Abstract
Background
Reduced folate carrier 1 (RFC1) gene is a candidate for susceptibility to nonsyndromic cleft lip with or without cleft palate (NSCL/P). Association between RFC1 A80G polymorphism and NSCL/P have been studied. The published results are conflicting.
Methods
A meta-analysis of the association between RFC1 A80G polymorphism and NSCL/P was carried out using Stata13.0. A systematic literature search was performed through the PubMed, EMBASE, the Cochrane Library, Web of Science, ScienceDirect, EBSCOhost, China Biology Medicine databases, China National Knowledge Infrastructure and the Wanfang databases. All relevant studies up to 9 September 2019 were identified.
Results
Nine case-control studies including 4,229 total participants (1,334 NSCL/P children, 1,515 healthy children, 656 mothers of the NSCL/P children, and 724 mothers of healthy control children) were included in this study. The meta-analysis revealed that two genetic models of RFC1 A80G polymorphism in NSCL/P children increased risk of NSCL/P: the homozygote model (GG vs. AA, OR =2.346, 95% CI: 1.127–4.884) and the recessive model (GG vs. AG + AA, OR =1.503, 95% CI: 1.049–2.152). Further sensitivity analysis indicated that the frequency of G allele and GG genotype in NSCL/P children was significantly higher than those in the control. However, there was no significant statistical differences after Bonferroni correction. Subgroup analyses indicated the presence of the association of all the model with NSCL/P risk in the Indian children. RFC1 A80G polymorphism in the maternal population of NSCL/P children was not significantly associated with children NSCL/P.
Conclusions
The RFC1 A80G polymorphism was a candidate for susceptibility to NSCL/P in the Indian pediatric population. More studies with larger samples are necessary to reach more conclusive outcomes.
Keywords: Reduced folate carrier 1 (RFC1), genetic polymorphism, nonsyndromic cleft lip, NSCL/P, meta-analysis
Introduction
Nonsyndromic cleft lip with or without cleft palate (NSCL/P), including cleft lip, cleft lip and palate, and cleft palate only (CPO), is a series of disorders affecting the lips and oral cavity (1). NSCL/P is the most common craniofacial malformation with an incidence of approximately 1/700–1,000 live births (2). Being a multifactorial disease, NSCL/P arises as a result of an interplay among remarkably variable factors, such as geographic origin, ethnicity, and socioeconomic status. Several studies have highlighted the role of folate/homocysteine pathway in preventing the risk of NSCL/P during early pregnancy and embryonic development (3), where folic acid and its derivatives play important roles in cell cycle regulation, DNA methylation, homocysteine remethylation, and “one carbon unit” transfer to purines and pyrimidines during DNA biosynthesis (4).
Reduced folate carrier 1 (RFC1) gene is considered a candidate for susceptibility to NSCL/P (1). This gene is also known as the solute carrier family gene 19 (SLC19A1) and mapped into 21q22.2-q22.3. The RFC1 gene encodes an integral membrane protein which delivers the metabolically active form of folate, the 5-methyltetrahydrofolate, into a variety of cells (5). Study by Mossey et al. indicated that RFC1 polymorphisms affected folate metabolism and maternal folate intake (1). The missense variation rs1051266 (A80G) in the exon 2 was proposed as a risk factor for NSCL/P. Conflicting results regarding association between allelic RFC1 A80G polymorphism and NSCL/P in diverse populations have been published (3,4,6,7). Kumari et al. found that RFC1 A80G polymorphism was associated with NSCL/P in the south India (6). Soghani et al. reported that the RFC1 A80G polymorphism was associated with the NSCL/P in Iranian population (8). In this study, we performed a meta-analysis to evaluate current evidence on the relationship between RFC1 A80G polymorphism and NSCL/P in NSCL/P children and their mothers.
Methods
This study is reported according to “Preferred Reporting Items for Systematic Reviews and Meta-analyses” (PRISMA) guidelines (Figure S1).
Figure S1.
PRISMA checklist.
Literature search
A systematic literature search was performed through the PubMed, EMBASE, the Cochrane Library, Web of Science, ScienceDirect, EBSCOhost, China Biology Medicine disc, China National Knowledge Infrastructure and the Wanfang databases. All relevant studies up to 9 September 2019 were identified. The medical subject headings (MeSH) “cleft lip”, “Cleft Palate”, “Orofacial Cleft 1”, “SLC19A1”, “Polymorphism, Single Nucleotide”, “Genotype, Alleles”, “Genetic Variation”, and the free-text words “Harelip”, “Orofacial Cleft”, “Cleft Lip with or without Cleft Palate”, “solute carrier family 19 member 1”, “folate transporter 1”, “IFC-1”, “RFC”, “intestinal folate carrier 1”, “placental folate transporter”, “reduced folate carrier protein”, “genetic polymorphism”, “genetic”, “genetic variant”, “genetic variants”, “SNP”, “mutation”, “variation”, “single nucleotide polymorphism”, “variant” were combined in search relevant literature. The full detailed search strategy and searching terms are shown in Table S1. Further, the search spectrum was expanded to the “related articles”. All retrieved studies were hand-searched and selected. Authors were contacted when necessary. No language restriction was imposed.
Table S1. The full detailed search strategy and searching terms.
| Search NO. | Query results | Items found |
|---|---|---|
| Search criterion of Medline (via PubMed, from inception to 9 September 2019) (n=19) | ||
| #18 | Search ((((((((((((((((("cleft lip"[MeSH Terms] OR “Cleft Palate”[MeSH Terms] OR "Orofacial Cleft 1" [Supplementary Concept])) OR ((“Cleft Lip”[All Fields] OR “Cleft Lips”[All Fields] OR Harelip[All Fields] OR Harelips[All Fields]))) OR ((Lip[All Fields] AND Cleft[All Fields]))) OR (Lips[All Fields] AND Cleft[All Fields])) OR (“Cleft Palate”[All Fields] OR “Cleft Palates” [All Fields])) OR (Palate[All Fields] AND Cleft[All Fields])) OR (Palates[All Fields] ] AND Cleft[All Fields])) OR (Cleft Palate[All Fields] AND Isolated[All Fields])) OR “Orofacial Cleft” [All Fields]) OR (Cleft Lip-Palate[All Fields] AND Nonsyndromic[All Fields])) OR (Cleft Lip Palate[All Fields] AND Nonsyndromic[All Fields])) OR (Cleft Lip with or without Cleft Palate[All Fields] AND Nonsyndromic[All Fields])) OR (Orofacial Cleft[All Fields] AND Nonsyndromic[All Fields])) OR OFC1[All Fields])) AND (SLC19A1/ OR CHMD/ OR FOLT/ OR IFC1/ OR REFC/ OR RFC1/ OR “folate transporter 1”/ OR IFC-1/ OR RFC/ OR “intestinal folate carrier 1”/ OR “placental folate transporter”/ OR “reduced folate carrier protein”/ OR “solute carrier family 19 member 1”/ OR “Reduced folate carrier”/)) AND ((((((((((((((((((((((("Polymorphism, Single Nucleotide" [Mesh]) OR "Genotype" [Mesh]) OR "Alleles" [Mesh]) OR polymorphism[Title/Abstract]) OR genetic variant[Title/Abstract]) OR genetic variants[Title/Abstract]) OR genetic polymorphism[Title/Abstract]) OR genetic[Title/Abstract]) OR genetic variant) OR genetic variants) OR "Genetic Variation" [Mesh]))))))) OR (((((SNP/) OR mutation/) OR variation/) OR single nucleotide polymorphism) OR variant/))))))) | 19 |
| #17 | Search (((((((((((((((((((((("Polymorphism, Single Nucleotide" [Mesh]) OR "Genotype" [Mesh]) OR "Alleles" [Mesh]) OR polymorphism[Title/Abstract]) OR genetic variant[Title/Abstract]) OR genetic variants[Title/Abstract]) OR genetic polymorphism[Title/Abstract]) OR genetic[Title/Abstract]) OR genetic variant) OR genetic variants) OR "Genetic Variation" [Mesh]))))))) OR (((((SNP/) OR mutation/) OR variation/) OR single nucleotide polymorphism) OR variant/)))))) | 1,901,876 |
| #16 | Search SLC19A1/ OR CHMD/ OR FOLT/ OR IFC1/ OR REFC/ OR RFC1/ OR “folate transporter 1”/ OR IFC-1/ OR RFC/ OR “intestinal folate carrier 1”/ OR “placental folate transporter”/ OR “reduced folate carrier protein”/ OR “solute carrier family 19 member 1”/ OR “Reduced folate carrier”/ | 2,736 |
| #15 | Search (((((((((((((((Cleft Lip[mesh]) OR "Orofacial Cleft 1" [Supplementary Concept]) OR ("cleft lip"[MeSH Terms] OR “Cleft Palate”[MeSH Terms] OR "Orofacial Cleft 1" [Supplementary Concept])) OR ((“Cleft Lip”[All Fields] OR “Cleft Lips”[All Fields] OR Harelip[All Fields] OR Harelips[All Fields]))) OR ((Lip[All Fields] AND Cleft[All Fields]))) OR (Lips[All Fields] AND Cleft[All Fields])) OR (“Cleft Palate”[All Fields] OR “Cleft Palates” [All Fields])) OR (Palate[All Fields] AND Cleft[All Fields])) OR (Palates[All Fields] AND Cleft[All Fields])) OR (Cleft Palate[All Fields] AND Isolated[All Fields])) OR “Orofacial Cleft” [All Fields]) OR (Cleft Lip-Palate[All Fields] AND Nonsyndromic[All Fields])) OR (Cleft Lip Palate[All Fields] AND Nonsyndromic[All Fields])) OR (Cleft Lip with or without Cleft Palate[All Fields] AND Nonsyndromic[All Fields])) OR (Orofacial Cleft[All Fields] AND Nonsyndromic[All Fields])) OR OFC1[All Fields] | 26,784 |
| #14 | Search OFC1[All Fields] | 70 |
| #13 | Search Orofacial Cleft[All Fields] AND Nonsyndromic[All Fields] | 69 |
| #12 | Search Cleft Lip with or without Cleft Palate[All Fields] AND Nonsyndromic[All Fields] | 878 |
| #11 | Search Cleft Lip Palate[All Fields] AND Nonsyndromic[All Fields] | 59 |
| #10 | Search Cleft Lip-Palate[All Fields] AND Nonsyndromic[All Fields] | 59 |
| #9 | Search “Orofacial Cleft” [All Fields] | 315 |
| #8 | Search Cleft Palate[All Fields] AND Isolated[All Fields] | 1,297 |
| #7 | Search Palates[All Fields] ] AND Cleft[All Fields] | 24,186 |
| #6 | Search Palate[All Fields] AND Cleft[All Fields] | 24,116 |
| #5 | Search “Cleft Palate”[All Fields] OR “Cleft Palates” [All Fields] | 21,173 |
| #4 | Search Lips[All Fields] AND Cleft[All Fields] | 16,321 |
| #3 | Search (Lip[All Fields] AND Cleft[All Fields]) | 16,456 |
| #2 | Search (“Cleft Lip”[All Fields] OR “Cleft Lips”[All Fields] OR Harelip[All Fields] OR Harelips[All Fields]) | 16,789 |
| #1 | Search "cleft lip"[MeSH Terms] OR “Cleft Palate”[MeSH Terms] OR "Orofacial Cleft 1" [Supplementary Concept] | 20,456 |
| Search criterion of Embase (from 1966 to 9 September 2019) (n=17) | ||
| #18 | #15 AND #16 AND #17 | 17 |
| #17 | 'polymorphism, single nucleotide'/exp OR 'polymorphism, single nucleotide' OR 'genotype'/exp OR 'genotype' OR 'alleles'/exp OR 'alleles' OR 'polymorphism':ab,ti OR 'genetic variant':ab,ti OR 'genetic variants':ab,ti OR 'genetic polymorphism':ab,ti AND 'genetic':ab,ti OR 'genetic variant':ab,ti OR 'genetic variants':ab,ti OR 'genetic variation'/exp OR 'genetic variation' OR 'snp' OR 'mutation'/exp OR 'mutation' OR 'variation' OR 'single nucleotide polymorphism'/exp OR 'single nucleotide polymorphism' OR 'variant' | 1,798,554 |
| #16 | SLC19A1/exp OR CHMD/exp OR FOLT/exp OR IFC1/exp OR REFC/exp OR RFC1/exp OR ‘folate transporter 1’/exp OR IFC-1/exp OR RFC/exp OR ‘intestinal folate carrier 1’/exp OR ‘placental folate transporter’/exp OR ‘reduced folate carrier protein’/exp OR ‘solute carrier family 19 member 1’/exp OR ‘Reduced folate carrier’/exp | 4,765 |
| #15 | #1 OR #2 OR #3 OR #4 OR #5 OR #6 OR #7 OR #8 OR #9 OR #10 OR #11 OR #12 OR #13 OR #14 | 34,321 |
| #14 | OFC1 | 22 |
| #13 | Orofacial Cleft AND Nonsyndromic | 189 |
| #12 | Cleft Lip with or without Cleft Palate AND Nonsyndromic | 712 |
| #11 | Cleft Lip Palate AND Nonsyndromic | 701 |
| #10 | Cleft Lip-Palate AND Nonsyndromic | 256 |
| #9 | 'Orofacial Cleft' | 359 |
| #8 | Cleft Palate AND Isolated | 1,687 |
| #7 | Palates AND Cleft | 1,167 |
| #6 | Palate AND Cleft | 30,399 |
| #5 | ‘Cleft Palate’ OR ‘Cleft Palates’ | 26,812 |
| #4 | Lips AND Cleft | 876 |
| #3 | (Lip AND Cleft) | 20,432 |
| #2 | (‘Cleft Lip’ OR ‘Cleft Lips’ OR Harelip OR Harelips) | 19,890 |
| #1 | ‘cleft lip’/de OR ‘Cleft Palate’/de OR ‘Orofacial Cleft 1’/de | 33,722 |
| Search criterion of Cochrane Library (Issue 5 of 12, September 2019) (n=0) | ||
| #1 | MeSH descriptor cleft lip | 22 |
| #2 | MeSH descriptor Cleft Palate | 31 |
| #3 | MeSH descriptor Orofacial Cleft 1 | 7 |
| #4 | Cleft Lip:ti,ab,kw or Cleft Lips:ti,ab,kw or Harelip:ti,ab,kw or Harelips:ti,ab,kw | 299 |
| #5 | Lip:ti,ab,kw and Cleft:ti,ab,kw | 296 |
| #6 | Lips:ti,ab,kw and Cleft:ti,ab,kw | 5 |
| #7 | Cleft Palate:ti,ab,kw or Cleft Palates:ti,ab,kw | 390 |
| #8 | Palate:ti,ab,kw and Cleft:ti,ab,kw | 386 |
| #9 | Palates:ti,ab,kw and Cleft:ti,ab,kw | 12 |
| #10 | Cleft Palate:ti,ab,kw and Isolated:ti,ab,kw | 14 |
| #11 | Orofacial Cleft:ti,ab,kw | 18 |
| #12 | Cleft Lip-Palate:ti,ab,kw and Nonsyndromic:ti,ab,kw | 2 |
| #13 | Cleft Lip Palate:ti,ab,kw and Nonsyndromic:ti,ab,kw | 10 |
| #14 | Cleft Lip with or without Cleft Palate:ti,ab,kw and Nonsyndromic:ti,ab,kw | 314 |
| #15 | Orofacial Cleft:ti,ab,kw and Nonsyndromic:ti,ab,kw | 1 |
| #16 | OFC1:ti,ab,kw | 0 |
| #17 | #1 or #2 or #3 or #4 or #5 or #6 or #7 or #8 or #9 or #10 or #11 or #12 or #13 or #14 or #15 o #16 | 514 |
| #18 | MeSH descriptor: [Polymorphism, Single Nucleotide] explode all trees | 1,345 |
| #19 | MeSH descriptor: [Alleles] explode all trees | 710 |
| #20 | MeSH descriptor: [Genotype] explode all trees | 4,739 |
| #21 | MeSH descriptor: [Genetic Variation] explode all trees | 5,388 |
| #22 | polymorphism:ti,ab,kw or genetic variant:ti,ab,kw or genetic variants:ti,ab,kw or genetic polymorphism:ti,ab,kw or genetic:ti,ab,kw or genetic variant:ti,ab,kw or genetic variants:ti,ab,kw | 10,215 |
| #23 | #18 or #19 or #20 or #21 or #22 | 12,971 |
| #24 | #17 and #23 | 9 |
| #25 | MeSH descriptor: [Reduced Folate Carrier Protein] explode all trees | 7 |
| #26 | SLC19A1:ti,ab,kw or CHMD:ti,ab,kw or FOLT:ti,ab,kw or IFC1:ti,ab,kw or REFC:ti,ab,kw or RFC1:ti,ab,kw or folate transporter 1:ti,ab,kw or IFC-1:ti,ab,kw or RFC:ti,ab,kw or intestinal folate carrier 1:ti,ab,kw or placental folate transporter:ti,ab,kw or reduced folate carrier protein:ti,ab,kw or solute carrier family 19 member 1:ti,ab,kw or Reduced folate carrier:ti,ab,kw | 56 |
| #27 | #25 or #26 | 56 |
| #28 | #24 and #27 | 0 |
| Search criterion of Web of Science (WOS) Database (from inception to September 2019) (n=39) | ||
| #1 | TS=((((((((((((((("cleft lip" OR “Cleft Palate” OR "Orofacial Cleft 1")) OR ((“Cleft Lip” OR “Cleft Lips” OR Harelip OR Harelips))) OR ((Lip AND Cleft))) OR (Lips AND Cleft)) OR (“Cleft Palate” OR “Cleft Palates”)) OR (Palate AND Cleft)) OR (Palates AND Cleft)) OR (Cleft Palate AND Isolated)) OR “Orofacial Cleft”) OR (Cleft Lip-Palate AND Nonsyndromic)) OR (Cleft Lip Palate AND Nonsyndromic)) OR (Cleft Lip with or without Cleft Palate AND Nonsyndromic)) OR (Orofacial Cleft AND Nonsyndromic)) OR OFC1) | 42,213 |
| #2 | TS=((((((((((((((((((((("Polymorphism, Single Nucleotide") OR "Genotype") OR "Alleles") OR polymorphism) OR genetic variant) OR genetic variants) OR genetic polymorphism) OR genetic) OR genetic variant) OR genetic variants) OR "Genetic Variation"))))))) OR (((((SNP) OR mutation) OR variation) OR single nucleotide polymorphism) OR variant))))) | 6,014,876 |
| #3 | #2 AND #1 | 7,532 |
| #4 | TS=(SLC19A1 OR CHMD OR FOLT OR IFC1 OR REFC OR RFC1 OR "folate transporter 1" OR IFC-1 OR RFC OR "intestinal folate carrier 1" OR "placental folate transporter" OR "reduced folate carrier protein" OR "solute carrier family 19 member 1" OR "Reduced folate carrier") | 5,678 |
| #5 | #4 AND #3 | 39 |
| Search criterion of ScienceDirect Database (from inception to September 2019) (n=145) | ||
| #1 | ((SLC19A1 OR CHMD OR FOLT OR IFC1 OR REFC OR RFC1 OR "folate transporter 1" OR IFC-1 OR RFC OR "intestinal folate carrier 1" OR "placental folate transporter" OR "reduced folate carrier protein" OR "solute carrier family 19 member 1" OR "Reduced folate carrier") AND ((((((((((((((("cleft lip" OR “Cleft Palate” OR "Orofacial Cleft 1")) OR ((“Cleft Lip” OR “Cleft Lips” OR Harelip OR Harelips))) OR ((Lip AND Cleft))) OR (Lips AND Cleft)) OR (“Cleft Palate” OR “Cleft Palates”)) OR (Palate AND Cleft)) OR (Palates AND Cleft)) OR (Cleft Palate AND Isolated)) OR “Orofacial Cleft”) OR (Cleft Lip-Palate AND Nonsyndromic)) OR (Cleft Lip Palate AND Nonsyndromic)) OR (Cleft Lip with or without Cleft Palate AND Nonsyndromic)) OR (Orofacial Cleft AND Nonsyndromic)) OR OFC1)) AND ((((((((((((((((((((("Polymorphism, Single Nucleotide") OR "Genotype") OR "Alleles") OR polymorphism) OR genetic variant) OR genetic variants) OR genetic polymorphism) OR genetic) OR genetic variant) OR genetic variants) OR "Genetic Variation"))))))) OR (((((SNP) OR mutation) OR variation) OR single nucleotide polymorphism) OR variant))))) | 145 |
| Search criterion of EBSCOhost Databases (from inception to September 2019) (n=27) | ||
| #1 | "(((((((((((((((("cleft lip" OR “Cleft Palate” OR "Orofacial Cleft 1")) OR ((“Cleft Lip” OR “Cleft Lips” OR Harelip OR Harelips))) OR ((Lip AND Cleft))) OR (Lips AND Cleft)) OR (“Cleft Palate” OR “Cleft Palates”)) OR (Palate AND Cleft)) OR (Palates AND Cleft)) OR (Cleft Palate AND Isolated)) OR “Orofacial Cleft”) OR (Cleft Lip-Palate AND Nonsyndromic)) OR (Cleft Lip Palate AND Nonsyndromic)) OR (Cleft Lip with or without Cleft Palate AND Nonsyndromic)) OR (Orofacial Cleft AND Nonsyndromic)) OR OFC1)) AND (((((((((((((((((((((("Polymorphism, Single Nucleotide") OR "Genotype") OR "Alleles") OR polymorphism) OR genetic variant) OR genetic variants) OR genetic polymorphism) OR genetic) OR genetic variant) OR genetic variants) OR "Genetic Variation"))))))) OR (((((SNP) OR mutation) OR variation) OR single nucleotide polymorphism) OR variant)))))) AND ((SLC19A1 OR CHMD OR FOLT OR IFC1 OR REFC OR RFC1 OR "folate transporter 1" OR IFC-1 OR RFC OR "intestinal folate carrier 1" OR "placental folate transporter" OR "reduced folate carrier protein" OR "solute carrier family 19 member 1" OR "Reduced folate carrier")) on 2016-05-19 11:24 PM" | 27 |
Inclusion and exclusion criteria
Inclusion criteria: (I) case-control studies evaluating the association between RFC1 A80G polymorphism and NSCL/P; (II) studies with sufficient data for calculating the odds ratios (ORs) and their 95% confidence intervals (CIs); and (III) studies contained at least two groups (NSCL/P group vs. control group). Exclusion criteria: (I) duplicated studies were removed but the latest article was kept; (II) review articles; (III) editorials or case reports; (IV) animal or cell line studies.
Data extraction
Two investigators independently and carefully extracted data from all eligible studies into a standard data extraction table in duplicate. If any conflict occurred, a discussion was launched to achieve consensus. The list of items extracted from each study included the name of authors, year of publication, country of author, source of patients, ethnicity, source of controls (population-based or hospital-based controls), matching criteria for controls, number of cases and controls, genotyping method, NSCL/P diagnostic method, genotype distribution of cases and controls, and Hardy-Weinberg equilibrium (HWE) tests in the control group. The authors were contacted for any missing data.
Quality assessment
Two investigators independently evaluated the quality of eligible studies using the Newcastle-Ottawa Scale (NOS) (9). If any conflict occurred, a discussion was initiated to achieve consensus.
Data synthesis and analysis
The meta-analysis was carried out using Stata13.0 (Stata Corporation, College Station, TX, USA). Allele frequencies for the RFC1 A80G polymorphisms from each study were determined by the allele counting method (Table 1). The genotype distributions of controls were used to estimate the frequency of the putative risk allele (80G) using the inverse variance method. A χ2 test determined whether the controls of each study conform to HWE. Meta-analyses were built on the following five genetic models: (I) allele model: G vs. A; (II) heterozygote model: AG vs. AA; (III) homozygote model: GG vs. AA; (IV) the dominant model (AG + GG) vs. AA; (V) the recessive model GG vs. (AG + AA). Heterogeneity across studies were measured by I2 statistics and Q tests. When I2<40% and P>0.1, the fixed-effects model was used. On the contrary, I2≥40% or P≤0.1, the meta-regression analysis was carried out to detect the source of heterogeneity, and stratification analyses were performed according to the outcome of meta-regression analysis. If heterogeneity across subgroup studies I2<40% and P>0.1, the fixed-effects model was applied, otherwise the random-effects model was used. Results are shown as odds ratios (ORs) with 95% confidence intervals (CIs), with two tailed P values and statistical significance set at P<0.05. When significant heterogeneity existed, sensitivity analysis was also performed by omitting each study in turn. The sensitivity analyses were conducted by excluding studies not in HWE or not in the population-based (PB) study. The funnel plots, Egger test, and Begg’s test were used to assess the publication bias, and P<0.05 was considered significant. The False Discovery Rate (FDR, Benjamini-Hochberg) method and Bonferroni method were applied for multiple comparisons (10,11).
Table 1. Main characteristics of included studies in the meta-analysis.
| Reference | Country (Ethnicity) | Sample type | Genotyping method | Source of controls | Mothers | Children | NOS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele frequencies (cases/controls) |
HWE | Allele frequencies (cases/controls) |
HWE | |||||||||
| A* | G* | A* | G* | |||||||||
| Shaw 2003 | America (Caucasian) | Blood | PCR-RFLP | PB | NA | NA | NA | 277/351 | 333/377 | No | 9 | |
| Pei 2006 | China (Asian) | Blood | PCR-RFLP | PB | 78/91 | 82/109 | Yes | 79/94 | 85/104 | Yes | 7 | |
| Mostowska 2006 | Poland (Caucasian) | Blood | PCR-RFLP | PB | 124/85 | 110/77 | Yes | NA | NA | NA | 6 | |
| Wang 2009 | China (Asian) | Blood | PCR-RFLP | HB | 103/118 | 89/88 | Yes | 73/94 | 119/112 | Yes | 6 | |
| Bufalino 2010 | Brazil (Mixed) | Tissue | PCR-RFLP | PB | 112/193 | 100/175 | Yes | NA | NA | NA | 9 | |
| Kumari 2013 | India (Caucasian) | Blood | PCR-RFLP | PB | NA | NA | NA | 338/417 | 596/521 | Yes | 7 | |
| Bezerra 2015 | Brazil (Mixed) | Blood | PCR-RFLP | PB | 233/296 | 47/54 | Yes | 212/254 | 68/96 | Yes | 8 | |
| Lakkakula 2015 | India (Caucasian) | Blood | PCR-RFLP | PB | NA | NA | NA | 84/122 | 162/160 | Yes | 6 | |
| Soghani 2017 | Iran (Caucasian) | Blood | PCR-RFLP | PB | NA | NA | NA | 96/269 | 146/59 | Yes | 8 | |
PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; PB, population-based; HB, hospital-based; NA, not available; HWE, Hardy-Weinberg equilibrium; NOS, Newcastle-Ottawa Scale. A: Major allele, the basic formulas of allele frequencies: 2× AA + AG. G: Minor allele, the basic formulas of allele frequencies: 2× GG + AG. *, number was calculated by the formula of transforming genotype frequency to gene frequency, not the original data in the studies.
Results
Selection and characterization of studies
Our search yielded 248 records, including 19 from PubMed, 17 from EMBASE, 39 from WOS, 145 from ScienceDirect, 27 from EBSCOhost, 3 from CBMdis, 7 from CNKI, 15 from Wanfang Databases. There was no record from the Cochrane Library. After excluding duplicates and irrelevances, we obtained 21 titles and abstracts. Among them, 9 records were deleted as they were reviews and had no numerical data. After further examination of full-text of the remaining 12 studies by two independent reviewers, 2 uncontrolled studies were excluded (4,12), and 2 studies overlapped in subjects (13,14). The article published earlier was excluded (13) and the recently published one was included (14). A detailed flowchart of the selection process is shown in Figure 1.
Figure 1.
Flowchart for selecting studies.
The major characteristics of the 9 case-control studies (3,5-8,14-17) were summarized in Table 1, including 4,229 total participants (1,334 NSCL/P children, 1,515 healthy children, 656 mothers of the NSCL/P children and 724 mothers of healthy control children). Two studies were conducted in Asian populations (14,17), 2 in Brazilian populations (3,7) and 5 in Caucasus populations (5,6,8,15,16). In terms of source of controls, 1 study recruited controls from hospital-base population (HB) (16) and 8 from general population-base population (PB) (3,5-8,14,15,17). Two studies demonstrated the affect of maternal folate intake (14,15), and other two studies reported the cleft palate only (CPO) and RFC1 A80G polymorphism (5,15). One study only reported the AA and GG genotypes in maternal population, ignoring the AG genotype (16). Polymerase chain reaction (PCR) was utilized as the genotyping method in all studies. All 9 studies were of high quality with an NOS score ≥6 (Table S2).
Table S2. Results of quality assessment using the Newcastle-Ottawa Scale for case-control studies.
| Study | Selection | Comparability | Exposure | Scores | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Adequate definition of cases | Representativeness of the cases | Selection of controls | Definition of controls | Control for important factora | Ascertainment of exposure | Same method of ascertainment for cases and controls | Non response rate | ||||
| Shaw 2003 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | ☆ | 9 | ||
| Pei 2006 | ☆ | ☆ | ☆ | – | ☆☆ | ☆ | ☆ | – | 7 | ||
| Mostowska 2006 | ☆ | ☆ | – | ☆ | ☆ | ☆ | ☆ | – | 6 | ||
| Wang 2009 | ☆ | ☆ | – | ☆ | ☆ | ☆ | ☆ | – | 6 | ||
| Bufalino 2010 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | ☆ | 9 | ||
| Kumari 2013 | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | – | 7 | ||
| Bezerra 2015 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | – | 8 | ||
| Lakkakula 2015 | ☆ | ☆ | ☆ | ☆ | – | ☆ | ☆ | – | 6 | ||
| Soghani 2017 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | – | 8 | ||
a, a maximum of 2 stars can be allotted in this category, one for age, the other for other controlled factors.
Association between RFC1 A80G polymorphism and NSCL/P risks in the population of children
The results of meta-analysis of the association between RFC1 A80G polymorphism and NSCL/P risks are summarized in Table 2. The meta-analysis revealed that NSCL/P risk was significantly increased in 2 genetic models: GG vs. AA (OR =2.346, 95% CI: 1.127–4.884), GG vs. (AG + AA) (OR =1.503, 95% CI: 1.049–2.152) (Figure 2). However, there was no statistical differences after Bonferroni correction. The meta-regression analysis to identify the source of heterogeneity revealed that it was not deviated in the published year, country, ethnicity, HWE, sample size, NOS score (Table S3). When stratified by country, a significantly increased risk was also found in all 5 genetic models in India (Table 2). When stratified by ethnicity, the recessive model did not exhibit an increased risk in Caucasian population (OR =1.890, 95% CI: 1.117–3.197; FDR =0.045 with P=0.090 in Bonferroni testing). When stratified by PB or HWE (yes), the recessive model exhibited a significantly increased risk (FDR =0.005 with P=0.005 in Bonferroni testing). In the stratified analysis by CPO, maternal folic acid use and maternal folic acid no-use of the children, non-significant associations were found when all studies were pooled with fixed or random-effects models for all these 5 genetic models.
Table 2. Meta-analysis of the association RFC1 A80G polymorphism with risk of cleft lip with or without cleft palate for children.
| Overall and subgroup | No. of trials (participants) | Heterogeneity | Model | Meta-analysis | Bonferroni | FDR | ||
|---|---|---|---|---|---|---|---|---|
| P | I2 (%) | OR (95% CI) | P for OR | |||||
| Total | ||||||||
| G vs. A | 7 (2,849) | <0.001 | 92.7 | R | 1.507 (0.988–2.300) | 0.057 | 0.285 | 0.071 |
| AG vs. AA | 7 (2,849) | <0.001 | 90.1 | R | 1.852 (0.944–3.631) | 0.073 | 0.365 | 0.073 |
| GG vs. AA | 7 (2,849) | <0.001 | 88.5 | R | 2.346 (1.127–4.884) | 0.023 | 0.115 | 0.065 |
| AG+GG vs. AA | 7 (2,849) | <0.001 | 91.5 | R | 2.008 (1.011–3.985) | 0.046 | 0.230 | 0.071 |
| GG vs. AG + AA | 7 (2,849) | 0.002 | 71.6 | R | 1.503 (1.049–2.152) | 0.026 | 0.130 | 0.065 |
| China | ||||||||
| G vs. A | 2 (380) | 0.245 | 25.9 | F | 1.159 (0.829–1.619) | 0.310 | 1.00 | 0.485 |
| AG vs. AA | 2 (380) | 0.338 | 0 | F | 1.349 (0.783–2.324) | 0.280 | 1.00 | 0.485 |
| GG vs. AA | 2 (380) | 0.204 | 38.0 | F | 1.399 (0.653–3.000) | 0.388 | 1.00 | 0.485 |
| AG+GG vs. AA | 2 (380) | 0.238 | 28.1 | F | 1.364 (0.739–2.518) | 0.321 | 1.00 | 0.485 |
| GG vs. AG + AA | 2 (380) | 0.412 | 0 | F | 1.130 (0.727–1.755) | 0.587 | 1.00 | 0.587 |
| India | ||||||||
| G vs. A | 2 (1,200) | 0.840 | 0 | F | 1.424 (1.208–1.678) | <0.001 | 0.005 | 0.001 |
| AG vs. AA | 2 (1,200) | 0.257 | 22.3 | F | 1.633 (1.163–2.292) | 0.005 | 0.025 | 0.005 |
| GG vs. AA | 2 (1,200) | 0.756 | 0 | F | 2.165 (1.519–3.087) | <0.001 | 0.005 | 0.001 |
| AG + GG vs. AA | 2 (1,200) | 0.425 | 0 | F | 1.839 (1.331–2.540) | <0.001 | 0.005 | 0.001 |
| GG vs. AG + AA | 2 (1,200) | 0.426 | 0 | F | 1.489 (1.174–1.889) | 0.001 | 0.005 | 0.001 |
| Caucasian | ||||||||
| G vs. A | 4 (2,154) | <0.001 | 95.7 | R | 1.972 (1.048–3.708) | 0.035 | 0.175 | 0.048 |
| AG vs. AA | 4 (2,154) | <0.001 | 94.0 | R | 2.741 (0.942–7.982) | 0.064 | 0.320 | 0.064 |
| GG vs. AA | 4 (2,154) | <0.001 | 93.5 | R | 3.921 (1.283–11.979) | 0.016 | 0.080 | 0.045 |
| AG + GG vs. AA | 4 (2,154) | <0.001 | 94.7 | R | 3.124 (1.063–9.181) | 0.038 | 0.190 | 0.048 |
| GG vs. AG + AA | 4 (2,154) | <0.001 | 79.5 | R | 1.890 (1.117–3.197) | 0.018 | 0.090 | 0.045 |
| PB | ||||||||
| G vs. A | 6 (2,650) | <0.001 | 93.9 | R | 1.532 (0.946–2.481) | 0.083 | 0.415 | 0.104 |
| AG vs. AA | 6 (2,650) | <0.001 | 91.7 | R | 1.869 (0.874–3.993) | 0.107 | 0.535 | 0.107 |
| GG vs. AA | 6 (2,650) | <0.001 | 90.4 | R | 2.414 (1.044–5.584) | 0.039 | 0.195 | 0.098 |
| AG + GG vs. AA | 6 (2,650) | <0.001 | 92.9 | R | 2.034 (0.939–4.405) | 0.072 | 0.360 | 0.104 |
| GG vs. AG + AA | 6 (2,650) | 0.001 | 76.4 | R | 1.428 (1.201–1.699) | <0.001 | 0.005 | 0.005 |
| HWE (Yes) | ||||||||
| G vs. A | 6 (2,180) | <0.001 | 93.4 | R | 1.590 (0.940–2.687) | 0.083 | 0.415 | 0.104 |
| AG vs. AA | 6 (2,180) | <0.001 | 91.5 | R | 2.040 (0.867–4.801) | 0.102 | 0.510 | 0.107 |
| GG vs. AA | 6 (2,180) | <0.001 | 89.2 | R | 2.703 (1.068–6.840) | 0.036 | 0.180 | 0.098 |
| AG + GG vs. AA | 6 (2,180) | <0.001 | 92.6 | R | 2.236 (0.932–5.363) | 0.072 | 0.360 | 0.104 |
| GG vs. AG + AA | 6 (2,180) | 0.002 | 73.4 | R | 1.548 (1.275–1.881) | <0.001 | 0.005 | 0.005 |
| Maternal Folic acid use | ||||||||
| G vs. A | 2 (460) | 0.002 | 89.3 | R | 0.660 (0.176–2.470) | 0.537 | 1.00 | 0.671 |
| AG vs. AA | 2 (460) | 0.017 | 82.4 | R | 0.339 (0.022–5.183) | 0.437 | 1.00 | 0.671 |
| GG vs. AA | 2 (460) | 0.004 | 87.9 | R | 0.304 (0.010–9.107) | 0.492 | 1.00 | 0.671 |
| AG + GG vs. AA | 2 (460) | 0.006 | 86.5 | R | 0.318 (0.015–6.558) | 0.458 | 1.00 | 0.671 |
| GG vs. AG + AA | 2 (460) | 0.053 | 73.2 | R | 0.808 (0.240–2.722) | 0.730 | 1.00 | 0.730 |
| Maternal Folic acid mouse | ||||||||
| G vs. A | 2 (383) | 0.114 | 60.1 | R | 1.214 (0.755–1.952) | 0.423 | 1.00 | 0.529 |
| AG vs. AA | 2 (383) | 0.397 | 0 | F | 1.492 (0.906–2.455) | 0.116 | 0.58 | 0.529 |
| GG vs. AA | 2 (383) | 0.103 | 62.4 | R | 1.516 (0.560–4.103) | 0.412 | 1.00 | 0.529 |
| AG + GG vs. AA | 2 (383) | 0.196 | 40.2 | R | 1.474 (0.784–2.768) | 0.228 | 1.00 | 0.529 |
| GG vs. AG + AA | 2 (383) | 0.178 | 44.8 | R | 1.106 (0.573–2.135) | 0.765 | 1.00 | 0.765 |
| CPO | ||||||||
| G vs. A | 2 (647) | 0.956 | 0 | F | 1.030 (0.789–1.344) | 0.829 | 1.00 | 0.878 |
| AG vs. AA | 2 (647) | 0.258 | 21.7 | F | 0.902 (0.561–1.449) | 0.669 | 1.00 | 0.878 |
| GG vs. AA | 2 (647) | 0.726 | 0 | F | 1.061 (0.645–1.745) | 0.815 | 1.00 | 0.878 |
| AG + GG vs. AA | 2 (647) | 0.383 | 0 | F | 0.967 (0.629–1.486) | 0.878 | 1.00 | 0.878 |
| GG vs. AG + AA | 2 (647) | 0.508 | 0 | F | 1.102 (0.737–1.646) | 0.636 | 1.00 | 0.878 |
CPO, cleft palate only; R, random; F, fixed.
Figure 2.
Forest plot of overall analysis in different genetic models. (A) Allele model: G vs. A; (B) heterozygote model: AG vs. AA; (C) homozygote model: GG vs. AA; (D) the dominant model: (AG + GG) vs. AA; (E) the recessive model respectively: GG vs. (AG + AA).
Table S3. Meta-regression analysis to detect the source of heterogeneity for the association between RFC1 A80G polymorphism and NSCL/P risks in the population of children.
| Genetic model | Variables | Coefficient | 95% CI | P | t |
|---|---|---|---|---|---|
| G vs. A | Year | 1.070 | 0.939, 1.219 | 0.239 | 1.34 |
| Country | 1.332 | 0.868, 2.044 | 0.146 | 1.72 | |
| Ethnicity | 0.984 | 0.305, 3.169 | 0.972 | −0.04 | |
| Source of control | 0.892 | 0.105, 7.540 | 0.896 | −0.14 | |
| NOS score | 1.220 | 0.679, 2.194 | 0.423 | 0.87 | |
| Sample size | 1.000 | 0.997, 1.003 | 0.820 | −0.24 | |
| HWE | 0.704 | 0.090, 5.483 | 0.678 | −0.44 | |
| AG vs. AA | Year | 1.106 | 0.872, 1.403 | 0.324 | 1.09 |
| Country | 1.579 | 0.722, 3.451 | 0.194 | 1.50 | |
| Ethnicity | 0.900 | 0.119, 6.777 | 0.898 | −0.13 | |
| Source of control | 0.942 | 0.022, 39.898 | 0.969 | −0.04 | |
| NOS score | 1.485 | 0.546, 4.040 | 0.357 | 1.01 | |
| Sample size | 1.000 | 0.995, 1.004 | 0.857 | −0.19 | |
| HWE | 0.564 | 0.016, 19.920 | 0.697 | −0.41 | |
| GG vs. AA | Year | 1.158 | 0.880, 1.526 | 0.228 | 1.37 |
| Country | 1.868 | 0.756, 4.617 | 0.136 | 1.77 | |
| Ethnicity | 1.028 | 0.084, 12.559 | 0.979 | 0.03 | |
| Source of control | 0.821 | 0.008, 78.378 | 0.916 | −0.11 | |
| NOS score | 1.549 | 0.444, 5.405 | 0.409 | 0.90 | |
| Sample size | 1.000 | 0.994, 1.005 | 0.832 | −0.22 | |
| HWE | 0.442 | 0.006, 34.083 | 0.650 | −0.48 | |
| GG + AG vs. AA | Year | 1.120 | 0.879, 1.427 | 0.282 | 1.21 |
| Country | 1.636 | 0.735, 3.641 | 0.174 | 1.58 | |
| Ethnicity | 0.926 | 0.113, 7.600 | 0.929 | −0.09 | |
| Source of control | 0.914 | 0.019, 44.783 | 0.955 | −0.06 | |
| NOS score | 1.469 | 0.511, 4.225 | 0.393 | 0.93 | |
| Sample size | 1.000 | 0.995, 1.005 | 0.853 | −0.19 | |
| HWE | 0.525 | 0.013, 21.510 | 0.675 | −0.45 | |
| GG vs. AG + AA | Year | 1.075 | 0.958, 1.206 | 0.170 | 1.60 |
| Country | 1.366 | 0.920, 2.028 | 0.098 | 2.03 | |
| Ethnicity | 1.100 | 0.330, 3.672 | 0.847 | 0.20 | |
| Source of control | 0.841 | 0.103, 6.892 | 0.841 | −0.21 | |
| NOS score | 1.176 | 0.648, 2.135 | 0.517 | 0.70 | |
| Sample size | 1.000 | 0.997, 1.002 | 0.780 | −0.29 | |
| HWE | 0.676 | 0.098, 4.687 | 0.626 | −0.52 |
NOS, Newcastle-Ottawa Scale; HWE, Hardy-Weinberg equilibrium; Source of control, the population source of the control group.
Sensitivity analysis on the association between RFC1 A80G polymorphism and NSCL/P risks in the population of children
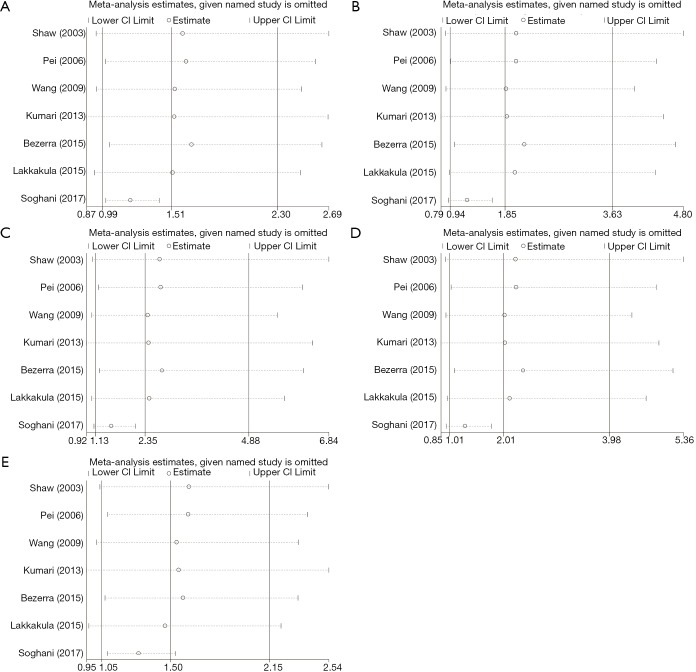
We performed sensitivity analysis by excluding the study by Soghani et al. because of the outlier data and publication bias (8) (Figure 3). The result showed that the GG genotype frequency (GG vs. AA, GG vs. AG + AA) in the NSCL/P patients were significantly higher than those in the controls (Table 3). The pooled ORs were not qualitatively changed in sensitivity analysis, compared with the overall meta-analysis.
Figure 3.
Sensitivity analysis in different genetic models. (A) Allele model: G vs. A; (B) heterozygote model: AG vs. AA; (C) homozygote model: GG vs. AA; (D) the dominant model: (AG + GG) vs. AA; (E) the recessive model respectively: GG vs. (AG + AA).
Table 3. Sensitivity analysis of the association RFC1 A80G polymorphism with risk of cleft lip with or without cleft palate for children.
| Overall | No. of trials (participants) | Heterogeneity | Model | Meta-analysis | Bonferroni | FDR | ||
|---|---|---|---|---|---|---|---|---|
| P | I2 (%) | OR (95% CI) | P for OR | |||||
| Children (Children with cleft lip with or without cleft palate) | ||||||||
| G vs. A | 6 (2,564) | 0.094 | 46.9 | R | 1.198 (1.013–1.416) | 0.034 | 0.170 | 0.057 |
| AG vs. AA | 6 (2,564) | 0.116 | 43.3 | R | 1.225 (0.915–1.641) | 0.173 | 0.865 | 0.173 |
| GG vs. AA | 6 (2,564) | 0.142 | 39.5 | F | 1.566 (1.241–1.976) | <0.001 | 0.005 | 0.005 |
| AG + GG vs. AA | 6 (2,564) | 0.040 | 57.0 | R | 1.299 (0.946–1.784) | 0.106 | 0.530 | 0.133 |
| GG vs. AG + AA | 6 (2,564) | 0.508 | 0 | F | 1.293 (1.088–1.536) | 0.003 | 0.015 | 0.008 |
CPO, cleft palate only; R, random; F, fixed.
Association between RFC1 A80G polymorphism in the maternal population and susceptibility to NSCL/P in children
The meta-analysis findings of the association between RFC1 A80G polymorphism in the maternal population and susceptibility to NSCL/P in children were summarized in Table 4. The results showed that all RFC1 A80G genetic models exhibited no significant association in overall comparisons between RFC1 A80G polymorphism in the maternal population and NSCL/P risks in children. In the succeeding stratified subgroup analysis including the Asian subgroup and the Caucasian subgroup, neither did we find any significant association (Table 4). No substantial alterations occurred in results during sensitivity analysis through omitting one study every time, implying the robustness of the results.
Table 4. Meta-analysis of the association RFC1 A80G polymorphism with risk of cleft lip with or without cleft palate for mothers of NSCP/L children.
| Overall and subgroup | No. of trials (participants) | Heterogeneity | Model | Meta-analysis | Bonferroni | FDR | ||
|---|---|---|---|---|---|---|---|---|
| P | I2 (%) | OR (95% CI) | P for OR | |||||
| Total | ||||||||
| G vs. A | 5 (1,380) | 0.892 | 0 | F | 1.015 (0.852–1.209) | 0.871 | 1 | 0.937 |
| AG vs. AA | 4 (984) | 0.352 | 8.3 | F | 1.022 (0.759–1.375) | 0.888 | 1 | 0.937 |
| GG vs. AA | 4 (984) | 0.640 | 0 | F | 1.052 (0.684–1.618) | 0.817 | 1 | 0.937 |
| AG + GG vs. AA | 4 (984) | 0.504 | 0 | F | 1.037 (0.781–1.378) | 0.800 | 1 | 0.937 |
| GG vs. AG + AA | 4 (984) | 0.496 | 0 | F | 1.015 (0.705–1.460) | 0.937 | 1 | 0.937 |
| Caucasian | ||||||||
| G vs. A | 3 (1,001) | 0.898 | 0 | F | 1.014 (0.813–1.265) | 0.901 | 1 | 0.901 |
| AG vs. AA | 2 (605) | 0.430 | 0 | F | 0.867 (0.597–1.259) | 0.454 | 1 | 0.897 |
| GG vs. AA | 2 (605) | 0.318 | 0 | F | 1.116 (0.616–2.023) | 0.717 | 1 | 0.897 |
| AG + GG vs. AA | 2 (605) | 0.497 | 0 | F | 0.935 (0.657–1.330) | 0.708 | 1 | 0.897 |
| GG vs. AG + AA | 2 (605) | 0.411 | 0 | F | 1.284 (0.764–2.159) | 0.345 | 1 | 0.897 |
| Asian | ||||||||
| G vs. A | 2 (379) | 0.344 | 0 | F | 1.016 (0.762–1.353) | 0.916 | 1 | 0.964 |
| AG vs. AA | 2 (379) | 0.435 | 0 | F | 1.365 (0.830–2.246) | 0.220 | 1 | 0.692 |
| GG vs. AA | 2 (379) | 0.435 | 0 | F | 0.986 (0.528–1.840) | 0.964 | 1 | 0.964 |
| AG + GG vs. AA | 2 (379) | 0.333 | 0 | F | 1.256 (0.778–2.027) | 0.351 | 1 | 0.692 |
| GG vs. AG + AA | 2 (379) | 0.686 | 0 | F | 0.809 (0.486–1.346) | 0.415 | 1 | 0.692 |
F, the fixed model.
Evaluation of publication bias
None of the studies included in the meta-analysis stated that genotyping was performed blinded to clinical status of the subjects. Funnel plot with egger recursive line seemed symmetric for each genetic model, showing no significant publication bias (Figure 4), which was confirmed with Egger’s test in the children and maternal population, respectively (Table S4).
Figure 4.
Funnel plot for NSCL/P risk and RFC1 A80G polymorphism in different genetic models. (A) Allele model: G vs. A; (B) heterozygote model: AG vs. AA; (C) homozygote model: GG vs. AA; (D) the dominant model: (AG+GG) vs. AA; (E) the recessive model respectively: GG vs. (AG + AA).
Table S4. Meta-analysis of the association RFC1 A80G polymorphism with risk of cleft lip with or without cleft palate: publication bias.
| Genetic model | Group | Studies | Begg’s test (P) | Egger test | |
|---|---|---|---|---|---|
| P | 95% CI | ||||
| Children | |||||
| G vs. A | Overall | 7 | 0.764 | 0.414 | (−5.096, 10.507) |
| AG vs. AA | Overall | 7 | 0.230 | 0.347 | (−5.716, 13.455) |
| GG vs. AA | Overall | 7 | 0.764 | 0.414 | (−5.096, 10.507) |
| AG + GG vs. AA | Overall | 7 | 0.230 | 0.931 | (−14.029, 15.066) |
| GG vs. AG + AA | Overall | 7 | 0.764 | 0.478 | (−3.264, 6.037) |
| Mother | |||||
| G vs. A | Overall | 5 | 1.000 | 0.861 | (−9.580, 10.802) |
| AG vs. AA | Overall | 4 | 0.734 | 0.509 | (−12.541, 18.247) |
| GG vs. AA | Overall | 4 | 0.308 | 0.231 | (−2.490, 5.754) |
| AG +GG vs. AA | Overall | 4 | 1.000 | 0.567 | (−11.499, 5.808) |
| GG vs. AG + AA | Overall | 4 | 0.734 | 0.538 | (−5.164, 7.295) |
Discussion
In recent years, several case-control studies were performed to identify the potential contribution of RFC1 to NSCL/P (3,5-7). However, the results are inconclusive. In this study, we investigated association between RFC1 A80G polymorphism and NSCL/P susceptibility using meta-analysis approach based on 9 case-control studies. We found that associations exist between the RFC1 A80G polymorphism and susceptibility to NSCL/P for all the 5 genetic models in pediatric populations of India even with multiple corrections. Subgroups analyses detected a significant association in recessive model for the PB pediatric population, but no association with Caucasian, Chinese, and Asian (Table 2). Sensitivity analyses showed the results were robust (Table 3). The Funnel plot and the egger recursive line seemed symmetric for each genetic model, suggesting there was no significant publication bias. Moreover, the results of HWE and source of controls indicated that studies out of HWE and studies with controls from hospital might be the source of bias.
RFC1 mediates delivery of 5-methyltetrahydrofolate into cytoplasm from endocytotic vesicles (18-20), which is one of the few identified mechanisms responsible for internalizing and transporting folate molecules in eukaryotic cells (21). When the concentration of 5-methyltetrahydrofolate is reduced, remethylation of homocysteine into methionine consequently become diminished, and fewer amount of the methyl group is available for DNA methylation. Hypomethylation can change the transcription and suppression of genes involved in formation of the lip, alveolus, and/or palate. Moreover, RFC1 had been proposed to be an organic anion exchanger in folic acid absorption and transports 5-methyltetrahydrofolate and thiamine monophosphate bi-directionally (22,23). The RFC1 A80G polymorphism results in the change of amino acid from glutamine (encoded by CAG) to arginine (encoded by CGG). The variants of RFC1 may result in lower levels of folate, potentially affecting NSCL/P risk.
The study by van Rooij et al. indicated that maternal periconceptional use of folic acid supplements was an independent preventive factor for NSCL/P (24). Consistent results were also obtained in other studies (14,15). It seems that periconceptional folic acid use may reduce incidence of NSCL/P of the offspring to some certain degree. In this regard, we conducted a subgroup analysis based on maternal intake of folate status in the current study. The results do not support that the association between RFC1 A80G polymorphism and the susceptibility to NSCL/P was affected by maternal folate status and dietary intake (Table 2). On the one hand, the sample size and number of the studies was very small, yielding imprecise risk estimates. On the other hand, study by Ray et al. showed that low-dose folic acid supplementation cannot protect against NSCL/P (25). Further, Tolarova et al. showed that only a very high dose of supplementary folic acid (10 mg/day) could reduce the risk of NSCL/P significantly (65% reduction was observed) (26). Meanwhile, study by Crider et al. suggested that serum/plasma folate concentrations increased 11.6% (95% CI: 8.4–14.9) for every 100 µg/day folic acid intake basis on regression analysis, and it is generally too late to prevent if folic acid consumption is only initiated after a woman learns she is pregnant (27). It seems that maternal serum/plasma folate concentrations decreased the risk for NSCL/P in a dose-dependent manner. Further studies should be carried out to clarify this issue.
In the current study, we also did not observe the association of RFC1 A80G polymorphism with CPO susceptibility from the two studies (5,15). The meta-analysis results didn’t support that associations exist between RFC1 A80G polymorphism of maternal populations and susceptibility of children’s NSCL/P for all genetic models (Table 4). This may be due to a small quantity of the sample size and number of the studies included in the current study.
Maternal RFC1 genotypes might be more important than those of the infant. Because the mother provides the environment for the embryo of development, the embryo is completely dependent on the mother's folic acid state. We further explored if the risk of delivering a NSCL/P child would increase, when the maternal 80GG genotype was present. It was likely that folic acid supplement use or dietary folate intake were not properly changed by the variants of the RFC1 genotypes in the maternal population.
NSCL/P is complex and heterogeneous as shown in its extensive involvement in craniofacial syndromes. Many genes are involved in the development of the primary and secondary palates which may be disrupted by genetic, environmental, or combined factors at any time point during developmental process, leading to NSCL/P. Those are candidate NSCL/P genes, encoding transcription factors (e.g., TBX22, MSX1), growth factors (e.g., TGFA, TGFB3), and adhesion molecules (e.g., PVRL1). More genes causing NSCL/P can be identified by the human genome-wide gene discovery, and positional cloning or positional candidate approaches. In addition, studies investigating the relative risk of NSCL/P contributed by changes of candidate genes such as SNPs can also provide insights into the mechanism of NSCL/P and lead to prevention of NSCL/P.
This meta-analysis was conducted following a canonical systematic process, included 7 case-control studies which confirmed the relationship between RFC1 A80G polymorphism and susceptibility to NSCL/P in children. In addition, the current study does not support the significant association between maternal RFC1 A80G polymorphism and susceptibility to NSCL/P in children based on 5 case-control studies. All studies included in the current meta-analysis were of high quality with NOS scores higher than 6, which suggests the data in our analysis is reliable. Our study is the exploratory meta-analysis focusing on the association between RFC1 A80G polymorphism and NSCL/P and CPO risk. We also considered the influence of maternal folate use status and stratified analysis according to the maternal folate use status, which included more valuable information on this topic. Moreover, we conducted eight subgroup meta-analyses based on characteristics of studies.
Although we have tried our best to retrieve more literatures, some limitations are inevitable. First, moderate heterogeneity was pooled in from some genetic models with the random-effects model. Sensitivity analysis was performed to evaluate the stability of the results. Second, the results of the Egger test and Begg’s test demonstrated that the publication biases may not affect the stability of positive results, but more studies are expected to be included in the future to make the results more precise. Additionally, our meta-analysis didn’t provide more advisories for the use of folate on prevention, diagnosis, and treatment of maternal NSCL/P.
In conclusion, our results supported that the RFC1 A80G polymorphism is a candidate for susceptibility to NSCL/P in the Indian pediatric population, especially for those conformed to the HWE. Because of the heterogeneity of our meta-analysis, a large number of homogeneous studies should be performed to evaluate the results in the future.
Acknowledgments
Funding: This research was supported by the National Natural Science Foundation of China (81902498), Natural Science Foundation of Hubei Province of China (2019CFB177), Natural Science Foundation of Hubei Provincial Department of Education (Q20182105), Chen Xiao-ping Foundation for the development of science and technology of Hubei Provincial (CXPJJH11800001-2018333), Natural Science Foundation of Hubei Province of China (2016CFB530) and Faculty Development Foundation of Hubei University of Medicine (2014QDJZR01), and National Students’ platform for innovation and entrepreneurship training program (201810929005, 201810929009, 201810929068, and 201813249010).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Mossey PA, Little J, Munger RG, et al. Cleft lip and palate. Lancet;374:1773-85. 10.1016/S0140-6736(09)60695-4 [DOI] [PubMed] [Google Scholar]
- 2.Sa J, Araujo L, Guimaraes L, et al. Dental anomalies inside the cleft region in individuals with nonsyndromic cleft lip with or without cleft palate. Med Oral Patol Oral Cir Bucal 2016;21:e48-52. 10.4317/medoral.20757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bezerra JF, Oliveira GHM, Soares CD, et al. Genetic and non-genetic factors that increase the risk of non-syndromic cleft lip and/or palate development. Oral Dis 2015;21:393-9. 10.1111/odi.12292 [DOI] [PubMed] [Google Scholar]
- 4.Girardi A, Martinelli M, Cura F, et al. RFC1 and non-syndromic cleft lip with or without cleft palate: an association based study in Italy. J Craniomaxillofac Surg 2014;42:1503-5. 10.1016/j.jcms.2014.04.021 [DOI] [PubMed] [Google Scholar]
- 5.Lakkakula B, Murthy J, Gurramkonda VB. Relationship between reduced folate carrier gene polymorphism and non-syndromic cleft lip and palate in Indian population. J Matern Fetal Neonatal Med 2015;28:329-32. 10.3109/14767058.2014.916677 [DOI] [PubMed] [Google Scholar]
- 6.Kumari P, Ali A, Sukla KK, et al. Lower incidence of nonsyndromic cleft lip with or without cleft palate in females: is homocysteine a factor? J Biosci 2013;38:21-6. 10.1007/s12038-013-9298-7 [DOI] [PubMed] [Google Scholar]
- 7.Bufalino A, Ribeiro Paranaiba LM, Nascimento de Aquino S, et al. Maternal polymorphisms in folic acid metabolic genes are associated with nonsyndromic cleft lip and/or palate in the Brazilian population. Birth Defects Res A Clin Mol Teratol 2010;88:980-6. 10.1002/bdra.20732 [DOI] [PubMed] [Google Scholar]
- 8.Soghani B, Ebadifar A, Khorram Khorshid HR, et al. The study of association between reduced folate carrier 1 (RFC1) polymorphism and non-syndromic cleft lip/palate in Iranian population. Bioimpacts 2017;7:263-8. 10.15171/bi.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603-5. 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 10.Benjamini Y, Drai D, Elmer G, et al. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001;125:279-84. 10.1016/S0166-4328(01)00297-2 [DOI] [PubMed] [Google Scholar]
- 11.Armstrong RA. When to use the Bonferroni correction. Ophthalmic Physiol Opt 2014;34:502-8. 10.1111/opo.12131 [DOI] [PubMed] [Google Scholar]
- 12.Vieira AR, Cooper ME, Marazita ML, et al. Reduced folate carrier 1 (RFC1) is associated with cleft of the lip only. Braz J Med Biol Res 2008;41:689-93. 10.1590/S0100-879X2008000800009 [DOI] [PubMed] [Google Scholar]
- 13.Pei LJ, Ren AG, Hao L, et al. Study on the association between reduced folate carrier gene polymorphism and congenital heart defects and cleft lip with or without cleft palate. Zhonghua Liu Xing Bing Xue Za Zhi 2004;25:1063-7. [PubMed] [Google Scholar]
- 14.Pei L, Zhu H, Zhu J, et al. Genetic variation of infant reduced folate carrier (A80G) and risk of orofacial defects and congenital heart defects in China. Ann Epidemiol 2006;16:352-6. 10.1016/j.annepidem.2005.02.014 [DOI] [PubMed] [Google Scholar]
- 15.Shaw GM, Zhu H, Lammer EJ, et al. Genetic variation of infant reduced folate carrier (A80G) and risk of orofacial and conotruncal heart defects. Am J Epidemiol 2003;158:747-52. 10.1093/aje/kwg189 [DOI] [PubMed] [Google Scholar]
- 16.Mostowska A, Hozyasz KK, Jagodzinski PP. Maternal MTR genotype contributes to the risk of non-syndromic cleft lip and palate in the Polish population. Clin Genet 2006;69:512-7. 10.1111/j.1399-0004.2006.00618.x [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Song X, Guo J, et al. Relationship between genetic polymorphisms of RFC1 A80G and nonsymdromic cleft lip with or without palate. Wei Sheng Yan Jiu 2009;38:276-9. [PubMed] [Google Scholar]
- 18.Chango A, Emery-Fillon N, de Courcy GP, et al. A polymorphism (80G->A) in the reduced folate carrier gene and its associations with folate status and homocysteinemia. Mol Genet Metab 2000;70:310-5. 10.1006/mgme.2000.3034 [DOI] [PubMed] [Google Scholar]
- 19.Dixon KH, Lanpher BC, Chiu J, et al. A novel cDNA restores reduced folate carrier activity and methotrexate sensitivity to transport deficient cells. J Biol Chem 1994;269:17-20. [PubMed] [Google Scholar]
- 20.Kamen BA, Wang MT, Streckfuss AJ, et al. Delivery of folates to the cytoplasm of MA104 cells is mediated by a surface membrane receptor that recycles. J Biol Chem 1988;263:13602-9. [PubMed] [Google Scholar]
- 21.Blanton SH, Henry RR, Yuan Q, et al. Folate pathway and nonsyndromic cleft lip and palate. Birth Defects Res A Clin Mol Teratol 2011;91:50-60. 10.1002/bdra.20740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao R, Gao F, Wang Y, et al. Impact of the reduced folate carrier on the accumulation of active thiamin metabolites in murine leukemia cells. J Biol Chem 2001;276:1114-8. 10.1074/jbc.M007919200 [DOI] [PubMed] [Google Scholar]
- 23.Zhao R, Seither R, Brigle KE, et al. Impact of overexpression of the reduced folate carrier (RFC1), an anion exchanger, on concentrative transport in murine L1210 leukemia cells. J Biol Chem 1997;272:21207-12. 10.1074/jbc.272.34.21207 [DOI] [PubMed] [Google Scholar]
- 24.van Rooij IA, Vermeij-Keers C, Kluijtmans LA, et al. Does the interaction between maternal folate intake and the methylenetetrahydrofolate reductase polymorphisms affect the risk of cleft lip with or without cleft palate? Am J Epidemiol 2003;157:583-91. 10.1093/aje/kwg005 [DOI] [PubMed] [Google Scholar]
- 25.Ray JG, Meier C, Vermeulen MJ, et al. Association between folic acid food fortification and congenital orofacial clefts. J Pediatr 2003;143:805-7. 10.1067/S0022-3476(03)00495-5 [DOI] [PubMed] [Google Scholar]
- 26.Tolarova M, Harris J. Reduced recurrence of orofacial clefts after periconceptional supplementation with high-dose folic acid and multivitamins. Teratology 1995;51:71-8. 10.1002/tera.1420510205 [DOI] [PubMed] [Google Scholar]
- 27.Crider KS, Devine O, Qi YP, et al. Systematic Review and Bayesian Meta-analysis of the Dose-response Relationship between Folic Acid Intake and Changes in Blood Folate Concentrations. Nutrients 2019. doi: . 10.3390/nu11010071 [DOI] [PMC free article] [PubMed] [Google Scholar]