Abstract
Background
The world-wide prevalence of carbapenem-resistant Klebsiella pneumoniae (CRKP) poses a threat to the public health. The objective of this study was to determine the epidemiological and molecular patterns of KPC-producing Klebsiella pneumoniae (K. pneumoniae) clinical isolates.
Methods
In this study, a total of 82 non-duplicated CRKP isolates were analyzed for the prevalence of resistant determinants including carbapenemase, extended spectrum β-lactamase (ESBLs), and AmpC as well as integrons and cassette regions by polymerase chain reaction (PCR) and DNA sequencing. The genetic relatedness was investigated by pulsed field gel electrophoresis (PFGE) and multi-locus sequencing typing (MLST).
Results
Overall, blaKPC-2 (n=75) was the predominant carbapenemase gene, followed by high prevalence of blaSHV (92.7%) and blaCTX-M (90.2%). PFGE and MLST analysis revealed that 65 out of 68 KPC-2-producing CRKP belonged to the ST11 clone and were distributed mainly in the department of neurology ICU. Moreover, first report on clonal dissemination of KPC-2-producing CRKP ST48 clone and NDM-5-producing CRKP ST337 clone was also identified. Class I integron were detected in 17 (20.7%) of 82 isolates with aadA2 being the most common cassette. And a novel cassette array of integron, aac(6')-II-blaCARB/PSE-1 was identified.
Conclusions
All in all, KPC-2-producing CRKP ST11 and ST48 clone were widely disseminated in multiple departments of our hospital, which triggers the need for active surveillance and implementation of infection control measures.
Keywords: Carbapenemases, Klebsiella pneumoniae (K. pneumoniae), KPC-2, NDM-5, clonal dissemination
Introduction
Klebsiella pneumoniae (K. pneumoniae) is one of the most opportunistic pathogen responsible for numerous infections, including respiratory tract infection, urinary tract infection, bacteremia, skin and soft tissue infections. Carbapenem has long been regarded as the last resort against infections caused by K. pneumoniae resistant to the 3rd or 4th cephalosporin. However, in recent years, the rapid emergence of CRKP isolates poses a great challenge to public health because of the limited therapeutic regimen, high mortality and healthcare costs (1). Up to date, clonal dissemination and outbreak caused by CRKP isolates have occurred worldwide due to the endemic clone spread and rapid transmission of carbapenemase encoding gene mediated by plasmids, integrons and transposons (2,3). It has been reported that the production of carbapenemase is the predominant mechanism leading to carbapemen resistance (4,5), in addition to the presence of extended-spectrum β-lactamases (ESBLs)/AmpC β-lactamases with porin loss combination and the overexpression of efflux pumps (6,7). The high-risk clone, ST11, has been reported to be the dominant sequence type (ST) in CRKP isolates which frequently caused clonal dissemination and outbreaks in healthcare settings in China (8-10). Recently, due to the rapid evolution of such strain under selective pressure, hypervirulent ST11 and ST11 co-producing NDM and KPC have also been consistently reported (11). Moreover, the emergence of new sequence non-ST11 isolates also poses a great challenge to clinicians and microbiologists.
Therefore, more information on the molecular characterization of such strains is needed to effectively control the transmission of CRKP and prevent the outbreaks. In this study, CRKP isolates responsible for clonal dissemination were analyzed for resistant determinants and genetic relatedness as well as integrons to provide data on CRKP isolates.
Methods
Bacterial isolates
In total, eighty-two non-duplicated CRKP isolates were collected from the Department of Laboratory Medicine of the Affiliated Hospital of Xuzhou Medical University from June, 2015 to August, 2016. The strains were obtained from sputum (n=61), blood (n=9), urine (n=6), pus (n=3), secretion (n=1), cerebrospinal fluid (n=1), and catheter (n=1). Species confirmation was performed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS, Bruker Daltonics, Bremen, Germany) and Vitek 2 Compact system (bioMérieux, France). Antimicrobial susceptibility testing towards tigecycline, meropenem, imipenem, cefazolin, cefoxitin, cefepime, ceftriaxone, aztreonam, amikacin, gentamicin, ciprofloxacin, levofoxacin, and piperacillin/tazobactam were also performed by the Vitek 2 Compact system according to the manufacturer’s instructions. Escherichia coli ATCC25922 was used as the quality control. The interpretation of results was based on the Clinical and Laboratory Standards Institute 2016 (12). The breakpoint of Food and Drug Administration (FDA) was used for tigecycline. Clinical characteristics on clinical features and laboratory tests were retrieved from the electronic record. The informed consent was granted by all patients and this study protocols were approved by the Ethics Committee of Affiliated Hospital of Xuzhou Medical University (XYFY2019-KL112-03).
The identification of infection and colonization bacteria were based on the clinical symptoms and signs in individual patients, imaging findings. Moreover, patients must have had fever >38 °C without other recognized cause, or abnormal white blood cell count [leukopenia (<4,000 WBC/mm3) or leukocytosis (≥12,000 WBC/mm3)], and at least two of the following: new onset of purulent sputum or change in the sputum characteristics, increased respiratory secretions or increased suctioning requirements, new onset or worsening of a cough or dyspnea or tachypnea, rales or bronchial breath sounds, or worsening gas exchange.
Detection of antimicrobial resistance determinants
DNA templates were extracted by absorption column method (Tiangen, China). All isolates that exhibited resistance to carbapenem (imipenem or meropenem) were screened for presence of the resistance genes including carbapenemase gene (blaKPC, blaSME, blaGES, blaNDM, blaVIM, blaIMP, blaOXA-48-like), extended spectrumβ-lactamase gene (blaSHV, blaTEM, blaCTX-M1 group, blaCTX-M2 group, blaCTX-M8 group, blaCTX-M9 group) and AmpC β-lactamase genes (blaACC, blaFOX, blaMOX, blaDHA, blaCIT/SPM, and blaEBC) by polymerase chain reaction (PCR) as previously described (10). All purified positive amplicons were sequenced by GENEWIZ Company (Suzhou, China) and subtypes of β-lactamase genes were aligned on blast database.
PCR detection of integrons and RPLP analysis of cassette regions
PCR was performed to screen the presence of integrons and integron cassette regions among the isolates using degenerate primers as described previously (13). Integrase products were digested with HinfI to identify classes of integrons. The amplicons of cassettes with the same HinfI pattern were considered to contain the same variable region. The representative amplicons were selected for DNA sequence by GENEWIZ Company (Suzhou, China). The results of sequencing were aligned in BLAST (http://blast.ncbi.nlm.nih.gov).
Pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis (PFGE) was used for molecular typing and analysis of clone relatedness. Plugs containing genomic DNA were prepared according to previous protocol by Pereira et al. (14). The DNA fragments digested with restriction endonuclease XbaI (TaKaRa Biotechnology, Dalian, China) were separated by PFGE on 1% SeaKem Gold agarose (Lonza, Rockland, ME, United States) in 0.5× TBE (Vicmad, China) buffer using the CHEF Mapper XA PFGE system (Bio-Rad, United States) and the electrophoresis conditions were as follows: running time for 18 h at 6V/cm, temperature at 14 °C, and electrophoretic switch times from 6 to 36 s. The similarity of PFGE patterns was calculated by Dice coefficients, and cluster analysis was performed by unweighted pair group method with arithmetic averages (UPGMA) by the BioNumerics software version 5.10. Isolates were considered to be of the same PFGE cluster when their dice similarity index was ≥80%.
Multi-locus sequence typing
MLST was performed according to the protocol shown on the Klebsiella pneumoniae MLST website (http://www.pasteur.fr/mlst/Kpneumoniae.html). Seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB and tonB) were amplified and sequenced for multilocus sequence typing of carbapenem-resistant Klebsiella pneumoniae (CRKP). Alleles and STs were identified using the MLST database.
Results
Clinical characteristics and antimicrobial susceptibility of CRKP isolates
The CRKP isolates were obtained from patients admitted to 15 wards in our hospital with majority being from neurology ICU (30.5%, 25/82), followed by emergency ICU (23.2%, 19/82), neurosurgery (11.0%, 9/82), critical medicine ICU (8.5%, 7/82), neurology (7.3%, 6/82), respiratory department (3.7%, 3/82), urology (3.7%, 3/82), and neonatal ICU (2.4%, 2/82). The other wards were department of bone marrow transplantation center (n=1), gastroenterology (n=1), orthopaedics (n=1), otorhinolaryngology (n=1), oncology (n=1), geriatric (n=1), and cardiology (n=1).
And all cases were identified as infected with CRKP strains.
All CRKP isolates exhibited resistance to penicillins, cefazolin, cefoxitin, cefepime, ceftriaxone, piperacillin/tazobactam, impenem and meropenem, whereas non-susceptible rates towards levofloxacin, ciprofloxacin, amikacin, and gentamicin were 96.6%, 97.7%, 74.7%, and 92.0% respectively. Furthermore, susceptibility of 100% to tigecycline was observed.
Prevalence of antibiotic resistance determinants
Carbapenemase encoding genes were identified in 80 out of 82 CRKP isolates. According to DNA alignment results, blaKPC-2 (n=75) was predominant followed by blaNDM-5 (n=4), and blaNDM-1 (n=1). 2 of 4 NDM-5-producing isolates were obtained from neonatal ICU while 2 were from department of respiratory and emergency ICU respectively. 1 isolate with blaNDM-1 gene was obtained from bone marrow transplantation center.
Analysis of ESBL genes revealed that blaCTX-M, blaSHV, and blaTEM were identified in 74, 76, and 65 isolates respectively. Of 74 blaCTX-M positive isolates, 55 isolates carried blaCTX-M-65, 16 blaCTX-M-15, 3 blaCTX-M-14, 2 blaCTX-M-27, and 1 blaCTX-M-9. Among 76 blaSHV isolates, the most prevalent ESBLs blaSHV gene was blaSHV-12 (n=47) followed by blaSHV-2a (n=8). The other non-ESBLs blaSHV genes including blaSHV-11 (n=18), blaSHV-1 (n=2) and blaSHV-28 (n=1) were also identified, all of which co-existed with ESBLs. Moreover, blaTEM-1 subtype was identified in all blaTEM isolates (n=65). Screening for AmpC β-lactamase genes revealed that blaDHA-1 (n=46) were predominant followed by blaCMY-2 (n=1) and blaCMY-42 (n=1). The blaSME, blaGES, blaVIM, blaIMP, blaOXA-48-like, blaCTX-M2 group, blaCTX-M8 group, blaACC, blaFOX, blaMOX, and blaEBC were not detected.
Seventy-seven isolates co-carried 2 or more resistant determinants with the combination of blaKPC-2, blaSHV-12, blaTEM-1, blaCTX-M-65 and/or blaDHA-1 being the most common type, accounting for 29.8%. Specifically, isolates co-production of blaKPC-2, blaSHV-12, blaTEM-1, blaCTX-M-65 were identified in 13 (56.5%) strains in neurology ICU, 5 (29.4%) in emergency ICU, 3 (37.5%) in neurosurgery.
Integron identification
No class II and class III were detected. Class I of integrons were detected in 17 isolates: 14 ST11, 2 ST37, and 1 ST15 isolate (Table 1). The length of amplicons of 17 fragments varied from 0.15 kb to 1.9 kb. The DNA sequence analysis of gene cassette arrays revealed 6 distinct profiles with the aadA2 (n=8) being the most prevalent array, and dfrA12-orfF-aadA2 (n=3), dfrA16-aadA2 (n=2), and dfrA17-aadA5 (n=1) were also identified. Of note, we found a novel cassette arrays of integron, aac(6')-II-blaCARB/PSE-1. Additionally, we found integron with the amplicon size at 0.15kb in which not any gene cassettes were present but 5' and 3' conserved segments of class I integron.
Table 1. The distribution of genotypes among 82 CRKP isolates.
| ST | No. | Genotypes [No.] | Integron | Ward | |||
|---|---|---|---|---|---|---|---|
| Carbapenemase | ESBLs | Non-ESBLs | AmpC | ||||
| ST 11 | 68 | KPC-2 [65], NDM-5 [2], NDM-1 [1] | SHV-12 [51], SHV-2a [9], CTX-M-15 [5], CTX-M-65 [60], CTX-M-14 [3], CTX-M-9 [1], CTX-M-27 [1] | SHV-11 [10], TEM-1 [70] | DHA-1 [50], CMY-42 [1] | aadA2 [8], dfrA16-aadA2 [2], dfrA17-aadA5 [1], dfrA12-orfF-aadA2 [1], aac [6’]-II-blaCRAB-1 [1], 5’CS-3’CS [1] | Neurology ICU [22], neurology [6], neurosurgery [8]; emergency ICU [15]; critical care medicine ICU [5]; Respiratory [3]; urinary surgery [3]; othersa [6] |
| ST 48 | 7 | KPC-2 [7] | CTX-M-15 [7] | SHV-11 [6], TEM-1 [6] | CMY-2 [1] | 0 | Neurology ICU [2]; critical care medicine ICU [2]; emergency ICU [2]; cardiology [1] |
| ST 337 | 2 | NDM-5 [2] | CTX-M-15 [2] | SHV-11 [2], TEM-1 [2] | 0 | 0 | Neonatal ICU [2] |
| ST 1 | 1 | 0 | CTX-M-15 [1] | TEM-1 [1] | 0 | 0 | Neurology [1] |
| ST 15 | 1 | KPC-2 [1] | SHV-12 [1], CTX-M-15 [1] | TEM-1 [1] | 0 | dfrA12-orfF-aadA2 [1] | Geriatrics [1] |
| ST 37 | 1 | 0 | SHV-2a [1], CTX-M-27 [1] | 0 | DHA-1 [1] | dfrA12-orfF-aadA2 [1], 5’CS-3’CS [1] | Emergency ICU [1] |
| ST 700 | 1 | KPC-2 [1] | CTX-M-65 [1] | SHV-1 [1], TEM-1 [1] | 0 | 0 | Neurosurgery [1] |
a, geriatrics (n=2), gastroenterology (n=1), otolaryngology (n=1), oncology (n=1), and bone marrow transplantation center (n=1) were included. CRKP, carbapenem-resistant Klebsiella pneumoniae; ESBLs, extended spectrum β-lactamase.
Molecular typing of CRKP isolates
Seven STs were identified among 82 clinical isolates, and ST11 was the most prevalent sequence type accounting for 82.9%, followed by ST48 (n=7) (Table 1). The remaining isolates were identified as ST337 (n=2), ST15 (n=1), ST700 (n=1), ST1 (n=1), ST37 (n=1), and 1 isolate could not be typed.
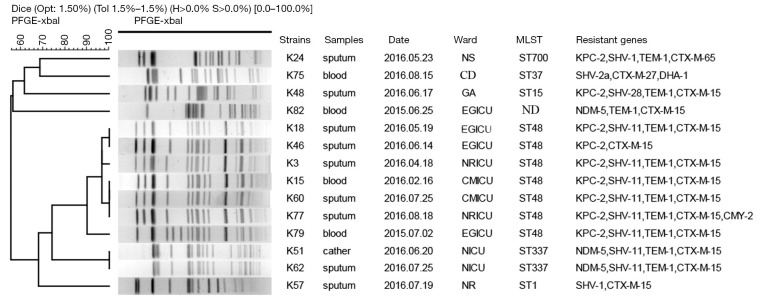
PFGE patterns of XbaI-digested genomic DNA of 82 CRKP isolates revealed 10 different clusters. A predominant cluster consisting of 65 KPC-2-producing CRKP ST11 clone isolates was identified. These isolates were mainly obtained from neurology ICU (n=22), emergency ICU (n=15), neurosurgery (n=8), critical care medicine ICU (n=5) and neurology (n=6) (Figure 1, Table 1). Furthermore, PFGE profiles of other 14 non-ST11 CRKP isolates displayed six different patterns (Figure 2). Among them, 7 KPC-2-producing CRKP ST48 clone displayed the same profiles. All ST48 CRKP isolates were found to harbor blaKPC-2 and blaCTX-M-15 genes indicating clonal dissemination of isolates from neurology ICU (n=2), critical care medicine ICU (n=2), emergency ICU (n=3). In addition, 2 NDM-5-producing CRKP ST337 clone also shared the same PFGE pattern, both of which were isolated from neonatal ICU.
Figure 1.
Dendrogram of PFGE profiles of XbaI-digested DNA restriction fragments from 68 carbapenem-resistant Klebsiella pneumoniae ST11 isolates. The UPGMA algorithm was performed to conduct dendrogram based on the Dice similarity coefficient. Isolates were categorized to be of the same cluster when their dice similarity index was ≥80%. NRICU, neurology ICU; EGICU, emergency ICU; NS, neurosurgery; CMICU, critical care medicine ICU; NR, neurology; RA, respiratory; UL, urology; BMTC, bone marrow transplantation center; GT, gastroenterology; OT, otolaryngology; OL, oncology; OR, orthopaedics; GA, geriatrics.
Figure 2.
Dendrogram of PFGE profiles of XbaI-digested DNA restriction fragments from 14 carbapenem-resistant Klebsiella pneumoniae non-ST 11 isolates. The UPGMA algorithm was performed to conduct dendrogram based on the Dice similarity coefficient. Isolates were categorized to be of the same cluster when their dice similarity index was ≥80%. NRICU, neurology ICU; EGICU, emergency ICU; CMICU, critical care medicine ICU; NICU, neonatal ICU; NS, neurosurgery; GA, geriatrics, NR, neurology; CD, cardiology.
Discussion
CRKP is of increasing clinical concern due to its high transmission capacity and pathogenicity (15). In this study, we described the clonal dissemination of KPC-2-producing CRKP ST11 and ST48 isolates in multiple departments and also provide the first report on clonal spread of NDM-5-producing CRKP ST337 clone.
All CRKP isolates exhibited resistance to all β-lactams and cephalosporins, in addition to high resistance rate to amikacin (>70%), which is consistent with a previous report in China (16). Moreover, amikacin has been reported to show a higher rate of microbiologic clearance than polymyxin B or tigecycline (17), which can still be considered for the infections caused by amikacin-susceptible CRKP isolates. A 100% of sensitivity toward tigecycline observed in our study is in accordance with previous study indicating that tigecycline may be considered as the basis of combination treatments for infections caused by such strains although tigecycline-resistant isolates have been reported (18). Tigecycline exhibited high susceptibility towards carbapenem-resistant K. pneumoniae, however, there is still some limitations of tigecycline therapy. Because of the wide distribution of tissues and low blood concentration, it is often used in combination with other antibiotics.
The high prevalence of blaKPC-2 gene among our isolates is in line with previous reports, suggesting that blaKPC-2 gene remains to be the most common enzyme among carbapenemase in K. pneumoniae isolates (15). The success of KPC is based on both gene and plasmid dissemination and on the clonal spread of K. pneumoniae ST258 and its variants (e.g., ST11). The dissemination of mobile elements may be attributed to high prevalence of blaKPC-2 gene due to frequently reports presence of different sizes of plasmids harboring blaKPC-2 gene (19). Moreover, the blaKPC gene is located on a highly mobile Tn3-related transposon, Tn4401, that can be carried by different transferable plasmids of various incompatibility groups (20). Previous studies demonstrated that the emergence of blaKPC-2 was characterized by two patterns of dispersion: the occurrence of K. pneumoniae harboring blaKPC-2 in the IncL/M transferable plasmid, and the clonal spread of K. pneumoniae harboring blaKPC-2 in Tn4401 different isoforms (21). In addition to KPC enzyme, New Delhi metallo-β-lactamase (NDM) was the only metallo-β-lactamase identified in our study, which included blaNDM-1 and blaNDM-5 subtypes. The NDM-1 enzyme, first identified in K. pneumoniae from Swedish patient with history of hospitalization in India, could hydrolyze all β-lactams besides monobactams (22). To our knowledge, blaNDM-1 gene has been frequently detected in different species of Enterobacteriaceae from multiple countries such as Spain, Dutch, Algeria, and Korea (15). However, the emergence of NDM variants that exhibit high resistance poses a great challenge to treatment of isolates with blaNDM-1 gene. For blaNDM-5, it has been reported that substitutions at positions 88 and 154 on blaNDM-1 resulted in increased resistance to carbapenems and broad-spectrum cephalosporins, moreover, it is also reported that blaNDM-5 gene were carried by higher virulent strain (23). Noteworthy, blaNDM-5 gene has been found to be co-carried with mcr-1 in Escherichia coli from Spain (24) and K. pneumoniae from China (25). Moreover, isolates co-production of NDM-5 and OXA-181 enzyme have also been identified in Escherichia coli from Egypt (26) and K. pneumoniae from Singapore (27). Thus, the prevalence of blaNDM-5 among K. pneumoniae in this study increases the awareness and urgency to implement surveillance on these strains to avoid the outbreaks, especially in the neonatal ICU.
It has been demonstrated that the production of ESBLs/AmpC β-lactamases in combination with porin loss and overexpression of efflux pumps contribute to carbapenem resistance. This might have happened in 2 of our isolates which co-carried blaSHV and blaCTX genes, although no carbapenemase encoding genes were detected. Of note, the blaCTX-M-65 gene was the dominant ESBLs gene in our study, which is inconsistent with other regions of China, where blaCTX-M-15 or blaCTX-M-14 genes were the predominant types of blaCTX-M variant (28,29). Although there is a low prevalence of blaCTX-M-65 gene in China, outbreak of infection caused by CTX-M-65-producing strains has been reported (9). Moreover, the prevalence of blaCTX-M-15 gene (18.2%) herein is higher than other regions of China (10,28), which is similar to those in Europe and America (30), indicating the rapid dissemination of blaCTX-M-15. The blaCTX-M-15 gene was frequently found to be associated with outbreaks caused by multidrug resistant K. pneumonia worldwide (31,32). Meanwhile, high prevalence of CRKP-ST11 isolates co-carrying blaKPC-2, blaSHV-12, blaTEM-1, blaCTX-M-65 and/or blaDHA-1 in our hospital was in line with previous studies (33), which may be mainly attributed to spread of elements such as plasmids or clonal dissemination of such strains. Altogether, multiple resistance determinants among CRKP isolates suggested that rapid spread of mobile genetic elements such as plasmids or transposons may play a key role in the resistant determinants under the selective pressure produced by the widely used antimicrobial agents in clinical therapy.
Furthermore, class I integron is the most common and widespread between different genera (34). However, this study found a quite lower prevalence of Class I integron in CRKP than that found in ESBL-producing Enterobacteriaceae in China (35). Existence of atypical integrons and regional differences may explain the imbalance of this phenomenon. A novel cassette arrays of integron, aac(6')-II-blaCARB/PSE-1, as far as we know, has not been identified.
Different from the high prevalence of ST258 in America, ST11 is the dominant sequence type in China which spreads rapidly around the healthcare settings (36). The ST11 clone is a single-locus variant (tonB) from ST258, which has been identified worldwide, especially in Asian regions such as Singapore, Korea, and Japan. Andrade et al. (11) reported that ST11 clone exhibited multidrug resistance phenotype with high prevalence of virulence factors favoring the colonization, biofilm formation, and defense against phagocytosis, which can explain the persistence and clonal spread successfully worldwide. Up to date, the outbreaks caused by ST11 have been reported in China and abroad, with cabapenemase coding gene being central to its rapid dissemination, especially blaKPC-2 and blaNDM (37). In our study, clonal dissemination of KPC-2-producing CRKP ST11 isolates were identified in multiple departments, mostly in the department of neurology and ICU, which is in line with previous studies (38). Evidence that neurology and ICU are the main departments where CRKP isolates spread rapidly, which may result from medical equipment used for invasive therapy, hence, high-risk wards such as neurology and ICU might be the focus of active surveillance.
Notably, this is the first report on clonal dissemination of KPC-2-producing CRKP ST48 clone. Albeit the CRKP ST48 clone has been identified in Korea and Thailand (39,40) and some of them exhibited tigecycline resistance (41), the clonal dissemination of ST48 CRKP isolates have never been reported. Furthermore, the clonal dissemination of such strains demonstrated that ST48 is a potential high-risk clone that needs close attention. Noteworthy, a minor clonal dissemination of NDM-5-producing ST337 clone isolates were also identified in neonatal ICU ward in our study, so far, this is the first report, indicating rapid evolution of blaNDM gene and CRKP isolates.
Some limitations of this study exist. Firstly, active surveillance was not performed during the study period, otherwise the isolation rate of CRKP could be higher than we found and the sample size of study could be expanded to further realize the clinical characteristics. Moreover, fecal samples from healthy carrier were not included in our study, which could provide an extensive description of clonal dissemination of such strains.
Conclusions
In summary, the clonal dissemination of KPC-2-producing CRKP ST11 clone was identified in multiple departments with neurology ICU being the most common, indicating extensive cross-transmission of CRKP isolates among high-risk departments in our hospital. This first report on the clonal dissemination of KPC-2 producing CRKP ST48 clone and a minor clonal dissemination of NDM-5-producing ST337 clone isolates hints at the potential occurrence of outbreak caused by such strains. Due to limited selective clinical treatment for infections caused by these strains in our hospital, active surveillance and implementation of infection control measures are therefore urgently needed.
Acknowledgments
Funding: This research was supported by the National Natural Science Foundation of China (81471994), Jiangsu Provincial Natural Science Foundation (BK20151154), Jiangsu Provincial Medical Talent (ZDRCA2016053), Six Talent Peaks Project of Jiangsu Province (WSN-135), Advanced Health Talent of Six-One Project of Jiangsu Province (LGY2016042), and Jiangsu Provincial Commission of Health and Family Planning Research Project (H201631).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The informed consent was granted by all patients and this study protocols were approved by the Ethics Committee of Affiliated Hospital of Xuzhou Medical University (XYFY2019-KL112-03).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Guh AY, Limbago BM, Kallen AJ. Epidemiology and prevention of carbapenem-resistant Enterobacteriaceae in the United States. Expert Rev Anti Infect Ther 2014;12:565-80. 10.1586/14787210.2014.902306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poulakou G, Lagou S, Karageorgopoulos DE, et al. New treatments of multidrug-resistant Gram-negative ventilator-associated pneumonia. Ann Transl Med 2018;6:423. 10.21037/atm.2018.10.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen P, Zhang Y, Li G, et al. Characterization of the genetic environment of the blaKPC-2 gene among Klebsiella pneumoniae isolates from a Chinese Hospital. Braz J Infect Dis 2016;20:384-8. 10.1016/j.bjid.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 2009;9:228-36. 10.1016/S1473-3099(09)70054-4 [DOI] [PubMed] [Google Scholar]
- 5.Qian H, Liu G, Chen Y, et al. Increasing clinical resistance rate of Shigella sonnei to cefotaxime in Jiangsu Province, China, between 2012 and 2015. Ann Transl Med 2018;6:207. 10.21037/atm.2018.05.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundmann H, Livermore DM, Giske CG, et al. Carbapenem-non-susceptible Enterobacteriaceae in Europe: conclusions from a meeting of national experts. Euro Surveill. 2010;15(46). 10.2807/ese.15.46.19711-en [DOI] [PubMed] [Google Scholar]
- 7.Ho PL, Cheung YY, Wang Y, et al. Characterization of carbapenem-resistant Escherichia coli and Klebsiella pneumoniae from a healthcare region in Hong Kong. Eur J Clin Microbiol Infect Dis 2016;35:379-85. 10.1007/s10096-015-2550-3 [DOI] [PubMed] [Google Scholar]
- 8.Liang Y, Yin X, Zeng L, et al. Clonal replacement of epidemic KPC-producing Klebsiella pneumoniae in a hospital in China. BMC Infect Dis 2017;17:363. 10.1186/s12879-017-2467-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou T, Zhang Y, Li M, et al. An outbreak of infections caused by extensively drug-resistant Klebsiella pneumoniae strains during a short period of time in a Chinese teaching hospital: epidemiology study and molecular characteristics. Diagn Microbiol Infect Dis 2015;82:240-4. 10.1016/j.diagmicrobio.2015.03.017 [DOI] [PubMed] [Google Scholar]
- 10.Cheng L, Cao XL, Zhang ZF, et al. Clonal dissemination of KPC-2 producing Klebsiella pneumoniae ST11 clone with high prevalence of oqxAB and rmtB in a tertiary hospital in China: results from a 3-year period. Ann Clin Microbiol Antimicrob 2016;15:1. 10.1186/s12941-015-0109-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrade LN, Vitali L, Gaspar GG, et al. Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), QnrS1-, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone. J Clin Microbiol 2014;52:2530-5. 10.1128/JCM.00088-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CLSI M100S. Performance Standards for Antimicrobial Suscrptibility Testing, 26th edn. Wayne, PA: CLSI, 2016. [Google Scholar]
- 13.Gu B, Tong M, Zhao W, et al. Prevalence and characterization of class I integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J Clin Microbiol 2007;45:241-3. 10.1128/JCM.01318-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira PS, Borghi M, de Araújo CF, et al. Clonal Dissemination of OXA-370-Producing Klebsiella pneumoniae in Rio de Janeiro, Brazil. Antimicrob Agents Chemother 2015;59:4453-6. 10.1128/AAC.04243-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 2014;20:821-30. 10.1111/1469-0691.12719 [DOI] [PubMed] [Google Scholar]
- 16.Bogan C, Kaye KS, Chopra T, et al. Outcomes of carbapenem-resistant Enterobacteriaceae isolation: matched analysis. Am J Infect Control 2014;42:612-20. 10.1016/j.ajic.2014.02.013 [DOI] [PubMed] [Google Scholar]
- 17.Satlin MJ, Kubin CJ, Blumenthal JS, et al. Comparative effectiveness of aminoglycosides, polymyxin B, and tigecycline for clearance of carbapenem-resistant Klebsiella pneumoniae from urine. Antimicrob Agents Chemother 2011;55:5893-9. 10.1128/AAC.00387-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taniguchi Y, Maeyama Y, Ohsaki Y, et al. Co-resistance to colistin and tigecycline by disrupting mgrB and ramR with IS insertions in a canine Klebsiella pneumoniae ST37 isolate producing SHV-12, DHA-1 and FosA3. Int J Antimicrob Agents 2017;50:697-8. 10.1016/j.ijantimicag.2017.09.011 [DOI] [PubMed] [Google Scholar]
- 19.Yang J, Ye L, Guo L, et al. A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin Microbiol Infect 2013;19:E509-15. 10.1111/1469-0691.12275 [DOI] [PubMed] [Google Scholar]
- 20.Cuzon G, Naas T, Truong H, et al. Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerg Infect Dis 2010;16:1349-56. 10.3201/eid1609.091389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez SA, Pasteran FG, Faccone D, et al. Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clin Microbiol Infect. 2011;17:1520-4. 10.1111/j.1469-0691.2011.03600.x [DOI] [PubMed] [Google Scholar]
- 22.Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 2009;53:5046-54. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mei YF, Liu PP, Wan LG, et al. Virulence and Genomic Feature of a Virulent Klebsiella pneumoniae Sequence Type 14 Strain of Serotype K2 Harboring blaNDM-5 in China. Front Microbiol 2017;8:335. 10.3389/fmicb.2017.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang RS, Feng Y, Lv XY, et al. Emergence of NDM-5- and MCR-1-Producing Escherichia coli Clones ST648 and ST156 from a Single Muscovy Duck (Cairina moschata). Antimicrob Agents Chemother 2016;60:6899-902. 10.1128/AAC.01365-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du H, Chen L, Tang YW, et al. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis 2016;16:287-8. 10.1016/S1473-3099(16)00056-6 [DOI] [PubMed] [Google Scholar]
- 26.Gamal D, Fernández-Martínez M, El-Defrawy I, et al. First identification of NDM-5 associated with OXA-181 in Escherichia coli from Egypt. Emerg Microbes Infect 2016;5:e30. 10.1038/emi.2016.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balm MN, La MV, Krishnan P, et al. Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin Microbiol Infect 2013;19:E421-3. 10.1111/1469-0691.12247 [DOI] [PubMed] [Google Scholar]
- 28.Yu J, Tan K, Rong Z, et al. Nosocomial outbreak of KPC-2- and NDM-1-producing Klebsiella pneumoniae in a neonatal ward: a retrospective study. BMC Infect Dis 2016;16:563. 10.1186/s12879-016-1870-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao SZ, Wang S, Wu WM, et al. The Resistance Phenotype and Molecular Epidemiology of Klebsiella pneumoniae in Bloodstream Infections in Shanghai, China, 2012-2015. Front Microbiol 2017;8:250. 10.3389/fmicb.2017.00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill 2008;13(47). [PubMed] [Google Scholar]
- 31.Valsdottir F, Elfarsdottir Jelle A, Gudlaugsson O, et al. Long-lasting outbreak due to CTX-M-15-producing Klebsiella pneumoniae ST336 in a rehabilitation ward: report and literature review. J Hosp Infect 2017;97:42-51. 10.1016/j.jhin.2017.04.002 [DOI] [PubMed] [Google Scholar]
- 32.Machuca J, López-Cerero L, Fernández-Cuenca F, et al. Characterization of an outbreak due to CTX-M-15-producing Klebsiella pneumoniae lacking the blaOXA-48 gene belonging to clone ST405 in a neonatal unit in southern Spain. J Antimicrob Chemother 2016;71:2353-5. 10.1093/jac/dkw137 [DOI] [PubMed] [Google Scholar]
- 33.Li H, Zhang J, Liu Y, et al. Molecular characteristics of carbapenemase-producing Enterobacteriaceae in China from 2008 to 2011: predominance of KPC-2 enzyme. Diagn Microbiol Infect Dis 2014;78:63-5. 10.1016/j.diagmicrobio.2013.10.002 [DOI] [PubMed] [Google Scholar]
- 34.Goudarzi M, Fazeli M, Azad M, et al. Carriage of Class 1 and Class 2 Integron in Multidrug Resistant Pseudomonas aeruginosa Isolated from Burn Patients in Tehran Hospitals, Iran. West Indian Med J 2015;65:32-9. [DOI] [PubMed] [Google Scholar]
- 35.Harada K, Shimizu T, Mukai Y, et al. Phenotypic and Molecular Characterization of Antimicrobial Resistance in Klebsiella spp. Isolates from Companion Animals in Japan: Clonal Dissemination of Multidrug-Resistant Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae. Front Microbiol 2016;7:1021. 10.3389/fmicb.2016.01021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi Y, Wei Z, Ji S, et al. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 2011;66:307-12. 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- 37.Savov E, Politi L, Spanakis N, et al. NDM-1 Hazard in the Balkan States: Evidence of the First Outbreak of NDM-1-Producing Klebsiella pneumoniae in Bulgaria. Microb Drug Resist 2018;24:253-9. 10.1089/mdr.2017.0230 [DOI] [PubMed] [Google Scholar]
- 38.Nouvenne A, Ticinesi A, Lauretani F, et al. Comorbidities and disease severity as risk factors for carbapenem-resistant Klebsiella pneumoniae colonization: report of an experience in an internal medicine unit. PLoS One 2014;9:e110001. 10.1371/journal.pone.0110001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin SY, Bae IK, Kim J, et al. Resistance to carbapenems in sequence type 11 Klebsiella pneumoniae is related to DHA-1 and loss of OmpK35 and/or OmpK36. J Med Microbiol 2012;61:239-45. 10.1099/jmm.0.037036-0 [DOI] [PubMed] [Google Scholar]
- 40.Netikul T, Kiratisin P. Genetic Characterization of Carbapenem-Resistant Enterobacteriaceae and the Spread of Carbapenem-Resistant Klebsiella pneumonia ST340 at a University Hospital in Thailand. PLoS One 2015;10:e0139116. 10.1371/journal.pone.0139116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahn C, Yoon SS, Yong TS, et al. The Resistance Mechanism and Clonal Distribution of Tigecycline-Nonsusceptible Klebsiella pneumoniae Isolates in Korea. Yonsei Med J 2016;57:641-6. 10.3349/ymj.2016.57.3.641 [DOI] [PMC free article] [PubMed] [Google Scholar]