| HOPX | Down | NS | 1.27 | 0.62 | – | – |
| CYBRD1 | Down | NS | 1.47 | 0.56 | – | – |
| UPB1 | Up | NS | 2.13 | 0.20 | – | – |
| OCLN | Down | NS | 1.35 | 0.76 | – | – |
| C21orf7 | NA | NA | NA | NA | NA | NA |
| HIST2H4B | Up | 0.02 | 3.75 | 0.12 | NS | – |
| TREM1 | Up | NS | 1.60 | 0.57 | – | – |
| HIST1H3H† | Up | NS | 1.50 | 0.52 | – | – |
| CDKN1A† | Up | 0.036 | 1.71 | 0.14 | NS | – |
| BTNL8 | Up | NS | 2.08 | 0.35 | – | – |
| HLA-DQB1 | Up | NS | 1.00 | 1.00 | – | – |
| CDKN1C | Up | NS | 1.02 | 0.95 | – | – |
| 2015, Kangelaris et al. | ||||||
| OLFM4 | Up | NS | 4.08 | 0.29 | – | – |
| MMP8 | Up | NS | 5.79 | 0.29 | – | – |
| LCN2 | Up | NS | 1.93 | 0.53 | – | – |
| CD24 | Up | NS | 2.37 | 0.29 | – | – |
| HP | Up | 0.05 | 3.30 | 0.16 | NS | – |
| BPI | Up | NS | 4.14 | 0.21 | – | – |
| RETN | Up | NS | 3.36 | 0.38 | – | – |
| TCN1 | Up | NS | 1.54 | 0.63 | – | – |
| SNORD1A | NA | NA | NA | NA | NA | NA |
| HCAR3‡ | Up | NS | 1.51 | 0.68 | – | – |
| RBP7‡ | Up | NS | 2.43 | 0.21 | – | – |
| MME‡ | Up | NS | 2.50 | 0.50 | – | – |
Genes shown in boldface are differentially expressed in present study with P<0.05 and FDR <0.25. †, genes that expressed in 2015 Kangelaris et al. study with FDR <0.25; ‡, a different alteration of gene expression in previous studies when compared between septic patients with hypoxemia versus without. NA, not included in the Affymetrix transcript cluster ID annotation; FDR, false discovery rate; NS, not significant.
Discussion
T lymphocytes, as part of adaptive immune response are influenced in septic process and their response characters vary in heterogeneous septic populations. Thus, it is important to further explore T lymphocytes involvement in clinically septic subsets. The objective of this study was to determine T lymphocytes gene expression in septic patients with distinct hypoxemic phenotypes and their potential associations.
Our study found septic patients with hypoxemic phenotype held higher incidence of lymphopenia with higher N/L ratio, serum lactate and creatine levels, indicating more severe illness, compared to those without hypoxemia and identified several candidate genes enriched in important T cell-related biological processes associated with hypoxemia in sepsis. After adjustment for age, sex, and lymphopenia, we identified CD300A, NLRP3 and STAT3 as increased expression in septic patients with hypoxemia, which has been described to be involved in the development of sepsis and hypoxia (12-16). Particularly, we compared the gene signatures related to T cell function and found the gene signatures of cell death and inhibitory receptors were up-regulated following hypoxemia phenotype, which might be consistent with the higher incidence of lymphopenia in septic patients with hypoxemia compared with those without.
We detected several novel up-regulated genes upon hypoxemia in sepsis, including PSMA2, PSMA5, CLEC4D, SOS1, ELF1, PRKD2 and then set up a cell culture system to validate the consistency of gene expression alterations in vitro. CD4+ T cells derived from mice spleen were cocultured with RAW246.7 as antigen presenting cells and anti-CD3/CD28 as the physiological activation. LPS or/and hypoxia of 5% were set as stimuli. The candidate genes showed altered expression after stimulation by LPS compared to controls while only the genes of NLPR3, STAT3, SOS1 and ELF1 held similar change trends of upregulation in hypoxia condition with those in vivo and the rest gene expressions of CD300A, CLEC4D, PRKD2, PSMA2 and PSMA5 held the opposite trends. The mechanism of their alterations was not completely clear either in vivo or ex vivo.
All the up-regulated genes were highly correlated, indicating the possibility of their coregulation in the pathogenesis of hypoxemia in early sepsis. The up-regulated genes were detected with negative correlation with PaO2/FiO2 except for CD300A while the CD300A gene express was found to be positively related to neutrophil and lymphocyte ratio. Several studies have shown the increased expression of CD300A under hypoxia condition in human monocytes, macrophages (17) or eosinophils (18), which was partly consistent with our results. The increased expression of CD300A could enhance the neutrophil recruitment and modulate T cell activation and T cell receptor signaling pathway as shown in GO analysis of Table 2 possibly via CD32a mediated signaling (19), which might explain its correlation with N/L ratio in septic patients with hypoxemia and the decreased expression in hypoxia condition with LPS stimulating might be attributed to the lack of monocytes or macrophages recruitment in ex vivo culture system . Additionally, CD300a is an inhibitory marker during degranulation processes and was reported to be up-regulated in patients following activation by formyl-methyinoyl-leucyl-phenylalanine (fMLP) and anti-FcɛI (20). Septic patients with hypoxemic phenotype held both higher levels of CD300A expression and serum creatine while no significant correlation was detected.
SOS1 and ELF1 were demonstrated with increased expression in septic patients with hypoxia compared to those without and were further confirmed ex vivo. SOS1 is involved in of mitotic spindle formation and proliferation (21) and was reported to be significantly upregulated in patients of high-altitude pulmonary edema (HAPE), followed by lymphocyte activation (22), which was in line with our results. ELF1 belongs to ETS family transcription factors. It has been proved to be expressed in T cells and regulate T cell activation and T cell specific cellular genes (IL-3 and granulocyte-macrophage colony-stimulating factor genes) (23,24). In the study related to HIV infection, a down-regulated expression of CD3ζ through ELF-1 inhibition was found on T cells, leading to T-cell anergy and lymphopenia (25). However, there remains lack of study regarding to its function in other infections or hypoxia condition.
Myeloid cells express members of the C-type lectin-like receptor (CTLR) family, including CLEC4D, which can function as pattern recognition receptors for several types of pathogens including fungi, bacteria and parasites, driving both innate and adaptive immunity. It was demonstrated that active tumor necrosis factor (TNF) and reactive oxygen species (ROS) production as well as phagocytosis followed CLEC4D triggering (26). Clec4d was detected on neutrophils and monocytes, but not be detectable on the surface of eosinophils, T cells, B cells, and natural killer (NK) cells (27), which might could partly explain the difference between ex vivo and in vivo parts. The increasing expression of CLEC4D shown in septic patients with hypoxemia compared to that of the group without hypoxemia may derive from that of peripheral neutrophils or monocytes, which were absent in ex vivo cell culture system. However, there is no relevant research about its change in hypoxia condition.
PRKD2 is one of isoforms among the calcium/calmodulin-dependent protein kinase superfamily. Our study demonstrated that PRKD2 was up-regulated upon hypoxemia in septic patients while its down-modulation was detected in ex vivo culture system. PRKD2 has been reported to induce HIF-1α accumulation in a low-oxygen environment, resulting in activation of NF-κB, implying its alteration and effect on immune cells in hypoxia condition (28). Of note, most present studies of PRKD2 are focus on its function in tumor cell survival, proliferation, migration, and angiogenesis (29) as well as the pathogenesis of hyperinsulinemia with its deficiency in exocrine and endocrine cells of the pancreas (28), indicating its expression on other cells rather than T cells. Whether its function related to T cells or the underlying molecular mechanism remains to be investigated.
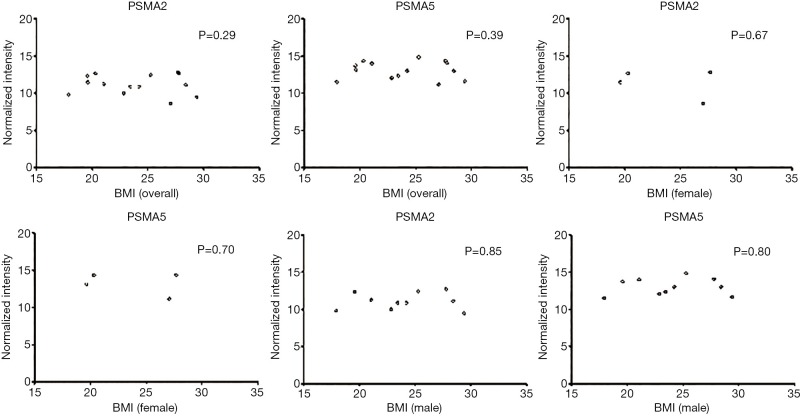
PSMA2 and PSMA5 are subunits of the proteasome family, the main proteolytic enzyme for the ubiquitin–proteasome system, regulation of the cell cycle, apoptosis, angiogenesis, cell adhesion and transcription (30), involving in several important biological processes including immune surveillance and tissue protein content maintenance (31). Our study showed that PSMA2 and PSMA5 mRNA was up-regulated among septic patients with hypoxemia while their expressions were decreased upon hypoxia ex vivo. Of interest, the expression of these two genes in peripheral blood was reported to be negatively correlated with body mass index (BMI) in 52 healthy men (30). However, there was no correlation detected of these two genes with BMI of the included population or in the gender stratification (Figure S1). The proteasome generates antigenic peptides that are presented on MHC class II molecules to CD4+ T lymphocytes and promotes lymphocyte activation to produce antibodies (32) while the mechanism of PSMA2 and PSMA5 during the sepsis with or without hypoxemia has not been reported.
Figure S1.
Correlation analysis of BMI with PSMA2 and PSMA5. BMI, body mass index; PSMA, proteasome subunit alpha.
We also validated several genes previously identified in microarray studies of sepsis-related ARDS by our study. As summarized in Table 4, some of the candidate genes identified in prior gene analysis studies could be confirmed in our study, supporting validity to their potential relevance in ARDS development during septic process as well as to the approach in this study. Not all the candidate genes in prior studies held the similar alterations between the two cohorts in the present study. There might be several potential explanations as followed. The characteristics of the prior studies were listed in Table S1. First, the included population was not completely the same. The hypoxemia cohort was sepsis induced ARDS (8-11). The control group was septic population except for Dolinay study (10), which included patients with SIRS as control settings. Besides, the objective of this study was to understand early changes of T cell-related genes following different clinical phenotype during sepsis process and we targeted on the immunocompetent septic patients by excluding the patients if they had tumors, hematological or immunological disease, or treatment with chemotherapy agents or corticosteroids within 6 months prior to hospitalization, which was not the case or reported in previous studies (8-11). More than 20% of the patients were cancer in both sepsis and sepsis with ARDS cohorts in Kangelaris study (8) and the proportion receiving the corticosteroids reached 65% in Howrylak study (9). These underlying effects on the peripheral blood gene expression should not be overlooked.
Table S1. Characteristics of prior studies with gene expressions in sepsis-induced ARDS or hypoxia.
| Study | Patients enrollment | Group setting | Factors that would affect the T cell related gene expression | Blood sample |
|---|---|---|---|---|
| 2009, Howrylak et al. (9) | Patients admitted to the MICU for 48 h or less who were intubated and receiving mechanical ventilation | Sepsis only (n=20) vs. sepsis and ALI (n=13) | No exclusion for past history of tumor, chemotherapy, hematology malignancy or medication that influence the immune system | At the time of enrollment in the study, whole blood was collected for RNA preservation |
| Patients were classified as having both sepsis and ALI if they met the criteria for sepsis as defined by the SCCM statement, and the criteria for ALI as defined by the AECC on ARDS | Patient characteristics | The GeneChip Human Genome U133A 2.0 Array (Affymetrix, Santa Clara, CA) was applied | ||
| Patients were classified as having sepsis only if they met only the above criteria for sepsis | • Cancer: 5% in ALI+ sepsis vs. 0 in sepsis; | |||
| Patients with sepsis and bilateral infiltrates who did not meet the AECC criteria for ALI were excluded in the study | • Immunosuppression: 15% in ALI+ sepsis vs. 5% in sepsis; | |||
| • Corticosteroids use: 62% in ALI+ sepsis vs. 65% in sepsis | ||||
| 2012, Dolinay et al. (10) | Patients were classified as SIRS, sepsis or sepsis with ARDS based on information available at the first 48 hours following MICU hospitalization. SIRS and Sepsis was identified according to the 2001 SCCM/ESICM/ACCP guidelines. ARDS was defined according to American-European Consensus Conference on ARDS guidelines | SIRS (n=19) vs. Sepsis+ ARDS (n=18) | No exclusion for past history of tumor, chemotherapy, hematology malignancy or medication that influence the immune system | Whole blood samples of 2.5 mL were collected within 48 h following MICU admission |
| No reports in patient characteristics of the past history of tumor, chemotherapy, hematology malignancy or medication that effected the immune system, or use of corticosteroids | The Illumina Human HT-12 v4 BeadChip arrays (Illumina) was applied | |||
| 2013, Chen et al. (11) | The same with the study of 2009, Howrylak et al. | |||
| 2015, Calfee et al. (8) | Critically ill patients admitted via the emergency department were enrolled at the time of triage to the ICU. Patients from this cohort who met consensus criteria for severe sepsis according to 2012 | Sepsis only (n=28) vs. Sepsis ARDS (n=29) | No exclusion for past history of tumor, chemotherapy, hematology malignancy or medication that influence the immune system; | Whole blood was collected within 24 h of ICU admission for the isolation of RNA |
| Patients were defined as having ARDS if they met criteria as defined by the Berlin definition of ARDS within 24 h of enrollment to the study | Patient characteristics | |||
| Patients were also excluded if whole blood suitable for RNA isolation was not collected | • Cancer: 21% in sepsis only vs. 52% in sepsis ARDS | The Affymetrix Human GeneChip Gene 1.0 ST array (Affymetrix, Santa Clara, CA) was applied |
The inconsistency of the in vivo and in vitro results suggested these candidate genes were expressed by other cells than T cells while they would drive both innate and adaptive immunity alterations. In spite of T cell-related genes directly activated or up-regulated in hypoxemic phenotype, the number of peripheral lymphocyte count was decreased and higher illness severity was observed, indicating the likely involvement of the candidate genes in T cells exhausting (33) and cell death pathways including apoptosis, necroptosis, necrosis, and pyroptosis (34). Our analysis supports the recent findings, namely that T cell-mediated alteration constitute an important pathophysiological mechanism of early sepsis, e.g., to transiently activation and subsequently to suppression (35,36).
Our study merges several strengths including enrollment of community-acquired sepsis and exclusion of the underlying condition that would influence the investigation of T cell-related genes alteration, prior clinical phenotyping of patients by hypoxemia, correlation of mRNA expression within candidate genes and with PaO2/FiO2, validation of candidate genes in ex vivo culture system and several genes identified in previous studies of sepsis with ARDS or hypoxemia, and analysis of potential mechanism of candidate genes. To our knowledge, this is the first study utilizing whole blood gene expression, focus on the T lymphocytes involved genes alteration following clinically hypoxemic phenotype in sepsis.
There are several limitations in our study as well. First, though we adopted the FDR <0.20, which seemed reasonable as prior literature suggested (8) and confirmed the difference seen in the candidate genes with the power calculation (Table S2), it requires further validation of candidate genes in a second study population. In addition, the sample size remains small and unconfirmed. Larger sample sizes would be needed to detect a gene signature upon hypoxemia in early sepsis. Third, genes detected by our approach are possible to be originated from circulating cells including the endothelial and epithelial cells and it remains investigation of their origination and interaction. Last, the causation between T cell-related gene expression alterations and hypoxemia in sepsis needs further investigation. Septic patients of hypoxemia phenotype held higher levels of illness severity and incidence of renal injury. The differentially expressed genes detected due to hypoxemia specifically, or rather due to multiple organ injury remains unclear. In addition, the timing of the hypoxemia occurrence could not be exactly captured in spite of the identification by P/F, which was also affected by the therapy strategy, like mechanical ventilating settings.
Table S2. The power calculation for the detected normalized intensity difference of candidate genes.
| Gene symbol | Non-hypoxemic | Hypoxemic | Mean difference | Alpha (significance level) |
Beta | Power (1-beta) | |||
|---|---|---|---|---|---|---|---|---|---|
| N | Normalized intensity (mean ± SD) | N | Normalized intensity (mean ± SD) | ||||||
| NLRP3 | 6 | 7.649±0.714 | 9 | 9.25±1.157 | 1.601 | 0.05 | 0.065 | 0.935 | |
| PSMA5 | 6 | 12.14±0.823 | 9 | 13.67±0.902 | 1.53 | 0.05 | 0.058 | 0.942 | |
| CLEC4D | 6 | 9.278±1.606 | 9 | 11.76±1.707 | 2.518 | 0.05 | 0.135 | 0.865 | |
| SOS1 | 6 | 6.216±1.43 | 9 | 7.976±1.039 | 1.76 | 0.05 | 0.223 | 0.777 | |
| CD300A | 6 | 9.462±1.354 | 9 | 10.91±0.7057 | 1.448 | 0.05 | 0.293 | 0.706 | |
| ELF1 | 6 | 9.278±1.077 | 9 | 11.05±0.9506 | 1.772 | 0.05 | 0.077 | 0.923 | |
| PRKD2 | 6 | 7.868±1.311 | 9 | 9.487±0.9846 | 1.619 | 0.05 | 0.225 | 0.775 | |
| STAT3 | 6 | 8.687±0.950 | 9 | 10.05±0.673 | 1.363 | 0.05 | 0.121 | 0.879 | |
| PSMA2 | 6 | 10.18±1.062 | 9 | 11.82±0.986 | 1.64 | 0.05 | 0.116 | 0.883 | |
The power calculation for the detected normalized intensity difference of candidate genes between groups with the present sample size was performed by Power Analysis and Sample Size (PASS) software (PASS 2008. NCSS, LLC. Kaysville, Utah, USA). For the current sample size of 9 in septic group with hypoxemia and 6 in that without and the detected normalized intensity difference of candidate genes, the detailed variables were set as above. Normalized intensity values were recorded from the microarray analysis. Mean difference = Mean (Hypoxemic) - Mean (Non-hypoxemic). The Alpha value of 0.05, as a level of significance was applied for estimation. The value of beta and respective power were obtained from power analysis of a non-Inferiority test of the difference of two means by PASS.
Conclusions
Our findings show septic patients with hypoxemic phenotype hold higher illness severity and incidence of lymphopenia as well as upregulated T lymphocytes function involved gene signatures of cell death and inhibitory receptors. The discovery suggests the potential association of T lymphocyte related genes alterations with distinct subphenotypes by hypoxemia during sepsis and their causal relationship would be worthy of further investigation in the future.
Acknowledgments
We would like to thank Mingyuan Gu, Xu Liu and Yuying Tang for clinical data collection. We would like to thank Lei Wang for his insightful readings and comments.
Funding: This work was supported by National Natural Science Foundations of China [grant number 81571874, 81372093 to H.Q.] and Jiangsu Provincial Key Medical Discipline [grant number ZDXKA2016025 to Y.Y.].
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The clinical study part was conducted in accordance with the amended Declaration of Helsinki. All the included subjects were from sepsis biological specimen bank from Department of Critical Care, Zhongda Hospital. The sepsis biological specimen bank was registered in 2017 by approval and supervise of Institutional Ethics Committee of Zhongda Hospital (Registration number: 2017ZDSYLL105). Written informed consent was obtained from all patients or their surrogate when possible. The Animal Care and Use Committee of Southeast University approved all experiments involving the use of animals.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Shankar-Hari M, Phillips GS, Levy ML, et al. Developing a New Definition and Assessing New Clinical Criteria for Septic Shock: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:775-87. 10.1001/jama.2016.0289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol 2013;13:862-74. 10.1038/nri3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen IJ, Sjaastad FV, Griffith TS, et al. Sepsis-induced T cell immunoparalysis: The ins and outs of impaired T cell immunity. J Immunol 2018;200:1543-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Englert JA, Bobba C, Baron RM. Integrating molecular pathogenesis and clinical translation in sepsis-induced acute respiratory distress syndrome. JCI Insight 2019. [Epub ahead of print]. 10.1172/jci.insight.124061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen PB, Hrobjartsson A, Nielsen DL, et al. Prevalence and prognosis of acutely ill patients with organ failure at arrival to hospital: A systematic review. PLoS One 2018;13:e0206610. 10.1371/journal.pone.0206610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.SRLF Trial Group Hypoxemia in the ICU: prevalence, treatment, and outcome. Ann Intensive Care 2018;8:82. 10.1186/s13613-018-0424-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westendorf AM, Skibbe K, Adamczyk A, et al. Hypoxia enhances immunosuppression by inhibiting CD4+ effector T cell function and promoting Treg activity. Cell Physiol Biochem 2017;41:1271-84. 10.1159/000464429 [DOI] [PubMed] [Google Scholar]
- 8.Kangelaris KN, Prakash A, Liu KD, et al. Increased expression of neutrophil-related genes in patients with early sepsis-induced ARDS. Am J Physiol Lung Cell Mol Physiol 2015;308:L1102-13. 10.1152/ajplung.00380.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howrylak JA, Dolinay T, Lucht L, et al. Discovery of the gene signature for acute lung injury in patients with sepsis. Physiol Genomics 2009;37:133-9. 10.1152/physiolgenomics.90275.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolinay T, Kim YS, Howrylak J, et al. Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am J Respir Crit Care Med 2012;185:1225-34. 10.1164/rccm.201201-0003OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Shi JX, Pan XF, et al. DNA microarray-based screening of differentially expressed genes related to acute lung injury and functional analysis. Eur Rev Med Pharmacol Sci 2013;17:1044-50. [PubMed] [Google Scholar]
- 12.Jones HD, Crother TR, Gonzalez-Villalobos RA, et al. The NLRP3 inflammasome is required for the development of hypoxemia in LPS/mechanical ventilation acute lung injury. Am J Respir Cell Mol Biol 2014;50:270-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palladino MA, Fasano GA, Patel D, et al. Effects of lipopolysaccharide-induced inflammation on hypoxia and inflammatory gene expression pathways of the rat testis. Basic Clin Androl 2018;28:14. 10.1186/s12610-018-0079-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Backert I, Koralov SB, Wirtz S, et al. STAT3 activation in Th17 and Th22 cells controls IL-22-mediated epithelial host defense during infectious colitis. J Immunol 2014;193:3779-91. 10.4049/jimmunol.1303076 [DOI] [PubMed] [Google Scholar]
- 15.Dang EV, Barbi J, Yang HY, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 2011;146:772-84. 10.1016/j.cell.2011.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karra L, Singh Gangwar R, Shamri R, et al. Leukocyte CD300a contributes to the resolution of murine allergic inflammation. J Immunol. 2018;201:2998-3005. 10.4049/jimmunol.1801000 [DOI] [PubMed] [Google Scholar]
- 17.Raggi F, Blengio F, Eva A, et al. Identification of CD300a as a new hypoxia-inducible gene and a regulator of CCL20 and VEGF production by human monocytes and macrophages. Innate Immun 2014;20:721-34. 10.1177/1753425913507095 [DOI] [PubMed] [Google Scholar]
- 18.Nissim Ben Efraim AH, Karra L, Ben-Zimra M, et al. The inhibitory receptor CD300a is up-regulated by hypoxia and GM-CSF in human peripheral blood eosinophils. Allergy 2013;68:397-401. 10.1111/all.12092 [DOI] [PubMed] [Google Scholar]
- 19.Alvarez Y, Tang X, Coligan JE, et al. The CD300a (IRp60) inhibitory receptor is rapidly up-regulated on human neutrophils in response to inflammatory stimuli and modulates CD32a (FcgammaRIIa) mediated signaling. Mol Immunol 2008;45:253-8. 10.1016/j.molimm.2007.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aljadi Z, Nopp A, Winqvist O, et al. Altered basophil function in patients with chronic kidney disease on hemodialysis. Clin Nephrol 2017;88:86-96. 10.5414/CN108992 [DOI] [PubMed] [Google Scholar]
- 21.Perucca S, Di Palma A, Piccaluga PP, et al. Mesenchymal stromal cells (MSCs) induce ex vivo proliferation and erythroid commitment of cord blood haematopoietic stem cells (CB-CD34+ cells). PLoS One 2017;12:e0172430. 10.1371/journal.pone.0172430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma M, Singh SB, Sarkar S. Genome wide expression analysis suggests perturbation of vascular homeostasis during high altitude pulmonary edema. PLoS One 2014;9:e85902. 10.1371/journal.pone.0085902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wurster AL, Siu G, Leiden JM, et al. Elf-1 binds to a critical element in a second CD4 enhancer. Mol Cell Biol 1994;14:6452-63. 10.1128/MCB.14.10.6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassuk AG, Barton KP, Anandappa RT, et al. Expression pattern of the Ets-related transcription factor Elf-1. Mol Med 1998;4:392-401. 10.1007/BF03401746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tumino N, Turchi F, Meschi S, et al. In HIV-positive patients, myeloid-derived suppressor cells induce T-cell anergy by suppressing CD3ζ expression through ELF-1 inhibition. AIDS 2015;29:2397-407. 10.1097/QAD.0000000000000871 [DOI] [PubMed] [Google Scholar]
- 26.Kerscher B, Willment JA, Brown GD. The Dectin-2 family of C-type lectin-like receptors: an update. Int Immunol 2013;25:271-7. 10.1093/intimm/dxt006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerscher B, Wilson GJ, Reid DM, et al. Mycobacterial receptor, Clec4d (CLECSF8, MCL), is coregulated with Mincle and upregulated on mouse myeloid cells following microbial challenge. Eur J Immunol 2016;46:381-9. 10.1002/eji.201545858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azoitei N, Fröhling S, Scholl C, et al. PRKD2: A two-pronged kinase crucial for the tumor-supporting activity of HSP90. Mol Cell Oncol 2015;2:e981444. 10.4161/23723556.2014.981444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao Y, Wang C, Chen JY, et al. Deficiency of PRKD2 triggers hyperinsulinemia and metabolic disorders. Nat Commun 2018;9:2015. 10.1038/s41467-018-04352-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakamoto K, Sato Y, Shinka T, et al. Proteasome subunits mRNA expressions correlate with male BMI: implications for a role in obesity. Obesity (Silver Spring) 2009;17:1044-9. 10.1038/oby.2008.612 [DOI] [PubMed] [Google Scholar]
- 31.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol 2006;17:1807-19. 10.1681/ASN.2006010083 [DOI] [PubMed] [Google Scholar]
- 32.Kisselev AF, Akopian TN, Woo KM, et al. The sizes of peptides generated from protein by mammalian 26 and 20S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J Biol Chem 1999;274:3363-71. 10.1074/jbc.274.6.3363 [DOI] [PubMed] [Google Scholar]
- 33.Mittal R, Ford ML, Coopersmith CM. Getting older can be exhausting. Crit Care 2014;18:465. 10.1186/s13054-014-0465-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Coopersmith CM. Dying as a Pathway to Death in Sepsis. Anesthesiology 2018;129:238-40. 10.1097/ALN.0000000000002271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hopp L, Loeffler-Wirth H, Nersisyan L, et al. Footprints of sepsis framed within community acquired pneumonia in the blood transcriptome. Front Immunol 2018;9:1620. 10.3389/fimmu.2018.01620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue M, Xie J, Liu L, et al. Early and dynamic alterations of Th2/Th1 in previously immunocompetent patients with community-acquired severe sepsis: a prospective observational study. J Transl Med 2019;17:57. 10.1186/s12967-019-1811-9 [DOI] [PMC free article] [PubMed] [Google Scholar]