Abstract
Background and Purpose
Growing evidence indicates targeting mitochondrial dynamics and biogenesis could accelerate recovery from renal ischemia–reperfusion (I/R) injury, but the underlying mechanisms remain elusive. Transcription factor forkhead box O1 (FOXO1) is a key regulator of mitochondrial homeostasis and plays a pathological role in the progression of renal disease.
Experimental Approach
A mouse model of renal I/R injury and a hypoxia/reoxygenation (H/R) injury model for human renal tubular epithelial cells were used.
Key Results
I/R injury up‐regulated renal expression of FOXO1 and treatment with FOXO1‐selective inhibitor AS1842856 prior to I/R injury decreased serum urea nitrogen, serum creatinine and the tubular damage score after injury. Post‐I/R injury AS1842856 treatment could also ameliorate renal function and improve the survival rate of mice following injury. AS1842856 administration reduced mitochondrial‐mediated apoptosis, suppressed the overproduction of mitochondrial ROS and accelerated recovery of ATP both in vivo and in vitro. Additionally, FOXO1 inhibition improved mitochondrial biogenesis and suppressed mitophagy. Expression of PPAR‐γ coactivator 1α (PGC‐1α), a master regulator of mitochondrial biogenesis, was down‐regulated in both I/R and H/R injury, which could be abrogated by FOXO1 inhibition. Experiments using integrated bioinformatics analysis and coimmunoprecipitation established that FOXO1 inhibited PGC‐1α transcription by competing with cAMP‐response element binding protein (CREB) for its binding to transcriptional coactivators CREBBP/EP300 (https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737).
Conclusion and Implications
These findings suggested that FOXO1 was critical to maintain mitochondrial function in renal tubular epithelial cells and FOXO1 may serve as a therapeutic target for pharmacological intervention in renal I/R injury.
Abbreviations
- BAX
BCL2‐associated X protein
- BCL2
B‐cell lymphoma 2
- BUN
blood urea nitrogen
- CBP
CREB binding protein
- CREB
cAMP‐response element binding protein
- DRP1
dynamin‐related protein‐1
- FOXO1
forkhead box O1
- H/R
hypoxia and reoxygenation
- HK2
human kidney cell line 2
- I/R
ischemia and reperfusion
- MFN1
mitofusin 1
- MFN2
mitofusin 2
- MMP
mitochondrial membrane potential
- mtDNA
mitochondrial DNA
- mtROS
mitochondrial ROS
- OPA1
optic atrophy 1
- P300
E1A binding protein p300
- PGC‐1α
PPAR‐γ coactivator‐1α
- PINK1
PTEN‐induced kinase 1
- TFAM
mitochondrial transcription factor A
What is already known
Mitochondrial damage contributes to the proximal tubular epithelial cell dysfunction in I/R injury.
What this study adds
FOXO1 mediated alteration of mitochondrial homeostasis induced by I/R in renal tubular epithelial cells.
What is the clinical significance
FOXO1 might serve as a therapeutic target for the prevention of acute kidney injury.
1. INTRODUCTION
Acute kidney injury (AKI) is associated with a high mortality rate and it predisposes to the progression of chronic kidney disease (Grams & Rabb, 2012; Khwaja, 2012). Ischemia and reperfusion (I/R) injury unavoidably occurs after surgical procedures is one of the most common cause of AKI (Ferenbach & Bonventre, 2015). However, no therapeutic strategy has been approved for AKI after I/R injury (McCurley et al., 2017). A novel pharmacological treatment for AKI is urgently required.
AKI induced by I/R injury is generally described as the injury of renal tubular epithelial and endothelial cells accompanied by the activation of inflammatory process (Andrade‐Oliveira et al., 2015). Recently, mitochondrial damage has been confirmed to be a major contributor to the proximal tubular epithelial cell dysfunction during I/R injury (Yang et al., 2016). The renal proximal tubular cells with high ATP demand, responssible for reabsorbing a bulk of the glomerular ultrafiltrate and contains more mitochondria than other renal cell types (Emma, Montini, Parikh, & Salviati, 2016). In I/R injury, the balance of mitochondrial homeostasis is disrupted, resulting in the mitochondrial fragmentation (Liu & Hajnóczky, 2011; Zhan, Brooks, Liu, Sun, & Dong, 2013). The fragmented mitochondria are potential source of reactive oxygen species (ROS), cytochrome C, mitochondrial DNA (mtDNA) and other potentially injurious molecules (Emma et al., 2016; Zhang et al., 2010). Researching into the effects of targeting mitochondrial dynamics and biogenesis has yielded consistent and exciting results, which suggests the pharmacological enhancement of mitochondrial mass or compensation of normal mitochondria might accelerate recovery from AKI (Tran & Parikh, 2014; Weinberg, 2011). Therefore, mitochondria protective strategies could benefit AKI. However, the mechanisms responsible for I/R injury‐induced mitochondrial dysfunction remain poorly understood.
Forkhead box protein O1 (FOXO1) is a member of the forkhead transcription factors family which is expressed relatively ubiquitously in mammals (Sanchez, Candau, & Bernardi, 2014). FOXO1 regulates the process of cell proliferation, apoptosis, autophagy, oxidative stress and energy metabolism by modulating the transcription of downstream target genes. Several lines of evidence indicate that FOXO1 plays a critical role in mitochondrial dynamics. It has been shown that FOXO1 promotes mitophagy through regulating transcription of PTEN‐induced kinase 1 (https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2161) and LC3 in a ROS‐dependent manner (Baldelli, Aquilano, & Ciriolo, 2014). Our previous study reveals that FOXO1 mediate alteration of mitochondrial dynamics by https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1503‐dynamin‐related protein‐1 (DRP1) pathway (Shi et al., 2018). Moreover, FOXO1 could activate the transcription of Bcl‐2‐like protein 11 (Bim), which triggers B‐cell lymphoma 2 (https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2844)‐associated X protein (BAX)‐mediated mitochondria‐dependent apoptosis (Shukla, Rizvi, Raisuddin, & Kakkar, 2014; Zhang et al., 2017). A great deal of information has been demonstrated that FOXO1 is crucially involved in different processes of renal diseases. However, the involvement of FOXO1 in renal I/R injury has not been determined.
In the current study, we used renal I/R injury mice model in vivo and cellular hypoxia and reoxygenation (H/R) injury model in vitro to evaluate the involvement of FOXO1 in the regulation of mitochondrial homeostasis using the FOXO1 inhbitor AS1842856 (5‐Amino‐7‐(cyclohexylamino)‐1‐ethyl‐6‐fluoro‐4‐oxo‐1,4‐dihydroquinoline‐3‐carboxylic acid) (Nagashima et al., 2010).
Our results demonstrate that inhibition of FOXO1 could prevent I/R‐induced renal injury and preserve mitochondrial homeostasis in renal tubular epithelial cells. The putative mechanisms include the increased transcription of PPAR‐γ coactivator 1α (PGC‐1α) by reducing the competitive binding of FOXO1 and p‐cAMP‐response element binding protein (CREB) to CREB binding protein (https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734)/E1A binding protein (https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737).
2. METHODS
2.1. Animal model
All animal care and experimental procedures complied with the Animals (Scientific procedures) Act 1986 and all procedures in this study were strictly conducted in accordance with the European Community guidelines for the use and care of laboratory animals and approved by the Biomedical Ethics Committee of Peking University (LA 2010–048). Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny, Browne, Cuthill, Emerson, & Altman, 2010) and with the recommendations made by the British Journal of Pharmacology. C57BL/6 male mice at 10 weeks of age, weighing 22–23 g, were purchased from the Animal Centre of Peking University. Mice were housed in open‐top conventional cages with usual bedding material and were kept in pathogen‐free conditions. A maximum of five mice were housed in a single cage. Animals were housed with a 12‐hr light and 12‐hr dark cycle under defined environmental conditions at 25 ± 2°C with a relative humidity of 50%. Water and food were available ad libitum. All efforts were made to prevent any animals suffering.
To establish renal I/R injury model, mice were anaesthetized with https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5480 (60 mg/kg, i.p.) and placed on a heating blanket to maintain body temperature at 37°C. The right kidney was removed and the left renal artery and vein were identified and with a non‐traumatic clamp. After 35 min the clamp was released and reperfusion was confirmed visually. To investigate if there was a protective effect of AS1842856 (Nagashima et al., 2010), mice were injected intraperitoneally with a dose of 1 or 10 mg·kg−1·day−1 of AS1842856 for 7 days before injury. Mice were then killed 24 hr after reperfusion. To investigate the effect of AS1842856 on the injury caused by reperfusion mice were injected intraperitoneally with 10 mg·kg−1·day−1 of AS1842856 for 7 days after the injury and then killed . Animals were killed by the administration of the anaesthetic pentobarbital (60 mg/kg, i.p.) followed by decapitation.
2.2. Randomization and blinding
In the present study animals were randomized for treatment. All the experiments were performed and analysed under blinded conditions.
2.3. Renal function test
Serum creatinine and blood urea nitrogen (BUN) were measured by commercial kits (Nanjing Jiancheng Bioengineering Institute). Serum collected 200 µl from mice was optimized to measure these two chemicals following the manufacturer's instructions.
2.4. Histology and immunohistochemistry
The immuno‐related procedures used comply with the recommendations made by the British Journal of Pharmacology (Alexander et al., 2017). Kidneys were fixed with 4% formaldehyde, embedded in paraffin, and sectioned into 5 mm thick. The sections were stained by haematoxylin and eosin for histological examination. Tubular damage was scored in a double‐blind manner method based on the percentage of injury included tubular dilation and intertubular haemorrhage: 0, no damage; 1, <25%; 2, 25–50%; 3, 50–75%; 4, >75%. For immunohistochemical staining, renal paraffin sections were deparaffinized, rehydrated and then incubated with anti‐FOXO1 antibody (Cell Signaling Technology) at 4°C overnight after heating‐induced antigen retrieval in 0.01‐M citrate buffer. Secondary antibody (Jackson ImmunoResearch Laboratories) was used to incubate for 45 min at room temperature. Then the diluted solution comprised Vector Nova Red chromogen (Vector Laboratories) was applied to incubate for 5 min. The sections were counterstained with haematoxylin and mounted. Properly diluted solutions of non‐immune bovine serum were used as negative control.
2.5. Cells
Human kidney proximal tubular cells (HK2s) were purchased from Cell Culture Centre, Institute of Basic Medical Science Chinese Academy of Medical Sciences (Beijing, China). HK2s were respectively maintained in DMEM (M&C Gene Technology), supplemented with 10% FBS (GIBCO), 100 U·ml−1 penicillin and 100 μg·ml−1 streptomycin, in a humidified atmosphere of 5% CO2 at 37°C. To establish H/R injury model HK2s were deprived of serum for 24 hr. Then the HKs were incubated in low‐glucose DMEM at 37°C under hypoxia for 12 hr and reoxygenation for 4 hr. AS1842856 was co‐incubated with HK2s in dose of 50, 100 and 200 nM, 24 hr prior to H/R injury. For https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 inhibition, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7529 (S7256, Selleck Chemicals) was co‐incubated with HK2s in dose of 50 and 100 nM, 24 hr prior to H/R injury. For CREB inhibition, a potent and selective CREB inhibitor, 666‐15 (HY‐101120, MedChemExpress), was co‐incubated with HK2s in dose of 20 μΜ, 24 hr prior to H/R injury. For FOXO1 knockdown, 30 nM of FOXO1 siRNA duplex or scramble siRNA with Lipofectamine RNAiMAX (Invitrogen) were used to transfect HK2s. Sequences corresponding to the siRNA were shown in Table S1. For FOXO1 overexpression, recombinant adenovirus expressing human FOXO1‐flag (Yingrun) and Ad‐EGFP was used to transfect HK2s. To determine the degradation rate of FOXO1, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5433 (HY‐12320, MedChemExpress), a reversible inhibitor of protein synthesis, was used. HK2s were incubated with 100‐μΜ cycloheximide for different times under control or H/R condition.
2.6. TUNEL assay
For TUNEL assay staining, renal tissues were embedded in Tissue‐Tek optimal cutting temperature compound (Sakura Finetek) and sectioned into 5 mm thick. The sections were stained by in situ Cell Death Detection kit (Promega). The process of HK2s was the same as renal tissues treated as indicated.
2.7. Mitochondrial function and morphology detection
For measuring ATP level in kidney and HK2s an ATP Assay Kit (Beyotime) was used. To investigate mitochondrial ROS (mtROS) level, 5‐μm sections of renal tissues embedded in Tissue‐Tek optimal cutting temperature compound and HK2s were stained by the fluorogenic probe MitoSOX Red (Thermofisher Scientific). The fluorescence images were identified and captured by inverted microscope (Olympus) and confocal microscope (Leica). For measuring mitochondrial membrane potential (MMP) a JC‐1 kit (Beyotime) was used. HK2s were planted on 96‐well plates and incubated with JC‐1. Quantitative analysis was carried out using ImageJ software. Mitochondrial morphology detection was performed as previously described (Shi et al., 2018). Briefly, HK2s were incubated with 2.5% glutaraldehyde solution and managed following dehydration, embedding and sectioning. Mitochondria were identified by a transmission electron microscope (Hitachi).
2.8. Bioinformatic analysis
The profile of GSE52004 was downloaded from the Gene Expression Omnibus Datasets (https://www.ncbi.nlm.nih.gov/geo/) and analysed by SangerBox (http://sangerbox.com/). Genes with a corrected P value less than .05 and log fold change >2 were considered as differentially expressed genes (DEGs) and revealed in the form of Volcano Plot. The gene ontology (GO) pathway and KEGG pathway enrichment of DEGs were performed by g:profiler online analyses (https://biit.cs.ut.ee/gprofiler/). The results were visualized by enrichment plugin of Cytoscape (http://www.cytoscape.org/) software. The DEGs expression products in renal I/R injury were constructed by the STRING database (http://string-db.org/). It was visualized and analysed by MCODE plugin and BINGO plugin of Cytoscape software. Bisogenet plugin of Cytoscape software was used to predict the specific interactions between FOXO1 and PGC‐1α.
2.9. Quantitative real‐time PCR
Total RNA was extracted from renal tissues and HK2s with TRIzol reagent (Thermofisher Scientific); 2‐μg RNA was used for reverse transcription by a RevertAid First Strand cDNA Synthesis Kit (Thermofisher Scientific). Real‐time quantitative PCR was performed using the Mx3005P system (Agilent Technologies) with SYBR Green Real‐Time PCR Master Mix (Promega). The specific primers used were shown in Table S1.
2.10. Western blot analysis
The Western blot analysis had been conducted according to BJP Guidelines (Alexander et al., 2017). Renal tissues and HK2s were homogenized in RIPA buffer (1% Triton X‐100, 20‐mM HEPES [pH 7.4], 100‐mM KCl, and 2‐mM EDTA) containing protease inhibitors (Calbiochem). Protein expression was analysed by Western blot analysis as previously described (Shi et al., 2018). The primary antibodies and HRP‐conjugated secondary antibody used were shown in Table S2. Blots were developed with Western Blotting Luminol Reagent (Santa Cruz Biotechnology). The bands were scanned with Epson scanning system and the staining intensity of bands were analysed by ImageJ software.
2.11. Coimmunoprecipitation
HK2s were lysed to extract proteins. Then the proteins were incubated with anti‐https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734 antibody (Cell Signaling Technology) or anti‐https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 antibody (Cell Signaling Technology) followed by precipitation with precleared Protein A/G Plus–agarose beads (Santa Cruz Biotechnology). The immunoprecipitates were detected by Western blot analysis using anti‐FOXO1 antibody (Cell Signaling Technology) or anti‐p‐CREB antibody (Santa Cruz Biotechnology).
2.12. Data and statistical analysis
The data and statistical analysis comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology (Curtis et al., 2018). Data are expressed as the means ± SEM. Based on our preliminary experiments the calculated minimum number of the sample size n required to achieve a difference with 95% confidence and 80% power was five, so at least five samples or independent experiments were performed with all the assays. Statistical analyses were conducted in GraphPad Prism 6 (RRID:SCR_002798). Kolmogorov–Smirnov's test was used to determine normality. The statistical differences between groups were evaluated by unpaired Student's t‐test. When more than two treatment groups were compared, ANOVA with the Tukey's test was used when the F statistic was significant and there was no significant variance inhomogeneity. Differences were considered to be statistically significant at P < .05.
2.13. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander et al., 2017).
2.14. Materials
SGC‐CBP30 (CAS # S7256) was purchased fromSelleck Chemicals. 666‐15 (CAS # HY‐101120)and cycloheximide (CAS # HY‐12320)were purchased from MedChemExpress. MitoSOX Red, RevertAid First StrandcDNA Synthesis Kit and TRIzol reagent were purchased from ThermofisherScientific. ATP Assay Kit and JC‐1 kit were purchased from Beyotime. Cell Death Detection kitand SYBR Green Real‐Time PCR Master Mix werepurchased from Promega. anti‐CBP antibody, anti‐P300 antibody and anti‐FOXO1 antibody were purchased from Cell Signaling Technology. LipofectamineRNAiMAX were purchased from Invitrogen. Protein A/G Plus‐agarose beads and anti‐p‐CREB antibody werepurchased from Santa Cruz Biotechnology.
3. RESULTS
3.1. Renal FOXO1 expression was induced by I/R
To investigate the specific role of FOXO1 in vivo, a mouse renal I/R injury model was established. After 24‐hr reperfusion following 35‐min ischemia, the expression of FOXO1 was significantly increased and the phosphorylation of FOXO1 at Ser256 was inhibited in both the renal cortex and medulla (Figure 1a), but I/R had no significant effect on the levels of mRNA for the other two isoforms—FOXO3 and FOXO4 (Figure 1b). We further examined the localization of FOXO1 using immunohistochemical staining. We found that FOXO1 was widely expressed in the mice glomerulus and tubules and was up‐regulated in renal tubular cells including proximal tubular cells following I/R (Figure 1c).
Figure 1.
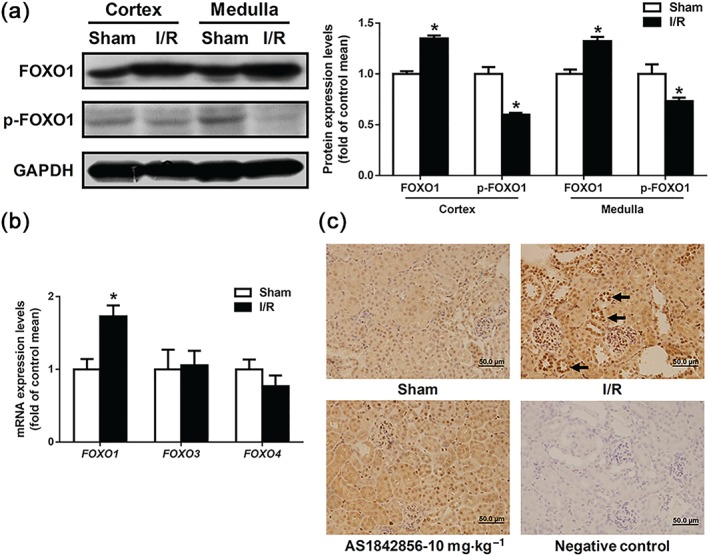
FOXO1 was specifically elevated in renal I/R injury. (a) FOXO1 and phosphorylated FOXO1 protein levels were respectively detected in renal cortex and medulla by Western blot analysis. Immunoblots of representative samples were shown. Data are presented as histograms and shown as mean ± SEM. n = 5 per group. * P < .05 versus sham. (b) The mRNA expression levels of FOXO1, FOXO3, and FOXO4 in renal cortex were analysed by qPCR. Data are shown as mean ± SEM. n = 6 per group, * P < .05 versus sham. (c) Immunohistochemical staining of FOXO1 in kidney of sham, I/R and AS1842856 pre‐treated mice after reperfusion for 24 hr. n = 5 per group. Black arrows: positive stains of FOXO1 in proximal tubular
3.2. FOXO1 inhibition ameliorated functional and histological renal injury induced by I/R
To investigate the effects of FOXO1 inhibition on I/R‐induced AKI, AS1842856, a selective inhibitor of FOXO1, was injected intraperitoneally at doses of 1 and 10 mg·kg−1 respectively for 7 days prior to the renal I/R operation (Figure 2a). In I/R mice, serum creatinine concentration and serum BUN levels were elevated; 1 and 10 mg·kg−1 AS1842856 pre‐treatment dose dependently decreased serum creatinine concentration and BUN levels (Figure 2b). I/R evoked severe tubular injury, characterized by tubular brush border dilatation and loss in proximal tubular. Pre‐treatment of AS1842856 reduced I/R‐induced tubular damage (Figure 2d). Kidney injury molecule‐1 and neutrophil gelatinase‐associated lipocalin, important biomarkers of renal injury, were up‐regulated in I/R group and had a significant decrease in AS1842856 pre‐treated groups (Figure 2c).
Figure 2.
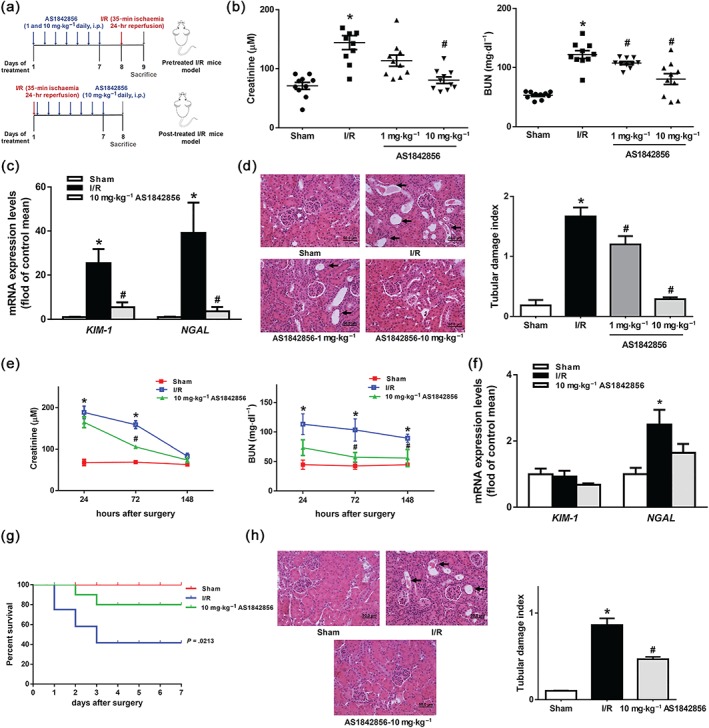
FOXO1 inhibition alleviated I/R‐induced AKI. (a) Two modes of AS1842856 administration were used. The specific operating time nodes of AS1842856 pre‐treated mice and post‐treated mice are shown. (b) Serum creatinine and serum BUN levels in sham, I/R and AS1842856 pre‐treated mice after reperfusion for 24 hr were monitored. Data are shown as mean ± SEM. n = 10 per group, * P < .05 versus sham, # P < .05 versus I/R. (c) The mRNA expression levels of KIM‐1 and NGAL in renal tissues in sham, I/R and AS1842856 pre‐treated mice were analysed by qPCR. Data are shown as mean ± SEM, n = 5 per group, * P < .05 versus sham, # P < .05 versus I/R. (d) Representative images of renal tissue with H&E staining in sham, I/R and AS1842856 pre‐treated mice after reperfusion for 24 hr (magnification 400×). Data are presented as histogram and expressed as mean ± SEM. n = 10 per group, * P < .05 versus sham, # P < .05 versus I/R. Black arrows: tubular brush border dilatation and loss. (e) The serum creatinine and serum BUN levels in sham, I/R and AS1842856 post‐treated mice were shown as line graphs. Data were expressed as mean ± SEM (n = 7 for each time period of serum creatinine and n = 5 for each time period of serum BUN). ** P < .05 versus sham, # P < .05 versus I/R. (f) The mRNA expression levels of KIM‐1 and NGAL in renal tissues in sham, I/R and AS1842856 post‐treated mice after reperfusion for 148 hr were analysed by qPCR. Data are shown as mean ± SEM, n = 5 per group, * P < .05 versus sham. (g) Survival rates in sham, I/R and AS1842856 post‐treated mice. Data were expressed as mean ± SEM. n = 10 for each group. P = .0213 versus sham. (h) Representative images of renal tissue with H&E staining in sham, I/R and AS1842856 post‐treated mice on Day 7 after reperfusion (magnification 400×). Data are presented as histograms. Data were expressed as mean ± SEM. n = 5 for each group. * P < .05 versus sham, # P < .05 versus I/R. Black arrows: tubular brush border dilatation and loss
We next examined whether post‐treatment of AS1842856 could protect against I/R injury. AS1842856 was injected with a 10 mg·kg−1 dose for seven consecutive days since the date of I/R (Figure 2a). Compared with sham group, mice died at Day 1 after I/R and the mortality was ≤60% 7 days following I/R, whereas post‐treatment of AS1842856 significantly improved survival from I/R (Figure 2g). Serum creatinine concentration increased 2.8‐ and 2.3‐fold at 24 and 72 hr after I/R and serum BUN levels increased 2.5‐, 2.4‐ and 2.0‐fold and at 24, 72, and 148 hr after I/R as compared to sham group. Post‐treatment of AS1842856 suppressed serum creatinine at 72 hr and serum BUN levels at 72 and 148 hr after I/R (Figure 2e). In addition post‐treatment of AS1842856 also reduced I/R‐induced neutrophil gelatinase‐associated lipocalin expression (Figure 2f) and tubular damage (Figure 2h).
3.3. FOXO1 inhibition prevented I/R‐induced renal tubular epithelial cells apoptosis
To investigate I/R‐induced tubular damage regulated by FOXO1, the incidence of apoptosis was assessed by TUNEL assay and activation of pro‐CASPASE‐3 and pro‐CASPASE‐7. Compared with sham group, I/R elevated the TUNEL‐positive staining in kidney and pre‐treatment of AS1842856 1 mg/kg/day or 10 mg/kg/day decreased the percentage of TUNEL‐positive cells (Figure 3a). I/R evoked the cleavage and activation of pro‐CASPASE‐3 and pro‐CASPASE‐7 compared with sham group. Pre‐treatment with AS1842856 inhibited its cleavage and restored the amount of pro‐CASPASE‐3 and pro‐CASPASE‐7 (Figure 3b). HK2s were subjected to acute H/R as an in vitro model of renal I/R. Hypoxia for 12 hr followed by reoxygenation for 4 hr significantly enhanced the TUNEL‐positive staining of HK2s. Co‐incubation with AS1842856 decreased the percentage of TUNEL‐positive cells (Figure 3c). AS1842856 abrogated cleavage and activation of pro‐CASPASE‐3 and pro‐CASPASE‐7 in H/R group (Figure 3d).
Figure 3.
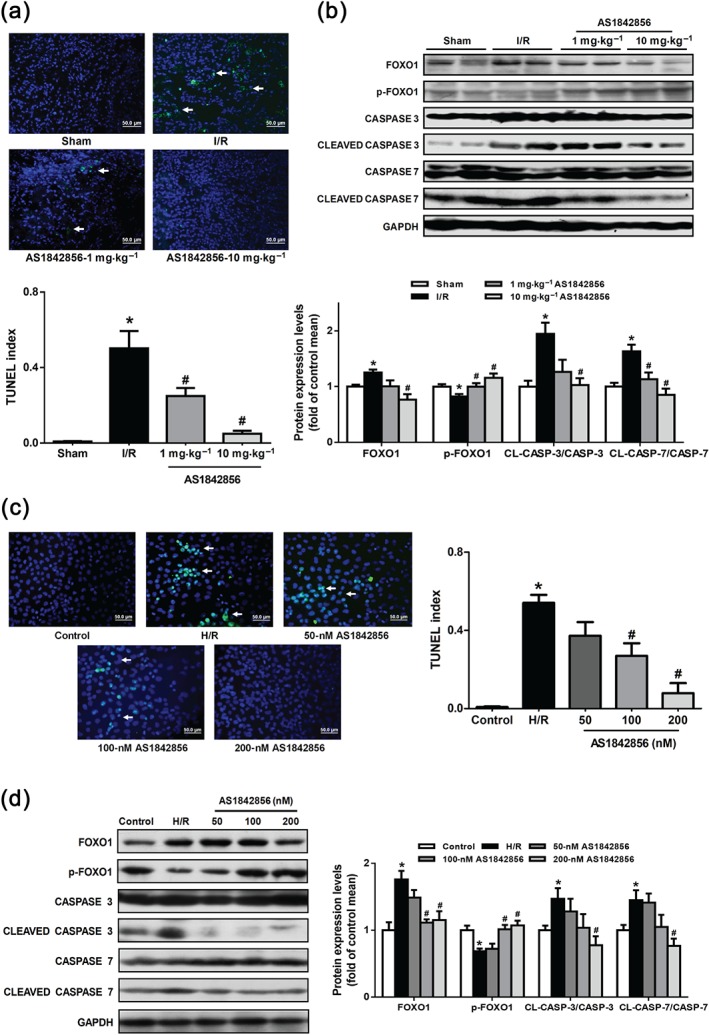
FOXO1 inhibition by AS1842856 pre‐treatment suppressed I/R‐induced apoptosis of tubular cells. (a) Representative images of TUNEL staining in kidneys of different groups (green fluorescence, magnification 400×). Scale bar: 50 μm. Quantitative analysis of TUNEL‐positive cells among groups. The number of TUNEL‐positive cells was expressed as a TUNEL positive index. Data are shown as mean ± SEM, n = 5 per group, * P < .05 versus sham, # P < .05 versus I/R. White arrows: TUNEL‐positive cells. (b) FOXO1, phosphorylated‐FOXO1, CASPASE 3, CLEAVED CASPASE 3, CASPASE 7 and CLEAVED CASPASE 7 protein levels among groups in renal cortex were monitored by Western blot analysis. Statistical data were shown as histograms. Data are shown as mean ± SEM. n = 6 per group. * P < .05 versus sham, # P < .05 versus I/R. (c) HK2s were subjected to TUNEL staining. Representative images of TUNEL staining in different groups. Scale bar: 50 μm. Data were expressed as mean ± SEM, n = 5 per group, * P < .05 versus control, # P < .05 versus H/R. White arrows: TUNEL‐positive cells. (d) The levels of FOXO1, phosphorylated‐FOXO1, CASPASE 3, CLEAVED CASPASE 3, CASPASE 7 and CLEAVED CASPASE 7 protein levels in HK2s among groups were detected by Western blot analysis. Data are shown as mean ± SEM, n = 5 per group. * P < .05 versus control, # P < .05 versus H/R
BAX as homologous binding partner of https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2844 can commit a cell to the mitochondrial apoptosis and subsequent initiation of the CASPASE cascade. We next determined the effect of FOXO1 inhibition on BAX, https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2844 and cytochrome c. I/R up‐regulated BAX and cytochrome c and down‐regulated https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2844 compared with sham group. Pre‐treatment with AS1842856 inhibited the up‐regulation of BAX/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2844 ratio and cytochrome c expression level (Figure 4a), which is consistent with the results in vitro (Figure 4b).
Figure 4.
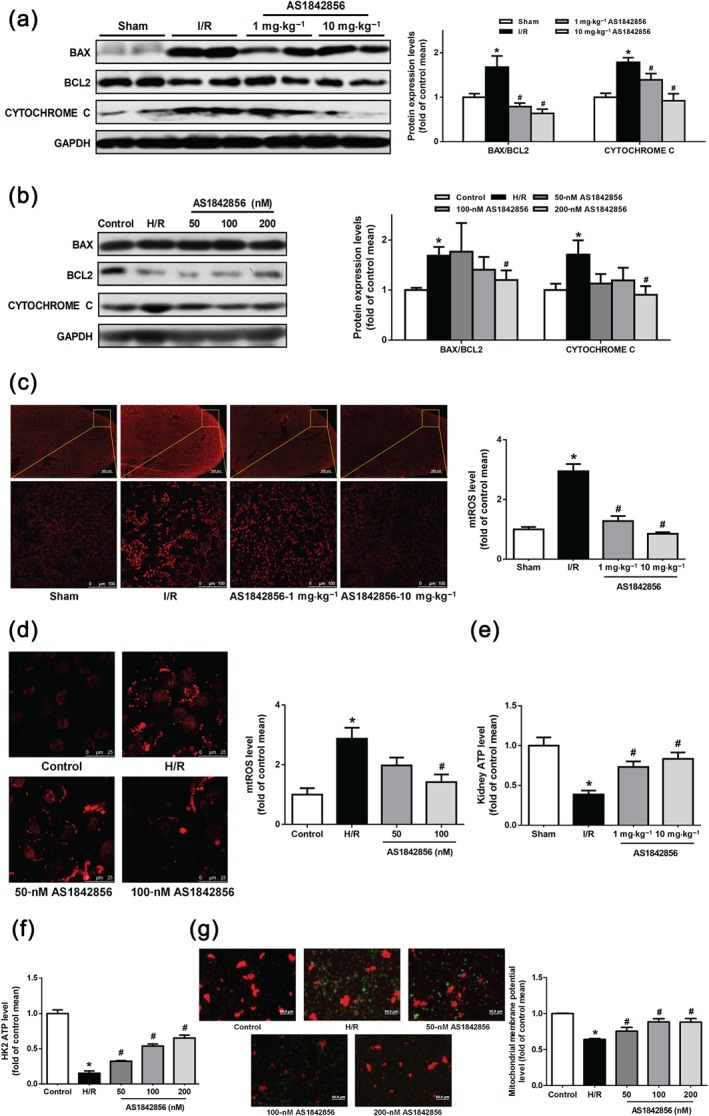
FOXO1 inhibition by AS1842856 pre‐treatment improved I/R‐impaired mitochondrial function in tubular cells. (a) The levels of cytochrome c expression and BAX/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2844 in renal cortex in different groups were monitored by Western blot analysis. Data were expressed as mean ± SEM. n = 6 per group. * P < .05 versus sham, # P < .05 versus I/R. (b) Cytochrome c and BAX/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2844 protein levels in HK2s in different groups were detected by Western blot analysis. Data are shown as mean ± SEM, n = 5 per group, * P < .05 versus control, # P < .05 versus H/R. (c) Renal mtROS level in different groups of mice was stained with MitoSOX Red. Representative sections of renal tissues (5 μm) were shown. Scale bar: 500 and 100 μm. Data of fluorescence measurement were presented as histograms and shown as mean ± SEM. n = 5 per group. * P < .05 versus sham, # P < .05 versus I/R. (d) Quantitative analysis of intracellular mtROS among groups. Scale bar: 25 μm. Data were expressed as mean ± SEM, n = 5 per group, * P < .05 versus control, # P < .05 versus H/R. (e) Quantitative analysis of renal ATP in different groups. Data were expressed as mean ± SEM and were shown as a percentage of the sham. n = 5 per group, * P < .05 versus sham, # P < .05 versus I/R. (f) Quantitative analysis of intracellular ATP among groups treated with or without AS1842856 in different concentrations. Data are shown as mean ± SEM and were shown as a percentage of the control. n = 5 per group, * P < .05 versus control, # P < .05 versus H/R. (g) Mitochondrial transmembrane potential levels in HK2s among groups. Bar diagram shows the ratio of red fluorescence to green fluorescence. Data were expressed as mean ± SEM and are shown as a percentage of the control. n = 5 per group, * P < .05 versus control, # P < .05 versus H/R
3.4. FOXO1 inhibition attenuated mitochondrial dysfunction induced by H/R
To determine whether FOXO1 affected mitochondrial function, mtROS generation, ATP content and MMP were detected. In vivo measurement demonstrated a 1.9‐fold increase in renal mtROS generation at 24 hr after I/R, which was prevented by pre‐treatment of AS1842856 (Figure 4c). In vitro FOXO1 inhibitor prevented mtROS generation induced by H/R (Figure 4d). In addition, ATP production was suppressed in response to either I/R or H/R. Pre‐treatment with AS1842856 enhance ATP content in a dose‐dependent manner (Figure 4e,f). MMP was dramatically decreased by 36% after H/R, which was potentiated by treatment with AS1842856 in a dose‐dependent manner (Figure 4g).
3.5. FOXO1 inhibition improved mitochondrial biogenesis and suppressed mitophagy
Alteration of mitochondrial dynamics and biogenesis has shown to be responsible for I/R‐induced mitochondrial dysfunction and ROS accumulation. We next determined effects of FOXO1 inhibition on mitochondrial dynamics and biogenesis. The expression of mitochondrial fission‐related protein DRP1 and mitophagy‐related proteins Parkin RBR E3 ubiquitin protein ligase and https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2161 were increased in kidney of I/R compared to sham. Pre‐treatment with AS1842856 suppressed the DRP1, Parkin RBR E3 ubiquitin protein ligase and https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2161 expression levels (Figure 5a,c). The expression of mitochondrial fusion‐ and biogenesis‐related optic atrophy 1 (OPA1), mitofusin 1 (MFN1), mitofusin 2 (MFN2), PGC‐1α and mitochondrial transcription factor A (TFAM) were down‐regulated. AS1842856 pre‐treatment potentiated MFN2, PGC‐1α and TFAM expression levels (Figure 5a,c). In HK2s, H/R resulted in down‐regulation of OPA1, PGC‐1α and TFAM and up‐regulation of https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2161. The changes were reversed by AS1842856 except for OPA1 (Figure 5b,d). Electron microscopy was used to evaluate morphological changes of mitochondria in HK2s. Broken mitochondria with cristae lost and more autophagosomes were found in H/R samples (Figure 5f). Formation of autophagosome was significantly decreased and mitochondria were intact and long with clear cristae in HK2s treated with AS1842856. Furthermore, mtDNA copy number decreased in I/R group and pre‐treatment with AS1842856 could significantly augment the mtDNA copy number (Figure 5g).
Figure 5.
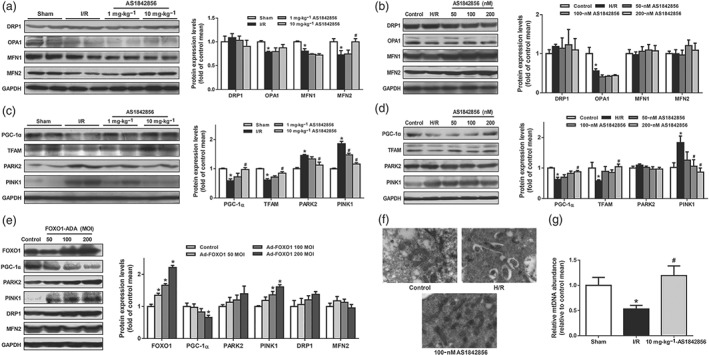
FOXO1 inhibition by AS1842856 pre‐treatment prevented I/R‐induced mitophagy and improves mitochondrial biogenesis. (a) DRP1, OPA1, MFN1, and MFN2 protein levels in renal cortex in different groups were monitored by Western blot analysis. Immunoblots of representative samples were shown. Data are shown as mean ± SEM, n = 6 per group. * P < .05 versus sham, # P < .05 versus I/R. (b) Quantitative analysis of intracellular DRP1, OPA1, MFN1 and MFN2 proteins in HK2s treated with or without different concentrations of AS1842856 was analysed by Western blot analysis. Data are shown as mean ± SEM. n = 5 per group, * P < .05 versus control. (c) Quantitative analysis of PGC‐1α, TFAM, PARK2, and https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2161 proteins in renal cortex were analysed by Western blot analysis. Data are shown as mean ± SEM. n = 6 per group, * P < .05 versus sham, # P < .05 versus I/R. (d) Quantitative analysis of intracellular PGC‐1α, TFAM, PARK2 and https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2161 proteins in HK2s. Data are shown as mean ± SEM. n = 5 per group, * P < .05 versus control, # P < .05 versus H/R. (e) The protein levels of FOXO1, PGC‐1α, PARK2, https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2161, DRP1 and MFN2 in HK2s transfected with Ad‐FOXO1 were monitored by Western blot analysis. Data are shown as mean ± SEM. n = 5 per group, * P < .05 versus control. (f) Morphology of mitochondria in HK2s was detected by transmission electron microscopy, n = 5 per group. (g) Relative mitochondrial DNA abundance in renal cortex of different groups were analysed by qPCR. Data sre shoen as mean ± SEM. n = 5 per group, * P < .05 versus sham, # P < .05 versus I/R
To further confirm the regulation of FOXO1 on PGC‐1α, recombinant adenovirus overexpressing FOXO1 (i.e. Ad‐FOXO1) was used. The expression level of https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2161 was up‐regulated and PGC‐1α expression was down‐regulated in response to FOXO1 adenovirus infection (Figure 5e). In addition, Ad‐FOXO1 transfection suppressed the PGC‐1α mRNA expression (Figure 6a). FOXO1 inhibition with siRNA knockdown could prevent the decreased mRNA expression of PGC‐1α induced by H/R (Figure 6b), indicating that FOXO1 could inhibit the transcription of PGC‐1α.
Figure 6.
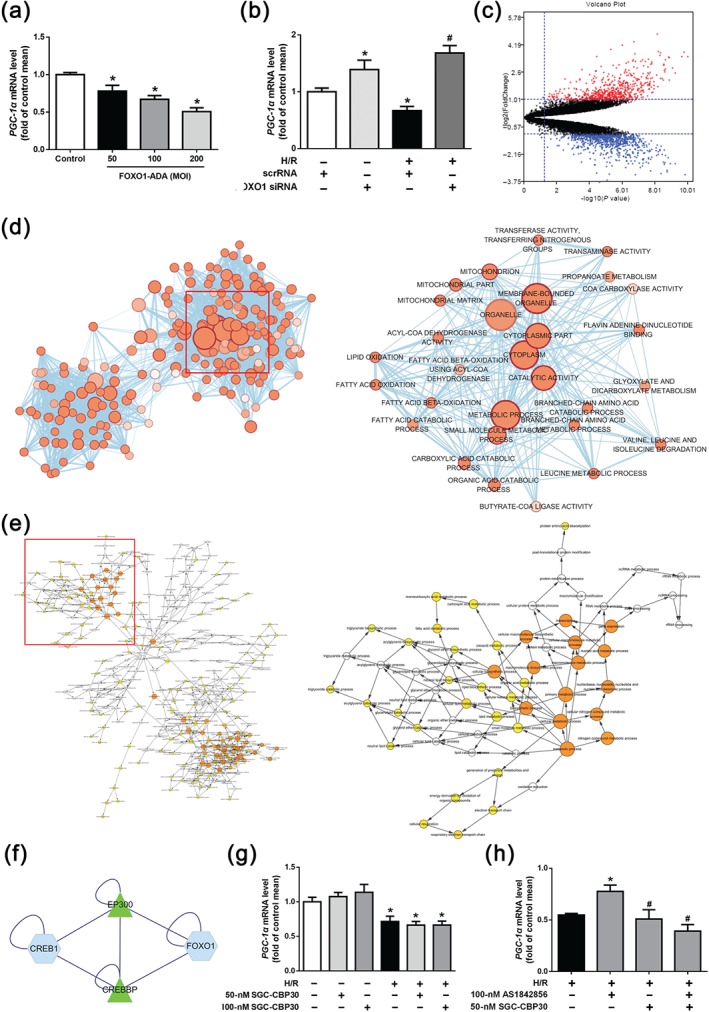
FOXO1 mediated mitochondrial biogenesis by regulating the transcription of PGC‐1α. (a) qPCR analysis of PGC‐1α mRNA level in HK2s treated with Ad‐FOXO1. Data are shown as mean ± SEM. n = 5 per group, * P < .05 versus control. (b) PGC‐1α mRNA level in HK2s under control or H/R condition treated with scrRNA or FOXO1 siRNA were monitored by qPCR. Data are shown as mean ± SEM. n = 5 per group, * P < .05 versus control, # P < .05 versus H/R. (c) Differential expression of genes in GSE52004 data. The up‐regulated genes which screened on the basis of|fold change| > 2.0 and a corrected P value <.05 were shown as red points. The blue points represent down‐regulated genes screened on the basis of|fold change| > 2.0 and a corrected P value <.05. The black points represent genes with no significant difference. (d) GO enrichment analysis of DEGs in renal I/R injury. (e) Significant pathway enrichment of DEGs which were related with FOXO1 and PGC‐1α. (f) PPI network between FOXO1 and CREB. (g) PGC‐1α mRNA level in HK2s under control or H/R condition treated with different concentrations of https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 inhibitor, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7529, were monitored by qPCR. Data are shown as mean ± SEM. n = 5 per group, * P < .05 versus control. (h) PGC‐1α mRNA level in HK2s under H/R condition treated with AS1842856 or https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7529 was monitored by qPCR. Data are shown as mean ± SEM. n = 5 per group, * P < .05 versus H/R, # P < .05 versus H/R with 100‐nM AS1842856
3.6. Identification of differentially expressed genes (DEGs) and signalling pathways in I/R by integrated bioinformatics analysis
It is well established that FOXO1 could bind to https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2161 with consequent up‐regulation of its expression (Baldelli et al., 2014), but the regulation between FOXO1 and PGC‐1α in HK2s has not been clearly demonstrated. Bioinformatics analysis was used to explore the important pathways in renal I/R injury and the regulation mechanism between FOXO1 and PGC‐1α. The profile of GSE52004 contained 45 samples, including 19 cases of sham samples and 26 cases of I/R samples exported from Gene Expression Omnibus Datasets. Samples from two groups were analysed by limma package (corrected P value <.05, log fold change >1) in SangerBox. After rank analysis, 1,015 DEGs were identified, with 404 up‐regulated genes and 611 down‐regulated genes (Figure 6c). Biological annotation of the DEGs in I/R identified from an integrated analysis of microarray data was performed by g:profiler online analysis tool and GO functional enrichments of up‐ and down‐regulated genes with a P value <.05 were obtained (Figure S1A). The enrichment results were mapped to construct a network by Cytoscape (Figure 6d). DEGs were mainly enriched in metabolic process, catalytic activity, cytoplasm and mitochondria‐related pathways, indicating that these pathways may play a major role in renal I/R injury. DEGs were submitted to KEGG analysis and visualized by Cytoscape. The signalling pathways of DEGs were mainly enriched in metabolic and mitochondria‐related pathways. This is similar to the results of GO enrichment analysis (Figure S1B). Using the STRING database (http://string-db.org) and Cytoscape, the DEGs expression products in renal I/R injury were filtered into the DEGs protein–protein interaction network complex containing 610 nodes and 1,555 edges (Figure S1C). Among the 610 nodes, 17 proteins interacted with FOXO1 were identified (Figure S1D) and further analysed utilizing Bingo plugin of Cytoscape. Pathway enrichment analysis showed that those 17 proteins were mainly associated with transcription process (Figure 6e). Results above indicated that FOXO1 and its related proteins may down‐regulate PGC‐1α in a transcriptional way. CREB (also known as CREB1) is an upstream transcription factor of PGC‐1α. It has been confirmed that phosphorylated CREB can initiate the transcription of PGC‐1α (Singh, Simpson, & Bennett, 2015), suggesting that FOXO1 may interact directly with CREB to regulate the transcription of PGC‐1α. To investigate the specific connection between FOXO1 and CREB, BisoGenet was used to construct a protein–protein interaction network. BisoGenet is a plugin of Cytoscape, used for visualization and analysis of biomolecular relationships (Martin et al., 2010). It showed that both FOXO1 and CREB could bind to https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737, a histone dimer in the nucleus (Figure 6f).
3.7. FOXO1 suppressed PGC‐1α transcription by competing with CREB to bind https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737
https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 is a pair of homologous histone acetyltransferases (HATs). They are known to interact with a broad range of transcription factors and function as transcriptional coactivators (Kalkhoven, 2004). It has been reported that https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 could interact with CREB to fully activate transcriptional initiation (Han et al., 2013). To further examine whether FOXO1 inhibited PGC‐1α transcription by competing with CREB for its binding to https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737, a selective inhibitor of https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7529 (Hammitzsch et al., 2015)was used. https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7529 did not affect the mRNA expression of PGC‐1α under normal or H/R conditions (Figure 6g). However, co‐incubation with https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7529 prevented FOXO1 inhibition‐induced increase of PGC‐1α mRNA expression in HK2s exposed to H/R (Figure 6h), indicating that https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 are involved in the regulation of FOXO1 on PGC‐1α. Furthermore, treatment with https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7529 suppressed the up‐regulation of MMP and decreased percentage of TUNEL‐positive cells by AS1842856 in response to H/R (Figure 7a,b).
Figure 7.
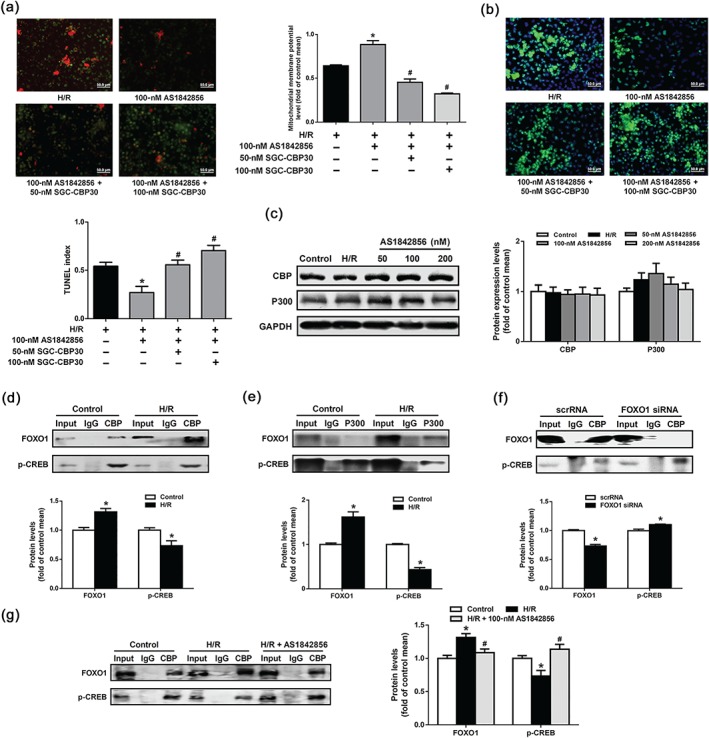
FOXO1 suppressed PGC‐1α transcription by competing with CREB to bind https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737. (a) Mitochondrial transmembrane potential levels in HK2s among groups. Data were expressed as mean ± SEM, n = 5 per group, * P < .05 versus H/R, # P < .05 versus H/R with 100‐nM AS1842856. (b) HK2s were subjected to TUNEL staining. Representative images of TUNEL staining in different groups. Scale bar: 50 μm. Data are shown as mean ± SEM, n = 5 per group, * P < .05 versus H/R, # P < .05 versus H/R with 100‐nM AS1842856. (c) https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734 and https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 protein levels in HK2s among groups were monitored by Western blot analysis. Data are presented as histograms. Data are shown as mean ± SEM. n = 6 per group. (d) Western blot analysis of a coimmunoprecipitation experiment in HK2s under control or H/R condition. The protein extracting solution of HK2s were immunoprecipitated with anti‐https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734 or IgG. FOXO1 and p‐CREB were visualized by Western blot analysis. Data are shown as mean ± SEM of five independent experiments. * P < .05 versus control. (e) Western blot analysis of a coimmunoprecipitation experiment in HK2s under control or H/R condition. The protein extracting solution of HK2s were immunoprecipitated with anti‐https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 or IgG. FOXO1 and p‐CREB were visualized by Western blot analysis. Data are shown as mean ± SEM of five independent experiments. * P < .05 versus control. (f) Western blot analysis of a coimmunoprecipitation experiment in HK2s under scrRNA and FOXO1 siRNA transfection. The protein extracting solution of HK2s were immunoprecipitated with anti‐https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734 or IgG. FOXO1 and p‐CREB were visualized by Western blot analysis. Data are shown as mean ± SEM of five independent experiments. * P < .05 versus scrRNA. (g) Western blot analysis of a coimmunoprecipitation experiment in HK2s under control‐, H/R‐, or AS1842856‐treated condition. The protein extracting solution of HK2s were immunoprecipitated with anti‐https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734 or IgG. FOXO1 and p‐CREB were visualized by Western blot analysis. Data are shown as mean ± SEM of five independent experiments. * P < .05 versus control, # P < .05 versus H/R with 100‐nM AS1842856
To further verify the competitive binding of FOXO1 and CREB to https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737, coimmunoprecipitation (CO‐IP) assays were used. H/R enhanced FOXO1 CO‐IP with https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734 or https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 and decreased phosphorylated CREB CO‐IP with https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734 or https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 (Figure 7d,e). Treatment with AS1842856 could significantly suppress FOXO1 CO‐IP with https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734 and enhance phosphorylated CREB CO‐IP with https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734 under H/R condition (Figure 7d,e). FOXO1 inhibition with siRNA knockdown could prevent FOXO1 CO‐IP with https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734 and increase phosphorylated CREB CO‐IP with https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734 (Figure 7f). These data indicate that FOXO1 decrease the transcription of PGC‐1α by competing with phosphorylated CREB for its binding to https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737. Furthermore, the expression levels of https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734, https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 and phosphorylated CREB were not significantly changed either in I/R mice and AS1842856 pre‐treated mice or in HK2 cells treated with H/R and AS1842856 (Figures S2D and 7c). This indicates that competitive binding of FOXO1 and CREB to https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 was the main reason of decreasing transcription of PGC‐1α.
4. DISCUSSION
The present study provides the first evidence that transcription factor FOXO1 is involved in the regulation of mitochondrial homeostasis in renal tubular epithelial cells. Selective inhibition of FOXO1 with AS1842856 can (a) prevent I/R‐induced renal injury and protect renal tubular epithelial cells from I/R‐induced apoptosis, (b) regulate mitochondrial homeostasis and improve mitochondrial function in renal tubular epithelial cells and (c) increase the transcription of PGC‐1α by reducing the competitive binding of FOXO1 and p‐CREB to https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737.
At present, four distinct isoforms of FOXO (FOXO1, FOXO3, FOXO4, and FOXO6) have been identified in mammals. All are found expressed in renal tissue except FOXO6, which is mainly restricted to brain and eye (Jacobs et al., 2003; Xin et al., 2018). In the present study, we provide evidence that I/R injury significantly enhanced renal FOXO1 mRNA and protein levels and decreased phosphorylation of FOXO1, whereas it had no effect on the mRNA levels of FOXO3 and FOXO4. Moreover, the t 1/2 of FOXO1 was decreased (Figure S2A) and protein level of FOXO1 was up‐regulated in HK2s exposed to H/R. The elevated protein levels were due to enhanced transcription of FOXO1. It has been reported that the protein expression of FOXO3 was attenuated in I/R renal tissue and or H/R HK2 cells (Tajima et al., 2019; Wu et al., 2016). Moreover, we only focused on FOXO1 function in renal I/R injury because AS1842856 is a selective inhibitor that blocks the transcription activity of FOXO1 (IC50: 33 nM; Nagashima et al., 2010). AS1842856 did not affect body weight, kidney and liver weight of mice (Table S3). Although the role of FOXO1 in renal I/R injury remains scant, FOXO1's involvements in I/R of other organs have been investigated. FOXO1 was up‐regulated in heart I/R injury of mice and it could increase sirtuin1 (https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2707) transcription by binding to its promoter region. https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2707 could deacetylate heart shock factor‐1 and promote heart shock proteins expression which inhibit I/R‐induced cardiomyocytes apoptosis (Cattelan et al., 2015). In hepatic I/R injury, increased FOXO1 expression and FOXO1 nuclear localization lead to severe liver cells apoptosis (Chen et al., 2017; Zhong et al., 2015). These findings suggest that FOXO1 serves as a critical mediator of I/R injury in various tissues.
Mitochondria is a dynamic organelle and supposed to be a potential therapeutic target for many diseases (Duann & Lin, 2017; Nunnari & Suomalainen, 2012; Yang et al., 2017). Mitochondrial dynamics and biogenesis are crucial processes underlying mitochondrial homeostasis (Mishra & Chan, 2016). It has been reported that impairment of mitochondrial homeostasis leads to the injury of renal tubular epithelial cells in I/R (Bhargava & Schnellmann, 2017). Our present results demonstrate that FOXO1 participates in mitophagy, biogenesis and mitochondria‐mediated apoptosis in I/R. We further provide evidence that FOXO1 is involved in the regulation of PGC‐1α, which is a key regulator of in mitochondrial biogenesis. The expression of PGC‐1α was primarily decreased at the early stage of I/R. Enforced overexpression of PGC‐1α in cultured proximal tubular epithelial cells compensate for the loss of mitochondrial number, respiratory capacity and mitochondrial proteins (Rasbach & Schnellmann, 2007; Wegrzyn et al., 2009; Zhang et al., 2007). Tran et al. (2016) developed an inducible tubular epithelial transgenic mouse model (iNephPGC1α), in which PGC‐1α was overexpressed specifically in tubular epithelial cells. They found that iNephPGC1α could protect against renal I/R injury, potentiate survival rates, preserve renal function and suppress tubular injury with increased mitochondrial abundance (Tran et al., 2016). However, the relation between FOXO1 and PGC‐1α remains unclear. It has been reported that FOXO1 could directly bind to the PGC‐1α promoter and trigger the transcription of PGC‐1α in liver and muscle tissues (Ropelle et al., 2009; Shute, Heesch, Zak, Kreiling, & Slivka, 2018). But up‐regulated FOXO1 was accompanied by a reduction of PGC‐1α, while PGC‐1α overexpression suppressed the activation of FOXO1 and FOXO3 in muscle (Kang & Ji, 2016). Hepatic PGC‐1α facilitated gluconeogenesis through multiple pathways served as a co‐activator for FOXO1 (Lee et al., 2017). Here, our results showed that elevated FOXO1 reduced PGC‐1α mRNA and protein levels, while FOXO1 inhibition increased transcription of PGC‐1α both in vivo and in vitro. The regulation between FOXO1 and PGC‐1α is complex between various tissue types and under different pathological conditions (Babu, Liu, & Gilbert, 2013; Mu et al., 2017).
It has been suggested that CREB regulates PGC‐1α transcription through interaction with https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 in the nucleus (Hussain et al., 2006; Rahnert, Zheng, Hudson, Woodworth‐Hobbs, & Price, 2016; Rui, 2014). https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734 and its paralogue https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 modulate locus‐specific transcription by separate mechanisms (Giotopoulos et al., 2016). https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 act as cofactors to modulate FOXO1 DNA‐binding capabilities and FOXO1‐mediated transcription (Senf, Sandesara, Reed, & Judge, 2011; van der Heide & Smidt, 2005). Here, we provide evidence that https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7529 elevated the mRNA expression of FOXO1 under normal condition and had no effect on the FOXO1 mRNA levels in HK2s exposed to H/R (Figure S2B). Neither https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7529 nor AS1842856 had effect on the mRNA expression of FOXO1 under H/R condition (Figure S2C). This observation is consistent with the fact that https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 participates in transcriptional regulation of FOXO‐1. FOXO1 competes with phosphorylated CREB for binding to https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 under H/R, resulting in the decrease of mitochondrial biogenesis by suppressing PGC‐1α expression. It has been reported that CREB regulated transcription coactivator 2 (CRTC2), which could also bind to https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 and form a transcriptional complex to promote transcription level of PGC‐1α (Rui, 2014). However, Rahnert and his colleagues reported that CRTC2 was not a central role for PGC‐1α transcription. Overexpression of CRTC2 was not sufficient to prevent the decrease in PGC‐1α mRNA or protein induced by muscle wasting (Rahnert et al., 2016). In the present study, we showed that CRTC2 did not change after I/R and FOXO1 inhibition decreased mRNA levels of CRTC2 under I/R (Figure S2B), indicating that CRTC2 had no significant effect on the transcription of PGC‐1α in I/R. Serving as histone acetyltransferase (HATs), https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734&https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 have a catalytic HAT domain and could acetylate some transcription factors (Wang, Marshall, & Ikura, 2013). https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737 could influence transcription of FOXO1 by regulation of its acetylation (Mortuza, Chen, Feng, Sen, & Chakrabarti, 2013; Senf et al., 2011). However, our current data did not address whether acetylated FOXO1 was involved in the process of renal I/R injury. It has been reported that acetylated FOXO1 is up‐regulated in I/R injury of different organs and results in an activation of BAX‐mediated apoptosis. In support of this possibility, https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2707‐dependent deacetylation of FOXO1 has been reported to protect from I/R injury (Cheng et al., 2016; Hsu et al., 2010; Pantazi et al., 2015). Therefore, further study is needed to elucidate the role of https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737‐mediated regulation of FOXO1 acetylation in renal I/R injury.
There is accumulating evidence that renal I/R injury could induce accumulation of long‐chain‐free fatty acids, long‐chain acylcarnitines, triglycerides and cholesterol. PPAR α and PPAR γ (PPARG) play an important role in the process (Corrales, Izquierdo‐Lahuerta, & Medina‐Gómez, 2018). Though we found that H/R could increase the transcription of PPARG and FOXO1, inhibition could affect the transcription of PPAR α and PPARG (Figure S2D). There was no significant changes in blood triglycerides and blood cholesterol in I/R group, compared to sham group (Figure S2A). I/R did not induce lipid accumulation in liver (Figure S2C). Results above indicate that lipid metabolism was not a major factor in the development of renal I/R injury. So we did not focus on the regulation between FOXO1 and PPARs in the present study.
In conclusion, the results of our study demonstrated that FOXO1 mediated alteration of mitochondrial homeostasis induced by I/R in renal tubular epithelial cells, at least in part, was by the p‐CREB‐https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2734/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2737‐PGC‐1α pathway. FOXO1 inhibition could attenuate mitochondrial dysfunction, ameliorate I/R‐induced renal injury and increase resistance of tubular epithelial cells to apoptosis (Figure 8). FOXO1 might serve as a potential target for the prevention and treatment of acute kidney injury.
Figure 8.
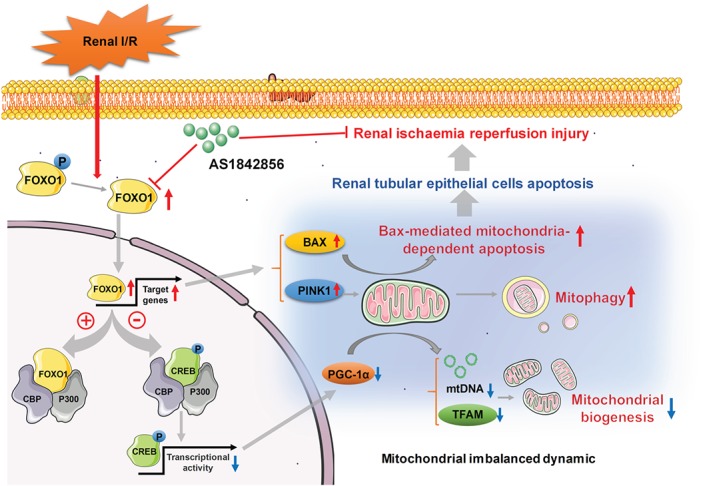
Schematic illustration of possible mechanisms contributing to mitochondrial dynamics regulated by FOXO1 in renal tubular epithelial cells. FOXO1 inhibition could suppress mitophagy and BAX‐mediated mitochondria‐dependent apoptosis, improve mitochondrial biogenesis and ameliorate impairment of I/R‐induced renal tubular injury in C57BL/6 male mice
4.1. Significance statement
Renal I/R injury‐induced acute kidney injury is associated with high morbidity and mortality and a lack of effective pharmacological treatment. Growing evidence indicates targeting mitochondrial dynamics and biogenesis could accelerate recovery from I/R injury, but the underlying mechanisms remain elusive. FOXO1, a member of the forkhead family of transcription factors essential for cell viability and metabolism, is considered to be critical for maintaining mitochondrial homeostasis. The present study found that FOXO1 suppresses mitochondrial biogenesis through inhibiting PGC‐1α transcription by competing CREB with its binding to CBP/P300, which is a new finding for FOXO1. Further, inhibition of FOXO1 by AS1842856 attenuated mitochondrial dysfunction, ameliorate I/R‐induced renal injury and increased resistance of tubular epithelial cells to apoptosis, indicating that FOXO1 may serve as a therapeutic target for pharmacological intervention in renal I/R injury.
AUTHOR CONTRIBUTIONS
L.T. designed research. D.W., Y.‐Q.W., and Y.‐D.S. performed experiments. D.W., X.‐T.Z., T.‐R.H.‐Y., and J.S. analysed the data. L.T., D.W., and Q.L. made the figures. L.T., Q.W., F.‐X.Z., and X.‐J.L. drafted and revised the paper. All authors approved the final version of the manuscript.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for https://bpspubs.onlinelibrary.wiley.com/doi/full/10.1111/bph.14207, https://bpspubs.onlinelibrary.wiley.com/doi/full/10.1111/bph.14208 and https://bpspubs.onlinelibrary.wiley.com/doi/full/10.1111/bph.14206, and as recommended by funding agencies, publishers and other organizations engaged with supporting research.
Supporting information
Table S1. Primer sequence used in the present study.
Table S2. The primary antibodies used in Western blot analysis.
Table S3. General characteristics of mice following AS1842856 treatment.
Figure S1. Identification of differentially expressed genes and signaling pathways in renal I/R injury by integrated bioinformatics analysis. (A) GO enrichment significance items of up‐regulated genes and down‐regulated genes in different functional groups. (B) KEGG pathway analysis of DEGs in renal I/R injury. The result was visualized by Cytoscape. Note: green arrows represented the signaling pathways, red circles represented up‐regulated genes, blue circles represented down‐regulated genes. (C) PPI network, analyzed by STRING database and visualized by Cytoscape. Note: Circles represented DEGs expression products, lines represented the interaction of proteins which were expressed by DEGs. Red circles represented up‐regulated proteins, blue circles represented down‐regulated proteins. (D) Identification of 17 proteins from PPI network which were interacted with FOXO1.
Figure S2. The interaction among FOXO1, CBP/P300 and CREB. (A) Cells were incubated with 100 μΜ CHX. At different time points after CHX‐treated, cells were harvested and the protein level of FOXO1 were detected by Western blot. Data were shown as line graphs. Data were expressed as mean ± SEM. n=5 for each time period. *P < 0.05 vs. control. (B) FOXO1 mRNA levels in HK2s under control or H/R condition treated with different concentrations of CBP/P300 inhibitor, SGC‐CBP30, were monitored by Quantified Real‐time PCR. Data were expressed as mean ± SEM. n=5 per group, *P < 0.05 vs. control. (C) FOXO1 mRNA level in HK2s under H/R condition treated with AS1842856 and SGC‐CBP30, were monitored by Quantified Real‐time PCR. Data were expressed as mean ± SEM. n=5 per group. (D) CBP, P300, phosphorylated‐CREB protein levels among groups in renal cortex were monitored by Western blot analysis. Statistical data were shown as histograms. Data were expressed as mean ± SEM. n=6 per group. (E) Western blot analysis of a co‐immunoprecipitation experiment in HK2s under control condition treated with or without CREB inhibitor, 666‐15 (20 μΜ). The protein extracting solution of HK2s were immunoprecipitated with anti‐CBP or IgG. FOXO1 and p‐CREB were visualized by Western blotting. Data were expressed as mean ± SEM of five independent experiments. *P < 0.05 vs. control.
Figure S3. Effects of FOXO1 inhibition on lipid metabolism. (A) Mice were intraperitoneally administered with corresponding vehicle or AS1842846 (10 mg/kg) daily for 7 days before surgery. Blood TG and blood CHO levels were monitored after reperfusion for 24 hours. Data were expressed as mean ± SEM. n=5 for each group. *P<0.05 vs. I/R. (B) Representative images of liver tissue with H & E staining in sham, I/R and AS1842856 pre‐treated mice after reperfusion for 24 hours (magnification 400×). n=5 per group. (C) Quantified Real‐time PCR analysis of CRTC2 mRNA level in renal cortex in sham, I/R and AS1842856 pre‐treated mice after reperfusion for 24 hours. Data were expressed as mean ± SEM. n=5 for each group. *P<0.05 vs. I/R. (D) Quantified Real‐time PCR analysis of PPARA and PPARG mRNA levels in HK2s in different groups. Data were expressed as mean ± SEM, n=5 per group. *P < 0.05 vs. control, #P<0.05 vs. H/R.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (Grants 81673486, 81974506, 81373405, and 30901803 to L.T., Grants 81874318, 81673453, and 81473235 to X.‐J.L.), Beijing Natural Science Foundation (Grant 7172119), Beijing Higher Education Young Elite Teacher Project(Grant YETP0053), Beijing Golden Bridge Seed Capital Project (Grant ZZ16019), Leading Academic Discipline Project of Beijing Education Bureau (Grant BMU20110254), and the Fund of Janssen Research Council China (Grant JRCC2011).
Wang D, Wang Y, Zou X, et al. FOXO1 inhibition prevents renal ischemia–reperfusion injury via cAMP‐response element binding protein/PPAR‐γ coactivator‐1α‐mediated mitochondrial biogenesis. Br J Pharmacol. 2020;177:432–448. 10.1111/bph.14878
REFERENCES
- Alexander, S. P. H. , Roberts, R. E. , Broughton, B. R. S. , Sobey, C. G. , George, C. H. , Stanford, S. C. , … Ahluwalia, A. (2017). Goals and practicalities of immunoblotting and immunohistochemistry: A guide for submission to the British Journal of Pharmacology. British Journal of Pharmacology, 3, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade‐Oliveira, V. , Amano, M. T. , Correa‐Costa, M. , Castoldi, A. , Felizardo, R. J. F. , de Almeida, D. C. , … Câmara, N. O. (2015). Gut bacteria products prevent AKI induced by ischemia‐reperfusion. J Am Soc Nephrol, 26, 1877–1888. 10.1681/ASN.2014030288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu, P. V. A. , Liu, D. , & Gilbert, E. R. (2013). Recent advances in understanding the anti‐diabetic actions of dietary flavonoids. The Journal of Nutritional Biochemistry, 24, 1777–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldelli, S. , Aquilano, K. , & Ciriolo, M. R. (2014). PGC‐1α buffers ROS‐mediated removal of mitochondria during myogenesis. Cell Death & Disease, 5, e1515–e1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava, P. , & Schnellmann, R. G. (2017). Mitochondrial energetics in the kidney. Nature Reviews. Nephrology, 13, 629–646. 10.1038/nrneph.2017.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattelan, A. , Ceolotto, G. , Bova, S. , Albiero, M. , Kuppusamy, M. , De Martin, S. , … Avogaro, A. (2015). NAD+‐dependent SIRT1 deactivation has a key role on ischemia‐reperfusion‐induced apoptosis. Vascular Pharmacology, 70, 35–44. 10.1016/j.vph.2015.02.004 [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Wang, J. , Yang, W. , Chen, J. , Meng, Y. , Geng, B. , … Yang, J. (2017). FAM3A mediates PPARγ's protection in liver ischemia‐reperfusion injury by activating Akt survival pathway and repressing inflammation and oxidative stress. Oncotarget, 8, 49882–49896. 10.18632/oncotarget.17805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Di, S. , Fan, C. , Cai, L. , Gao, C. , Jiang, P. , … Yang, Y. (2016). SIRT1 activation by pterostilbene attenuates the skeletal muscle oxidative stress injury and mitochondrial dysfunction induced by ischemia reperfusion injury. Apoptosis, 21, 905–916. 10.1007/s10495-016-1258-x [DOI] [PubMed] [Google Scholar]
- Corrales, P. , Izquierdo‐Lahuerta, A. , & Medina‐Gómez, G. (2018). Maintenance of kidney metabolic homeostasis by PPAR γ. International Journal of Molecular Sciences, 19, E2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M. J. , Alexander, S. , Cirino, G. , Docherty, J. R. , George, C. H. , Giembycz, M. A. , … Ahluwalia, A. (2018). Experimental design and analysis and their reporting II: Updated and simplified guidance for authors and peer reviewers. British Journal of Pharmacology, 7, 987–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duann, P. , & Lin, P. H. (2017). Mitochondria damage and kidney disease. Advances in Experimental Medicine and Biology, 982, 529–551. 10.1007/978-3-319-55330-6_27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emma, F. , Montini, G. , Parikh, S. M. , & Salviati, L. (2016). Mitochondrial dysfunction in inherited renal disease and acute kidney injury. Nature Reviews. Nephrology, 12, 267–280. 10.1038/nrneph.2015.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenbach, D. A. , & Bonventre, J. V. (2015). Mechanisms of maladaptive repair after AKI leading to accelerated kidney ageing and CKD. Nature Reviews. Nephrology, 11, 264–276. 10.1038/nrneph.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giotopoulos, G. , Chan, W. I. , Horton, S. J. , Ruau, D. , Gallipoli, P. , Fowler, A. , … Huntly, B. J. (2016). The epigenetic regulators CBP and p300 facilitate leukemogenesis and represent therapeutic targets in acute myeloid leukemia. Oncogene, 35, 279–289. 10.1038/onc.2015.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grams, M. E. , & Rabb, H. (2012). The distant organ effects of acute kidney injury. Kidney International, 81, 942–948. 10.1038/ki.2011.241 [DOI] [PubMed] [Google Scholar]
- van der Heide, L. P. , & Smidt, M. P. (2005). Regulation of FoxO activity by CBP/p300‐mediated acetylation. Trends in Biochemical Sciences, 30, 81–86. 10.1016/j.tibs.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Hammitzsch A, Tallant C, Fedorov O, et al. (2015). CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. Proceedings of the National Academy of Sciences, 201501956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Du Y, Song C, et al. (2013) Degradation of CREB‐binding protein and modulation of type I interferon induction by the zinc finger motif of the porcine reproductive and respiratory syndrome virus nsp1α subunit. Virus Research, 172(1–2: 54–65. [DOI] [PubMed] [Google Scholar]
- Harding S D, Sharman J L, Elena F, et al. (2018). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Research, 46(D1): D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, C. P. , Zhai, P. , Yamamoto, T. , Maejima, Y. , Matsushima, S. , Hariharan, N. , … Sadoshima, J. (2010). Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation, 122, 2170–2182. 10.1161/CIRCULATIONAHA.110.958033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, M. A. , Porras, D. L. , Rowe, M. H. , West, J. R. , Song, W. J. , Schreiber, W. E. , & Wondisford, F. E. (2006). Increased pancreatic beta‐cell proliferation mediated by CREB binding protein gene activation. Molecular and Cellular Biology, 26, 7747–7759. 10.1128/MCB.02353-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, F. M. , van der Heide, L. P. , Wijchers, P. J. , Burbach, J. P. , Hoekman, M. F. , & Smidt, M. P. (2003). FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. The Journal of Biological Chemistry, 278, 35959–35967. 10.1074/jbc.M302804200 [DOI] [PubMed] [Google Scholar]
- Kalkhoven, E. (2004). CBP and p300: HATs for different occasions. Biochemical Pharmacology, 68, 1145–1155. 10.1016/j.bcp.2004.03.045 [DOI] [PubMed] [Google Scholar]
- Kang, C. , & Ji, L. L. (2016). PGC‐1α overexpression via local transfection attenuates mitophagy pathway in muscle disuse atrophy. Free Radical Biology & Medicine, 93, 32–40. 10.1016/j.freeradbiomed.2015.12.032 [DOI] [PubMed] [Google Scholar]
- Khwaja, A. (2012). KDIGO clinical practice guidelines for acute kidney injury. Nephron. Clinical Practice, 120, c179–c184. 10.1159/000339789 [DOI] [PubMed] [Google Scholar]
- Kilkenny, C. , Browne, W. , Cuthill, I. C. , Emerson, M. , & Altman, D. G. (2010). Animal research: Reporting in vivo experiments: The ARRIVE guidelines. British Journal of Pharmacology, 160(7), 1577–1579. 10.1111/j.1476-5381.2010.00872.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Salazar Hernández, M. A. , Auen, T. , Mucka, P. , Lee, J. , & Ozcan, U. (2017). PGC‐1α functions as a co‐suppressor of XBP1s to regulate glucose metabolism. Molecular Metabolism, 7, 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , & Hajnóczky, G. (2011). Altered fusion dynamics underlie unique morphological changes in mitochondria during hypoxia‐reoxygenation stress. Cell Death and Differentiation, 18, 1561–1572. 10.1038/cdd.2011.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, A. , Ochagavia, M. E. , Rabasa, L. C. , Miranda, J. , Fernandez‐de‐Cossio, J. , & Bringas, R. (2010). BisoGenet: A new tool for gene net work building, visualization and analysis. BMC Bioinformatics, 11, 91–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurley, A. , Alimperti, S. , Campos‐Bilderback, S. B. , Sandoval, R. M. , Calvino, J. E. , Reynolds, T. L. , … Crackower, M. A. (2017). Inhibition of αvβ5 integrin attenuates vascular permeability and protects against renal ischemia‐reperfusion injury. Journal of the American Society of Nephrology, 28, 1741–1752. 10.1681/ASN.2016020200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, P. , & Chan, D. C. (2016). Metabolic regulation of mitochondrial dynamics. The Journal of Cell Biology, 212, 379–387. 10.1083/jcb.201511036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortuza, R. , Chen, S. , Feng, B. , Sen, S. , & Chakrabarti, S. (2013). High glucose induced alteration of SIRTs in endothelial cells causes rapid aging in a p300 and FOXO regulated pathway. PLoS ONE, 8, e54514–e54514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu, X. , Qi, W. , Liu, Y. , Zhou, J. , Li, Y. , Rong, X. , & Lu, L. (2017). IGF‐II‐mediated downregulation of peroxisome proliferator‐activated receptor‐γ coactivator‐1α in myoblast cells involves PI3K/Akt/FoxO1 signaling pathway. Molecular and Cellular Biochemistry, 432, 199–208. 10.1007/s11010-017-3010-4 [DOI] [PubMed] [Google Scholar]
- Nagashima, T. , Shigematsu, N. , Maruki, R. , Urano, Y. , Tanaka, H. , Shimaya, A. , … Shibasaki, M. (2010). Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: Improvement of fasting glycemia in diabetic db/db mice. Molecular Pharmacology, 78, 961–970. [DOI] [PubMed] [Google Scholar]
- Nunnari, J. , & Suomalainen, A. (2012). Mitochondria: In sickness and in health. Cell, 148, 1145–1159. 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazi, E. , Zaouali, M. A. , Bejaoui, M. , Folch‐Puy, E. , Ben Abdennebi, H. , Varela, A. T. , … Roselló‐Catafau, J. (2015). Sirtuin 1 in rat orthotopic liver transplantation: an IGL‐1 preservation solution approach. World Journal of Gastroenterology, 21, 1765–1774. 10.3748/wjg.v21.i6.1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahnert, J. A. , Zheng, B. , Hudson, M. B. , Woodworth‐Hobbs, M. E. , & Price, S. R. (2016). Glucocorticoids alter CRTC‐CREB signaling in muscle cells: Impact on PGC‐1α expression and atrophy markers. PLoS ONE, 11, e0159181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasbach, K. A. , & Schnellmann, R. G. (2007). Signaling of mitochondrial biogenesis following oxidant injury. The Journal of Biological Chemistry, 282, 2355–2362. 10.1074/jbc.M608009200 [DOI] [PubMed] [Google Scholar]
- Ropelle, E. R. , Pauli, J. R. , Cintra, D. E. , Frederico, M. J. , de Pinho, R. A. , Velloso, L. A. , & de Souza, C. T. (2009). Acute exercise modulates the Foxo1/PGC‐1α pathway in the liver of diet‐induced obesity rats. The Journal of Physiology, 587, 2069–2076. 10.1113/jphysiol.2008.164202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui, L. (2014). Energy metabolism in the liver. Comprehensive Physiology, 4, 177–197. 10.1002/cphy.c130024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, A. M. , Candau, R. B. , & Bernardi, H. (2014). FoxO transcription factors: Their roles in the maintenance of skeletal muscle homeostasis. Cellular and Molecular Life Sciences, 71, 1657–1671. 10.1007/s00018-013-1513-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senf, S. M. , Sandesara, P. B. , Reed, S. A. , & Judge, A. R. (2011). p300 Acetyltransferase activity differentially regulates the localization and activity of the FOXO homologues in skeletal muscle. American Journal of Physiology. Cell Physiology, 300, C1490–C1501. 10.1152/ajpcell.00255.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , Fan, S. , Wang, D. , Huyan, T. , Chen, J. , Chen, J. , … Tie, L. (2018). FOXO1 inhibition potentiates endothelial angiogenic functions in diabetes via suppression of ROCK1/Drp1‐mediated mitochondrial fission. Biochimica et Biophysica Acta ‐ Molecular Basis of Disease, 1864, 2481–2494. 10.1016/j.bbadis.2018.04.005 [DOI] [PubMed] [Google Scholar]
- Shukla, S. , Rizvi, F. , Raisuddin, S. , & Kakkar, P. (2014). FoxO proteins' nuclear retention and BH3‐only protein Bim induction evoke mitochondrial dysfunction‐mediated apoptosis in berberine‐treated HepG2 cells. Free Radical Biology & Medicine, 76, 185–199. 10.1016/j.freeradbiomed.2014.07.039 [DOI] [PubMed] [Google Scholar]
- Shute, R. J. , Heesch, M. W. , Zak, R. B. , Kreiling, J. L. , & Slivka, D. R. (2018). Effects of exercise in a cold environment on transcriptional control of PGC‐1α. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 314, R850–R857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. , Simpson, R. L. , & Bennett, R. G. (2015). Relaxin activates peroxisome proliferator‐activated receptor γ (PPARγ) through a pathway involving PPARγ coactivator 1α (PGC1α). The Journal of Biological Chemistry, 290, 950–959. 10.1074/jbc.M114.589325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, T. , Yoshifuji, A. , Matsui, A. , Itoh, T. , Uchiyama, K. , Kanda, T. , … Itoh, H. (2019). β‐hydroxybutyrate attenuates renal ischemia‐reperfusion injury through its anti‐pyroptotic effects. Kidney International, 95, 1120–1137. 10.1016/j.kint.2018.11.034 [DOI] [PubMed] [Google Scholar]
- Tran, M. , & Parikh, S. M. (2014). Mitochondrial biogenesis in the acutely injured kidney. Nephron. Clinical Practice, 127, 42–45. 10.1159/000363715 [DOI] [PubMed] [Google Scholar]
- Tran, M. T. , Zsengeller, Z. K. , Berg, A. H. , Khankin, E. V. , Bhasin, M. K. , Kim, W. , … Parikh, S. M. (2016). PGC1α drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature, 531, 528–532. 10.1038/nature17184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Marshall, C. B. , & Ikura, M. (2013). Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: Structural and functional versatility in target recognition. Cellular and Molecular Life Sciences, 70, 3989–4008. 10.1007/s00018-012-1254-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn, J. , Potla, R. , Chwae, Y.‐J. , Sepuri, N. B. V. , Zhang, Q. , Koeck, T. , … Larner, A. C. (2009). Function of mitochondrial Stat3 in cellular respiration. Science, 323, 793–797. 10.1126/science.1164551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg, J. M. (2011). Mitochondrial biogenesis in kidney disease. Journal of the American Society of Nephrology, 22, 431–436. [DOI] [PubMed] [Google Scholar]
- Wu, H. , Huang, T. , Ying, L. , Han, C. , Li, D. , Xu, Y. , … Dong, Z. (2016). MiR‐155 is involved in renal ischemia‐reperfusion injury via direct targeting of FoxO3a and regulating renal tubular cell pyroptosis. Cellular Physiology and Biochemistry, 40, 1692–1705. 10.1159/000453218 [DOI] [PubMed] [Google Scholar]
- Xin, Z. , Ma, Z. , Hu, W. , Jiang, S. , Yang, Z. , Li, T. , … Yang, Y. (2018). FOXO1/3: Potential suppressors of fibrosis. Ageing Research Reviews, 41, 42–52. 10.1016/j.arr.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Yang, S. , Han, Y. , Liu, J. , Song, P. , Xu, X. , Zhao, L. , … Sun, L. (2017). Mitochondria: A novel therapeutic target in diabetic nephropathy. Current Medicinal Chemistry, 24, 3185–3202. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Song, M. , Liu, Y. , Liu, H. , Sun, L. , Peng, Y. , … Dong, Z. (2016). Renoprotective approaches and strategies in acute kidney injury. Pharmacology & Therapeutics, 163, 58–73. 10.1016/j.pharmthera.2016.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, M. , Brooks, C. , Liu, F. , Sun, L. , & Dong, Z. (2013). Mitochondrial dynamics: Regulatory mechanisms and emerging role in renal pathophysiology. Kidney International, 83, 568–581. 10.1038/ki.2012.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Gao, P. , Fukuda, R. , Kumar, G. , Krishnamachary, B. , Zeller, K. I. , … Semenza, G. L. (2007). HIF‐1 inhibits mitochondrial biogenesis and cellular respiration in VHL‐deficient renal cell carcinoma by repression of C‐MYC activity. Cancer Cell, 11, 407–420. 10.1016/j.ccr.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Raoof, M. , Chen, Y. , Sumi, Y. , Sursal, T. , Junger, W. , … Hauser, C. J. (2010). Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature, 464, 104–107. 10.1038/nature08780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. Q. , Dong, J. J. , Cai, T. , Shen, X. , Zhou, X. J. , & Liao, L. (2017). High glucose induces apoptosis via upregulation of Bim expression in proximal tubule epithelial cells. Oncotarget, 8, 24119–24129. 10.18632/oncotarget.15491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C. , Pu, L. Y. , Fang, M. M. , Gu, Z. , Rao, J. H. , & Wang, X. H. (2015). Retinoic acid receptor α promotes autophagy to alleviate liver ischemia and reperfusion injury. World Journal of Gastroenterology, 21, 12381–12391. 10.3748/wjg.v21.i43.12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequence used in the present study.
Table S2. The primary antibodies used in Western blot analysis.
Table S3. General characteristics of mice following AS1842856 treatment.
Figure S1. Identification of differentially expressed genes and signaling pathways in renal I/R injury by integrated bioinformatics analysis. (A) GO enrichment significance items of up‐regulated genes and down‐regulated genes in different functional groups. (B) KEGG pathway analysis of DEGs in renal I/R injury. The result was visualized by Cytoscape. Note: green arrows represented the signaling pathways, red circles represented up‐regulated genes, blue circles represented down‐regulated genes. (C) PPI network, analyzed by STRING database and visualized by Cytoscape. Note: Circles represented DEGs expression products, lines represented the interaction of proteins which were expressed by DEGs. Red circles represented up‐regulated proteins, blue circles represented down‐regulated proteins. (D) Identification of 17 proteins from PPI network which were interacted with FOXO1.
Figure S2. The interaction among FOXO1, CBP/P300 and CREB. (A) Cells were incubated with 100 μΜ CHX. At different time points after CHX‐treated, cells were harvested and the protein level of FOXO1 were detected by Western blot. Data were shown as line graphs. Data were expressed as mean ± SEM. n=5 for each time period. *P < 0.05 vs. control. (B) FOXO1 mRNA levels in HK2s under control or H/R condition treated with different concentrations of CBP/P300 inhibitor, SGC‐CBP30, were monitored by Quantified Real‐time PCR. Data were expressed as mean ± SEM. n=5 per group, *P < 0.05 vs. control. (C) FOXO1 mRNA level in HK2s under H/R condition treated with AS1842856 and SGC‐CBP30, were monitored by Quantified Real‐time PCR. Data were expressed as mean ± SEM. n=5 per group. (D) CBP, P300, phosphorylated‐CREB protein levels among groups in renal cortex were monitored by Western blot analysis. Statistical data were shown as histograms. Data were expressed as mean ± SEM. n=6 per group. (E) Western blot analysis of a co‐immunoprecipitation experiment in HK2s under control condition treated with or without CREB inhibitor, 666‐15 (20 μΜ). The protein extracting solution of HK2s were immunoprecipitated with anti‐CBP or IgG. FOXO1 and p‐CREB were visualized by Western blotting. Data were expressed as mean ± SEM of five independent experiments. *P < 0.05 vs. control.
Figure S3. Effects of FOXO1 inhibition on lipid metabolism. (A) Mice were intraperitoneally administered with corresponding vehicle or AS1842846 (10 mg/kg) daily for 7 days before surgery. Blood TG and blood CHO levels were monitored after reperfusion for 24 hours. Data were expressed as mean ± SEM. n=5 for each group. *P<0.05 vs. I/R. (B) Representative images of liver tissue with H & E staining in sham, I/R and AS1842856 pre‐treated mice after reperfusion for 24 hours (magnification 400×). n=5 per group. (C) Quantified Real‐time PCR analysis of CRTC2 mRNA level in renal cortex in sham, I/R and AS1842856 pre‐treated mice after reperfusion for 24 hours. Data were expressed as mean ± SEM. n=5 for each group. *P<0.05 vs. I/R. (D) Quantified Real‐time PCR analysis of PPARA and PPARG mRNA levels in HK2s in different groups. Data were expressed as mean ± SEM, n=5 per group. *P < 0.05 vs. control, #P<0.05 vs. H/R.


