Abstract
Influenza A virus in swine (IAV-S) is an important pathogen in pigs in the United States, in addition to posing a potential risk to humans through zoonotic events. Intervention strategies continue to be explored to better control virus circulation. Improved surveillance efforts has led to significantly increased sequence data available on circulating strains, vastly improving our understanding of the genetic and antigenic diversity of IAV-S. IAV-S in North America is characterized by repeated spillover events of human viruses into pigs followed by genetic and antigenic diversification. An important gap that needs to be addressed is our understanding of the role that various vaccine platforms have on efficacy against antigenically heterologous challenge. Currently licensed vaccines often update their components to adapt to a dynamic antigenic landscape and newly developed technologies are continue to be approved. Hence, it remains critical to test the performance of vaccines against challenge with antigenically distinct viruses. We tested the level of protection conferred by an alphavirus-vectored hemagglutinin (HA) subunit vaccine, delivered as a monovalent or bivalent formulation, against challenge with IAV-S. Monovalent alphavirus-vectored HA vaccines provided efficient protection against challenge with viruses with matched and mismatched HA, although in one mismatched HA challenge group there was a trend for reduced protection. A bivalent vaccine, in which two HA’s were simultaneously delivered, was effective in producing antibody response against both antigens and provided protection against challenge. The alphavirus platform is a promising new technology available to swine producers to help reduce the burden of disease caused by IAV-S.
Keywords: influenza, replicon particle, alphavirus, subunit vaccine
1. Introduction
Influenza A virus (IAV) is an important pathogen that causes disease in humans, livestock and poultry, posing a risk to public health as well as being a burden to the agricultural economy. There are two biological features of IAV that make it challenging to develop efficacious vaccines: the first, a polymerase complex with a high rate of mutation facilitates the emergence of antigenically distinct hemagglutinin (HA) that can evade antibodies in a process called antigenic drift [1]. The second, co-infection of a single cell by two distinct viruses can result in progeny virions that carry novel gene constellations through gene reassortment in a process called antigenic shift [2]. The HA is a glycoprotein on the surface of the virion that mediates virus entry into susceptible cells [3]. HA is a main target for antibody responses in vaccine efforts to induce antibodies that can block receptor-binding interactions and therefore neutralize virus infection. Amino acid positions in the head of the HA (HA1) have been associated with monoclonal antibody escape [4, 5], and some positions have been associated with antigenic evolution of human, avian and swine IAV [6-10]. Despite these advances in understanding the molecular determinants of the antigenicity of IAV, viruses continue to persist and efforts to prevent sustained circulation are performed in hindsight of the emergence of antigenically distinct viruses. In the biological arms race to better control the burden of disease caused by IAV, it has become evident that improved intervention strategies are required [11].
Several strategies have been proposed to improve vaccines including but not limited to: targeting conserved epitopes in the HA stalk region, targeting non-HA proteins that are more conserved (matrix protein, nucleoprotein), sequential prime-boost regimens altering vaccine components or platform, and use of vaccine platforms that elicit an innate immune response in addition to an adaptive response [12, 13]. Among the vaccine platforms that elicit an innate immune response are subunit vaccines that are delivered with an alphavirus vector [14]. One alphavirus-vectored vaccine uses Venezuelan equine encephalitis (VEE) virus where the genome is genetically modified so that viral structural genes are replaced by the transgene of choice, rendering them defective in producing infectious virus unless grown in a cell line that provides the structural genes in trans [15, 16]. An important biological feature of the system is that the RNA is self-replicating and therefore transgenes are expressed to high levels [16, 17]. The vaccine platform, when generated in a helper cell line that provides the structural proteins, consists of a propagation-defective replicon particle (RP) that can undergo a single-cycle of replication that does not require an adjuvant to induce a robust antibody response.
The alphavirus-vectored platform has been explored for use in humans as a vaccine to prevent influenza infection [18, 19], but there is yet to be a commercially available alphavirus-based vaccine for IAV in humans. In swine, an alphavirus-vectored RP influenza vaccine has been developed [20-23] and is licensed for use in the United States [24]. Use of the RP vaccine in swine has been shown to reduce viral replication and lung lesions against challenge with a virus encoding a homologous HA for both H1 and H3 subtype viruses, although the platform was not able to overcome maternally derived antibodies [20-22]. The RP platform is flexible and allows for introduction of any of the viral genes, and an RP expressing the nucleoprotein (NP) as a vaccine has been previously tested in pigs and significantly reduced levels of viral titers in the upper and lower respiratory tract when challenged with a virus encoding a genetically distinct NP [22]. While both the HA and NP have been used as antigens in the RP platform, a bivalent vaccine that employs co-administration of two RP vaccines expressing distinct viral proteins has not been reported. In this study, we tested vaccine efficacy of monovalent and bivalent RP vaccines against matched and mismatched HA challenge strains.
2. Materials and methods
2.1. Viruses and vaccines
The two challenge viruses used in the study were wild-type A/swine/Iowa/A01480656/2014 H3N2 (IA/14; GenBank accession KJ635928 for HA gene) and wild-type A/swine/New York/A01104005/2011 H3N2 (NY/11; GenBank accession JN940422 for HA gene). Challenge viruses were chosen from the previously described phylogenetic clade IV-A to represent distinct antigenic clusters termed red and green [6, 8]. All viruses were propagated in Madin-Darby Canine Kidney (MDCK) cells. Commercial RP vaccines (Harrisvaccines, IA, USA) [20-22, 25] were obtained based on the IA/14 and NY/11 sequences requested. RP-GFP expressed green fluorescent protein, and was used as a control; RP-red expressed an HA protein with high homology to the NY/11 (differed in the HA1 at amino acid position 273; H3 numbering); and RP-green expressed the IA/14 HA protein.
2.2. Virus antigenic characterization
Hemagglutination inhibition (HI) assays were performed with a subset of a previously described reference antisera panel [8]. Prior to HI testing, sera were treated with receptor-destroying enzyme (Sigma-Aldrich, MO, USA), heat inactivated at 56°C for 30 min, and adsorbed with 50% turkey red blood cells (RBC) to remove nonspecific inhibitors of hemagglutination. Serial 2-fold dilutions starting at 1:10 were tested for the ability to inhibit the agglutination of 0.5% turkey RBC with 8 HAU of IA/14 and NY/11.
2.3. Animal study design
All pigs were cared for in compliance with the Institutional Animal Care and Use Committee of the National Animal Disease Center. Pigs were treated with ceftiofur crystalline free acid and tulathromycin (Zoetis Animal Health, Florham Park, NJ) and were shown to be seronegative to IAV antibodies by a commercial ELISA kit (Swine Influenza Virus Ab Test, IDEXX, Westbrook, ME) prior to the start of the study. The various vaccine challenge groups are outlined in Table 1.
Table 1.
Animal study design.
| Groups | Vaccine | Challenge virus (antigenic cluster) |
Number of pigs per group |
|---|---|---|---|
| RP-GFP->NC | RP-GFP | No challenge | 5 |
| RP-GFP->NY/11 | RP-GFP | NY/11 (red) | 10 |
| RP-red->NY/11 | RP-red | NY/11 (red) | 10 |
| RP-green->NY/11 | RP-green | NY/11 (red) | 9 |
| RP-red+RP-green->NY/11 | RP-red + RP-green | NY/11 (red) | 10 |
| RP-GFP->IA/14 | RP-GFP | IA/14 (green) | 10 |
| RP-red->IA/14 | RP-red | IA/14 (green) | 10 |
| RP-green->IA/14 | RP-green | IA/14 (green) | 8 |
| RP-red+RP-green->IA/14 | RP-red + RP-green | IA/14 (green) | 9 |
NC, non-challenged.
Eighty-one 5 week-old pigs were vaccinated with 2 ml of RP delivered intramuscularly on the right side of the neck (1 × 108 alphavirus replicon particles). Three weeks post vaccination a booster dose was administered (2 ml intramuscularly, 1 × 108 alphavirus particles). Pigs were challenged three weeks post booster dose at 11 weeks of age (6 weeks after the first dose of vaccine was delivered).
The challenge virus was delivered to pigs intratracheally (2 ml) and intranasally (1 ml) with 1 × 105 TCID50/ml under anesthesia, using an intramuscular injection of a cocktail of ketamine (8 mg/kg of body weight; Phoenix, St. Joseph, MO), xylazine (4 mg/kg; Lloyd Inc., Shenandoah, IA), and Telazol (6 mg/kg; Zoetis Animal Health, Florham Park, NJ). Pigs were humanely euthanized with a lethal dose of pentobarbital (Fatal Plus, Vortech Pharmaceuticals, Dearborn, MI) at 5 dpi, when lungs were evaluated and bronchoalveolar lavage fluid (BALF), trachea and right cardiac or affected lung lobe were collected.
2.4. Serology
HI assays were performed with 0.5% turkey RBCs according to standard techniques as described above in section 2.2. [26]. Results were reported as geometric mean antibody titers.
Isotype specific (IgA and IgG) enzyme-linked immunosorbent assays (ELISA) were performed with using IA/14 and NY/11 as antigens after BALF samples were treated with 10mM Dithiothreitol (DTT; Sigma-Aldrich, St. Louis, MO) and diluted to 1:4, as previously described [27]. Results were reported as average optical density (O.D.) levels of duplicate wells for each sample.
2.5. Pathological examination and virus detection
At necropsy, the percent of lung surface affected with pneumonia was calculated as previously described [28, 29]. Formalin-fixed trachea and lung tissue samples were routinely processed and stained with hematoxylin and eosin. Lung and trachea microscopic lesions were scored according to previously described parameters [30] and individual composite scores for each pig were computed.
Nasal swabs (NS; Fisherbrand Dacron swabs, Fisher Scientific, Pittsburg, PA) were collected on 0, 3, and 5 dpi to be used for virus isolation as previously described [27]. Virus isolation-positive NS and BALF were titrated in MDCK cells as previously described [27, 31] and TCID50/ml virus titers were calculated for each sample by the Reed and Muench method [32].
2.6. Statistical analysis
Results were analyzed by analysis of variance (ANOVA), with P≤0.05 considered significant (Prism software; GraphPad, La Jolla, CA) and variables with significant effects by treatment group were subjected to pairwise mean comparisons using the Tukey-Kramer test.
3. Results
3.1. Antibody response to alphavirus-vectored HA subunit vaccine and mucosal response after infection
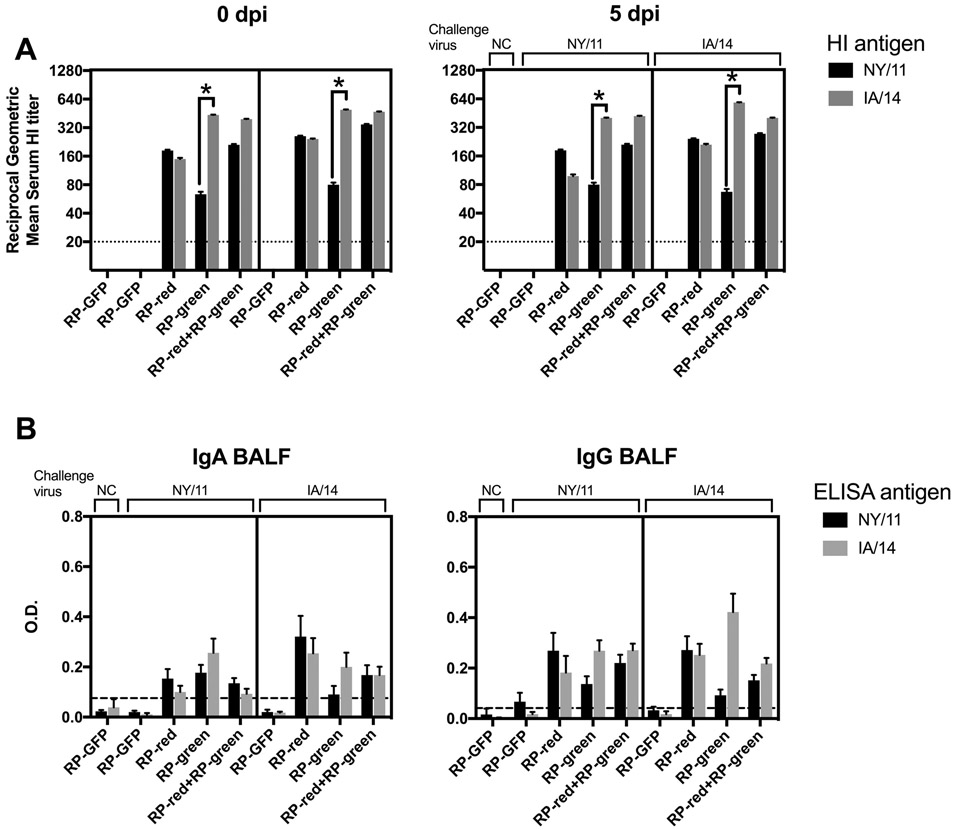
In the HA1, NY/11 (red) and IA/14 (green) only differed at two amino acid positions (145 and 289), yet exhibit significantly reduced cross-HI activity when using monovalent whole-inactivated virus antisera (Table 2). Pigs vaccinated with RP-red HA demonstrated an HI-specific antibody response against NY/11 and a comparable response against IA/14 (Fig. 1A). In contrast, while pigs vaccinated with RP-green HA generated an HI-antibody response against IA/14 there was a titer reduction greater than 2-fold in cross-HI antibodies against NY/11. Pigs that were administered two doses of the bivalent RP-red and the RP-green HA subunit vaccine had a comparable HI response against NY/11 and IA/14 when compared to the monovalent vaccine, suggesting that there was no antigen competition between the two HA antigens. No significant changes in HI titer were observed after challenge at 5 dpi, regardless of whether the challenge virus encoded a matched or mismatched HA.
Table 2.
Cross-HI titers reported as geometrical mean reciprocal titers.
Paired anti-sera generated in swine by delivering two doses of whole-inactivated virus.
Fig. 1. Antibody responses in pigs against NY/11 and IA/14 induced by monovalent and bivalent RP vaccines pre- and post-infection.
(A) Serum hemagglutination inhibition (HI) geometric mean titers against IA/14 and NY/11 at 0 and 5 dpi from all vaccinated groups and the sham vaccinated pigs (RP-GFP). (B) Whole virus IgA and IgG ELISA’s were performed using BALF from 5 dpi with IA/14 and NY/11 as antigens. Groups of pigs are labeled according to vaccine use. HI titers that differed by more than two serial dilutions within 0 or 5 dpi are marked with an asterisk (A). Data presented as mean optical density (O.D.) or geometric mean titers ± standard error of the mean. Cut-off values (dotted line) for ELISA data was calculated as the mean of blank wells plus three times the standard deviation. A black line within each graph separates groups according to challenge strain (NY/11 on the left, IA/14 on the right).
A potential advantage of using the alphavirus-vectored platform is that despite delivery by the parenteral route, there is an induction of a mucosal response to produce IgA antibodies [33, 34]. To assess this, we analyzed the presence of IgA and IgG-specific antibodies in BALF at 5 dpi that reacted with whole-virus antigen (Fig. 1B). Despite low levels, there were detectable IgA and IgG antibodies in the lungs of all vaccinated groups.
3.2. RP HA vaccine provides protection against antigenically HA-matched virus
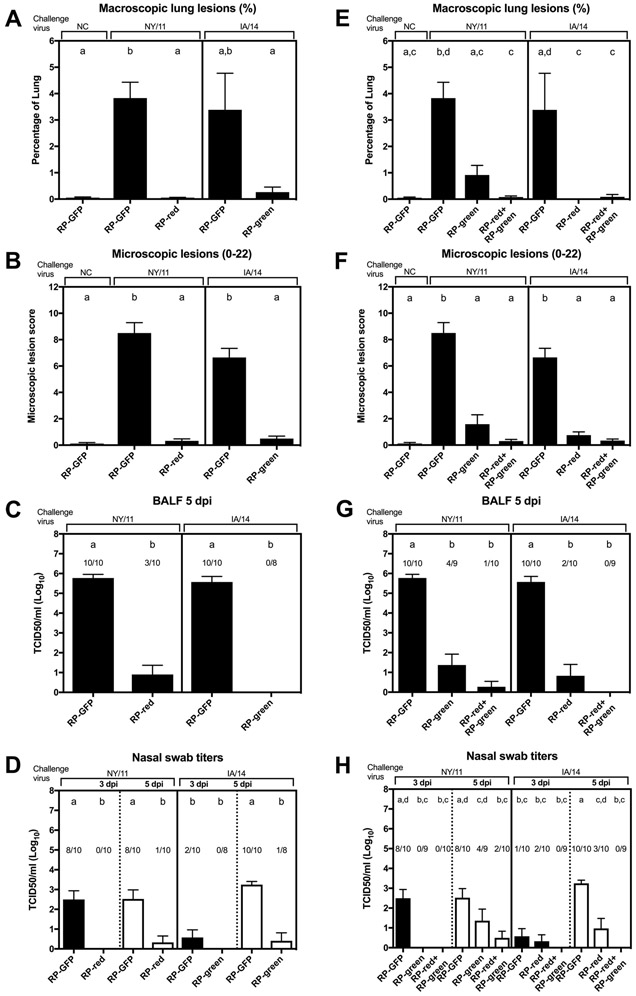
Mock-vaccinated pigs (RP-GFP) challenged with NY/11 and IA/14 exhibited mild macroscopic and microscopic lung lesions, high viral titers in BALF and infectious virus was isolated from nasal swab samples at 3 and 5 dpi (Figure 2A-D). The kinetics of virus shedding in the upper respiratory tract in pigs challenged with IA/14 was delayed, in which minimal virus shedding was observed at 3 dpi although robust virus titers were detected at 5 dpi (Fig. 2D). RP-red HA vaccinated pigs challenged with NY/11 demonstrated negligible macroscopic lung lesions, significantly reduced viral titers in BALF and in nasal swabs at 5 dpi. Similarly, RP-green HA vaccinated pigs challenged with IA/14 had reduced macroscopic lung lesions, no detectable virus in BALF and significantly reduced titers in nasal swab samples.
Fig. 2. Protection in pigs vaccinated with monovalent or bivalent RP vaccine.
Results from monovalent HA-matched (A-D) and HA-mismatched or bivalent groups (E-H). The percentage of macroscopic (A/E) and microscopic (B/F) lung lesions was evaluated at 5 dpi, and viral titers were measured in bronchoalveolar lavage fluid (BALF) at 5 dpi (C/G) and in nasal swabs at 3 and 5 dpi (D/H). Groups of pigs are labeled with the vaccine used followed by the challenge strain. NC, not challenged. Data presented as mean ± standard error of the mean. The number of pigs with a positive virus titer/total number of pigs are indicated above the bars (C-D, G-H). Treatment group means with statistically significant differences (P ≤ 0.05) are identified by different lowercase letters. A black line within each graph separates groups according to challenge strain (NY/11 on the left, IA/14 on the right). A dotted line separates nasal swab titers at 3 (black bars) and 5 dpi (white bars) within challenged groups.
3.3. Monovalent and bivalent RP HA vaccine provides broad protection against antigenically HA-mismatched viruses
In the HA-mismatched groups there was evidence of protection although there was a trend for greater protection in the RP-red HA vaccinated pigs challenged with IA/14 (Fig. 2E-H). In the RP-green HA vaccinated pigs challenged with NY/11 there were significantly reduced lung lesions, viral titers in the lung and viral titers in nasal swabs at 3 dpi, but no statistical difference in viral titers in nasal swabs at 5 dpi in comparison to GFP-vaccinated pigs (Fig. 2H). In the RP-red HA vaccinated pigs challenged with IA/14 there was minimal lung lesions detected and significantly reduced viral titers in the lung and in nasal swab samples on both days. Pigs that received the bivalent vaccine, a simultaneous dose of the RP-green and a dose of the RP-red, demonstrated minimal lung lesions, low to no detectable infectious virus in the lungs and nasal swab samples in both challenge groups.
4. Discussion
The level of antigenic diversity of IAV-S co-circulating in the United States continues to increase and this poses problems to effectively reduce the burden of disease through vaccines. Antigenic studies of contemporary H3 IAV-S revealed at least three major antigenic clusters co-circulate in the United States [8, 35]. In the face of such large antigenic diversity, there is a need for more broadly protective vaccines that can be flexible to the dynamic antigenic evolution of IAV-S.
In the present study we examined the efficacy of a recently licensed vaccine platform, alphavirus-vectored HA subunit vaccine. While products based on traditional vaccine platforms such as whole-inactivated virus and live-attenuated influenza virus are still produced and used, several virus vectors have been explored as delivery agents for vaccine purposes to protect against influenza infection [14]. Alphavirus vectors have been designed for potential use in humans with Semliki Forest virus, Sindbis virus and Venezuelan equine encephalitis virus as vectors [16, 36, 37]. Although, there is no commercially available alphavirus-based influenza vaccine for use in humans despite several existing candidates [38]. The same alphavirus-based influenza vaccine tested here has also been approved for use in avian species and an H5 HA vaccine has been stockpiled as a preventative measure for future highly-pathogenic avian influenza virus outbreaks [39].
We found that the RP vaccine produced robust HI titers in pigs, when delivered both as a monovalent and bivalent vaccine. In pigs challenged with an HA-matched virus, effective protection was observed with minimal lung lesions and no or minimal infectious virus detected in upper and lower respiratory tract. In pigs vaccinated with a monovalent vaccine and challenged with an HA-mismatched virus, there was significant protection for RP-red vaccinated pigs challenged with IA/14, but less so with the RP-green. In particular there was no significant difference between mean viral titers in the nasal swab samples at 5 dpi between the negative control pigs (RP-GFP) and the RP-green vaccinated pigs challenged with NY/11. Importantly, neither of the mismatched RP monovalent vaccines was associated with vaccine-associated enhanced respiratory disease (VAERD) that has been reported with adjuvanted whole inactivated influenza vaccines in similar experimental models [27, 30, 40, 41]. Additional studies with different permutations of vaccination timing and IAV strains are needed to confirm the absence of VAERD. The best level of protection, regardless of the challenge strain, was observed when the RP-green and RP-red were delivered together as a bivalent vaccine. Importantly, NY/11 and IA/14 only differed at two amino acid positions in the HA1 sequence, and given the observed efficacy of the bivalent vaccine, multivalent vaccines that include HA’s representative of the endemic antigenic diversity may be a more effective strategy.
One of the advantages of the alphavirus-based technology licensed for use in swine is that HA sequences can be readily replaced. This allows customers to either obtain an off the shelf, previously made RP vaccine or request a new RP vaccine to better match the HA of interest. Furthermore, as presented here, there is the potential to vaccinate with more than one RP particle and therefor deliver a multivalent vaccine that could be comprised of not only multiple HA’s but also a combination of HA with other viral proteins such as the NA or NP. There has been an increased interest and evidence to support the importance of anti-NA antibodies to reduce morbidity [42, 43], which makes it a strong candidate to provide NA in addition to the HA antigen to broaden efficacy.
In addition to implementing a multivalent vaccine, various prime-boost strategies can be explored to enhance duration and increase breadth of protection. In both cases, exploring distinct prime-boost strategies may improve vaccine protection. A recent paper has reported that priming with an RP vaccine followed by a whole-inactivated virus vaccine in turkeys conferred the best long-term protection [44]. Due to the genetic diversity of avian species, responses to vaccines can vary by host and caution should be used when extrapolating findings [45, 46]. Nevertheless, further experiments are warranted to examine the duration of protection in pigs vaccinated with RP’s. Furthermore, implementing a heterologous prime-boost could be explored as well to broaden protection as there is evidence that this strategy is effective at broadening protective antibodies when using whole-inactivated virus [47].
Swine producers now have various vaccine platforms at their disposal to better combat the burden of disease caused by IAV-S. One area where our understanding is lacking is how to best implement vaccine use to protect against a virus landscape that we know continues to diversify genetically and antigenically. Better vaccines, or improved prime-boost strategies, that broaden protection can help reduce the burden of disease caused by IAV-S on swine production systems. Further, improved vaccines may reduce the chances of inter-species transmission from swine to humans. Our findings indicate that multivalent RP’s may be one approach that can provide broader protection, although factors such as duration of protection and different prime-boost regimens should be explored to identify additional effective approaches.
Highlights.
Monovalent alphavirus-vectored HA vaccines provided efficient protection against challenge with viruses with matched HA
Monovalent alphavirus-vectored HA vaccines provided partial protection against challenge with viruses with mismatched HA.
A bivalent alphavirus-vectored HA vaccine was effective in producing antibody response against both antigens and provided protection against challenge.
Acknowledgements
The authors thank Michelle Harland and Gwen Nordholm for technical assistance and Jason Huegel, Justin Miller, Aaron Hebeisen and Keiko Sampson for assistance with animal studies. We thank Dr. Mark Mogler of Harrisvaccines (now Merck) for supplying the alphavirus vaccine used in this study.
Funding
This study was supported by USDA-ARS and by an NIH-National Institute of Allergy and Infectious Diseases (NIAID) interagency agreement (R21AI098079) associated with Center of Research in Influenza Pathogenesis, an NIAID funded Center of Excellence in Influenza Research and Surveillance (HHSN272201400008C). EJA was supported in part by an appointment to the ARS-USDA Research Participation Program administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and USDA under contract number DE-AC05-06OR23100. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Government. USDA is an equal opportunity provider and employer.
Footnotes
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Drake JW. Rates of spontaneous mutation among RNA viruses. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:4171–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Desselberger U, Nakajima K, Alfino P, Pedersen FS, Haseltine WA, Hannoun C, et al. Biochemical evidence that “new” influenza virus strains in nature may arise by recombination (reassortment). Proceedings of the National Academy of Sciences of the United States of America. 1978;75:3341–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–73. [DOI] [PubMed] [Google Scholar]
- [4].Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–8. [DOI] [PubMed] [Google Scholar]
- [5].Gerhard W, Yewdell J, Frankel ME, Webster R. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature. 1981;290:713–7. [DOI] [PubMed] [Google Scholar]
- [6].Abente EJ, Santos J, Lewis NS, Gauger PC, Stratton J, Skepner E, et al. The Molecular Determinants of Antibody Recognition and Antigenic Drift in the H3 Hemagglutinin of Swine Influenza A Virus. Journal of virology. 2016;90:8266–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342:976–9. [DOI] [PubMed] [Google Scholar]
- [8].Lewis NS, Anderson TK, Kitikoon P, Skepner E, Burke DF, Vincent AL. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine. Journal of virology. 2014;88:4752–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lewis NS, Daly JM, Russell CA, Horton DL, Skepner E, Bryant NA, et al. Antigenic and genetic evolution of equine influenza A (H3N8) virus from 1968 to 2007. Journal of virology. 2011;85:12742–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–6. [DOI] [PubMed] [Google Scholar]
- [11].Paules CI, Sullivan SG, Subbarao K, Fauci AS. Chasing Seasonal Influenza - The Need for a Universal Influenza Vaccine. The New England journal of medicine. 2018;378:7–9. [DOI] [PubMed] [Google Scholar]
- [12].Nachbagauer R, Krammer F. Universal influenza virus vaccines and therapeutic antibodies. Clin Microbiol Infect. 2017;23:222–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Houser K, Subbarao K. Influenza vaccines: challenges and solutions. Cell host & microbe. 2015;17:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tripp RA, Tompkins SM. Virus-vectored influenza virus vaccines. Viruses. 2014;6:3055–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kamrud KI, Coffield VM, Owens G, Goodman C, Alterson K, Custer M, et al. In vitro and in vivo characterization of microRNA-targeted alphavirus replicon and helper RNAs. Journal of virology. 2010;84:7713–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pushko P, Parker M, Ludwig GV, Davis NL, Johnston RE, Smith JF. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology. 1997;239:389–401. [DOI] [PubMed] [Google Scholar]
- [17].Rayner JO, Dryga SA, Kamrud KI. Alphavirus vectors and vaccination. Rev Med Virol. 2002;12:279–96. [DOI] [PubMed] [Google Scholar]
- [18].Bernstein DI, Reap EA, Katen K, Watson A, Smith K, Norberg P, et al. Randomized, double-blind, Phase 1 trial of an alphavirus replicon vaccine for cytomegalovirus in CMV seronegative adult volunteers. Vaccine. 2009;28:484–93. [DOI] [PubMed] [Google Scholar]
- [19].Hubby B, Talarico T, Maughan M, Reap EA, Berglund P, Kamrud KI, et al. Development and preclinical evaluation of an alphavirus replicon vaccine for influenza. Vaccine. 2007;25:8180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bosworth B, Erdman MM, Stine DL, Harris I, Irwin C, Jens M, et al. Replicon particle vaccine protects swine against influenza. Comp Immunol Microbiol Infect Dis. 2010;33:e99–e103. [DOI] [PubMed] [Google Scholar]
- [21].Vander Veen RL, Loynachan AT, Mogler MA, Russell BJ, Harris DL, Kamrud KI. Safety, immunogenicity, and efficacy of an alphavirus replicon-based swine influenza virus hemagglutinin vaccine. Vaccine. 2012;30:1944–50. [DOI] [PubMed] [Google Scholar]
- [22].Vander Veen RL, Mogler MA, Russell BJ, Loynachan AT, Harris DL, Kamrud KI. Haemagglutinin and nucleoprotein replicon particle vaccination of swine protects against the pandemic H1N1 2009 virus. The Veterinary record. 2013;173:344. [DOI] [PubMed] [Google Scholar]
- [23].Vander Veen R, Kamrud K, Mogler M, Loynachan AT, McVicker J, Berglund P, et al. Rapid development of an efficacious swine vaccine for novel H1N1. PLoS Curr. 2009;1:RRN1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Center for Veterinary Biologics. Notice N. 17–01. 2017.
- [25].Kamrud KI, Custer M, Dudek JM, Owens G, Alterson KD, Lee JS, et al. Alphavirus replicon approach to promoterless analysis of IRES elements. Virology. 2007;360:376–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].World Health Organization. WHO Manual on Animal Influenza Diagnosis and Surveillance. 2nd ed. Geneva: 2002. [Google Scholar]
- [27].Vincent AL, Ma W, Lager KM, Richt JA, Janke BH, Sandbulte MR, et al. Live attenuated influenza vaccine provides superior protection from heterologous infection in pigs with maternal antibodies without inducing vaccine-associated enhanced respiratory disease. Journal of virology. 2012;86:10597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gauger PC, Vincent AL, Loving CL, Lager KM, Janke BH, Kehrli ME Jr., et al. Enhanced pneumonia and disease in pigs vaccinated with an inactivated human-like (delta-cluster) H1N2 vaccine and challenged with pandemic 2009 H1N1 influenza virus. Vaccine. 2011;29:2712–9. [DOI] [PubMed] [Google Scholar]
- [29].Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, et al. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Veterinary pathology. 1995;32:648–60. [DOI] [PubMed] [Google Scholar]
- [30].Gauger PC, Vincent AL, Loving CL, Henningson JN, Lager KM, Janke BH, et al. Kinetics of lung lesion development and pro-inflammatory cytokine response in pigs with vaccine-associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Veterinary pathology. 2012;49:900–12. [DOI] [PubMed] [Google Scholar]
- [31].Gauger PC, Vincent AL. Serum virus neutralization assay for detection and quantitation of serum-neutralizing antibodies to influenza A virus in swine In: Spackman E, editor. Animal Influenza Virus. New York, NY: Springer; 2014. p. 313–24. [DOI] [PubMed] [Google Scholar]
- [32].Reed lJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. 1938;27:493–7. [Google Scholar]
- [33].Thompson JM, Whitmore AC, Konopka JL, Collier ML, Richmond EM, Davis NL, et al. Mucosal and systemic adjuvant activity of alphavirus replicon particles. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3722–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Khalil SM, Tonkin DR, Snead AT, Parks GD, Johnston RE, White LJ. An alphavirus-based adjuvant enhances serum and mucosal antibodies, T cells, and protective immunity to influenza virus in neonatal mice. Journal of virology. 2014;88:9182–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Feng Z, Gomez J, Bowman AS, Ye J, Long LP, Nelson SW, et al. Antigenic characterization of H3N2 influenza A viruses from Ohio agricultural fairs. Journal of virology. 2013;87:7655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Schultz-Cherry S, Dybing JK, Davis NL, Williamson C, Suarez DL, Johnston R, et al. Influenza virus (A/HK/156/97) hemagglutinin expressed by an alphavirus replicon system protects chickens against lethal infection with Hong Kong-origin H5N1 viruses. Virology. 2000;278:55–9. [DOI] [PubMed] [Google Scholar]
- [37].Tsuji M, Bergmann CC, Takita-Sonoda Y, Murata K, Rodrigues EG, Nussenzweig RS, et al. Recombinant Sindbis viruses expressing a cytotoxic T-lymphocyte epitope of a malaria parasite or of influenza virus elicit protection against the corresponding pathogen in mice. Journal of virology. 1998;72:6907–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Scorza FB, Pardi N. New Kids on the Block: RNA-Based Influenza Virus Vaccines. Vaccines (Basel). 2018;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].USDA Adds Avian Influenza Vaccine Doses to the National Veterinary Stockpile. https://www.aphis.usda.gov/aphis/newsroom/stakeholder-info/sa_by_date/sa_2015/sa_10/ct_ai_vaccines.
- [40].Gauger PC, Loving CL, Khurana S, Lorusso A, Perez DR, Kehrli ME Jr., et al. Live attenuated influenza A virus vaccine protects against A(H1N1)pdm09 heterologous challenge without vaccine associated enhanced respiratory disease. Virology. 2014;471–473:93–104. [DOI] [PubMed] [Google Scholar]
- [41].Souza CK, Rajao DS, Loving CL, Gauger PC, Perez DR, Vincent AL. Age at Vaccination and Timing of Infection Do Not Alter Vaccine-Associated Enhanced Respiratory Disease in Influenza A Virus-Infected Pigs. Clin Vaccine Immunol. 2016;23:470–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, et al. Influenza Infection in Humans Induces Broadly Cross-Reactive and Protective Neuraminidase-Reactive Antibodies. Cell. 2018;173:417–29 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, et al. Evaluation of Antihemagglutinin and Antineuraminidase Antibodies as Correlates of Protection in an Influenza A/H1N1 Virus Healthy Human Challenge Model. mBio. 2016;7:e00417–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Santos JJS, Obadan AO, Garcia SC, Carnaccini S, Kapczynski DR, Pantin-Jackwood M, et al. Short- and long-term protective efficacy against clade 2.3.4.4 H5N2 highly pathogenic avian influenza virus following prime-boost vaccination in turkeys. Vaccine. 2017;35:5637–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bertran K, Balzli C, Lee DH, Suarez DL, Kapczynski DR, Swayne DE. Protection of White Leghorn chickens by U.S. emergency H5 vaccination against clade 2.3.4.4 H5N2 high pathogenicity avian influenza virus. Vaccine. 2017;35:6336–44. [DOI] [PubMed] [Google Scholar]
- [46].Bertran K, Sa ESM, Pantin-Jackwood MJ, Swayne DE. Protection against H7N3 high pathogenicity avian influenza in chickens immunized with a recombinant fowlpox and an inactivated avian influenza vaccines. Vaccine. 2013;31:3572–6. [DOI] [PubMed] [Google Scholar]
- [47].Van Reeth K, Gracia JCM, Trus I, Sys L, Claes G, Versnaeyen H, et al. Heterologous prime-boost vaccination with H3N2 influenza viruses of swine favors cross-clade antibody responses and protection. NPJ Vaccines. 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]