Abstract
Yersinia enterocolitica, a zoonotic foodborne pathogen, is able to withstand low temperatures. This psychrotrophic ability allows it to multiply in food stored in refrigerators. However, little is known about the Y. enterocolitica cold response. In this study, isolate-specific behavior at 4°C was demonstrated and the cold response was investigated by examining changes in phenotype, gene expression, and the proteome. Altered expression of cold-responsive genes showed that the ability to survive at low temperature depends on the capacity to acclimate and adapt to cold stress. This cold acclimation at the transcriptional level involves the transient induction and effective repression of cold-shock protein (Csp) genes. Moreover, the resumption of expression of genes encoding other non-Csp is essential during prolonged adaptation. Based on proteomic analyses, the predominant functional categories of cold-responsive proteins are associated with protein synthesis, cell membrane structure, and cell motility. In addition, changes in membrane fluidity and motility were shown to be important in the cold response of Y. enterocolitica. Isolate-specific differences in the transcription of membrane fluidity- and motility-related genes provided evidence to classify strains within a spectrum of cold response. The combination of different approaches has permitted the systematic description of the Y. enterocolitica cold response and gives a better understanding of the physiological processes underlying this phenomenon.
Keywords: Yersinia enterocolitica, cold response, proteome, isolates specific, motility, fluidity
Introduction
Yersinia enterocolitica, the third most commonly reported foodborne zoonotic pathogen in the European Union, can cause serious diseases, including gastroenteritis, mesenteric lymphadenitis, reactive arthritis, erythema nodosum, and pseudoappendicitis (Ostroff et al., 1994; Horisaka et al., 2004; European Food Safety Authority and European Centre for Disease Prevention and Control, 2016). It occurs ubiquitously in the natural environment and is widespread in animal populations (Benembarek, 1994; Robins-Browne, 2013). Furthermore, it can be isolated frequently from a variety of foods, including milk and milk products, pork, poultry, eggs, and produce (Bari et al., 2011).
Yersinia enterocolitica is capable of growing at temperatures approaching and even below 0°C (Tudor et al., 2008; Divya and Varadaraj, 2013). Therefore, even refrigeration temperatures (0–4°C) can allow significant bacterial growth over time. Several studies have reported growth of Y. enterocolitica in food products stored at refrigeration temperatures: e.g., on raw beef, with increased cell counts of up to 2 log CFU/ml within 4 days (Tudor et al., 2008) and in pasteurized milk, reaching levels of 5–7 log CFU/ml after 7 days (with an initial inoculum of 1–3 log CFU/ml) (Amin and Draughon, 1987).
One of the most prominent cold responses is the induction of cold-shock proteins (Csps) in all psychrotrophs, mesophiles, and thermophiles (Polissi et al., 2003; Phadtare, 2004). As model systems, Escherichia coli and Bacillus subtilis have been studied in detail regarding cold response and Csps (Phadtare et al., 1999; Ermolenko and Makhatadze, 2002; Weber and Marahiel, 2003). The role of polynucleotide phosphorylase (PNPase, encoded by the pnp gene) in regulating cold response is also well described (Goverde et al., 1998; Yamanaka and Inouye, 2001; Cordin et al., 2006; Matos et al., 2009; Phadtare, 2011). This enzyme with the 3’- to 5’-exonucleolytic activities involved mostly in mRNA decay and ribosomes release (Coburn and Mackie, 1998; Polissi et al., 2003) is used to help repress the generation of Csps and relieve growth arrest (Neuhaus et al., 2003; Zhao et al., 2016). Meanwhile, in psychrotrophic bacteria such as Arthrobacter globiformis and Pseudomonas fragi, some cold-responsive proteins are synthesized at relatively moderate levels and prolonged in response to continuous growth at low temperatures (Berger et al., 1996; Michel et al., 1997). These proteins are of particular importance since they differentiate psychrotrophs from mesophiles, and they are probably one of the key determinants that allow survival at low temperature (Hébraud and Potier, 1999). Additionally, the ability to cope with temperature downshift must be accompanied by a number of changes in response to alterations of physical and biochemical parameters, including solubility, membrane fluidity, protein conformation and stability, and changes in gene expression (Hébraud and Potier, 1999; Vorachek-Warren et al., 2002; Albanesi et al., 2004; Phadtare, 2004; Cao-Hoang et al., 2010; Barria et al., 2013). Therefore, the biochemical and physiological effects allowing bacteria to adapt to temperature changes are likely to be complex, involving a number of cellular processes.
As a psychrotrophic bacterium, Y. enterocolitica has two well reported csp homolog genes (cspA and cspB), which are strongly expressed during the cold response. The cold-shock exoribonuclease PNPase and pnp gene have also been reported (Goverde et al., 1998; Phadtare, 2011). Additionally, a previous study has reported that genes involved in various functions (regulation, motility, virulence, and metabolism) are upregulated after a temperature downshift from optimal (30°C) to suboptimal (10°C) conditions in Y. enterocolitica (Bresolin et al., 2006). However, the effects of these genes and the cold response on protein expressional levels are not clarified in Y. enterocolitica.
Recently, advances in proteomics and bioinformatics technologies provide clear information on protein expression in response to cold and other stresses. High-throughput comparative proteomics with label-free quantification enabled the parsing of various potential mechanisms and regulatory networks of stress response in E. coli, B. subtilis, Pseudomonas putida, and Yersinia ruckeri (Delumeau et al., 2011; Stefanopoulou et al., 2011; Herbst et al., 2015; Kumar et al., 2016).
However, to our knowledge, the global proteomic profiles of Y. enterocolitica under the influence of low temperature have not been reported. Considerable research on Y. enterocolitica cold response has been limited to few proteins or genes and to single time points. The aim of this study is to describe the physiological processes of cold response in Y. enterocolitica via comparisons of growth ability, expression of cold-responsive genes and proteins, as well as cell motility and membrane fluidity of selected strains upon exposure to cold conditions.
Materials and Methods
Growth Profile at Low Temperature
In order to test the growth ability of Y. enterocolitica at low temperatures (4°C), 55 isolates were collected from different matrices, representing different serotypes and biotypes (details are given in Table 1). Isolates were incubated on Plate Count agar (PC agar, Merck, Darmstadt, Germany) at 28°C for 24 h. Single colonies were transferred to 3 ml of Brucella broth (BB, BD Franklin Lakes, NJ, United States) and incubated at 28°C for 20 h. Enriched cultures were serially diluted 1:106 in BB to reach a cell concentration of about 101–102 CFU/ml as the initial value. Growth abilities of 55 strains were tested based on cell concentration in BB after incubating at 4°C for 168 h. For growth profile investigation, cell concentration of the selected isolates (II7D, 8081, and 44B) was measured under cold stress for 0, 24, 48, 72, 144, and 168 h respectively. The experiment was carried out in six biological replicates (with two technical duplicates each).
TABLE 1.
Characteristics and growth ability of Y. enterocolitica strains at 4°C for 168 h.
| Isolates | Median | Median norm. | Serotype | Biotype | Matrix |
| 44B | 1.86E + 03 | 2.86E + 02 | O:5,27 | 1A | Food |
| IP566/82 | 4.60E + 03 | 8.85E + 02 | O:8 | n. a. | n. a. |
| 4780 | 5.90E + 05 | 5.98E + 04 | O:8 | 1B | Human |
| 96/10 | 1.50E + 05 | 1.01E + 05 | O:8 | 1B | n. a. |
| 39/91 | 5.00E + 06 | 2.87E + 05 | O:8 | 1 | Human |
| 21/08 | 4.20E + 07 | 3.41E + 06 | O:8 | 1A | n. a. |
| 8081 | 2.70E + 07 | 6.85E + 06 | O:8 | 1B | Human |
| 78/90 | 1.07E + 08 | 7.96E + 06 | O:8 | 1B | Human |
| 25 Ia | 1.25E + 08 | 1.22E + 07 | O:3 | 4 | Food |
| 96B | 2.60E + 08 | 2.23E + 07 | O:5,27 | 3 | Animal |
| 177B | 2.40E + 08 | 2.37E + 07 | O:5,27 | 2 | Human |
| 207 IIa | 3.60E + 08 | 2.64E + 07 | O:9 | 3 | Animal |
| 207 Ia | 3.40E + 08 | 3.56E + 07 | O:9 | 3 | Animal |
| 25/13 | 8.10E + 08 | 4.60E + 07 | O:5 | 1A | Food |
| 56/14 | 5.90E + 08 | 4.66E + 07 | O:5 | 1A | Food |
| 57/14 | 7.40E + 08 | 4.76E + 07 | O:9 | 2 | Food |
| 54/13 | 6.45E + 08 | 4.93E + 07 | O:8 | 1A | Food |
| 28/07 | 1.00E + 09 | 5.03E + 07 | O:9 | 3 | Animal |
| 04/13 | 4.90E + 08 | 5.05E + 07 | O:5 | 1A | Food |
| 32/07 | 4.30E + 08 | 5.77E + 07 | O:3 | 4 | Animal |
| 05/13 | 5.80E + 08 | 5.79E + 07 | O:5 | 1A | Food |
| 44/07 | 1.14E + 09 | 5.92E + 07 | O:3 | 4 | Food |
| III15D | 7.00E + 08 | 5.93E + 07 | O:5 | 1A | Food |
| 24/14 | 4.50E + 08 | 5.97E + 07 | O:5 | 1A | Food |
| 29/07 | 8.30E + 08 | 5.99E + 07 | O:9 | 3 | Animal |
| 09/11 | 1.37E + 09 | 6.12E + 07 | O:9 | 2 | Food |
| 37/12 | 5.40E + 08 | 6.18E + 07 | O:5 | 1A | Food |
| 65/14 | 1.03E + 09 | 6.29E + 07 | O:5 | 1A | Food |
| 47/13 | 5.50E + 08 | 6.31E + 07 | O:5 | 1A | Food |
| 77/14 | 6.40E + 08 | 6.46E + 07 | O:9 | 2 | Food |
| I15C | 6.80E + 08 | 6.92E + 07 | O:3 | 3 | Animal |
| 20/07 | 1.42E + 09 | 7.17E + 07 | O:9 | 3 | Human |
| 11/07 | 1.07E + 09 | 7.32E + 07 | O:3 | 4 | Human |
| 38/12 | 5.60E + 08 | 7.35E + 07 | O:5 | 1A | Food |
| 58/07 | 1.13E + 09 | 7.73E + 07 | O:3 | 4 | Animal |
| 31/13 | 7.60E + 08 | 7.86E + 07 | O:5,27 | 2 | food |
| 45/14 | 1.21E + 09 | 7.97E + 07 | O:5,27 | 2 | Food |
| 03/13 | 7.00E + 08 | 8.01E + 07 | O:8 | 1A | Food |
| 387/09 | 8.20E + 08 | 8.06E + 07 | O:9 | n. a. | Animal |
| 06/13 | 4.30E + 08 | 8.36E + 07 | O:8 | 1B | Food |
| 18/07 | 1.30E + 09 | 8.45E + 07 | O:9 | 3 | Human |
| 15/12 | 9.40E + 08 | 8.92E + 07 | O:5,27 | 2 | Food |
| 19/07 | 1.07E + 09 | 9.62E + 07 | O:3 | 4 | Human |
| 61/07 | 1.12E + 09 | 1.00E + 08 | O:3 | 4 | Animal |
| 13/14 | 1.12E + 09 | 1.01E + 08 | O:3 | 4 | Food |
| 12/07 | 8.80E + 08 | 1.02E + 08 | O:3 | 4 | Human |
| 30/14 | 9.50E + 08 | 1.04E + 08 | O:5,27 | 2 | Food |
| 33/07 | 4.50E + 08 | 1.09E + 08 | O:3 | 4 | Animal |
| 25/14 | 8.80E + 08 | 1.12E + 08 | O:5,27 | 2 | Food |
| 11/09 | 1.07E + 09 | 1.14E + 08 | O:5,27 | 2 | Food |
| 14/07 | 1.45E + 09 | 1.15E + 08 | O:3 | 4 | Human |
| 46/14 | 1.56E + 09 | 1.21E + 08 | O:5,27 | 2 | Food |
| 89/14 | 1.42E + 09 | 1.27E + 08 | O:3 | 4 | Food |
| 17/07 | 9.70E + 08 | 1.27E + 08 | O:9 | 3 | Human |
| II7D | 1.02E + 09 | 1.27E + 08 | O:5 | 1A | Food |
Median norm.: relative median value normalized with the initial concentration, respectively.
RNA Extraction Under Cold Stress
Yersinia enterocolitica isolates were selected for RNA extraction. Pre-culture was prepared in 12 ml BB at 28°C (as incubation temperature) for 24 h. The suspension was diluted in BB to 0.05 OD600 value and then incubated at 28°C for 2 h to reach an OD600 value between 0.1 and 0.2. After centrifugation, the bacteria were suspended into 10 ml cooled BB and incubated at 4°C for different time periods (5 min, 30 min, 2 h, 4 h, 24 h, and 48 h). The pellet suspended in BB at room temperature was used as control. Cold-shock stop mix solution (5% Roti-Aqua-phenol, 95% ethanol, Carl Roth, Karlsruhe, Germany) was added and samples were processed as described elsewhere (Blomberg et al., 1990). All samples were frozen at −80°C until further use.
RNA was extracted with Roti-Aqua-Phenol (Carl Roth). RNA quality of samples was tested by gel electrophoresis. The ratio of absorbance A260/A280 and A260/A230 were used to assess the purity of RNA photometrically with NanoDropTM 2000/2000c Spectrophotometers (Thermo Fisher Scientific). A ratio of ∼2.0 is generally accepted of A260/A280 and the expected A260/A230 values are set in the range of 2.0–2.2. Reverse transcription was performed with Maxima H Minus First Strand cDNA Synthesis Kit (Fermentas, St. Leon-Rot, Germany). The cDNA samples were diluted 1: 5 with nuclease-free water for RT-qPCR investigation.
Expressional Analysis of Cold-Responsive Genes
Real-time quantitative PCR (RT-qPCR) was used to test the transcription level of cold-responsive genes of Y. enterocolitica. Eight genes, which were reported to have enhanced at transcriptional levels at 10°C (Bresolin et al., 2006), were tested in this study. These genes cover the functions of regulation, metabolism, and motility (Supplementary Table S2 lists target genes and used primers). The SsoFast EvaGreen Supermix (SYBR-green, Bio-Rad, Munich, Germany) was used for RT-qPCR assays. The expression of the genes was normalized to the reference gene polA (DNA polymerase I) (Townsend et al., 2008). The results of RT-qPCR were visualized and evaluated by CFX software (Bio-Rad).
Whole Cell Protein Extraction
Three isolates (Y. enterocolitica strains II7D, 8081, and 44B) were subjected to incubation at 4°C for 0, 5 min, 2 h, and 24 h. The cells were harvested and the pellet was washed with PBS. Cell pellets were reconstituted with 300 μl distilled water and inactivated by addition of 900 μl ethanol. After the centrifugation and evaporation, the final pellet was reconstituted with 250 μl 20 mM HEPES (pH 7.4) and subjected to sonication for 1 min (cycle, 1.0; amplitude, 100%) with a sonicator (UP100H; Hielscher Ultrasound Technology, Teltow, Germany). Supernatants were collected and the concentration was measured using modified Bradford’s method with Coomassie PlusTM Protein Assays (Thermo Fisher Scientific, Rockford, IL, United States) and the samples were stored at -20°C for further analysis. Each strain was tested six times independently.
In-Solution Trypsin Digestion
The in-solution trypsin digestion of proteins was performed as described previously (Wareth et al., 2016). Briefly, 10 μg protein was used for acetone precipitation. The resultant peptides were then reconstituted with 20 μl denaturation buffer containing 6 M urea/2 M thiourea in 10 mM HEPES (pH 8.0) and reduced with 10 mM dithiothreitol in 50 mM of ammonium bicarbonate (ABC, Sigma, Germany). The alkylation was carried out with 55 mM iodacemtamide and subsequently 0.5 μg/μl LysC solution was added. The urea concentration was decreased by 0.5 μg/μl trypsin and the trypsin digestion was stopped by 5% acetonitrile/3% trifluoroacetic acid.
Liquid Chromatography-Electrospray Ionization-Tandem Mass Spectrometry (LC-ESI-MS/MS) Measurements
Liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS/MS) measurements were carried out as described elsewhere (Wareth et al., 2016). Resultant peptides of trypsin digestion were desalted by solid phase extraction and the peptides were separated using Dionex Ultimate 3000 nanoLC (Dionex/Thermo Fisher Scientific, Idstein, Germany) on fritless silica micro-columns with an inner diameter of 100 μm. Mass spectrometry measurements were carried out using LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). The LTQ-Orbitrap was operated in the positive mode to simultaneously measure full scan MS spectra in the range of m/z 300–1700 in the Orbitrap analyzer at a resolution of R = 60,000. After that, isolation and fragmentation of the 20 most intense ions in the LTQ part were carried out by collision-induced dissociation.
The raw mass spectra were processed using label-free quantification algorithm of the MaxQuant version 1.3.0.5 (Max Planck Institute of Biochemistry, Martinsried, Germany) (Tyanova et al., 2016) and protein identification was carried out by searching against protein sequence FASTA file of Y. enterocolitica strain YE02/02 (Proteome ID: UP000069750, protein count: 4760) with a wide range of homologous strains downloaded from UniProt database. The following parameters were set for protein identification: Initial maximum precursor—7 ppm, fragment mass deviations—0.5 Da; variable modification—methionine oxidation/acetylation of peptide N-termini; fixed modification—carbamidomethylation; enzymes—LysC and trypsin, both with a maximum of two missed cleavages; minimum peptide length—seven amino acids, and target-decoy-based false discovery rate (FDR) for peptide and protein identification—1%.
The statistical analysis was performed using the Perseus software version 1.4.1.3 (Max Planck Institute of Biochemistry, Martinsried, Germany) (Rudolph and Cox, 2019). The LFQ intensities of proteins were imported and transformed to logarithmic scale with base two. The Student’s t-test and Benjamini–Hochberg procedure FDR corrections of the significant p-values (p < 0.05) were applied for identification of differentially expressed proteins.
Motility Assay
Motility was tested as described for Y. enterocolitica (Bresolin et al., 2008). Three strains II7D, 8081, and 44B were assessed by measuring diameters of migration zone at 4°C with motility agar plates (0.3% agar, 0.5% NaCl, and 1% tryptone). Strains were incubated on PC agar plates overnight at 28°C. Single colonies were transferred onto motility agar plates and incubated initially at 37°C for 2 h to start the assay with non-motile bacteria. Plates were subsequently incubated at 28°C (for 21 h) and 4°C (for 44 h).
Fluidity Assay
Membrane fluidity of Y. enterocolitica was measured by a fluorescence polarization or anisotropy value, which corresponds to the reaction to polarized light of a fluorescent probe inside the membrane (Zaritsky et al., 1985; Aricha et al., 2004; Mykytczuk et al., 2007). Briefly, three isolates (Y. enterocolitica strains II7D, 8081, and 44B) were prepared and incubated at 4°C for 0, 2, 24, and 48 h with the method described above. Cultured cells were harvested and washed twice with PBS (10 mM, pH 7.4, Merck) and then incubated with 5 μM 1,6-diphenyl-1,3,5-hexatriene (DPH, Sigma–Aldrich, St. Louis, MO, United States) at 37°C for 1 h. Unlabeled cells were used as a scattering reference. The fluorescence polarization was measured using a Cary Eclipse Fluorescence spectrophotometer with Manual Polarizer (Agilent, Santa Clara, CA, United States) at 360 nm excitation and 430 nm emission. Fluorescence anisotropy was calculated by the formula A = [IVV − IVH (IHV/IHH)]/[IVV + 2IVH (IHV/IHH)], where I is the corrected fluorescence intensity, and the subscripts V and H indicate the values obtained with vertical or horizontal orientations, respectively. The emission polarized filter was set either in the vertical (IVV) or horizontal (IVH) position. Decrease in fluorescence anisotropy reflected increases in the fluidity of the lipid bilayer, which controls or alters the mobility of DPH in the membrane.
Bioinformatics and Statistical Analysis
Cell counts of the growth assays were expressed as the median with range for all the isolates (CFU/ml) and other quantitative data were expressed as the mean with the standard error of the mean. Paired sample t-tests were applied to determine differences in growth profile, gene expression, and fluidity. GraphPad Prism 6 was used to carry out the analyses cited above.
The Gene Ontology (GO) database1 and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database2 were used to classify proteins and related pathways of proteins (Kanehisa et al., 2016). The Clusters of Orthologous Groups (COGs) functional categories of differentially expressed proteins were assigned by BLAST and searched with the COG database3 referring to other research (Tatusov et al., 2001; Galperin et al., 2014).
Results and Discussion
Growth Profiles of Isolates at Low Temperatures
Altogether, 55 isolates of Y. enterocolitica collected from food, humans, and animals were tested for their growth profiles at 4°C after 168 h (end-point analysis). Diverse growth abilities at 4°C among the isolates were observed. Most of the isolates displayed enhanced growth rates at 4°C over 168 h, up to 108 CFU/ml (23.63%) and 107 CFU/ml (61.81%), while a minority of strains (14.6%) showed a slighter increase, up to 102–106 CFU/ml (Table 1). More than 85% of tested strains exhibited enhanced growth rates, which indicated a general survival and growth ability of Y. enterocolitica at low temperatures. This result is consistent with the observations of high levels of this bacterium in food products; e.g., meat, milk, cheese, and oysters (Peixotto et al., 1979; Greenwood et al., 1985; Amin and Draughon, 1987; Wang et al., 2009), and natural environmental conditions; e.g., soil and aqueous at low temperature (Asadishad et al., 2013). However, significant differences in growth ability among the tested isolates were observed at 4°C, which demonstrates the growth specificity of isolates at low temperature. Similar observations (specific behavior of strains under low temperature) were found in Y. enterocolitica previously. For example, strains with various serotypes survived differently at 4°C in soil and river water (Tashiro et al., 1991). The impact of low temperatures on the survival of Y. enterocolitica strains differs when inoculated on raw pork samples at 4 and −20°C for 90 days (Iliev and Najdenski, 2008).
To investigate the cold response of Y. enterocolitica, three isolates [44B (1A/O:5,27), 8081 (1B/O:8), and II7D (1A/O:5)] representing low, medium, and high growth ability at 4°C, respectively, were selected for further analysis (Supplementary Table S1).
Transcriptional Changes of Cold-Responsive Genes at Low Temperature
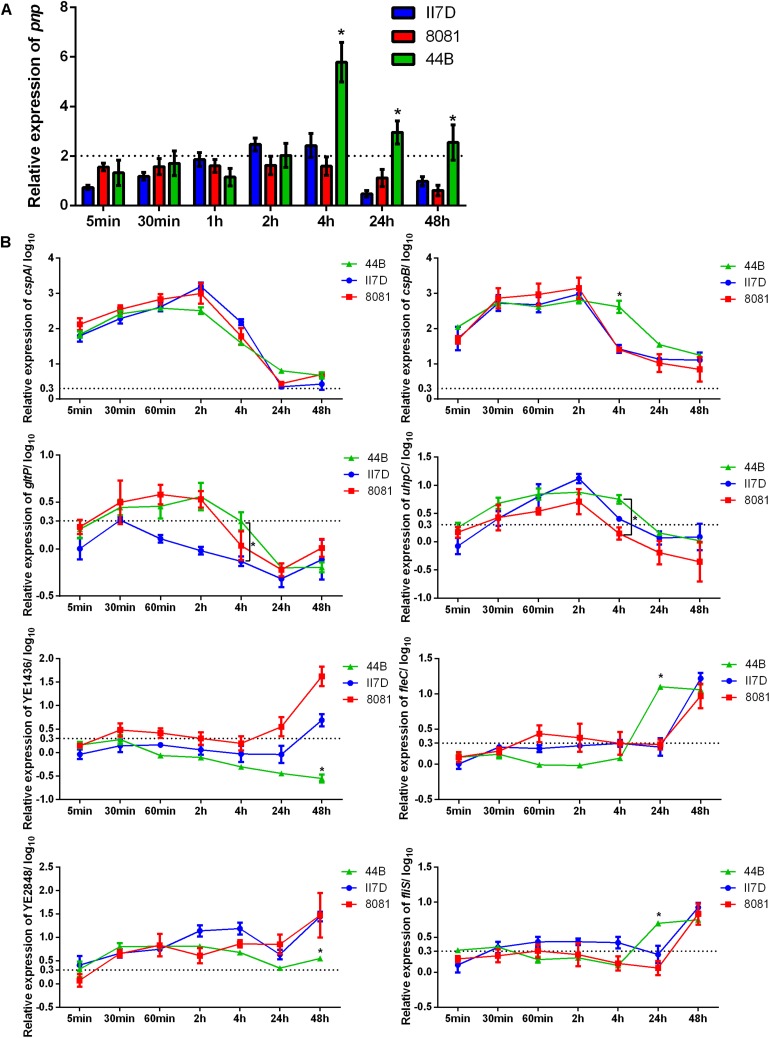
In order to better understand the cold response in Y. enterocolitica, the correlation between growth ability and transcriptional changes was investigated in the three isolates. It has been mentioned that pnp gene played an indispensable role in the cold response of Y. enterocolitica (Goverde et al., 1998) and other bacteria (Mathy et al., 2001; Hu et al., 2014; Briani et al., 2016). In our study, during a cold response, an increased expression of pnp gene was detected (Figure 1A). When exposed to 4°C for 5 min to 2 h, the pnp expression of the three isolates exhibited no significant difference. After 4 h of exposure at 4°C, the expression of pnp in 44B increased continuously and significantly exceeded that of II7D and 8081. The results indicated that different changes of pnp expression were found among tested isolates with various growth ability, which verified the essentiality of the pnp gene in cold adaptation. The continuous high expression of pnp gene implies the higher demand of PNPase and pnp in 44B.
FIGURE 1.
Expressional changes of the cold-responsive genes in II7D, 44B, and 8081. Three Y. enterocolitica strains were incubated at 4°C over time (from 5 min to 48 h) to show the expressional changes in cold response. Gene expression was normalized to the reference genes polA. (A) Expressional changes of pnp gene of three isolates at 4°C. Specific values are shown as the means ± SEM of the relative expression in four independent experiments. (B) Expressional changes of cspA, cspB, gltP, uhpC, YE1436, fliS, fleC, and YE284 genes at 4°C in three isolates. Specific values of relative gene expression are shown in log10 as the means ± SEM of four independent experiments. Statistically significant difference compared with the control according to multiple comparisons (∗p < 0.05). The line parallel to the x-axis represents a biologically relevant induction at 2 (fold-change) and 0.3 (log10 fold-change).
Based on the role of PNPase (encoded by the pnp gene) in repressing the generation of Csps and relieving growth arrest (Neuhaus et al., 2003; Zhao et al., 2016), the changes in related genes were investigated. RT-qPCR was performed with eight genes, which were reported to have increasing peaks or steady enhancement in gene expression after temperature downshift (Bresolin et al., 2006). The RNA used for this analysis was extracted from isolate cultures kept at 4°C from 5 min to 48 h and the related genes in response to cold with various functions are listed in Supplementary Table S2 accordingly.
As Figure 1B shown, the expression of the genes cspA, cspB, gltP, and uhpC increased rapidly after a cold stimulation and then decreased over time, which is consistent with the result from previous study regarding changes of the cold-shock genes (Bresolin et al., 2006; Horn et al., 2007). Based on the expression of these cold-shock genes, the expression decreased rapidly in strains II7D and 8081 after the transcriptional peak. However, in strain 44B, the expression of these genes decreased slowly and the relative expression of cspB, gltP, and uhpC was higher than that of II7D and 8081 at the end of 4 h after cold stress. Since the function of PNPase was RNA degradation and the higher expression of pnp was observed in 44B (Figure 1A), the repression of Csp generation might not be accomplished in 44B.
As reported previously, after the repression of Csp production, the growth reinitiated at the end of the acclimation phase (Yamanaka and Inouye, 2001). Therefore, the RNA degradation of Csps by PNPase was indispensable for cold acclimation and growth resumption. Similar cold acclimation was also found in E. coli, in which the synthesis of Csps transiently increases and the control of mRNA stability and translatability plays a major role in the adaptive response to cold temperature (Phadtare et al., 1999; Briani et al., 2016).
A different cold response was detected on transcriptional levels of YE1436, fleC, fliS, and YE2848, which did not show increased peaks but mostly increased under cold stress over prolonged growth. According to the expression of genes YE1436 and YE2848, the transcriptional levels increased over time and the upward tendencies in II7D and 8081 are more obvious than that in 44B (even no obvious uptrend of YE1436 gene expression). After 48 h of cold stress, the relative expression of YE1436 and YE2848 was significantly lower in 44B compared with II7D and 8081. Considering the worse growth ability of 44B at low temperature, the transcriptional regulation of gene YE1436 and YE2848 might be necessary for cold response. As it was mentioned in other studies, one of the psychrotrophic abilities in bacteria was to produce several non-Csps and allow growth during prolonged low temperatures in cold adaptation (Berger et al., 1996; Hébraud and Potier, 1999; Wouters et al., 2000; Phadtare, 2004). After the cold acclimation, the expression of non-cold shock genes has not been resumed in 44B, arresting the transition from acclimation to cell growth.
In addition, after 48 h of cold stress, the expression of fleC and fliS genes increased in 44B while their expression did not increase until 24 h in II7D and 8081. Since the genes fleC and fliS are associated with bacterial motility, the regulation of motility might contribute to cold adaptation as well.
Consequently, the transcriptional changes in cold-responsive genes play an important role in both cold acclimation and prolonged adaptation. The isolate-specific ability to survive under cold stress depends on the capacity of enabling transient induction and effective repression of cold-shock gene in cold acclimation. Meanwhile, the resumption of the non-cold shock gene expression was also required in prolonged cold adaptation.
Global Proteomic Analysis of the Cold-Responsive Proteins at Low Temperature
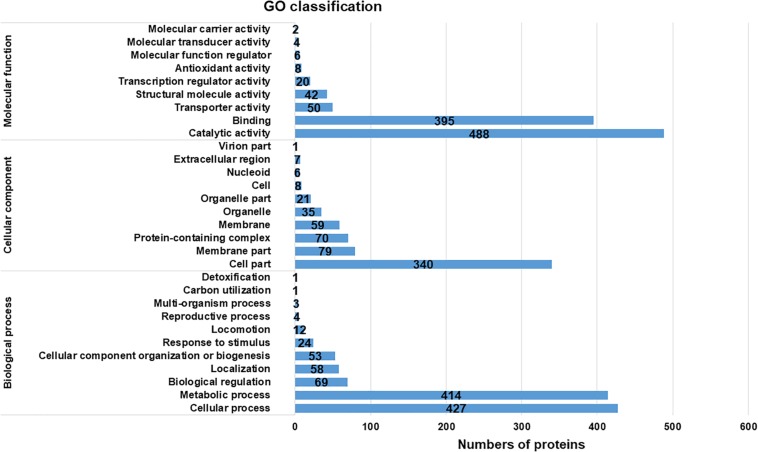
Three isolates (II7D, 8081, and 44B) with various growth abilities were used for the proteomic analysis to further investigate the underlying processes of cold response. A total of 1526 proteins were identified using label-free quantification analysis in six biological replicates. Among these, 809 proteins which expressed differentially under cold stress (at 4 versus 28°C) for 5 min, 2 h, and 24 h in three strains were identified. Functional classification and annotation indicated that 715 and 790 uniproteins were assigned to 30 GO annotations and 138 KEGG functional pathways, respectively.
The proteins assigned to GO functional groups were classified into three categories: “biological process,” “molecular function,” and “cellular component” (Figure 2). Various biological processes were involved in cold response. The most predominant processes were cellular and metabolic process; other major process categories were biological regulation, localization, and cellular component organization or biogenesis. These results indicated that the effects of cold response on protein level in Y. enterocolitica were involved in multiple processes. Furthermore, the predominant molecular functions of expressed proteins were associated with catalytic activity and binding; molecular functions of transporter and structural molecule activity were also involved in. Additionally, the most predominant cellular components were located cell and membrane parts. These results implied that the metabolism of the bacteria changed severely after cold response and it might lead to the alterations of cell and membrane components. Considerable groups of temperature-associated proteins were also reported previously in many other studies. For example, the periplasmic proteins associated with cellular component organization are strongly altered in Yersinia pestis in response to temperature changes (Pieper et al., 2008). The proteins involved in metabolic processes highly expressed at 4°C in Listeria monocytogenes (Cacace et al., 2010). During an abrupt temperature downshift in E. coli, expressional alterations occurred in the proteins associated with transport and binding (Kocharunchitt et al., 2014).
FIGURE 2.
Gene Ontology classification of the total assembled uniproteins.
For further investigation, the KEGG database was used and the expressed proteins were identified in four categories: “metabolism,” “genetic information processing,” “environmental information processing,” and “cellular processes” (Figure 3). It displayed that “Metabolism” with seven subcategories was the most enriched, which verified the active metabolic changes after a cold response. Among these subcategories, more proteins were enriched in metabolic related pathways: carbohydrate metabolism, nucleotide metabolism, amino acid biosynthesis, and translation. Similar pathways involved in cold response were also described in L. monocytogenes and E. coli (Cacace et al., 2010; Kocharunchitt et al., 2014).
FIGURE 3.
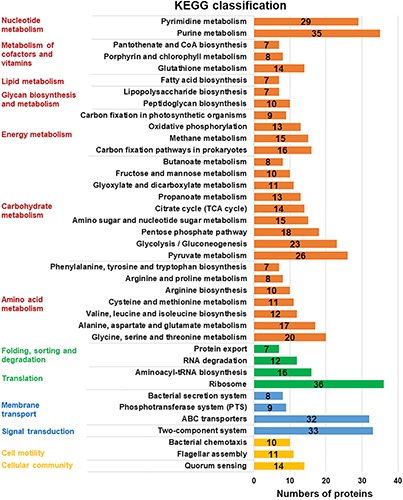
KEGG pathway clusters of assembled uniproteins. Metabolic pathways in different functional groups involved in cold response were classified with KEGG database in four related categories (protein numbers of each group higher than 10): “metabolism” in red, “genetic information processing” in green, “environmental information processing” in blue, and the “cellular processes” in yellow. The subcategory titles were also represented.
Analysis of Differentially Expressed Proteins at Low Temperature
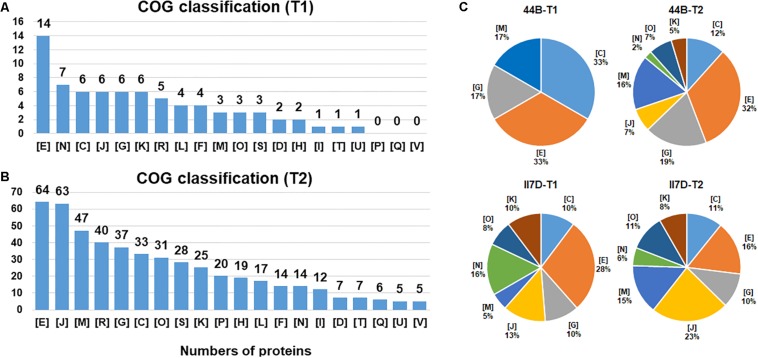
To investigate the alteration of metabolism related to growth profile under cold temperature over time, differentially expressed proteins were investigated at different time points in two isolates, 44B and II7D (with low and high growth ability at 4°C). Differentially expressed proteins of 44B and II7D under cold stress for 2 h (early stage, T1) and 24 h (late stage, T2) were compared (Figure 4).
FIGURE 4.
Functional category distribution of proteins identified from Y. enterocolitica cells at low temperature. (A) The number of identified proteins belong to each functional category is shown under cold stress after 2 h (T1). (B) The number of identified proteins belong to each functional category is shown under cold stress after 24 h (T2). (C) The percentage of differential expressed proteins that belong to each functional category in II7D and 44B is shown under cold stress in two time (T1 and T2). COG categories are as follows: C: energy production and conversion; D: cell division and chromosome partitioning; E: amino acid transport and metabolism; F: nucleotide transport and metabolism; G: carbohydrate transport and metabolism; H: coenzyme metabolism; I: lipid metabolism; J: translation, ribosomal structure and biogenesis, K: transcription; L: DNA replication, recombination and repair; M: cell wall/membrane/envelope biogenesis; N: cell motility; O: post translational modification, protein turnover, chaperones; P: inorganic ion transport and metabolism; Q: secondary metabolites biosynthesis, transport and catabolism; R: general function prediction only; S: function unknown; T: signal transduction mechanisms; U: intracellular trafficking, secretion, and vesicular transport; V: defense mechanisms.
Differentially expressed proteins were classified into 20 COGs functional groups with a relative fold change [log2 (FC) > 1.2 and log2 (FC) < -0.8, p < 0.05]. The expressed protein response to the early stage of the cold response (T1) was enriched into 17 functional clusters. Of these, the most predominant categories were “amino acid transport and metabolism,” “translation, ribosomal structure, and biogenesis,” “carbohydrate transport and metabolism,” “cell motility,” and also “transcription.” For the late stage of the cold response (T2), a high abundance of proteins was observed for categories of “amino acid transport and metabolism,” “translation, ribosomal structure, and biogenesis,” “cell wall/membrane/envelope biogenesis,” and “carbohydrate transport and metabolism.” In addition, a high abundance of the proteins belonging to “general function prediction only” was also found in both stages (Figures 4A,B). Throughout the whole testing course (T1–T2), proteins in specific functions of amino acid transport and metabolism (E), translation, ribosomal structure and biogenesis (J), carbohydrate transport and metabolism (G), and energy production and conversion (C) had higher enrichment in both T1 and T2. Hence, the proteins are involved mostly in metabolism in response to cold. Similarly, the high abundances of proteins regarding metabolism-related pathways and metabolic process were also investigated in the KEGG and GO analysis. Hence, we assume that the major cold-responsive proteins participate in the metabolic regulation of cells.
However, differences in protein abundance were observed between T1 and T2. Especially, protein abundance existed mostly in the clusters of cell motility (N) and transcription (K) in T1 while cell wall/membrane/envelope biogenesis (M) and post-translational modification, protein turnover, chaperones (O) in T2. This result indicates that the effects of the cold response on protein levels differ in the early and late stages. The time-dependent differences in protein categories were also found in E. coli in response to temperature and water-activity changes and were closely related to the cultivability after the temperature downshift (King et al., 2016). Different phases including adaptation and re-growth phases could be divided based on clustering analyses. Additionally, various protein categories were involved such as energy metabolism, DNA repair system, amino acid biosynthetic pathways, and carbohydrate catabolism (King et al., 2016).
According to the COG classification, eight protein clusters with the most protein abundance in T1 or T2 were chosen to compare the differences between strain 44B and II7D (Figure 4C). Compared with the protein abundance in the other three pie charts, protein clusters of (K), (O), (J), and (N) were undetectable in the early stage of 44B (T1) and the proportions of these proteins in all selected proteins in 44B (T2) were lower than those in II7D (T1) and II7D (T2). This result demonstrates that the biogenesis of responding proteins in 44B lags behind II7D under cold stress. Meanwhile, the proteins in clusters of (K), (O), and (J) represent key processes of protein biosynthesis. Hence, lower abundances of these proteins in 44B (T1) and 44B (T2) suggested that synthesis of general proteins might be inhibited in 44B compared to strain II7D. As was shown in many bacteria (e.g., E. coli), the arrest of cell growth upon temperature downshift is caused by the severe inhibition of general protein synthesis (Phadtare, 2004). The inhibition of general proteins in 44B (both in T1 and T2) might be the reason for low growth ability at low temperature. Considering the expressional repression of cold acclimation genes in 44B (Figure 1B), the inhibition might be involved in synthesis of cold acclimation proteins, which are essential for cold adaptation during prolonged growth.
In addition, a lower abundance of protein cluster (N) related to the “cell motility” was also mentioned in 44B. Base on the proteomic results, some cold-responsive proteins related to flagella and chemotaxis were detected in II7D but not in 44B (data not shown). For example, the Flg family, used for flagellar assembly and motility, are temperature-dependent in E. coli and other bacteria (Phadtare, 2012; Osterman et al., 2015). The chemotaxis protein, Che family is essential for motility and cold response (Burkart et al., 1998; Liu et al., 2014). According to the transcriptional analysis in Figure 1B, the correlation between motility and growth ability was demonstrated due to the different expressional changes of motility-related genes fleC (homologous to fliC and encoding Flagellin), fliS (putative cytoplasmic chaperone), and YE2848 (putative chemotaxis methyl-accepting transducer) in three isolates. Meanwhile, the Flagellin was detectable only in II7D but not in 44B in proteomic analysis (other related genes were not found). Since it is critical in motility and cold response in Salmonella enterica (Elhadad et al., 2015; Michaux et al., 2017), the involvement of motility in cold response might be confirmed.
On the other hand, the percentages of clusters of energy production and conversion (C), carbohydrate transport and metabolism (G), and cell wall/membrane/envelope biogenesis (M) in 44B (T1) are higher than those in 44B (T2), II7D (T1), and II7D (T2). Considering the high abundance of proteins related to carbohydrate metabolism and cell wall/membrane/envelope biogenesis, but low enrichment of proteins related to functional protein synthesis in strain 44B (T1), we might assume that 44B uses a high rate of energy for the cell wall structure, instead of initial growth at cold response. As an important protective structure against adverse environmental conditions, the cell membrane plays an important role in stress response. Previously, it was extensively discussed that membrane lipopolysaccharide, cell membrane, and the membrane fluidity contribute to temperature adaptation in bacteria (Carty et al., 1999; Phadtare, 2004; Storz and Hengge, 2010).
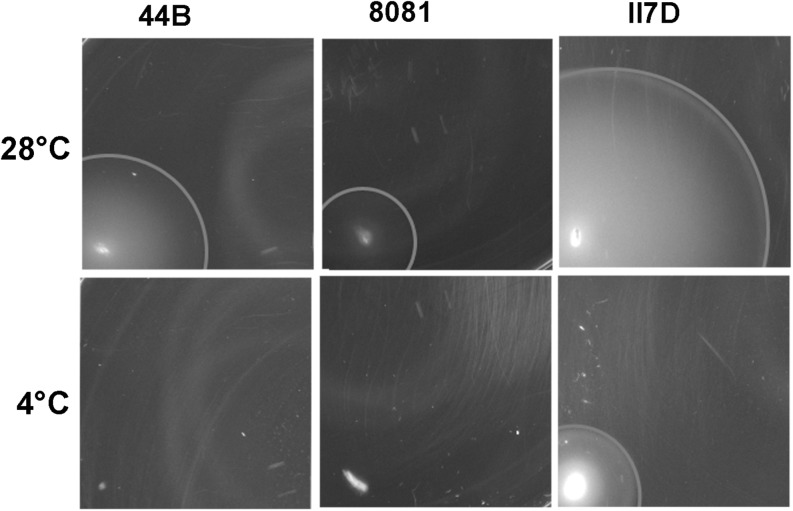
Motility at Low Temperature
To investigate the physiological changes in cold response, motility assays at low temperatures were performed on three isolates (44B, 8081, and II7D) with different growth profiles at low temperatures. All three strains showed motility at 28°C; however, at the temperature of 4°C, only II7D was motile (Figure 5).
FIGURE 5.
Motility of Y. enterocolitica strains at low temperature. Motility was tested on motility agar plate. Single colonies were stabbed on to motility agar plates and incubated initially for 2 h at 37°C to start the assay with non-motile bacteria. The plates were subsequently incubated at 28°C (for 21 h) and 4°C (for 44 h). The motility was assessed by measuring the diameters of migration zone of strains. Values of the migration zone diameters (means ± SEM) in each strains were tested of six independent experiments: 24.92 ± 0.239 cm (44B at 28°C), 11.58 ± 0.201 cm (8081 at 28°C), 49.00 ± 0.966 cm (II7D at 28°C), and 17.83 ± 0.105 cm (II7D at 4°C). No migration zone was detectable in 44B or 8081 at 4°C.
As shown in our transcriptional analysis, the expression of motility-related genes (fliS and fleC) increased under cold stress at 4°C and their expression was increased earlier in strain 44B than in the other two strains (Figure 1B). Meanwhile, a lower abundance of proteins was present in the “cell motility” group in 44B, which was consistent with the lower growth ability in 44B than II7D. Moreover, the differential growth ability correlates with motility in the three strains (only the strain with high growth ability was motile) at low temperature. Based on the results from transcriptional and proteomic analysis, the different induction of the motility-related genes and proteins among isolates with different growth behaviors indicated the close link between cell motility and growth ability, which has been described previously in Y. enterocolitica (Kapatral et al., 1996). However, due to the wide range of factors with complex mechanisms in regulating motility, how cell motility was affected by or contributed to the growth ability after cold response remains unclear (Young et al., 1999; Mukherjee et al., 2013; Xu et al., 2014).
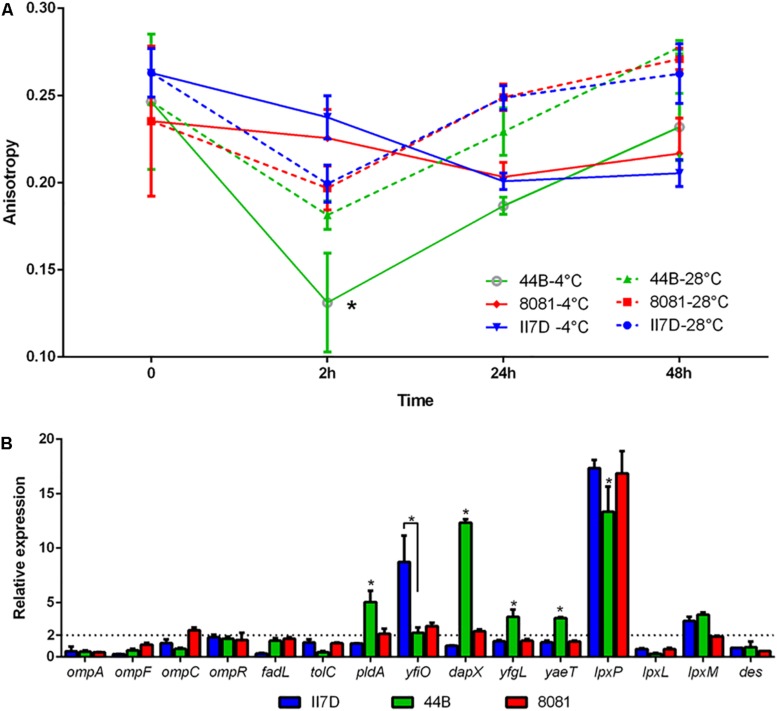
Cell Membrane and Fluidity at Low Temperature
To test other factors corresponding to membrane activity in cold response, fluidity assays were performed at 4°C on three isolates (44B, 8081, and II7D). All three tested strains showed stable fluidity under the temperature of 28°C in 48 h, while the membrane fluidity of 44B increased significantly at 4°C at 2 h and decreased to the normal level at 4°C after 24 h (Figure 6A). According to the results of fluidity, the membrane fluidity maintained at the normal level in both strains 8081 and II7D, but not in 44B. These results indicated that the balance of the membrane fluidity was changed in response to cold stress in 44B at 2 h. This finding might be correlated to the high protein abundance in the functional cluster of cell wall/membrane/envelope biogenesis in 44B at T1. Therefore, the differences in growth abilities at low temperature might be related to the maintenance of cell fluidity. The similar roles of membrane fluidity have been demonstrated in cold and other stresses in many bacteria (Yoon et al., 2015; Eberlein et al., 2018). However, the fluidities are regulated by various mechanisms in different bacteria; e.g., E. coli (Carty et al., 1999), B. subtilis (Aguilar et al., 2001), and Salmonella (Wollenweber et al., 1983; Ricke et al., 2018).
FIGURE 6.
Membrane fluidity and transcriptional changes of membrane-related genes in Y. enterocolitica strains at low temperature. (A) Fluidity assays in II7D, 44B, and 8081 under cold stress for 2, 24, and 48 h. Anisotropy value represented the membrane fluidity (higher anisotropy means lower fluidity). (B) Expressional changes of the membrane related genes were detected using RT-qPCR and were normalized to the reference gene polA. Specific values of relative gene expression are shown as the means ± SEM of four independent experiments. The line parallel to the x-axis represents a biologically relevant induction at 2 (fold-change). Statistically significant difference compared with the control according to multiple comparisons (∗p < 0.05).
To investigate the regulatory factors of membrane fluidity in Y. enterocolitica, groups of genes regarding outer membrane proteins and lipid A biosynthesis were selected according to previous studies (Dekker, 2000; Nikaido, 2003; Barria et al., 2013; Hussain and Bernstein, 2018; Robinson, 2019). Transcriptional changes in these genes were investigated under cold stress for 2 h in three isolates with the primers listed in Supplementary Table S3. In strain 44B, significantly higher expression of yaeT, yfgL, dapX, and pldA and lower expression of yfiO and lpxP was observed compared to the other isolates (Figure 6B). The four outer membrane protein assembly factors (encoded by yaeT, yfgL, dapX, and yfiO) were found in proteomic analysis, in which, BamC encoded by dapX was upregulated significantly in 44B. The differential expression of these genes and proteins might be involved in fluidity regulation in response to cold. Similar functions of the outer membrane protein YaeT, DapX, and YfgL (homologous to insert β-barrel proteins in E. coli) were shown in previous research in response to cold (Macintyre and Henning, 1990; Onufryk et al., 2005; Wu et al., 2005; Begic and Worobec, 2006; Sklar et al., 2007; Rollauer et al., 2015). Outer membrane phospholipase A (encoded by pldA), which is activated under various stress conditions, presents in the outer membrane of Gram-negative bacteria. Its possible role is maintaining the cell envelope integrity and permeabilization, which is related to temperature (Dekker, 2000; Belosludtsev et al., 2014). The different expressions of pldA gene and protein among isolates suggested the possible involvement of outer membrane phospholipase A in fluidity maintenance under cold stress. However, their functions in cold response still remain to be elucidated in Y. enterocolitica.
Des and LpxP (encoded by des and lpxP genes) are two fluidity-generated enzymes in B. subtilis and E. coli. In B. subtilis, upon a drop in temperature, the Des protein is synthesized and desaturates the acyl chains of membrane phospholipids to increase the membrane fluidity (Aguilar et al., 2001; Albanesi et al., 2004). Furthermore, in E. coli, cold-induced acyltransferase LpxP helps to attach more unsaturated fatty acids (palmitoleate instead of laurate attached at normal temperature by LpxL) to lipid A, thus increasing membrane fluidity and lowering its phase transition temperature, counteracting the effect of low temperature (Vorachek-Warren et al., 2002). In our study, no significant difference in the expression of des gene was observed among the three isolates and the Des protein did not induced, which implied that the regulation of Des in Y. enterocolitica might not be as important as that in B. subtilis. Meanwhile, expression of the cold induced gene lpxP was significantly lower in 44B and the related protein were induced differently between II7D and 44B according to proteomic analysis (Supplementary Table S4). Therefore, membrane fluidity related to growth ability in Y. enterocolitica might be regulated by LpxP in cold adaptation, which is identical to E. coli.
Proteomic Overview of the Cold Response in Y. enterocolitica
Cold response involved a series of complex and significant changes in the abundance of proteins in many processes and pathways rather than a simple increase or decrease in a specific category. To present an overview of the cold response of Y. enterocolitica, the upregulated proteins under cold stress were selected according to the main COG functional categories mentioned in this study. These particular proteins probably represented the key determinants that allow life at low temperature. Top KEGG pathways (including the BRITE hierarchies) were selected according to the proteins in COG categories (listed in Supplementary Table S4). Proteomic overview and the predicted regulation in cold response are presented in Figure 7.
FIGURE 7.
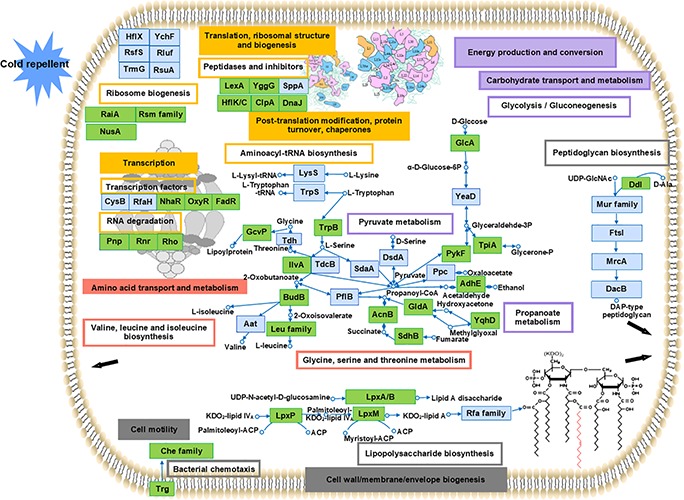
Overview of the cold response of Y. enterocolitica at proteomic level. Proteins were selected in the main COG functional categories and distinguished. Boxes with colored border represented the related pathways (including KEGG BRITE hierarchies). The main KEGG pathways reported in each category were presented: transcription factors, RNA degradation, ribosome biogenesis, peptidases and inhibitor, and aminoacyl-tRNA biosynthesis (in yellow); glycine, serine, and threonine metabolism and valine, leucine, and isoleucine biosynthesis (in red); lipopolysaccharide biosynthesis, peptidoglycan biosynthesis, and bacterial chemotaxis (in gray), glycolysis/gluconeogenesis, pyruvate metabolism, and propanoate metabolism (in purple). The related COG categories were marked with the same colors in boxes accordingly. The proteins were displayed in blue boxes representing the involvement in the related functional categories and pathways in this study. The green boxes were used for the proteins also clarified in cold response in other researches. Detailed information is listed in Supplementary Table S4.
First, a high abundance of proteins was observed associated with protein biosynthetic processes, such as transcriptional, translational, and ribosomal, and post-translational processes (related COG categories are shown in yellow boxes). These proteins were involved predominate in transcription factors, RNA degradation, peptidases and inhibitors, ribosome biogenesis, and aminoacyl-tRNA biosynthesis. Numbers of proteins related ribosome biogenesis [such as the transcription termination/anti-termination protein NusA, ribosome-associated inhibitor A (encoded by raiA), and ribosomal RNA small subunit methyltransferase B (encoded by rsmB)] were induced in response to cold. These related protein associated with cold stress was also reported in E. coli and other bacteria (Burakovsky et al., 2012; Di Pietro et al., 2013). Meanwhile, functions of the proteins induced in this study like ribosomal silencing factor RsfS, GTPase HflX, and ribosome-binding ATPase YchF under cold stress have not been clarified previously. Since some of them are involved in other stress response like heat, oxidative, and nutrient stress (Starosta et al., 2014; Hannemann et al., 2016; Dey et al., 2018), their potential roles under cold stress should be investigated. The proteins associated with peptidases and inhibitor such as LexA repressor, lipoprotein (encoded by yggG), and Protein HflK were induced under cold stress in this study. The cold-responsive function of these proteins was also found previously in other research (Phadtare and Inouye, 2004; Burakovsky et al., 2012; Jian et al., 2015). Besides, some transcription factors (encoded by nhaR, oxyR, fadR, cysB, and rfaH) were also involved in regulation of cold response. Since the essential cold-responsive roles of these transcription factors (encoded by nhaR, oxyR, and fadR) were investigated in E. coli (White-Ziegler et al., 2008), Vibrio vulnificus (Limthammahisorn et al., 2008), and Moraxella catarrhalis (Spaniol et al., 2013), transcription factors should also be focused on in Y. enterocolitica cold response.
A number of proteins involved in specific amino acids biosynthesis may reflect their importance in mediating survival under cold stress. In our research, the induced proteins participated in biosynthesis of various amino acids under cold stress (related COG categories shown in red boxes). These proteins are associated with the biosynthesis and metabolism of glycine, serine, and threonine (encoded by trpB, gcvP, ilvA, tdh, etc.), and valine, leucine, and isoleucine (encoded by leuA, leuB, leuC, leuD, budB, etc.). Similar amino acids have been demonstrated in previous studies to aid tolerance under cold stress conditions in many other bacteria (Fonseca et al., 2011; King et al., 2016).
Second, proteins associated with cell membrane and motility were identified (related COG categories shown in gray boxes). Certain proteins were identified in pathways, such as lipopolysaccharide biosynthesis (encoded by lpxA/B/P/M and rfaC/Q), peptidoglycan biosynthesis (encoded by murC/E, ddl, dacB, etc.), and bacterial chemotaxis (encoded by trg and cheB/D/Z). The cold-responsive functions have been reported previously in many proteins mentioned in Supplementary Table S4. However, although the genes related to motility (fliS and YE2848) were detected in our transcriptional, the induction of them cannot be detected in our proteomic results. According to the proteomic data, almost all the proteins related to flagellar assembly were downregulated. The cold-responsive effect of flagella on cell motility at the protein level is unknown.
Meanwhile, valine, leucine, and isoleucine, as the branched-chain amino acids and the precursors for biosynthesis of iso- and anteiso-branched-chain fatty acids, were utilized to regulate the membrane fluidity in response to cold in certain bacteria (Grau and de Mendoza, 1993; Annous et al., 1997; Klein et al., 1999). Levels of isoleucine and leucine significantly increase under cold stress in E. coli (Jozefczuk et al., 2010), and the expression of related genes (leuA/B/C/D and ilvB/C/D/E/H) was also elevated in Thermoanaerobacter tengcongensis (Liu et al., 2014). Based on our proteomic results, the induction of leuA/B/C/D encoding proteins was only detected in 44B, which implied the indispensable regulation of these branched-chain amino acids. However, the growth ability under cold stress of 44B was detected worse than II7D, which suggested that multiple pathways related to motility might be applied in cold response.
The considerable involvement of proteins has been detected and the transcriptional and physiological investigation associated with motility and fluidity contributes to our understanding of cold-response regulation of motility and membrane fluidity.
Additionally, certain pathways in energy production and conversion and carbohydrate transport and metabolism were also involved in this study (listed in Supplementary Table S4). The complex processes and pathways in cold response of Y. enterocolitica and the specific functions of other individual proteins predicted in the proteomic results are required to be investigated during cold adaptation.
This study demonstrates the strain-specific cold response of Y. enterocolitica at 4°C, which is time-dependent, including cold acclimation and adaptation. The transcriptional analysis revealed the importance of the induction and repression of cold-shock genes in cold acclimation as well as the resumption of the non-cold shock genes in prolonged cold adaptation. Meanwhile, the time-dependent response at protein level was also found and the cold-responsive proteins identified in proteomic analysis were closely related to protein synthesis, cell membrane parts and cell motility. Additionally, the physiological processes in cell fluidity and motility might be responsible for differential growth abilities at low temperatures. By combining different approaches, cold response was described systematically, providing a better understanding of the significant physiological processes involved in cold stress of Y. enterocolitica.
Data Availability Statement
All datasets generated for this study are included in the article and Supplementary Material.
Author Contributions
CL contributed to designing, carrying out the experiment, and writing the manuscript. JM provided assistance for the proteome experiment and reviewed the manuscript. CT provided assistance for the experiment. TA reviewed the manuscript and gave advice. CR reviewed the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was financially supported by the Chinese Scholarship Council.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03037/full#supplementary-material
References
- Aguilar P. S., Hernandez-Arriaga A. M., Cybulski L. E., Erazo A. C., de Mendoza D. (2001). Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 20 1681–1691. 10.1093/emboj/20.7.1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanesi D., Mansilla M. C., de Mendoza D. (2004). The membrane fluidity sensor desk of Bacillus subtilis controls the signal decay of its cognate response regulator. J. Bacteriol. 186 2655–2663. 10.1128/jb.186.9.2655-2663.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin M. K., Draughon F. A. (1987). Growth-characteristics of Yersinia-enterocolitica in pasteurized skim milk. J. Food Protect. 50 849–852. 10.4315/0362-028x-50.10.849 [DOI] [PubMed] [Google Scholar]
- Annous B. A., Becker L. A., Bayles D. O., Labeda D. P., Wilkinson B. J. (1997). Critical role of anteiso-C15: 0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 63 3887–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricha B., Fishov I., Cohen Z., Sikron N., Pesakhov S., Khozin-Goldberg I., et al. (2004). Differences in membrane fluidity and fatty acid composition between phenotypic variants of Streptococcus pneumoniae. J. Bacteriol. 186 4638–4644. 10.1128/Jb.186.14.4638-4644.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadishad B., Ghoshal S., Tufenkji N. (2013). Role of cold climate and freeze–thaw on the survival, transport, and virulence of Yersinia enterocolitica. Environ. Sci. Technol. 47 14169–14177. 10.1021/es403726u [DOI] [PubMed] [Google Scholar]
- Bari M. L., Hossain M. A., Isshiki K., Ukuku D. (2011). Behavior of Yersinia enterocolitica in foods. J. Pathog. 2011:420732. 10.4061/2011/420732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barria C., Malecki M., Arraiano C. M. (2013). Bacterial adaptation to cold. Microbiology 159 2437–2443. 10.1099/mic.0.052209-0 [DOI] [PubMed] [Google Scholar]
- Begic S., Worobec E. A. (2006). Regulation of Serratia marcescens ompF and ompC porin genes in response to osmotic stress, salicylate, temperature and pH. Microbiology 152(Pt 2), 485–491. 10.1099/mic.0.28428-0 [DOI] [PubMed] [Google Scholar]
- Belosludtsev K., Belosludtseva N., Kondratyev M., Agafonov A., Purtov Y. (2014). Interaction of phospholipase A of the E. coli outer membrane with the inhibitors of eucaryotic phospholipases A(2) and their effect on the Ca2+-induced permeabilization of the bacterial membrane. J. Membr. Biol. 247 281–288. 10.1007/s00232-014-9633-4 [DOI] [PubMed] [Google Scholar]
- Benembarek P. K. (1994). Presence, detection and growth of Listeria-monocytogenes in Seafoods - a review. Int. J. Food Microbiol. 23 17–34. 10.1016/0168-1605(94)90219-4 [DOI] [PubMed] [Google Scholar]
- Berger F., Morellet N., Menu F., Potier P. (1996). Cold shock and cold acclimation proteins in the psychrotrophic bacterium Arthrobacter globiformis SI55. J. Bacteriol. 178 2999–3007. 10.1128/jb.178.11.2999-3007.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg P., Wagner E. G., Nordström K. (1990). Control Of Replication Of Plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 9 2331–2340. 10.1002/j.1460-2075.1990.tb07405.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresolin G., Neuhaus K., Scherer S., Fuchs T. M. (2006). Transcriptional analysis of long-term adaptation of Yersinia enterocolitica to low-temperature growth. J. Bacteriol. 188 2945–2958. 10.1128/JB.188.8.2945-2958.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresolin G., Trcek J., Scherer S., Fuchs T. M. (2008). Presence of a functional flagellar cluster Flag-2 and low-temperature expression of flagellar genes in Yersinia enterocolitica W22703. Microbiology 154(Pt 1), 196–206. 10.1099/mic.0.2007/008458-0 [DOI] [PubMed] [Google Scholar]
- Briani F., Carzaniga T., Deho G. (2016). Regulation and functions of bacterial PNPase. Wiley Interdiscip. Rev. RNA 7 241–258. 10.1002/wrna.1328 [DOI] [PubMed] [Google Scholar]
- Burakovsky D. E., Prokhorova I. V., Sergiev P. V., Milon P., Sergeeva O. V., Bogdanov A. A., et al. (2012). Impact of methylations of m2G966/m5C967 in 16S rRNA on bacterial fitness and translation initiation. Nucleic Acids Res. 40 7885–7895. 10.1093/nar/gks508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart M., Toguchi A., Harshey R. M. (1998). The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95 2568–2573. 10.1073/pnas.95.5.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacace G., Mazzeo M. F., Sorrentino A., Spada V., Malorni A., Siciliano R. A. (2010). Proteomics for the elucidation of cold adaptation mechanisms in Listeria monocytogenes. J. Proteomics 73 2021–2030. 10.1016/j.jprot.2010.06.011 [DOI] [PubMed] [Google Scholar]
- Cao-Hoang L., Dumont F., Marechal P. A., Gervais P. (2010). Inactivation of Escherichia coli and Lactobacillus plantarum in relation to membrane permeabilization due to rapid chilling followed by cold storage. Arch. Microbiol. 192 299–305. 10.1007/s00203-010-0555-y [DOI] [PubMed] [Google Scholar]
- Carty S. M., Sreekumar K. R., Raetz C. R. H. (1999). Effect of cold shock on lipid A biosynthesis in Escherichia coli. Induction at 12 degrees C of an acyltransferase specific for palmitoleoyl-acyl carrier protein. J. Biol. Chem. 274 9677–9685. 10.1074/jbc.274.14.9677 [DOI] [PubMed] [Google Scholar]
- Coburn G. A., Mackie G. A. (1998). Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 62 55–108. 10.1016/s0079-6603(08)60505-x [DOI] [PubMed] [Google Scholar]
- Cordin O., Banroques J., Tanner N. K., Linder P. (2006). The DEAD-box protein family of RNA helicases. Gene 367 17–37. 10.1016/j.gene.2005.10.019 [DOI] [PubMed] [Google Scholar]
- Dekker N. (2000). Outer-membrane phospholipase A: known structure, unknown biological function. Mol. Microbiol. 35 711–717. 10.1046/j.1365-2958.2000.01775.x [DOI] [PubMed] [Google Scholar]
- Delumeau O., Lecointe F., Muntel J., Guillot A., Guédon E., Monnet V., et al. (2011). The dynamic protein partnership of RNA polymerase inBacillus subtilis. Proteomics 11 2992–3001. 10.1002/pmic.201000790 [DOI] [PubMed] [Google Scholar]
- Dey S., Biswas C., Sengupta J. (2018). The universally conserved GTPase HflX is an RNA helicase that restores heat-damaged <em>Escherichia coli</em> ribosomes. J. Cell Biol. 217 2519–2529. 10.1083/jcb.201711131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro F., Brandi A., Dzeladini N., Fabbretti A., Carzaniga T., Piersimoni L., et al. (2013). Role of the ribosome-associated protein PY in the cold-shock response of Escherichia coli. Microbiol. Open 2 293–307. 10.1002/mbo3.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divya K. H., Varadaraj M. C. (2013). Behavioral pattern of native food isolates of Yersinia enterocolitica under simulated time-temperature combinations of the food chain. Food Nutr. Sci. 4 365–375. 10.4236/fns.2013.44047 [DOI] [Google Scholar]
- Eberlein C., Baumgarten T., Starke S., Heipieper H. J. (2018). Immediate response mechanisms of Gram-negative solvent-tolerant bacteria to cope with environmental stress: cis-trans isomerization of unsaturated fatty acids and outer membrane vesicle secretion. Appl. Microbiol. Biotechnol. 102 2583–2593. 10.1007/s00253-018-8832-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhadad D., Desai P., Rahav G., McClelland M., Gal-Mor O. (2015). Flagellin is required for host cell invasion and normal Salmonella pathogenicity island 1 expression by Salmonella enterica serovar Paratyphi A. Infect. Immun. 83 3355–3368. 10.1128/IAI.00468-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko D. N., Makhatadze G. I. (2002). Bacterial cold-shock proteins. Cell. Mol. Life Sci. 59 1902–1913. 10.1007/Pl00012513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority, and European Centre for Disease Prevention, and Control (2016). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 14:4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca P., Moreno R., Rojo F. (2011). Growth of Pseudomonas putida at low temperature: global transcriptomic and proteomic analyses. Environ. Microbiol. Rep. 3 329–339. 10.1111/j.1758-2229.2010.00229.x [DOI] [PubMed] [Google Scholar]
- Galperin M. Y., Makarova K. S., Wolf Y. I., Koonin E. V. (2014). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43 D261–D269. 10.1093/nar/gku1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goverde R. L. J., Huis in’t Veld J. H. J., Kusters J. G., Mooi F. R. (1998). The psychrotrophic bacterium Yersinia enterocolitica requires expression of pnp, the gene for polynucleotide phosphorylase, for growth at low temperature (5°C). Mol. Microbiol. 28 555–569. 10.1046/j.1365-2958.1998.00816.x [DOI] [PubMed] [Google Scholar]
- Grau R., de Mendoza D. (1993). Regulation of the synthesis of unsaturated fatty acids by growth temperature in Bacillus subtilis. Mol. Microbiol. 8 535–542. 10.1111/j.1365-2958.1993.tb01598.x [DOI] [PubMed] [Google Scholar]
- Greenwood M. H., Coetzee E. F., Ford B. M., Gill P., Hooper W. L., Matthews S. C., et al. (1985). The microbiology of cooked prawns and shrimps on retail sale. J. Hyg. 94 319–326. 10.1017/s0022172400061544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannemann L., Suppanz I., Ba Q., MacInnes K., Drepper F., Warscheid B., et al. (2016). Redox activation of the universally conserved ATPase YchF by Thioredoxin 1. Antioxid. Redox Signal. 24 141–156. 10.1089/ars.2015.6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébraud M., Potier P. (1999). Cold shock response and low temperature adaptation in psychrotrophic bacteria. J. Mol. Microbiol. Biotechnol. 1 211–219. [PubMed] [Google Scholar]
- Herbst F.-A., Danielsen H. N., Wimmer R., Nielsen P. H., Dueholm M. S. (2015). Label-free quantification reveals major proteomic changes in Pseudomonas putida F1 during the exponential growth phase. Proteomics 15 3244–3252. 10.1002/pmic.201400482 [DOI] [PubMed] [Google Scholar]
- Horisaka T., Fujita K., Iwata T., Nakadai A., Okatani A. T., Horikita T., et al. (2004). Sensitive and specific detection of Yersinia pseudotuberculosis by loop-mediated isothermal amplification. J. Clin. Microbiol. 42 5349–5352. 10.1128/Jcm.42.11.5349-5352.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn G., Hofweber R., Kremer W., Kalbitzer H. R. (2007). Structure and function of bacterial cold shock proteins. Cell. Mol. Life Sci. 64 1457. 10.1007/s00018-007-6388-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., McCormick R. J., Means W. J., Zhu M. J. (2014). Polynucleotide phosphorylase is required for Escherichia coli O157:H7 growth above refrigerated temperature. Foodborne Pathog. Dis. 11 177–185. 10.1089/fpd.2013.1632 [DOI] [PubMed] [Google Scholar]
- Hussain S., Bernstein H. D. (2018). The Bam complex catalyzes efficient insertion of bacterial outer membrane proteins into membrane vesicles of variable lipid composition. J. Biol. Chem. 293 2959–2973. 10.1074/jbc.RA117.000349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev M., Najdenski H. (2008). Monitoring of plasmid dissociation and pathogenic potential among Yersinia enterocolotica and Yersinia pseudotuberculosis during storage of refrigerated pork meat. Ann. Microbiol. 58 623–632. 10.1007/bf03175567 [DOI] [Google Scholar]
- Jian H., Xiong L., He Y., Xiao X. (2015). The regulatory function of LexA is temperature-dependent in the deep-sea bacterium Shewanella piezotolerans WP3. Front. Microbiol. 6:627. 10.3389/fmicb.2015.00627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozefczuk S., Klie S., Catchpole G., Szymanski J., Cuadros-Inostroza A., Steinhauser D., et al. (2010). Metabolomic and transcriptomic stress response of Escherichia coli. Mol. Syst. Biol. 6:364. 10.1038/msb.2010.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. (2016). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45 D353–D361. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapatral V., Olson J. W., Pepe J. C., Miller V. L., Minnich S. A. (1996). Temperature-dependent regulation of Yersinia enterocolitica Class III flagellar genes. Mol. Microbiol. 19 1061–1071. 10.1046/j.1365-2958.1996.452978.x [DOI] [PubMed] [Google Scholar]
- King T., Kocharunchitt C., Gobius K., Bowman J. P., Ross T. (2016). Physiological response of Escherichia coli O157:H7 Sakai to dynamic changes in temperature and water activity as experienced during carcass chilling. Mol. Cell. Proteomics 15 3331–3347. 10.1074/mcp.M116.063065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein W., Weber M. H., Marahiel M. A. (1999). Cold shock response of Bacillus subtilis: isoleucine-dependent switch in the fatty acid branching pattern for membrane adaptation to low temperatures. J. Bacteriol. 181 5341–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocharunchitt C., King T., Gobius K., Bowman J. P., Ross T. (2014). Global genome response of Escherichia coli O157: H7 Sakai during dynamic changes in growth kinetics induced by an abrupt downshift in water activity. PLoS One 9:e90422. 10.1371/journal.pone.0090422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G., Hummel K., Ahrens M., Menanteau-Ledouble S., Welch T. J., Eisenacher M., et al. (2016). Shotgun proteomic analysis of Yersinia ruckeri strains under normal and iron-limited conditions. Vet. Res. 47:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limthammahisorn S., Brady Y. J., Arias C. R. (2008). Gene expression of cold shock and other stress-related genes in Vibrio vulnificus grown in pure culture under shellstock temperature control conditions. J. Food Prot. 71 157–164. 10.4315/0362-028x-71.1.157 [DOI] [PubMed] [Google Scholar]
- Liu B., Zhang Y., Zhang W. (2014). RNA-Seq-based analysis of cold shock response in Thermoanaerobacter tengcongensis, a bacterium harboring a single cold shock protein encoding gene. PLoS One 9:e93289. 10.1371/journal.pone.0093289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre S., Henning U. (1990). The role of the mature part of secretory proteins in translocation across the plasma-membrane and in regulation of their synthesis in Escherichia-Coli. Biochimie 72 157–167. 10.1016/0300-9084(90)90141-3 [DOI] [PubMed] [Google Scholar]
- Mathy N., Jarrige A. C., Robert-Le Meur M., Portier C. (2001). Increased expression of Escherichia coli polynucleotide phosphorylase at low temperatures is linked to a decrease in the efficiency of autocontrol. J. Bacteriol. 183 3848–3854. 10.1128/JB.183.13.3848-3854.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos R. G., Barbas A., Arraiano C. M. (2009). RNase R mutants elucidate the catalysis of structured RNA: RNA-binding domains select the RNAs targeted for degradation. Biochem. J. 423 291–301. 10.1042/Bj20090839 [DOI] [PubMed] [Google Scholar]
- Michaux C., Holmqvist E., Vasicek E., Sharan M., Barquist L., Westermann A. J., et al. (2017). RNA target profiles direct the discovery of virulence functions for the cold-shock proteins CspC and CspE. Proc. Natl. Acad. Sci. U.S.A. 114 6824–6829. 10.1073/pnas.1620772114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel V., Lehoux I., Depret G., Anglade P., Labadie J., Hebraud M. (1997). The cold shock response of the psychrotrophic bacterium Pseudomonas fragi involves four low-molecular-mass nucleic acid binding proteins. J. Bacteriol. 179 7331–7342. 10.1128/jb.179.23.7331-7342.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Babitzke P., Kearns D. B. (2013). FliW and FliS function independently to control cytoplasmic flagellin levels in Bacillus subtilis. J. Bacteriol. 195 297–306. 10.1128/JB.01654-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytczuk N., Trevors J., Leduc L., Ferroni G. (2007). Fluorescence polarization in studies of bacterial cytoplasmic membrane fluidity under environmental stress. Prog. Biophys. Mol. Biol. 95 60–82. 10.1016/j.pbiomolbio.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Neuhaus K., Anastasov N., Kaberdin V., Francis K. P., Miller V. L., Scherer S. (2003). The AGUAAA motif in cspA1/A2 mRNA is important for adaptation of Yersinia enterocolitica to grow at low temperature. Mol. Microbiol. 50 1629–1645. 10.1046/j.1365-2958.2003.03795.x [DOI] [PubMed] [Google Scholar]
- Nikaido H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67 593–656. 10.1128/mmbr.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onufryk C., Crouch M. L., Fang F. C., Gross C. A. (2005). Characterization of six lipoproteins in the sigma(E) regulon. J. Bacteriol. 187 4552–4561. 10.1128/Jb.187.13.4552-4561.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterman I. A., Dikhtyar Y. Y., Bogdanov A. A., Dontsova O. A., Sergiev P. V. (2015). Regulation of flagellar gene expression in Bacteria. Biochemistry 80 1447–1456. 10.1134/S000629791511005X [DOI] [PubMed] [Google Scholar]
- Ostroff S. M., Kapperud G., Hutwagner L. C., Nesbakken T., Bean N. H., Lassen J., et al. (1994). Sources of sporadic Yersinia-enterocolitica infections in Norway - a prospective case-control study. Epidemiol. Infect. 112 133–141. 10.1017/s0950268800057496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixotto S. S., Finne G., Hanna M. O., Vanderzant C. (1979). Presence, growth and survival of Yersinia-enterocolitica in oysters, shrimp and crab. J. Food Protect. 42 974–981. 10.4315/0362-028x-42.12.974 [DOI] [PubMed] [Google Scholar]
- Phadtare S. (2004). Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6 125–136. [PubMed] [Google Scholar]
- Phadtare S. (2011). Unwinding activity of cold shock proteins and RNA metabolism. RNA Biol. 8 394–397. 10.4161/rna.8.3.14823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadtare S. (2012). Escherichia coli cold-shock gene profiles in response to over-expression/deletion of CsdA, RNase R and PNPase and relevance to low-temperature RNA metabolism. Genes Cells 17 850–874. 10.1111/gtc.12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadtare S., Alsina J., Inouye M. (1999). Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 2 175–180. 10.1016/S1369-5274(99)80031-9 [DOI] [PubMed] [Google Scholar]
- Phadtare S., Inouye M. (2004). Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J. Bacteriol. 186 7007–7014. 10.1128/JB.186.20.7007-7014.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper R., Huang S. T., Clark D. J., Robinson J. M., Parmar P. P., Alami H., et al. (2008). Characterizing the dynamic nature of the Yersinia pestis periplasmic proteome in response to nutrient exhaustion and temperature change. Proteomics 8 1442–1458. 10.1002/pmic.200700923 [DOI] [PubMed] [Google Scholar]
- Polissi A., De Laurentis W., Zangrossi S., Briani F., Longhi V., Pesole G., et al. (2003). Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 154 573–580. 10.1016/S0923-2508(03)00167-0 [DOI] [PubMed] [Google Scholar]
- Ricke S. C., Dawoud T. M., Kim S. A., Park S. H., Kwon Y. M. (2018). Salmonella cold stress response: mechanisms and occurrence in foods. Adv. Appl. Microbiol. 104 1–38. 10.1016/bs.aambs.2018.03.001 [DOI] [PubMed] [Google Scholar]
- Robins-Browne R. M. (2013). “Chapter 14: Yersinia enterocolitica,” in Food Microbiology: Fundamentals and Frontiers, 4th Edn, ed. Doyle L. B. M. (Washington, DC: Amercian Society for Microbiology; ). [Google Scholar]
- Robinson J. A. (2019). Folded synthetic peptides and other molecules targeting outer membrane protein complexes in gram-negative bacteria. Front. Chem. 7:45. 10.3389/fchem.2019.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollauer S. E., Sooreshjani M. A., Noinaj N., Buchanan S. K. (2015). Outer membrane protein biogenesis in Gram-negative bacteria. Philos. Trans. R. Soc. B Biol. Sci. 370:20150023. 10.1098/Rstb.2015.0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J. D., Cox J. (2019). A network module for the perseus software for computational proteomics facilitates proteome interaction graph analysis. J. Proteome Res. 18 2052–2064. 10.1021/acs.jproteome.8b00927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar J. G., Wu T., Gronenberg L. S., Malinverni J. C., Kahne D., Silhavy T. J. (2007). Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 104 6400–6405. 10.1073/pnas.0701579104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaniol V., Wyder S., Aebi C. (2013). RNA-Seq-based analysis of the physiologic cold shock-induced changes in Moraxella catarrhalis gene expression. PLoS One 8:e68298. 10.1371/journal.pone.0068298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starosta A. L., Lassak J., Jung K., Wilson D. N. (2014). The bacterial translation stress response. FEMS Microbiol. Rev. 38 1172–1201. 10.1111/1574-6976.12083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanopoulou M., Kokoschka M., Sheldrick W. S., Wolters D. A. (2011). Cell response of Escherichia coli to cisplatin-induced stress. Proteomics 11 4174–4188. 10.1002/pmic.201100203 [DOI] [PubMed] [Google Scholar]
- Storz G., Hengge R. (2010). Bacterial Stress Responses. Washington, DC: American Society for Microbiology Press. [Google Scholar]
- Tashiro K., Kubokura Y., Kato Y., Kaneko K.-I., Ogawa M. (1991). Survival of Yersinia enterocolitica in soil and water. J. Vet. Med. Sci. 53 23–27. 10.1292/jvms.53.23 [DOI] [PubMed] [Google Scholar]
- Tatusov R. L., Natale D. A., Garkavtsev I. V., Tatusova T. A., Shankavaram U. T., Rao B. S., et al. (2001). The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29 22–28. 10.1093/nar/29.1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend M. K., Carr N. J., Iyer J. G., Horne S. M., Gibbs P. S., Prüß B. M. (2008). Pleiotropic phenotypes of a Yersinia enterocolitica flhD mutant include reduced lethality in a chicken embryo model. BMC Microbiol. 8:12. 10.1186/1471-2180-8-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor L., Togoe I., Pop A., Mitranescu E. (2008). The Yersinia enterocolitica species tolerance to temperature. Rom. Biotechnol. Lett. 13 17–22. 10.1089/fpd.2009.0526 [DOI] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Cox J. (2016). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11:2301. 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- Vorachek-Warren M. K., Carty S. M., Lin S., Cotter R. J., Raetz C. R. (2002). An Escherichia coli mutant lacking the cold shock-induced palmitoleoyltransferase of lipid A biosynthesis: absence of unsaturated acyl chains and antibiotic hypersensitivity at 12 degrees C. J. Biol. Chem. 277 14186–14193. 10.1074/jbc.M200408200 [DOI] [PubMed] [Google Scholar]
- Wang X., Cui Z., Jin D., Tang L., Xia S., Wang H., et al. (2009). Distribution of pathogenic Yersinia enterocolitica in China. Eur. J. Clin. Microbiol. Infect. Dis. 28 1237–1244. 10.1007/s10096-009-0773-x [DOI] [PubMed] [Google Scholar]
- Wareth G., Eravci M., Weise C., Roesler U., Melzer F., Sprague L. D., et al. (2016). Comprehensive identification of immunodominant proteins of Brucella abortus and Brucella melitensis using antibodies in the sera from naturally infected hosts. Int. J. Mol. Sci. 17:E659. 10.3390/ijms17050659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M. H., Marahiel M. A. (2003). Bacterial cold shock responses. Sci. Prog. 86(Pt 1–2), 9–75. 10.3184/003685003783238707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Ziegler C. A., Um S., Perez N. M., Berns A. L., Malhowski A. J., Young S. (2008). Low temperature (23 degrees C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology 154(Pt 1), 148–166. 10.1099/mic.0.2007/012021-0 [DOI] [PubMed] [Google Scholar]
- Wollenweber H. W., Schlecht S., Lüderitz O., Rietschel E. T. (1983). Fatty acid in lipopolysaccharides of Salmonella species grown at low temperature: identification and position. Eur. J. Biochem. 130 167–171. 10.1111/j.1432-1033.1983.tb07132.x [DOI] [PubMed] [Google Scholar]
- Wouters J. A., Rombouts F. M., Kuipers O. P., De Vos W. M., Abee T. (2000). The role of cold-shock proteins in low-temperature adaptation of food-related bacteria. Syst. Appl. Microbiol. 23 165–173. 10.1016/s0723-2020(00)80001-6 [DOI] [PubMed] [Google Scholar]
- Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T. J., Kahne D. (2005). Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121 235–245. 10.1016/j.cell.2005.02.015 [DOI] [PubMed] [Google Scholar]
- Xu S., Peng Z., Cui B., Wang T., Song Y., Zhang L., et al. (2014). FliS modulates FlgM activity by acting as a non-canonical chaperone to control late flagellar gene expression, motility and biofilm formation in Yersinia pseudotuberculosis. Environ. Microbiol. 16 1090–1104. 10.1111/1462-2920.12222 [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Inouye M. (2001). Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J. Bacteriol. 183 2808–2816. 10.1128/Jb.183.9.2808-2816.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y., Lee H., Lee S., Kim S., Choi K.-H. (2015). Membrane fluidity-related adaptive response mechanisms of foodborne bacterial pathogens under environmental stresses. Food Res. Int. 72 25–36. 10.1016/j.foodres.2015.03.016 [DOI] [Google Scholar]
- Young G. M., Smith M. J., Minnich S. A., Miller V. L. (1999). The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 181 2823–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky A., Parola A. H., Abdah M., Masalha H. (1985). Homeoviscous adaptation, growth rate, and morphogenesis in bacteria. Biophys. J. 48 337–339. 10.1016/S0006-3495(85)83788-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Liu Q., Xiao K., Hu Y., Liu X., Li Y., et al. (2016). Identification of the crp gene in avian Pasteurella multocida and evaluation of the effects of crp deletion on its phenotype, virulence and immunogenicity. BMC Microbiol. 16:125. 10.1186/s12866-016-0739-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article and Supplementary Material.