Abstract
Background
Epithelial-mesenchymal transition (EMT) of the retinal pigment epithelial (RPE) cells is a critical step in the pathogenesis of proliferative vitreoretinopathy (PVR). Some microRNAs (miRNAs) participate in regulating RPE cell EMT as post-transcriptional regulators. However, the function of miR-194 in RPE cell EMT remains elusive. Here, the role of miR-194 in PVR was investigated.
Methods
Retinal layers were obtained using laser capture microdissection (LCM). Gene expression at the mRNA and protein level in the tissues and cells was examined using quantitative reverse transcription (RT)-polymerase chain reaction and Western blotting, respectively. The related protein expression was observed by immunostaining. The effect of miR-194 on RPE cell EMT was examined by gel contraction, wound healing, and cell migration assays. RNAseq was performed in ARPE-19 with transfection of pSuper-scramble and pSuper-miR-194. The target gene of miR-194 was identified and confirmed via bioinformatics analysis and dual-luciferase reporter assay. ARPE-19 (adult retinal pigment epithelium-19) cells were treated with transforming growth factor (TGF)-β1 in the same fashion as the in vitro RPE cell EMT model. A PVR rat model was prepared by intravitreous injection of ARPE-19 cells with plasma-rich platelets.
Results
miR-194 was preferentially expressed in the RPE cell layer compared with the outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell layer in rat retina. RNAseq analysis indicated that miR-194 overexpression was involved in RPE cell processes, including phagocytosis, ECM-receptor interaction, cell adhesion molecules, and focal adhesion. miR-194 overexpression significantly inhibited the TGF-β1-induced EMT phenotype of RPE cells in vitro. Zinc finger E-box binding homeobox 1 (ZEB1), a key transcription factor in EMT, was confirmed as the direct functional target of miR-194. Knockdown of ZEB1 attenuated TGF-β1-induced α-smooth muscle actin expression in ARPE-19 cells, and overexpression of miR-194 could significantly reduce the expression of some genes which were up-regulated by ZEB1. Exogenous miR-194 administration in vivo effectively suppressed PVR in the rat model, both functionally and structurally.
Conclusions
Our findings demonstrate for the first time that miR-194 suppresses RPE cell EMT by functionally targeting ZEB1. The clinical application of miR-194 in patients with PVR merits further investigation.
Keywords: miR-194, ZEB1, epithelial-mesenchymal transition (EMT)
Introduction
Proliferative vitreoretinopathy (PVR) complicates 5–10% of rhegmatogenous retinal detachment (RRD) cases and is the major cause of surgical failure (1). The risk factors for PVR occurrence include longer RRD duration, greater extent of detachment, vitreous hemorrhage, intraocular inflammation, increased retinal tear size, extensive cryopexy, and laser retinopexy, failure to close retinal breaks, along with perioperative scleral perforation and hemorrhage (2). PVR is characterized by the formation of an abnormal contractile epiretinal membrane (ERM), and surgical management of PVR may require extensive ERM peeling. Despite recent improvements in surgery, isolating a standard treatment has remained elusive, while preventive measures against PVR are still not effective (2).
Currently, the etiology of PVR is not completely understood. What is known, however, is that the dislocation of cells to the vitreous cavity during retinal detachment is widely believed to be a contributing factor. The ERM is composed of extracellular matrix (ECM) and various cell types in the vitreous cavity and on the epiretinal surface (3). The cells in the ERM are retinal pigment epithelial (RPE) cells, fibroblasts, glial cells, and to a much lesser extent, macrophages (4-7). RPE cells are considered to have a significant role in PVR. They can trigger PVR development by dedifferentiating and migrating through a retinal break and proliferating on the retinal surface (8). Epithelial-mesenchymal transition (EMT) of RPE cells directly mediates the pathogenesis of PVR.
EMT is considered to be a highly dynamic process, where epithelial cells lose their polarity and intercellular junctions to become mesenchymal cells with migratory and invasive properties (9-11). It is an essential process in numerous developments in the human body, including mesoderm and neural tube formation, wound healing in organ fibrosis, and the initiation of metastasis for tumor progression. RPE cells undergoing EMT are transformed into fibroblast-like cells. They proliferate and produce ECM components, and participate in those fibrotic sequelae constituting the major pathological processes of PVR (12). Targeting RPE cell EMT may provide a promising strategy for developing potential prophylactic or therapeutic agents of PVR.
MicroRNAs (miRNAs) are short, single-stranded non-coding RNAs (~22 nucleotides in length) that can negatively regulate gene expression by binding to complementary sites in the target mRNA at the RNA-induced silencing complex (RISC) and inducing mRNA degradation or translation inhibition (13). Some miRNAs, such as miR-182, miR-21, miR-29b, miR-200, miR-204/211, and miR-124, have been recognized as molecular regulators and potential therapeutic targets in PVR, and all functionally target the EMT-inducer receptors or downstream signaling molecules of EMT (14-20). Moreover, the miRNA expression profile of RPE cells under oxidative stress conditions has revealed the regulatory functions of several miRNAs in retinitis pigmentosa (RP), a genetic disease-inducing progressive death in RPE cells and photoreceptors (21).
Myriad studies suggest that miR-194 plays a critical role in chondroitin sulfate biosynthesis (22), hepatic cell function (23), psoriasis (24), and tumor biology (25-29). However, miR-194 functions in tumor cell EMT remain controversial. Most studies support the premise that miR-194 functions as a tumor suppressor and plays a pivotal inhibitory role in EMT in some cancer cells (25-27), whereas two reports have provided opposing evidence indicating that miR-194 is a tumor promoter (28,29). Interestingly, a recent study demonstrated that miR-194 upregulation prevented glutamate excitotoxicity in developing chick retina (30). However, the function of miR-194 in RPE cell EMT remains unknown.
Vertebrates have two homologous zinc finger E-box binding homeobox (ZEB) proteins with zinc finger DNA-binding motifs as complex transcription factors. ZEB1 (ZFHX1A/TCF8/δEF1/Nil-2α) and ZEB2 (ZFHX1B/SIP1), which are ZEB family members, are CDH1 (E-cadherin) repressors that have been implicated in EMT, tumorigenesis, and metastasis (31). ZEB1 has a distinct common domain architecture consisting of the p300-binding domain and four zinc finger domains in the N-terminal region, including the SMAD-binding domain, homeodomain, and the COOH terminal-binding protein (CtBP)-binding domain in the middle region, along with three zinc finger domains in the C-terminal region (31). The structural complexity regulates EMT via the TGF-β or bone morphogenetic protein (BMP) signaling pathway, the NF-κB pathway, and the Notch signaling pathway (32).
In the present study, we examined the miR-194 expression pattern in the retina and retinal cells, its effects on in vitro and in vivo PVR models, and explored the mechanism of miR-194 protection in PVR.
Methods
Chemicals and reagents
All cell culture reagents without special specifications were purchased from Life Company. All chemicals were purchased from Sigma. TRIzol for RNA isolation and PrimeScript® RT Master Mix for reverse transcription (RT) were acquired from Takara Biotechnology (Dalian, China). The real-time quantitative RT-polymerase chain reaction (qRT-PCR) reagents were purchased from Tiangen Biotech (Beijing, China). The antibodies against GAPDH (ab8245), nectin-1 (ab66985), ZO1 (ab96587), OCLN (ab216327), goat anti-rabbit immunoglobulin G (IgG) H&L (Cy3®) preabsorbed (ab6939), goat anti-rabbit IgG H&L [fluorescein isothiocyanate (FITC)] preabsorbed (ab7086), were acquired from Abcam, and those against α-smooth muscle actin (α-SMA, 14395-1-AP), and ZEB1 (21544-1-AP) were acquired from Proteintech. Transforming growth factor β1 (TGF-β1, HZ-1011) was purchased from Sino Biologicals, and an optimum cutting temperature (OCT) compound was purchased from Sakura Finetechnical. pSUPER vector (VEC-PBS-0002) was purchased from OligoEngine (Seattle, WA, USA), AgomiR-194 was purchased from Tuoran Biotech (China), and XhoI and NotI were purchased from NEB Biolab. DH5α competent cells, Real Universal fluorescent quantitative premixed reagent (SYBR Green), and DNA agarose gel recovery kit were purchased from Tiangen Biotech. T4 DNA ligase was purchased from TaKaRa Bio Inc. (Shiga, Japan), while the LipoFilter transfection kit was purchased from Hanbio Biotech Co. Ltd.
Animal models
The male Sprague-Dawley (SD) rats and Dark Agouti (DA) rats (2–4 months old) used in this study were purchased from Bikai Biotech (Shanghai, China). They were bred on a 12/12-h light/dark cycle. All surgical procedures were performed after the animals were anesthetized with an intraperitoneal injection of pentobarbital (40 mg/kg body weight). The rats were sacrificed with sodium pentobarbital overdose.
Laser capture microdissection (LCM) of the rat retina
LCM was performed according to a previous report (33) with some modifications. Briefly, rat eyecups were freshly enucleated, and cryostat sections (10 µm) were prepared on PEN membrane slides (Leica, Wetzlar, Germany). All procedures were performed under RNase-free conditions. The RPE layer, inner nuclear layer (INL), and outer nuclear layer (ONL) were microdissected with a Leica AS LMD system. The excised layers were separately collected into tubes under gravity, minimizing sample damage and ensuring a contamination-free process. The separated retinal layers were then collected for subsequent total RNA extraction.
PVR rat model preparation
Experimental PVR models were prepared by intravitreal injection of platelet-rich plasma (PRP) containing human adult RPE-19 (ARPE-19) cells (34). Briefly, whole blood was collected from the tail vein into EDTA-treated tubes and then centrifuged at 180 ×g for 5 min. The supernatant was PRP. For PVR model preparation, SD rats were intravitreally injected with 8 µL PRP containing 3×106 ARPE-19 cells; the normal control was intravitreally injected with an equal volume of PBS. The intervention group was injected with ARPE-19 cells + PRP + agomiR-194 (0.1 nmol/eye). Color fundus photography was performed using APS-AER (Kanghuaruiming S&T, Chongqing, China) at 2 weeks post-injection.
Cell culture
The ARPE-19 cells were purchased from American Type Cell Culture (Manassas, VA, USA). Passage (P) 22–28 ARPE-19 cells were used for the experiments. The cells were maintained in Dulbecco’s modified Eagle’s minimum essential medium (DMEM)/F-12 supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. All cultures were maintained in a humidified incubator at 37 °C with an atmosphere of 5% CO2 and 95% air.
miR-194 overexpression construct
The miR-194 hairpin was formed after annealing forward (gatcccctgtaacagcaactccatgtggattcaagagatccacatggagttgctgttacatttttggaaa) and reverse (agcttttccaaaaatgtaacagcaactccatgtggatctcttgaatccacatggagttgctgttacaggg) primers and ligated into BglII/HindIII-digested pSUPER vector according to the manufacturer’s instructions. All constructs used for transfection were confirmed by sequencing before they were prepared using Plasmid Maxi Prep (Tiangen).
Isolation of total RNAs and real-time qRT-PCR quantification of gene expression
Total RNA from cells and tissues was isolated with TRIzol (Takara Biotechnology) according to the manufacturer’s instructions. RT was performed using the PrimeScript® RT Master Mix. The changes in the target mRNA expression levels were quantified by qPCR using Super PreMix Plus (SYBR Green) kit (Tiangen). The PCR cycling conditions were 95 °C for 15 min and 40 cycles with 95 °C for 15 sec, 60 °C for 20 sec, and 72 °C for 40 sec. The relative gene expression in the samples was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels using the comparative threshold cycle (2–ΔΔCT) method.
miRNA was quantified using stem-loop-based RT-PCR, as previously reported (35). Briefly, total RNA was extracted from cultured cells and retinal samples. The miRNA levels were measured using qRT-PCR. A 45-bp universal stem-loop sequence and 6 bp at the 3' end complementary to the miR-194 3' end sequence was used as the primer to reverse-transcribe miR-194 from rat complementary DNA (cDNA) (gtcgtatccagtgcagggtccgaggtattcgcactggatacgactgtaac) using RevertAid Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA). miR-194-specific forward primer (gccgcctaatgcccctaaaaatcc) and a commonly used reverse primer (gtgcagggtccgaggt) were used for PCR. U6 (forward primer: cttcggcagcacatatactaaaa, reverse primer: gaatttgcgtgtcatccttg) was used for the internal control. Amplifications were performed in a Bio-Rad cycler using FastStart Universal SYBR Green Master Mix (Tiangen). For the negative control, water was used in place of cDNA. The ZEB1 small interfering RNA (si-ZEB1) sequence was shown in Table S1 and synthesized from Tuoran Biotech.
Table S1. Sequences of siRNAs.
| Oligo name | Sequence |
|---|---|
| Human siZEB1#1-forward | CCUCUCUGAAAGAACACAUUA |
| Human siZEB1#1-reverse | UAAUGUGUUCUUUCAGAGAGG |
| Human siZEB1#2-forward | GCUGCCAAUAAGCAAACGAUU |
| Human siZEB1#2-reverse | AAUCGUUUGCUUAUUGGCAGC |
| Human siZEB1#3-forward | GCUGUUGUUCUGCCAACAGUU |
| Human siZEB1#3-reverse | AACUGUUGGCAGAACAACAGC |
Transfection of pSUPER-miR-194 into ARPE-19 cells
ARPE-19 cells were seeded in 6-well dishes at 70–80% density per well 24 h before transfection. The cells were transfected with pSuper-miR-194 or empty vector as a control, using Lipofectamine 2000 (Invitrogen) (2 µg per well) according to the manufacturer’s instruction.
RNA sequencing (RNAseq) analysis
Total RNA samples extracted from miR-194 overexpression and scramble groups were used for RNAseq using the BGISEQ-500 platform. Subsequent bioinformatics analysis was performed by BGI (Beijing, China). The RNAseq mapping statistics are presented in Table S2.
Table S2. RNA-Seq mapping statistics.
| Mapping type | Control | miR-194 | |||
|---|---|---|---|---|---|
| Reads number | Percentage (%) | Reads number | Percentage (%) | ||
| Total clean reads | 24,054,199 | 99.66 | 24,028,309 | 99.55 | |
| Total mapped reads | 20,682,738 | 85.98 | 21,282,186 | 88.57 | |
| Perfect match | 19,191,436 | 79.78 | 19,725,930 | 82.09 | |
| Mismatch | 1,491,302 | 6.2 | 1,556,256 | 6.48 | |
| Unique match | 18,334,354 | 76.22 | 18,993,903 | 79.05 | |
| Multi-position match | 2,348,384 | 9.76 | 2,288,283 | 9.52 | |
| Total unmapped reads | 3,371,461 | 14.02 | 2,746,123 | 11.43 | |
Wound healing assay
ARPE-19 cells transfected with miR-194 or empty vector were seeded into 24-well plates and scratched with 200-µL pipette tips after 8-h incubation. To remove floating cells, the cells were washed twice with phosphate-buffered saline (PBS). Thereafter, a fresh serum-free medium with and without 10 ng/mL TGF-β1 was added to the cells. At 0, 24, and 48 h after wounding, the width of the scratch was measured and compared among the groups under an inverted microscope.
Immunostaining and microscopy
ARPE-19 cells were seeded on coverslips in culture medium at a density of 1×105 cells per well in 24-well plates. After treatment, ARPE-19 cells or rat retina were fixed with 4% paraformaldehyde (PFA) in PBS for 24 h. The rat retinas were embedded in OCT compound after dehydration in gradient sucrose, and 8-µm thick sections were prepared. Before immunostaining, the cells and retinas were permeabilized in 0.25% Triton X-100 for 10 min (cells) or 0.5% Triton X-100 for 30 min (retinas). After 3 rinses in PBS, the samples were blocked with 1% bovine serum albumin (BSA) for 30 min at room temperature and incubated with primary antibodies overnight at 4 °C. The detailed information of the primary antibodies used in immunofluorescence staining is listed in Table S3. The samples were next incubated with Cy3/FITC-conjugated secondary antibodies for 1 h at room temperature after they had been washed 3 times with PBS. The nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). The staining was visualized and captured by Nikon A1R confocal microscopy.
Table S3. The information of primary antibodies used in western blotting and immunofluorescence staining.
| Primary antibody | Company | Product catalog No. | Dilution ratio | |
|---|---|---|---|---|
| Western blotting | Immunofluorescence staining | |||
| GAPDH | Abcam | ab8245 | 1:2,000 | – |
| Nectin-1 | Abcam | ab66985 | 1:1,000 | 1:100 |
| ZO1 | Abcam | ab96587 | 1:1,000 | 1:50 |
| Occludin | Abcam | ab216327 | – | 1:100 |
| α-SMA | Proteintech | 14395-1-AP | 1:4,000 | 1:100 |
| ZEB1 | Proteintech | 21544-1-AP | 1:1,000 | 1:100 |
| SNAI1 | Proteintech | 13099-1-AP | 1:1,000 | – |
Collagen gel contraction assay
Briefly, 24-well plates were coated with 1 mL 1% BSA for 1 h at 37 °C. ARPE-19 cells transfected with miR-194 or empty vector were harvested and resuspended in serum-free DMEM/F-12 medium. Collagen I, at a final concentration of 2 mg/mL (Thermo Fisher Scientific) with cell suspension (final cell density, 2.5×105/well), sterile double-distilled water (ddH2O), and sterile 1 N NaOH (0.025-fold of the volume of collagen) were prepared and mixed on ice. After the gel had been polymerized, serum-free DMEM/F-12 medium (0.5 mL) containing 10 ng/mL TGF-β1 was added on top of the gel. Photographs were taken after 24 or 48 h for quantitation of the ratio of the gel contraction area using ImageJ software [version 1.46r; National Institutes of Health (NIH), Bethesda, MD, USA].
Transwell migration assay
To measure cell migration, 8-µm pore size culture inserts (Transwell, Costar) were placed into the wells of 24-well culture plates with separate upper and lower chambers. ARPE-19 cells transfected with miR-194 or empty vector were harvested by trypsinization and washed once in PBS, and 5×104 cells were added to the upper chamber. In the lower chamber, 400 µL DMEM/F12 medium containing 10% FBS was added. After 24 h incubation at 37 °C with 5% CO2, the cells that had migrated through the pores were stained with crystal violet, and 5 fields of vision were counted under a Zeiss microscope using a 20× objective.
Western blotting
The samples were lysed in radioimmunoprecipitation assay buffer (Beyotime, Shanghai, China) supplemented with proteinase inhibitors and phosphatase inhibitors (Selleck, Houston, TX, USA), and placed on ice for 30 min (cells) or 60 min (tissue). Then, the protein concentrations were measured using a bicinchoninic acid protein assay reagent kit (Pierce, Rockford, IL, USA). Equal amounts of protein (30 µg) were loaded and separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). After blocking in 5% non-fat milk, the membranes were immunoblotted with primary antibodies at 4 °C overnight. After washing in PBS-T (PBS containing 0.1% Tween-20) 3 times (10 min per wash), the membranes were incubated with horseradish peroxidase-labeled secondary antibodies based on the source of their primary antibodies for 2 h at room temperature. Immunopositive bands were visualized using an enhanced chemiluminescence detection kit (Pierce) using Tanon 500S (Shanghai, China). The band intensities were analyzed using ImageJ version 1.41 (NIH) and normalized to GAPDH. More detailed information, including the dilution ratio of the primary antibodies used in Western blotting, is listed in Table S3.
Bioinformatics analysis of miR-194 target genes
The interaction between the miR-194 seed sequence and the 3' untranslated region (3' UTR) sequence of the target genes was predicted using the web-based miRNA target prediction software PicTar (pictar.mdc-berlin.de) and TargetScan (www.targetscan.org).
Dual-luciferase reporter assay
To verify the miR-194 sequence targeting the ZEB1 3' UTR, dual-luciferase reporter assay was performed according to a previous report (36) with slight modifications. The human ZEB1 sequence was amplified using human ARPE-19 cDNA as the template and primers with XhoI and NotI restriction sites (forward primer: atcgctcgagcttctataaacagtgttgggaac, reverse primer: aagcggccgcgatgaatcaaatgcaggatgaca). The PCR amplicon and psiCHECK2 vector (Promega, Madison, WI, USA) were digested with XhoI and NotI at 37 °C for 4 h and then separated by 1% agarose gel electrophoresis. The digested psiCHECK2 vector and PCR amplicon were recovered from the gel with a DNA agarose gel recovery kit (Tiangen) and ligated at 16 °C for 1 h. Then, the ligation reaction was transformed into competent DH5α cells. After growth in an ampicillin agar plate overnight, single colonies were seeded in 5-mL ampicillin-Luria-Bertani medium at 37 °C for 16 h and sequenced. The recombinant psiCHECK2-3'UTR-ZEB1 with the correct sequence was used for subsequent co-transfection experiments. HEK293 cells were plated at a density of 5×105 cells per well in 24-well plates. A mixture of 1.25 µL 20 µM agomir, mutant, or scramble, and 500 ng psiCHECK2-3'UTR-ZEB1 using 2 µL LipoFilter in OptiMEM was incubated for 6 h at 37 °C, and then 1 mL complete DMEM/F12 medium with FBS was added to each well and incubated for 48 h in a humidified atmosphere of 5% CO2 in air. Subsequently, the cells were washed twice with PBS and lysed by Passive Lysis Buffer (Promega). Luciferase activity was measured using a dual-luciferase assay kit (Promega) and a GloMax luminometer (Promega). Renilla luciferase activity was measured and normalized to firefly luciferase activity using the dual-luciferase assay kit (Promega). At least 3 independent experiments per assay were performed.
Statistical analysis
All statistical analysis was performed in GraphPad Prism 6.0 using one-way analysis of variance with Tukey’s honestly significant difference post hoc test or unpaired Student’s t-test. All numerical data are presented as the mean ± SEM; P<0.05 was considered statistically significant. All cell experiments were repeated at least 3 times.
Results
miR-194 was abundant in mammalian RPE cells
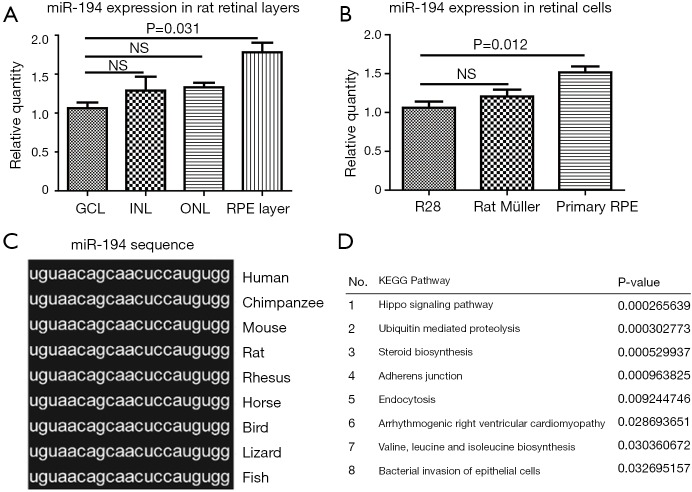
To reveal the role of miR-194 in the adult mammalian retina, we first examined its expression pattern in rat retina. LCM-assisted qRT-PCR (33) showed that miR-194 was expressed in all retinal cell layers, with expression being highest in RPE-Bruch’s membrane-choroid complex, where the RPE monolayer was involved (Figure 1A). Remarkably, miR-194 expression levels were higher in DA rat primary RPE cells compared to R28 cells, which is a retinal precursor cell line with markers of both glial and neuronal origin, and primary Müller cell lines (Figure 1B). Bioinformatics analysis showed that miR-194 was highly conserved among vertebrate species and even in bilaterian animals, with a completely identical seed sequence (Figure 1C). By DIANA (DNA Intelligent Analysis) pathway analysis, the Hippo signaling pathway relevant to miR-194 was ranked the highest (Figure 1D), followed by ubiquitin-mediated proteolysis. Among the pathways, the adherens junction pathway and bacterial invasion of the epithelial pathway also had higher scores. However, the true link between these pathways and the RPE function is missing.
Figure 1.
miR-194 expression pattern and bioinformatics analysis. (A) LCM combined with qRT-PCR showing that miR-194 expression was highest in the RPE layer compared to other retinal layers in rat models (P=0.031). (B) In rat retinal cells, the primary RPE cells had higher miR-194 expression levels than R28 cells and rat Müller cells (P=0.012). (C) The miR-194 sequence was highly conserved from fish to humans. (D) DIANA pathway analysis showing that miR-194 was involved in the top 10 pathways based on published articles. The data are the mean ± SEM (n=3). The one-way analysis of variance with Tukey’s honestly significant difference post hoc test was used for statistical analysis. LCM, laser capture microdissection; GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer.
miR-194 overexpression in ARPE-19 cells affected multiple pathways in the cells
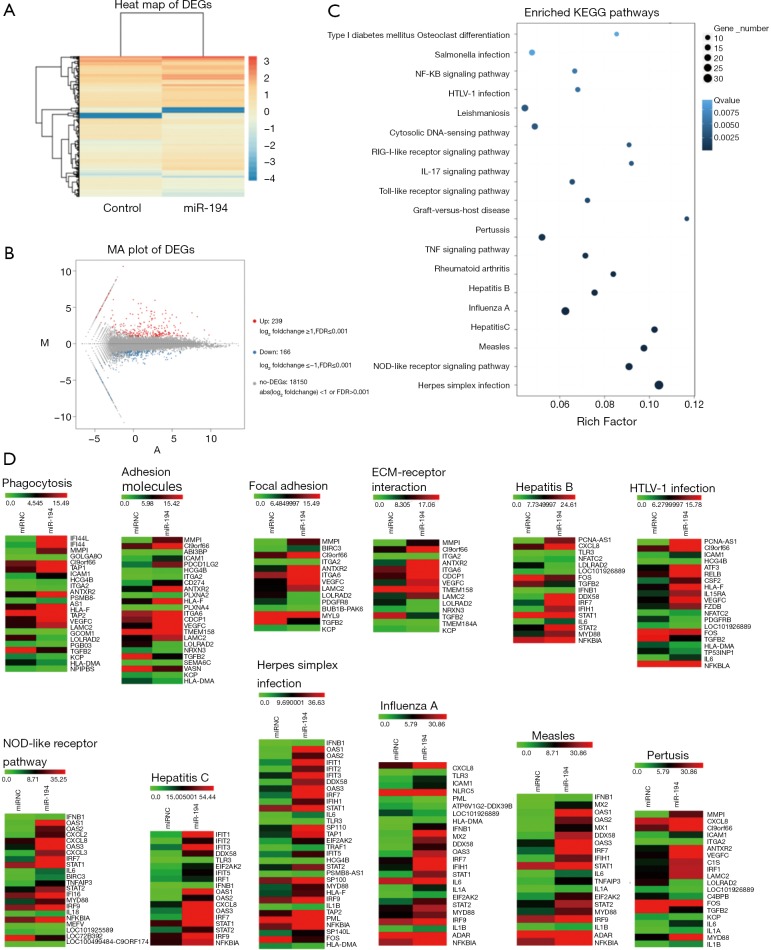
The above pathway prediction for miR-194 was based on non-RPE cell studies, most of which investigated cancer (Figure 1D). However, we wanted to determine the role of miR-194 in RPE cells. Accordingly, we investigated the effect of miR-194 overexpression in ARPE-19 cells on the global gene expression profiles via RNAseq at 1-week post-transfection. The heat map of the RNAseq for control and miR-194 group (Figure 2A) and the M-versus-A plot (MA plot) of the differentially expressed genes (DEGs) for both groups (Figure 2B) showed that the expression of 239 genes was upregulated while the expression for 166 genes was downregulated when miR-194 overexpressed in ARPE-19 cells.
Figure 2.
RNAseq was showing that miR-194 overexpression affected multiple signaling pathways in ARPE-19 cells. (A) Bioinformatics analysis based on RNAseq data showing the DEG profile in miR-194 overexpression ARPE-19 cells by heat map; (B) DEG numbers indicated in MA plots. (C) KEGG pathway enrichment analysis showing the top 20 pathways with the highest significant scores. (D) Heat maps for DEGs involved in the specific pathways of interest. DEGs, differentially expressed genes; MA plot, M-versus-A plot; M, minus; A, add.
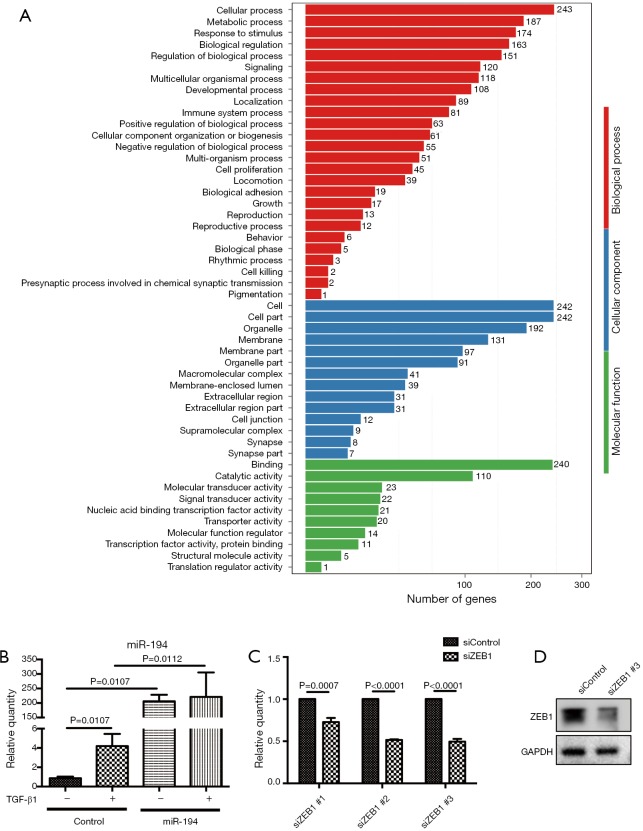
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis for DEGs was also conducted. Compared with the empty vector (control) group, the most significantly affected pathways of miR-194 overexpression in ARPE-19 cells were related to infections (herpes simplex infection, influenza A, hepatitis B and C, pertussis, HTLV-1 (human T-cell lymphotropic virus) infection, leishmaniosis, measles, salmonella infection), inflammation (NOD-like receptor signaling pathway, Toll-like receptor signaling pathway, nuclear factor (NF)-κB pathway, tumor necrosis factor (TNF) pathway, RIG-1-like receptor signaling pathway) (Figure 2C). Moreover, considering the specific functions of RPE cells, we performed a heat map analysis of those affected pathways closely related to RPE cell functions, including phagocytosis, cell adhesion molecules, focal adhesion, and ECM-receptor interaction, etc. (Figure 2D). Additionally, Gene Ontology analysis was performed to gain further insight into the possible functional implications of the gene expression alterations (Figure S1A).
Taken together, the RNAseq data indicated that miR-194 overexpression might play an important role in RPE cells by mediating inflammatory cytokine production, tight junction, cell adhesion, phagocytosis, and ECM-receptor interaction, all of which are related to EMT in RPE cells. We, therefore, speculated that miR-194 might play a key role in EMT. By analyzing the intersection between all downregulated DEGs in the miR-194 overexpression group and the predicted targets by TargetScan, we selected ZEB1, a key EMT-promoting transcription factor with a 0.5-fold decrease compared to the control group (Table S4) and which appeared in the TargetScan predicted gene list, for subsequent study . There was no big fold (0.5-fold) change of ZEB1 expression in miR-194 overexpressing RPE cells from RNAseq data. This might be partially due to the high abundance of miR-194 and low expression of ZEB1in normal ARPE-19, which was supported by a ZEB1 Ct value of 28 with a standard 20 ng input of cDNA.
Table S4. RNAseq data regarding ZEB1.
| Sample name | Control expression | miR-194 expression |
|---|---|---|
| Value | 2.02 | 1.18 |
miR-194 overexpression inhibited TGF-β1-induced ARPE-19 cell EMT
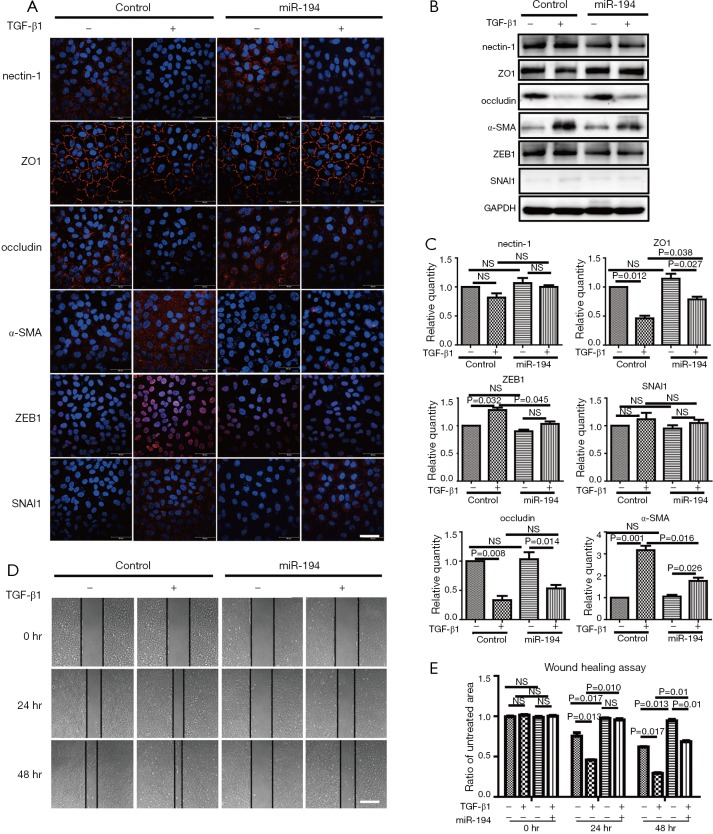
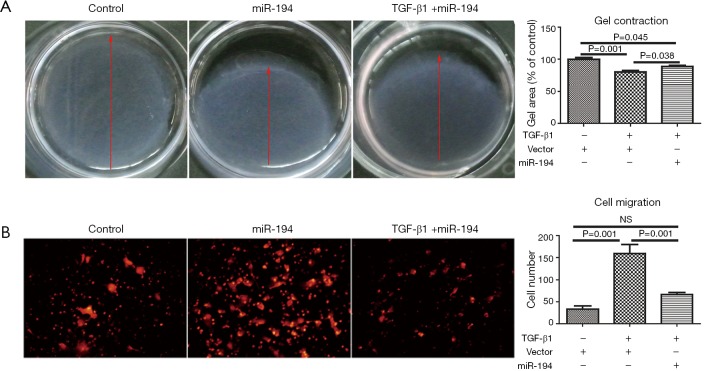
Based on the above data, we were interested to know whether miR-194 can suppress TGF-β1-induced ARPE-19 cell EMT. For this purpose, ARPE-19 cells were transfected with pSUPER-miR-194, and the transfection efficiency was observed (Figure S1B). TGF-β1 was administered 48 h post-transfection. After 48-h treatment, the EMT-related proteins were examined by immunostaining and western blotting. miR-194 overexpression without TGF-β1 increased nectin-1 expression and redistributed OCLN to the perinuclear region (Figure 3A). Loss of nectin-1 and OCLN expression along with disrupted ZO1 immunoreactivity were observed after TGF-β1 treatment. Meanwhile, α-SMA expression was induced, and significant ZEB1 immunoreactivity was observed in the nucleus. miR-194 overexpression greatly attenuated the immunoreactivity of ZEB1 and α-SMA induced by TGF-β1 (Figure 3A). The immunoreactivity of SNAI1, a key transcription factor, did not change significantly after TGF-β1 treatment (Figure 3A). Western blotting confirmed the altered expression of the EMT-related markers (Figure 3B,C). After normalization to GAPDH, the relative protein expression levels of occludin, nectin-1, and ZO1 were decreased, while ZEB1 and α-SMA expression levels were significantly increased (P<0.05) after TGF-β1 treatment; however, nectin-1 remained unchanged. miR-194 overexpression partially inhibited the upregulation of ZEB1 and α-SMA along with the downregulation of ZO-1 (Figure 3B,C). miR-194 suppression of cell migration and proliferation was confirmed by wound healing assay (Figure 3D,E), collagen gel contraction assay (Figure 4A), and transwell migration assay (Figure 4B).
Figure 3.
miR-194 overexpression suppressed TGF-β1-induced EMT in ARPE-19 cells. (A) ARPE-19 cells transfected with empty plasmid or miR-194 were treated with 10 ng/µL TGF-β1 for 24 h. Immunofluorescence staining showing partially disrupted ZO1 staining, diminished occludin signal, and no apparent significant change to nectin-1 compared to the untreated groups; miR-194 improved or maintained ZO1 integrity and altered occludin redistribution to the membrane. α-SMA and ZEB1 in the TGF-β1 treatment group had stronger immunoreactivity compared to the untreated group; miR-194 attenuated α-SMA and ZEB1 expression patterns. (B,C) Western blotting results were consistent with that of immunofluorescence staining. (D) Wound healing assay supported the premise that miR-194 inhibited cell migration ability. NS, no significance. Scale bar =50 µm (A), 200 µm (D). The data were the mean ± SEM. Cell experiments were repeated at least 3 times. The one-way analysis of variance with Tukey’s honestly significant difference post hoc test was used to calculate the statistical significance.
Figure 4.
miR-194 attenuated gel contraction and cell migration of ARPE-19 cells. (A) At 24 h after miR-194 transfection, ARPE-19 cells were treated with 10 ng/mL TGF-β1 for an additional 48 h, and the gel contraction area was quantified. (B) miR-194-treated ARPE-19 cells were treated with 10 ng/mL TGF-β1 for 48 h and then seeded in a Transwell culture system for 24 h. The migrated cells were visualized by crystal violet, and 5 random fields of vision were counted under a 20× objective. The data were the mean ± SEM (n=3). The one-way analysis of variance with Tukey’s honestly significant difference post hoc test was used for statistical analysis. Scale bar =50 µm.
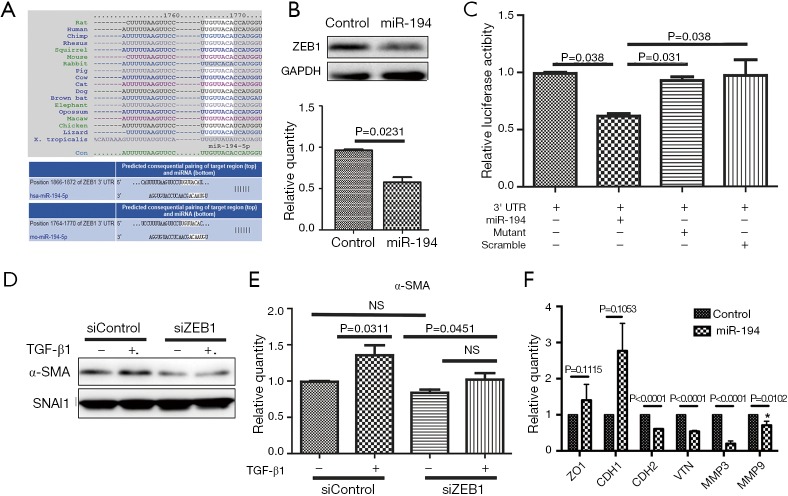
ZEB1 was identified as a direct target of miR-194
As miR-194 significantly suppressed EMT in the in vitro models, we were very interested in identifying the potential target of miR-194 in RPE cells. Based on bioinformatics analysis with DIANA (Figure 1D), we focused on the potential candidate targets of miR-194 that might also be closely correlated with EMT. As shown in Figure 5A, two regions of the ZEB1 3' UTR contained a 7-mer complementary to the miR-194 seed region, which is highly conserved from lizards to humans (Figure 5A). Although ZEB1 was predicted to be a potential target of miR-194 in both rats and humans (Figure 5A), there has been no previous relevant report on miR-194 and ZEB1. Western blotting was used to confirm the downregulated ZEB1 from RNAseq analysis. When miR-194 was overexpressed in ARPE-19 cells for 48 h, ZEB1 protein levels were significantly decreased (Figure 5B). To examine the interaction between miR-194 and the ZEB1 3' UTR, HEK293 cells were transfected with psiCHECK2-3'UTR-ZEB1 and miR-194 wild-type/mutant forms, and the scramble was used as the negative control. At 36 h post-transfection, the dual-luciferase assay was performed. AgomiR-194, but not the scramble and mutant miR-194, markedly inhibited luciferase activity (P<0.05) (Figure 5C), indicating the discovery of a post-transcriptional regulatory mechanism of ZEB1 by miR-194.
Figure 5.
ZEB1 was the functional target of miR-194. (A) TargetScan showed that the ZEB1 3' UTR among different species was highly conserved and that a conserved region of the ZEB1 3' UTR was complementary to the seed sequence of miR-194. (B) At 48 h after miR-194 transfection, ZEB1 protein levels were decreased compared with the empty vector group (P=0.0231). (C) Luciferase activity was significantly decreased following co-transfection with psiCHECK2-3'UTR-Zeb1 and agomir-194 in HEK293T cells (P=0.038). (D,E) At 48 h after siZEB1 transfection, knockdown of ZEB1 mRNA attenuated the expression of α-SMA at the protein level. (F) At post-transfection 48 h in ARPE-19, overexpression of miR-194 has no significant effect on ZO1 (P=0.1115) and CDH1(P=0.1053) while the mRNA expression of VTN (P<0.0001), CDH2 (P<0.0001), MMP3 (P<0.0001) and MMP9 (P=0.0102) were significantly decreased, respectively. All cell experiments were repeated at least 3 times. The unpaired t-test and the one-way analysis of variance with Tukey’s honestly significant difference post hoc test were used for statistical analysis. ZEB1, zinc finger E-box binding homeobox 1; ZO1, tight junction protein 1; CDH1, E-cadherin; CDH2, N-cadherin; VTN, vitronectin; MMP3, matrix metallopeptidase 3; MMP9, matrix metallopeptidase 9.
Moreover, we also wanted to know whether ZEB1 knockdown in ARPE-19 cells can affect the expression of α-SMA. The knockdown efficiency of siZEB1 was examined (Figure S1C,D), and the siZEB1 with the highest knockdown efficiency was transfected into ARPE-19 cells. At 48 h after TGF-β1 treatment, α-SMA protein expression levels decreased as expected (Figure 5D,E). However, ZEB1 knockdown did not affect SMAD2/3 phosphorylation (data not shown), indicating that ZEB1 knockdown attenuated TGF-β1-induced EMT without affecting the classic TGF-β1 pathway. Furthermore, we wanted to determine if miR-194 could affect the expression of some targets of ZEB1. Unsurprisingly, the expressions of factors inhibited by ZEB1, including ZO1 and CDH1, were relatively upregulated, but not significantly so, while CDH2 (N-cadherin), VTN (Vitronectin), MMP3 (matrix metallopeptidase 3) and MMP9 (matrix metallopeptidase 9), which are up-regulated by ZEB1, was downregulated significantly when miR-194 was overexpressed (Figure 5F), further illustrating the regulatory function of miR-194 on ZEB1.
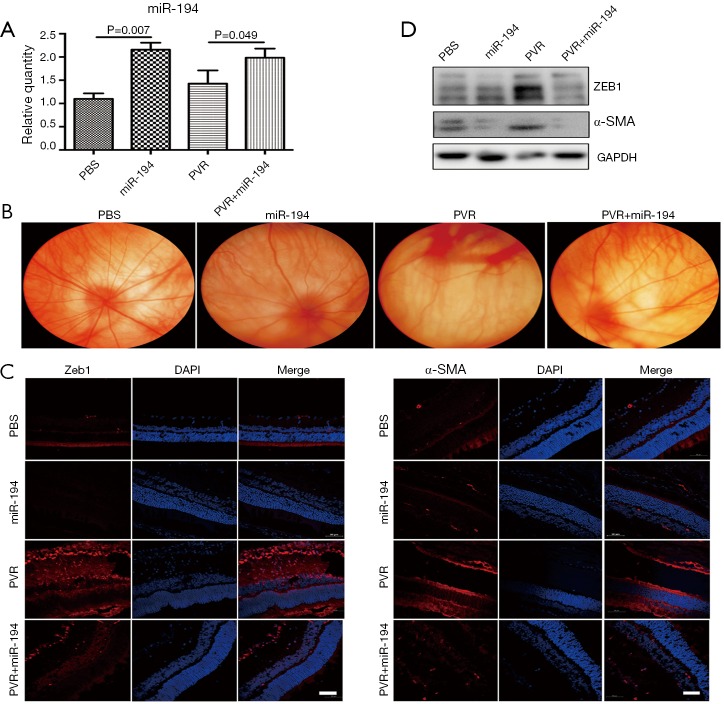
AgomiR-194 significantly suppressed experimental PVR in the rat model
Based on the miR-194 expression pattern in the retinal cells and the RNAseq data analysis of miR-194 overexpression in the ARPE-19 cells, we examined whether exogenous miR-194 could suppress EMT in RPE cells. To test our hypothesis, a PVR rat model was constructed by vitreous injection of PRP containing ARPE-19 cells. AgomiR-194 was mixed with PRP containing ARPE-19 cells and delivered intravitreally. AgomiR-194 (0.1 nmol/eye) was delivered, and an equal amount of scramble was used as the control. The intervention effect was evaluated by fundus photography. The expression of miR-194 was increased after miR-194 intravitreal injection in a rat PVR model (Figure 6A). Retinal fold-like structures and tortuous vessels were observed in the rat PVR model group, whereas the fundus structure in the agomiR-194 intervention group was improved at 2 weeks post-injection (Figure 6B). Morphologically, α-SMA and ZEB1 immunoreactivity were very strong, and ERM-like structures with positive α-SMA immunostaining were formed at 2 weeks post-injection in the PVR group. Consistent with our expectation, agomiR-194 significantly suppressed the formation of the ERM-like α-SMA-positive structures and attenuated ZEB1 immunostaining in the nucleus (Figure 6C). The bands for ZEB1 and α-SMA were much weaker under the intervention of miR-194 (Figure 6D), which was consistent with the result of immunofluorescence staining. Collectively, these findings support the premise that the exogenous administration of miR-194 suppresses EMT effectively in the PVR rat model.
Figure 6.
Exogenous administration of agomir-194 suppressed PVR effectively in the rat model. (A) The expression of miR-194 was increased after the miR-194 intravitreal injection in the rat PVR model. (D) Western blotting showed significantly increased ZEB1 and α-SMA protein levels in PVR retina (P<0.05) compared with the PBS group, and miR-194 intervention reduced this phenomenon. (B) Fundus photography showed the typical retinal fold in the PVR group; with miR-194 intervention, the fundus appeared better, i.e., without retinal fold and with relatively straight vessels. (C) Immunofluorescence staining showed that ZEB1 and α-SMA immunoreactions were very strong in the PVR group; under miR-194 intervention, their signal was significantly attenuated, which was consistent with the Western blotting results. The data were the mean ± SEM (n=6). The one-way analysis of variance with Tukey’s honestly significant difference post hoc test was used for statistical analysis. Scale bar =50 µm.
Discussion
Here, we describe, for the first time, ZEB1acting as a functional target of miR-194, an abundant miRNA in RPE cells. miR-194 overexpression suppressed EMT effectively in RPE cells mainly by negatively regulating ZEB1 expression both in the TGF-β1-induced in vitro model and the in vivo model of an intravitreal injection of PRP containing ARPE-19 cells. These findings are thus evidence of a novel therapeutic target in PVR.
Recently, the involvement of miRNAs in PVR pathogenesis has been elucidated, particularly in RPE cell EMT. However, little is known about the function of miR-194 in PVR or in the retina, and we, therefore, wanted to clarify the function of miR-194 in RPE cell EMT. qRT-PCR showed that miR-194 expression was highest in primary RPE cells and the RPE layer compared with other retinal cells and other microdissected retinal layers, respectively. To further understand the effect of miR-194 on the global gene expression profile in RPE cells, miR-194 overexpression in ARPE-19 cells and empty vector-transfected controls were subjected to RNAseq. The most significantly affected pathways of the miR-194overexpression ARPE-19 cells were the various inflammation and infection pathways. Gene Ontology analysis was performed to gain further insight into possible functional implications of the gene expression alterations (Figure S1A). Unexpectedly, the epithelial marker genes, i.e., those for NECTIN1, CDH1, CDH2, and OCLN, did not appear in the DEGs lists. Interestingly, previously identified target genes of miR-194 in tumor biologies, such as WEE1, SOCS2, BMI1, CUL4B, and KDM5B (25-29), were not detected among the ARPE-19 cell DEGs, indicating that the novel molecules in the ARPE-19 cells might be targets of miR-194. Analysis of the intersection between all downregulated DEGs in the miR-194 overexpression group and the TargetScan-predicted targets led to the selection of ZEB1 for our functional study of miR-194. Immunostaining, Western blotting, gel contraction assay, wound healing assay, and cell migration assay, in vitro, all support the notion that miR-194 overexpression significantly attenuates the phenotype of the EMT cell model. Moreover, the dual-luciferase activity assay confirmed the interaction of miR-194 with the ZEB1 3' UTR. In vivo, miR-194 suppressed PVR phenotypes and decreased ZEB1 expression levels in the rat PVR model. These findings collectively demonstrate that miR-194 suppresses PVR effectively by downregulating ZEB1 expression in a PVR rat model.
The function of ZEB1 protein, which can synergize with SMAD proteins to activate transcription and induce cell growth arrest, leads to TGF-β-dependent gene transcription, while ZEB2 has the opposite effect (37). Meanwhile, TGF-β signaling also reciprocally induces the expression of ZEB proteins during EMT (38), which in our results was indicated as ZEB1 upregulation after TGF-β treatment (Figure 3). ZEB1 protein then interacts with SMAD3 and directly represses the expression of epithelial marker genes such as CDH1 (E-cadherin) and ZO1, while expression of mesenchymal marker genes like CDH2 (N-cadherin) is increased (39). E-cadherin repression is the most reported signaling pathway of ZEB1 in inducing EMT (40); however, N-cadherin, but not E-cadherin, is the most abundant cadherin expressed by RPE cells (41,42). In our study, miR-194 overexpression unsurprisingly reduced CDH2 expression levels while increasing CDH1 expression levels in ARPE-19 cells, which further confirms the relationship between miR-194 and ZEB1 (Figure 5F). ZEB1 knockdown downregulated α-SMA expression levels in the TGF-β1-treated ARPE-19 cells (Figure 5D,E), supporting the premise that decreased ZEB1, either by siRNA knockdown or miR-194 as the post-transcriptional mechanism, attenuates the interaction between ZEB1 and SMAD3 in the TGF-β pathway in RPE cells, and finally represses EMT. Whether there is synergistic interaction between ZEB1 and the non-canonical TGF-β pathway in RPE cell EMT warrants further exploration.
In recent years, glyoxalase 1 (GLO1), a ubiquitous cellular enzyme involved in methylglyoxal detoxification, has attracted research attention. GLO1 can control EMT via miR-101, MG-H1-AP, and TGF-β/SMAD signaling, sustaining the metastatic phenotype of prostate cancer (43). Furthermore, a case-control study revealed that two analyzed GLO1 polymorphisms might be associated with RP, an inherited retinal degenerative disease (44), suggesting that GLO1 might be a promising target for further exploration of ZEB1 and the non-canonical TGF-β pathway in RPE cell EMT.
Conclusions
In summary, the present study demonstrates for the first time that ZEB1 is a novel functional target of miR-194. As a negative regulator of ZEB1, miR-194 suppresses EMT in RPE cells, and therefore miR-194 could potentially serve as a novel therapeutic target in PVR. It is well established that miRNAs control the cellular expression machinery via a single miRNA/multiple targets or multiple miRNAs/single target mechanism. More functional targets of miR-194 need to be identified. Comprehensive and extensive studies of the biology of miR-194 will shed new light on their roles in normal homeostasis and the pathology of retinal diseases.
Figure S1.
The verification of miR-194 overexpression post-transfection, GO analysis after overexpression of miR-194 in ARPE-19 and the sequence selection of siZEB1. (A) Gene ontology analysis of empty vector (control) group and miR-194 over expression group in ARPE-19 cells; (B) the expression level of miR-194 were significantly increased in miR-194 overexpression group compared to the control group with or without TGF-β1 treatment; (C) detection of ZEB1-knockdown efficiency in ARPE-19 cells using different siRNAs by qRT-PCR; (D) detection of ZEB1-knockdown efficiency in ARPE-19 cells using siZEB1 #3 by western blotting. The data are the mean ± SEM, n=3. The one-way analysis of variance with Tukey’s honestly significant difference post hoc test and 2-tailed Student t-test were used for statistical analysis.
Acknowledgments
Funding: This work was supported by grants from the Ministry of Science and Technology of China (2017YFA0104100, 2015CB964601, 2016YFA0101302), National Natural Science Foundation (81670867, 81372071, 81770942), Shanghai Municipal Commission of Health and Family Planning project (201640229), Shanghai Science and Technology Committee Grant (17ZR1431300), as well as from Shanghai East Hospital (ZJ2014-ZD-002).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The animal protocols were approved by the Tongji University Committee of Experimental Animals Ethics (Permit Number: TJmed-010-32) and were carried out in accordance with the ARVO (Association for Research in Vision and Ophthalmology) Statement for the Use of Animals in Ophthalmic and Vision Research. The study involving human participants was approved by the Ethics Committee of Shanghai Ninth People’s Hospital affiliated to Shanghai Jiaotong University and was in compliance with the Declaration of Helsinki.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Sadaka A, Giuliari GP. Proliferative vitreoretinopathy: current and emerging treatments. Clin Ophthalmol 2012;6:1325-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nemet A, Moshiri A, Yiu G, et al. A Review of Innovations in Rhegmatogenous Retinal Detachment Surgical Techniques. J Ophthalmol 2017;2017:4310643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamiya S, Liu L, Kaplan HJ. Epithelial-mesenchymal transition and proliferation of retinal pigment epithelial cells initiated upon loss of cell-cell contact. Invest Ophthalmol Vis Sci 2010;51:2755-63. 10.1167/iovs.09-4725 [DOI] [PubMed] [Google Scholar]
- 4.Banerjee S, Savant V, Scott RA, et al. Multiplex bead analysis of vitreous humor of patients with vitreoretinal disorders. Invest Ophthalmol Vis Sci 2007;48:2203-7. 10.1167/iovs.06-1358 [DOI] [PubMed] [Google Scholar]
- 5.Baudouin C, Fredj-Reygrobellet D, Brignole F, et al. Growth factors in vitreous and subretinal fluid cells from patients with proliferative vitreoretinopathy. Ophthalmic Res 1993;25:52-9. 10.1159/000267221 [DOI] [PubMed] [Google Scholar]
- 6.Canataroglu H, Varinli I, Ozcan AA, et al. Interleukin (IL)-6, interleukin (IL)-8 levels and cellular composition of the vitreous humor in proliferative diabetic retinopathy, proliferative vitreoretinopathy, and traumatic proliferative vitreoretinopathy. Ocul Immunol Inflamm 2005;13:375-81. 10.1080/09273940490518900 [DOI] [PubMed] [Google Scholar]
- 7.Asaria RH, Kon CH, Bunce C, et al. Silicone oil concentrates fibrogenic growth factors in the retro-oil fluid. Br J Ophthalmol 2004;88:1439-42. 10.1136/bjo.2003.040402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim IK, Arroyo JG. Mechanisms in proliferative vitreoretinopathy. Ophthalmol Clin North Am 2002;15:81-6. 10.1016/S0896-1549(01)00008-6 [DOI] [PubMed] [Google Scholar]
- 9.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420-8. 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong D, Li Y, Wang Z, et al. Cancer Stem Cells and Epithelial-to-Mesenchymal Transition (EMT)-Phenotypic Cells: Are They Cousins or Twins? Cancers (Basel) 2011;3:716-29. 10.3390/cancers30100716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phua YL, Martel N, Pennisi DJ, et al. Distinct sites of renal fibrosis in Crim1 mutant mice arise from multiple cellular origins. J Pathol 2013;229:685-96. 10.1002/path.4155 [DOI] [PubMed] [Google Scholar]
- 12.Garweg JG, Tappeiner C, Halberstadt M. Pathophysiology of proliferative vitreoretinopathy in retinal detachment. Surv Ophthalmol 2013;58:321-9. 10.1016/j.survophthal.2012.12.004 [DOI] [PubMed] [Google Scholar]
- 13.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010;79:351-79. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Dong F, Reinach PS, et al. MicroRNA-182 Suppresses HGF/SF-Induced Increases in Retinal Pigment Epithelial Cell Proliferation and Migration through Targeting c-Met. PLoS One 2016;11:e0167684. 10.1371/journal.pone.0167684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Usui-Ouchi A, Ouchi Y, Kiyokawa M, et al. Upregulation of Mir-21 Levels in the Vitreous Humor Is Associated with Development of Proliferative Vitreoretinal Disease. PLoS One 2016;11:e0158043. 10.1371/journal.pone.0158043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Li H, Liu X, et al. MicroRNA-29b regulates TGF-beta1-mediated epithelial-mesenchymal transition of retinal pigment epithelial cells by targeting AKT2. Exp Cell Res 2016;345:115-24. 10.1016/j.yexcr.2014.09.026 [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Ye S, Xiao W, et al. Differentially expressed microRNAs in TGFbeta2-induced epithelial-mesenchymal transition in retinal pigment epithelium cells. Int J Mol Med 2014;33:1195-200. 10.3892/ijmm.2014.1688 [DOI] [PubMed] [Google Scholar]
- 18.Adijanto J, Castorino JJ, Wang ZX, et al. Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J Biol Chem 2012;287:20491-503. 10.1074/jbc.M112.354761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jun JH, Joo CK. MicroRNA-124 Controls Transforming Growth Factor beta1-Induced Epithelial-Mesenchymal Transition in the Retinal Pigment Epithelium by Targeting RHOG. Invest Ophthalmol Vis Sci 2016;57:12-22. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko H, Terasaki H. Biological Involvement of MicroRNAs in Proliferative Vitreoretinopathy. Transl Vis Sci Technol 2017;6:5. 10.1167/tvst.6.4.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donato L, Bramanti P, Scimone C, et al. miRNAexpression profile of retinal pigment epithelial cells under oxidative stress conditions. FEBS Open Bio 2018;8:219-33. 10.1002/2211-5463.12360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu B, Xu C, Tian Y, et al. Inflammatory microRNA-194 and -515 attenuate the biosynthesis of chondroitin sulfate during human intervertebral disc degeneration. Oncotarget 2017;8:49303-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morimoto A, Kannari M, Tsuchida Y, et al. An HNF4alpha-microRNA-194/192 signaling axis maintains hepatic cell function. J Biol Chem 2017;292:10574-85. 10.1074/jbc.M117.785592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu X, An J, Hua Y, et al. MicroRNA-194 regulates keratinocyte proliferation and differentiation by targeting Grainyhead-like 2 in psoriasis. Pathol Res Pract 2017;213:89-97. 10.1016/j.prp.2016.11.020 [DOI] [PubMed] [Google Scholar]
- 25.Bao J, Zou JH, Li CY, et al. miR-194 inhibits gastric cancer cell proliferation and tumorigenesis by targeting KDM5B. Eur Rev Med Pharmacol Sci 2016;20:4487-93. [PubMed] [Google Scholar]
- 26.Zhang X, Wei C, Li J, et al. MicroRNA-194 represses glioma cell epithelialtomesenchymal transition by targeting Bmi1. Oncol Rep 2017;37:1593-600. 10.3892/or.2017.5376 [DOI] [PubMed] [Google Scholar]
- 27.Li P, Yang Y, Liu H, et al. MiR-194 functions as a tumor suppressor in laryngeal squamous cell carcinoma by targeting Wee1. J Hematol Oncol 2017;10:32. 10.1186/s13045-017-0402-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das R, Gregory PA, Fernandes RC, et al. MicroRNA-194 Promotes Prostate Cancer Metastasis by Inhibiting SOCS2. Cancer Res 2017;77:1021-34. 10.1158/0008-5472.CAN-16-2529 [DOI] [PubMed] [Google Scholar]
- 29.Mi J, Zou Y, Lin X, et al. Dysregulation of the miR-194-CUL4B negative feedback loop drives tumorigenesis in non-small-cell lung carcinoma. Mol Oncol 2017;11:305-19. 10.1002/1878-0261.12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Bo Q, Wu W, et al. alpha-Melanocyte-stimulating hormone prevents glutamate excitotoxicity in developing chicken retina via MC4R-mediated down-regulation of microRNA-194. Sci Rep 2015;5:15812. 10.1038/srep15812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci 2009;66:773-87. 10.1007/s00018-008-8465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurahara H, Takao S, Maemura K, et al. Epithelial-mesenchymal transition and mesenchymal-epithelial transition via regulation of ZEB-1 and ZEB-2 expression in pancreatic cancer. J Surg Oncol 2012;105:655-61. 10.1002/jso.23020 [DOI] [PubMed] [Google Scholar]
- 33.Lu L, Neff F, Dun Z, et al. Gene expression profiles derived from single cells in human postmortem brain. Brain Res Brain Res Protoc 2004;13:18-25. 10.1016/j.brainresprot.2003.12.003 [DOI] [PubMed] [Google Scholar]
- 34.Zheng XZ, Du LF, Wang HP. An immunohistochemical analysis of a rat model of proliferative vitreoretinopathy and a comparison of the expression of TGF-beta and PDGF among the induction methods. Bosn J Basic Med Sci 2010;10:204-9. 10.17305/bjbms.2010.2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Zhang J, Chen X, et al. miR-365 promotes diabetic retinopathy through inhibiting Timp3 and increasing oxidative stress. Exp Eye Res 2018;168:89-99. 10.1016/j.exer.2017.11.006 [DOI] [PubMed] [Google Scholar]
- 36.Donato L, Scimone C, Rinaldi C, et al. Stargardt Phenotype Associated With Two ELOVL4 Promoter Variants and ELOVL4 Downregulation: New Possible Perspective to Etiopathogenesis? Invest Ophthalmol Vis Sci 2018;59:843-57. 10.1167/iovs.17-22962 [DOI] [PubMed] [Google Scholar]
- 37.Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J 2003;22:2443-52. 10.1093/emboj/cdg225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma L, Young J, Prabhala H, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 2010;12:247-56. 10.1038/ncb2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong W, Yang H, He L, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol 2008;28:6773-84. 10.1128/MCB.00941-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez-Tillo E, Lazaro A, Torrent R, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 2010;29:3490-500. 10.1038/onc.2010.102 [DOI] [PubMed] [Google Scholar]
- 41.Marrs JA, Andersson-Fisone C, Jeong MC, et al. Plasticity in epithelial cell phenotype: modulation by expression of different cadherin cell adhesion molecules. J Cell Biol 1995;129:507-19. 10.1083/jcb.129.2.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Aken EH, De Wever O, Van Hoorde L, et al. Invasion of retinal pigment epithelial cells: N-cadherin, hepatocyte growth factor, and focal adhesion kinase. Invest Ophthalmol Vis Sci 2003;44:463-72. 10.1167/iovs.01-1096 [DOI] [PubMed] [Google Scholar]
- 43.Antognelli C, Cecchetti R, Riuzzi F, et al. Glyoxalase 1 sustains the metastatic phenotype of prostate cancer cells via EMT control. J Cell Mol Med 2018;22:2865-83. 10.1111/jcmm.13581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donato L, Scimone C, Nicocia G, et al. GLO1 gene polymorphisms and their association with retinitis pigmentosa: a case-control study in a Sicilian population. Mol Biol Rep 2018;45:1349-55. 10.1007/s11033-018-4295-4 [DOI] [PubMed] [Google Scholar]