Abstract
Background
Investigation of the myocardial strain characteristics of the left ventricular non-compaction (LVNC) phenotype with cardiovascular magnetic resonance (CMR) feature tracking.
Methods
CMR cine balanced steady-state free precession data sets of 59 retrospectively identified LVNC phenotype patients (40 years, IQR: 28–50 years; 51% male) and 36 healthy subjects (39 years, IQR: 30–47 years; 44% male) were evaluated for LV volumes, systolic function and mass. Hypertrabeculation in patients and healthy subjects was evaluated against established CMR diagnostic criteria. Global circumferential strain (GCS), global radial strain (GRS) and global longitudinal strain (GLS) were evaluated with feature-tracking software. Subgroup analyses were performed in patients (n = 25) and healthy subjects (n = 34) with normal LV volumetrics, and with healthy subjects (n = 18) meeting at least one LVNC diagnostic criteria.
Results
All LVNC phenotype patients, as well as a significant proportion of healthy subjects, met morphology-based CMR diagnostic criteria: non-compacted (NC): compacted myocardial diameter ratio > 2.3 (100% vs. 19.4%), NC mass > 20% (100% vs. 44.4%) and > 25% (100% vs. 13.9%), and NC mass indexed to body surface area > 15 g/m2 (100% vs. 41.7%). LVNC phenotype patients demonstrated reduced GRS (26.4% vs. 37.1%; p < 0.001), GCS (− 16.5% vs. -20.5%; p < 0.001) and GLS (− 14.6% vs. -17.1%; p < 0.001) compared to healthy subjects, with statistically significant differences persisting on subgroup comparisons of LVNC phenotype patients with healthy subjects meeting diagnostic criteria. GCS also demonstrated independent and incremental diagnostic value beyond each of the morphology-based CMR diagnostic criteria.
Conclusions
LVNC phenotype patients demonstrate impaired strain by CMR feature tracking, also present on comparison of subjects with normal LV volumetrics meeting diagnostic criteria. The high proportion of healthy subjects meeting morphology-based CMR diagnostic criteria emphasizes the important potential complementary diagnostic value of strain in differentiating LVNC from physiologic hypertrabeculation.
Keywords: Isolated noncompaction of the ventricular myocardium, Cardiovascular magnetic resonance, Myocardial strain, Feature tracking, Ventricular dysfunction
Background
Left ventricular non-compaction cardiomyopathy (LVNC) is a rare cardiomyopathy that can occur in isolation or in association with other congenital and acquired cardiac pathologies [1]. It is characterised by two distinct layers of the left ventricular (LV) myocardium – an endocardial layer of heavily hypertrabeculated myocardium with deep inter-trabecular recesses, and an abnormally thin epicardial layer of compacted myocardium [2]. Despite controversies regarding the development of the myocardial phenotype of LVNC, the abnormal morphology characteristically involves the final segments to undergo compaction during embryogenesis – the apical inferior and lateral segments, with variable basal extension [3, 4].
Previously considered a rare anomaly, the widespread availability and advances in image quality of echocardiography and cardiovascular magnetic resonance (CMR) imaging and increased awareness have led to increasingly frequent recognition of the LVNC phenotype [1]. However, variable clinical manifestations and a wide spectrum of natural history have been observed, ranging from an asymptomatic imaging finding to life-threatening conditions including heart failure, arrhythmia and systemic thromboembolism [5].
Multiple validated morphology-based echocardiographic diagnostic criteria have evolved since the early descriptions of LVNC [2, 6–8] with a similar development of diagnostic criteria using CMR [9–11]. However, despite increasingly sophisticated methods of describing the myocardial phenotype and quantifying the degree of non-compaction, significant limitations in the clinical utility of morphology-based diagnostic criteria persist. Importantly, there is a tendency to over-diagnose LVNC among healthy subjects [12], without significant adverse outcomes on long-term follow-up [13]. Indeed, it is critical to distinguish patients with physiologic remodelling of the myocardium from those with pathologic remodelling or LVNC [14].
In order to expand beyond pure morphologic features and global volumetric parameters of cardiac function in the diagnosis and assessment of ischemic and non-ischemic cardiomyopathies, quantitative assessment of myocardial deformation by strain analysis has developed over recent years. Speckle-tracking echocardiography (STE) has demonstrated abnormal patterns of LV myocardial strain and torsion in LVNC including decreased radial, circumferential and longitudinal strain, decreased LV twist and abnormal patterns of ventricular rotation [15–18]. Strain analysis by CMR feature tracking (FT) using routine cine balanced steady-state free precession (bSSFP) data sets can reproducibly quantify myocardial deformation across multiple parameters [19]. However, there is limited data available on the characteristics of myocardial deformation in LVNC assessed by CMR FT. In light of the limitations of current morphology-based diagnostic criteria, this study investigated functional parameters including myocardial strain characteristics among LVNC phenotype patients and healthy subjects with CMR FT to explore potential complementary diagnostic utility. Ancillary aims included investigating whether potential differences in myocardial strain persist independent of LV volumetrics, parameters known to affect strain [20], between LVNC phenotype patients and healthy subjects, including subjects with and without features of ‘physiologic’ hypertrabeculation.
Methods
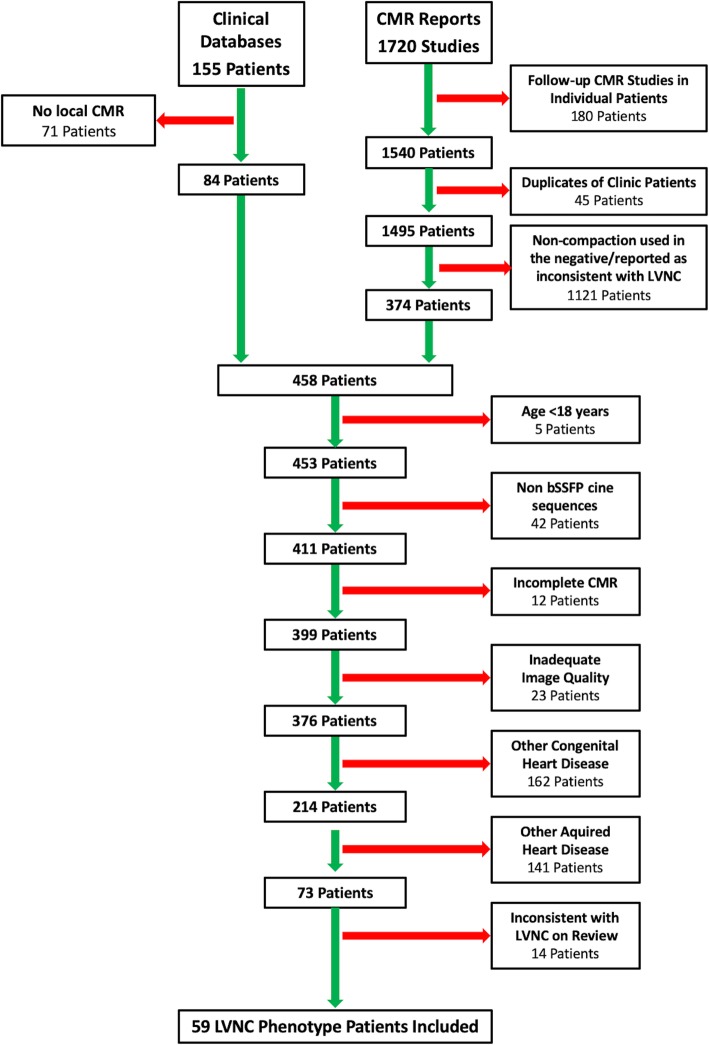
Study population
This single centre retrospective cohort study identified patients with isolated LVNC phenotype from heart failure and adult congenital cardiac disease clinic databases, as well as a Boolean search of CMR reports between June 2008 and July 2017. The keyword search of CMR reports included the terms ‘non-compaction’ and/or ‘hypertrabeculation’, and returned cases were qualitatively reviewed by two readers (JGD and BJW) to confirm findings consistent with the LVNC phenotype. Patient exclusion criteria included age < 18 years, incomplete cine bSSFP coverage of the LV, CMR studies with limited image quality, and any concurrent congenital or acquired heart disease with the exception of idiopathic dilated cardiomyopathy with hypertrabeculation. Patient identification and study inclusion is summarized in Fig. 1. Healthy volunteers with no known history of cardiovascular disease that were originally recruited as part of the EMBRACE-MRI (clinicaltrials.gov: NCT02306538) and MAFIO (clinicaltrials.gov: NCT02090699) studies were included as a healthy control population [21]. LVNC phenotype patients and healthy subjects with normal LV volumetrics were separated for subgroup analysis and labelled subgroup A and B, respectively. Healthy subjects were further subdivided into those meeting none of the CMR diagnostic criteria for LVNC (subgroup C) and those meeting one or more criteria (subgroup D). The following subgroup comparisons were performed: subgroup A vs. B; A vs. C; A vs. D; and C vs. D. The study protocol conformed to the Declaration of Helsinki and was approved by the institutional research ethics board (University Health Network Research Ethics Board). Written informed consent was obtained from all healthy subjects and was waived for the patient cohort.
Fig. 1.
Retrospective Patient Identification. CMR = cardiovascular magnetic resonance; LVNC = left ventricular non-compaction cardiomyopathy; bSSFP = balanced steady-state free precession
CMR technique
CMR scans were performed on one of four scanners: Magnetom Skyrafit/Verio (3 T) or Avanto/Avantofit (1.5 T) (Siemens Healthineers, Erlangen, Germany). All studies included retrospectively gated LV 2-, 3- and 4-chamber long-axis (LAX) single-slice and short-axis oblique (SAO) base-to-apex stack cine bSSFP sequences with 6–8 mm slice thickness, 2 mm slice gap, pixel size ≤1.8 × 1.8 mm2, and a temporal resolution < 50 ms.
CMR analysis
Analysis of LV volumes, systolic function, mass and strain was performed using commercially post-processing software (cvi42, Circle Cardiovascular Imaging, Calgary, Canada) by a single cardiac-fellowship trained radiologist with 2 years of cardiac imaging experience (JGD) blinded to pre-existing clinical and radiological information.
LV Volumetrics
Abnormality of LV volumetrics (i.e. dilation, impaired systolic function and hypertrophy) was assessed against gender-specific reference ranges validated for cine bSSFP sequences [22]. Definitions for males and females respectively included: LV dilation as an EDVi (end-diastolic volume (EDV) indexed to BSA) > 112 ml/m2 and > 99 ml/m2; impaired LV systolic function as an ejection fraction (EF) < 55 and < 54%; LV hypertrophy as a MASSi (mass indexed to BSA) > 83 g/m2 and > 67 g/m2.
CMR diagnostic criteria for LVNC
The following validated CMR diagnostic criteria for LVNC were assessed in all patients and volunteers: non-compacted (NC): compacted (C) myocardium diameter ratio > 2.3 in end-diastole [9], NC > 20% of LV mass [10], NC > 25% of LV mass, and NC indexed mass > 15 g/m2 [11].
Measurements of NC and C thickness for the calculation of the NC:C ratio were performed on end-diastolic SAO and/or LAX cine images with the myocardium orthogonal to the plane of measurement.
A previously validated technique was employed to measure the NC myocardial mass [10, 11]. A semi-automated threshold-based technique was used to contour the end-diastolic endocardial border of the NC myocardium along with standard epicardial contours to measure the ‘global myocardial mass’, without accounting for the intervening blood pool within the inter-trabecular recesses of the NC myocardium. The NC mass was then calculated by subtracting both the C myocardial mass (measured with standard endo- and epicardial contours) and papillary muscle mass from the global myocardial mass.
Methods of assessment of the NC myocardium are illustrated in Fig. 2.
Fig. 2.
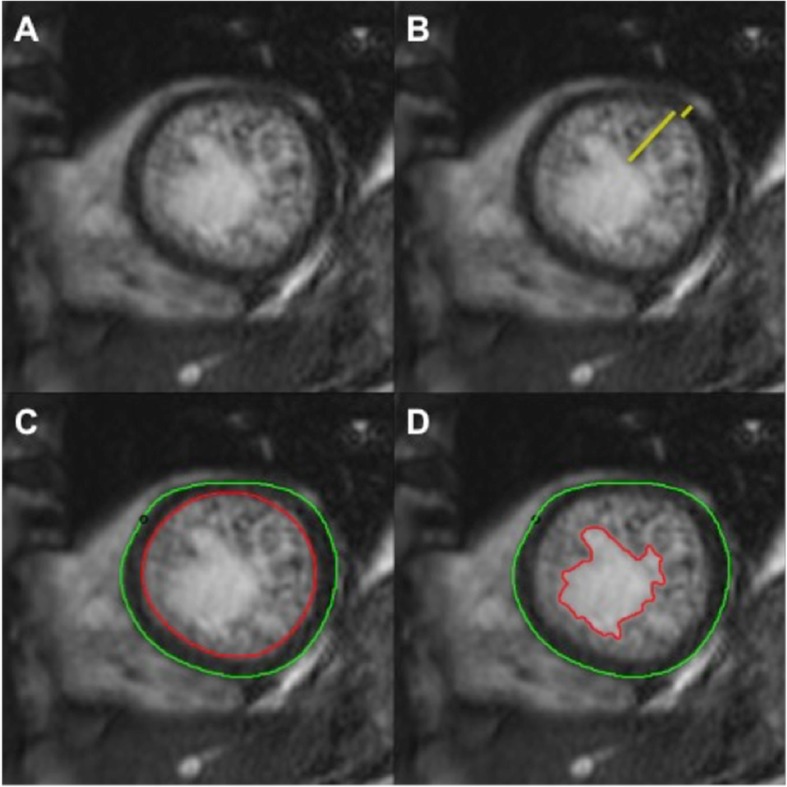
CMR Diagnostic Criteria for LVNC. a CMR short-axis cine bSSFP image of the apical LV demonstrating severely thickened myocardium with prominent trabeculations and deep intertrabecular recesses in a 49-year-old female with a history of ventricular tachycardia and LVNC phenotype. b Measurement of the maximum non-compacted (NC): compacted myocardium diameter ratio. c Standard epicardial contours and endocardial contours at the border of the compacted and NC myocardium for measurement of the compacted myocardial mass. d Endocardial contours at the border of the NC myocardium and non-trabeculated cavity for measurement of the global myocardial mass (compacted and non-compacted myocardial mass). CMR = cardiovascular magnetic resonance; LVNC = left ventricular non-compaction cardiomyopathy; bSSFP = balanced steady-state free precession; LV = left ventricle
CMR Strain Analysis
Radial, circumferential and longitudinal strain
Radial and circumferential strain were measured across the full thickness of the myocardium from the SAO cine bSSFP stack following application of end-diastolic endo- and epicardial contours with superior and inferior insertion point markers to three selected slices at representative basal, mid-ventricular and apical levels. The selected basal slices included circumferentially complete myocardium throughout the cardiac cycle, the selected apical slices avoided obliquely oriented myocardium, and the selected mid-ventricular slices were equidistant between the selected basal and apical slices at the level of the papillary muscles. Global radial strain (GRS) and global circumferential strain (GCS) were calculated from the average of the peak strain of the three selected SAO slices.
Longitudinal strain was measured across the full thickness of the myocardium from the LV 2-, 3- and 4-chamber cine bSSFP single-slice images by applying end-diastolic endo- and epicardial contours with a T-bar defining the mitral valve plane and LV apex. Global longitudinal strain (GLS) was calculated from the average of the peak strain of the three LAX slices.
Twist and torsion
Twist was measured as the rotational circumferential displacement (measured in degrees) at the selected basal and apical SAO slices. Torsion was calculated as the difference between basal and apical rotation (i.e. twist) divided by the distance between the two slices along the long axis of the LV.
Statistical analysis
All statistical analysis was performed using STATA v14.1 (StataCorp, College Station, Texas) and statistical significance was defined as a two-tailed p-value of < 0.05. Categorical variables are described as number and percentage and continuous variables are presented as median and interquartile range (IQR). Testing for normal distribution was performed using the Shapiro-Wilk test. Comparison of categorical variables was performed using Fisher’s exact test, normally distributed continuous variables with the independent samples t-test, and non-normally distributed continuous variables with the Wilcoxon rank sum test. Global strain values were evaluated with receiver operating characteristic (ROC) analyses.
To assess whether strain parameters are independent of EF in the subgroup A vs. D comparison, individual bivariable logistic regression models were fitted with respective strain measures and EF, and to assess for incremental diagnostic value, the likelihood ratio test was used to compare nested models with EF alone versus a model with GCS added. To assess the independence and incremental diagnostic value of GCS against each of the morphologic CMR diagnostic criteria for LVNC in differentiating all healthy volunteers and LVNC phenotype patients, individual bivariable logistic regression models were fitted with GCS and each diagnostic criteria, and the likelihood ratio test was used to compare nested models with each diagnostic criteria alone versus models with GCS added.
Results
Study population
One hundred fifty-five study candidates were identified from the clinical databases and 1720 exams from the CMR report database. A total of 59 LVNC phenotype patients were included following elimination of duplicate patients and CMR reports using keywords in the negative (e.g. “no evidence of non-compaction”), application of exclusion criteria, and review of CMR data sets studies by the two readers. All 59 (100%) of the patients met each of the four CMR diagnostic criteria for LVNC. LVNC phenotype patients included 30 males (51%) with a median age of 40 years (IQR: 28–50 years). The control group of 36 healthy subjects included 16 males (44%) with a median age of 39 years (IQR: 30–47 years). Demographic characteristics are detailed in Table 1. There were no significant differences in sex (p = 0.673), age (p = 0.866) or BSA (p = 0.863) between all healthy subjects and LVNC phenotype patients. All healthy subjects (n = 36/36) were scanned at 1.5 T. Of the LVNC phenotype patients, 61% (n = 36/59) were scanned at 1.5 T and 39% (n = 23/59) at 3.0 T.
Table 1.
Demographic Characteristics and LV Volumetrics
| All patients and volunteers | Subgroup A | Subgroup B | Subgroup C | Subgroup D | A vs. B | A vs. C | A vs. D | C vs. D | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All LVNC phenotype patients | All healthy subjects | p-values | LVNC phenotype patients (normal LV volumetrics) | Healthy subjects (normal LV volumetrics) | Healthy subjects (normal LV volumetrics, not meeting any LVNC criteria) | Healthy subjects (normal LV volumetrics, meeting one or more LVNC criteria) | p-values | p-values | p-values | p-values | |
| (n = 59) | (n = 36) | (n = 25) | (n = 34) | (n = 16) | (n = 18) | ||||||
| Malea | 30 (51%) | 16 (44%) | 0.673 | 9 (36%) | 15 (44%) | 4 (25%) | 11 (61%) | 0.598 | 0.513 | 0.130 | 0.045 |
| Age (years)b | 40.3 (28.4, 50.5) | 39.0 (30.2, 46.6) | 0.866 | 33.2 (22.7, 42.5) | 39.0 (30.4, 47) | 44.7 (38.0, 57.9) | 34.3 (30.0, 40.8) | 0.089 | 0.017 | 0.588 | 0.012 |
| BSA (m2)c | 1.87 (1.64, 2.08) | 1.82 (1.63, 2.02) | 0.863 | 1.69 (1.55, 1.87) | 1.82 (1.63, 1.99) | 1.72 (1.63, 1.89) | 1.94 (1.71, 2.05) | 0.118 | 0.527 | 0.045 | 0.198 |
| Heart Rate (bpm)b | 65 (56, 74) | 62 (59, 706) | 0.466 | 66 (57, 72) | 63 (59, 71) | 64 (60, 70) | 62 (56, 71) | 0.545 | 0.820 | 0.445 | 0.469 |
| EDV (ml)b | 173 (143, 203) | 152 (122, 180) | 0.054 | 143 (127, 155) | 145 (122, 176) | 137 (120, 155) | 171 (122, 197) | 0.439 | 0.669 | 0.102 | 0.079 |
| EDVI (ml/m2)b | 93.0 (80.7, 102.9) | 84.2 (72.5, 95.6) | 0.036 | 83.0 (74.8, 91.7) | 83.8 (71.4, 92.6) | 78.4 (69.3, 85.5) | 88.3 (81.1, 102.1) | 0.902 | 0.181 | 0.153 | 0.032 |
| ESV (ml)b | 79 (58, 103) | 54 (45, 69) | < 0.001 | 58 (53, 70) | 53 (44, 68) | 50 (40, 56) | 68 (48, 74) | 0.390 | 0.019 | 0.431 | 0.027 |
| ESVI (ml/m2)b | 42.4 (33.6, 53.1) | 30.2 (25.7, 37.8) | < 0.001 | 34.1 (31.1, 39.2) | 30.1 (25.6, 36.0) | 29.8 (22.8, 30.6) | 33.9 (25.8, 39.6) | 0.051 | 0.002 | 0.787 | 0.019 |
| SV (ml)b | 86 (76, 107) | 95 (77, 118) | 0.121 | 80 (74, 91) | 93 (77, 116) | 87 (795, 99) | 103 (76, 122) | 0.051 | 0.219 | 0.046 | 0.178 |
| SVI(ml/m2)c | 49.2 (41.3, 52.7) | 54.1 (46.8, 59.0) | 0.796 | 49.5 (43.9, 52.6) | 53.3 (46.7, 58.3) | 50.1 (46.6, 55.2) | 56.5 (47.0, 59.9) | 0.033 | 0.205 | 0.008 | 0.125 |
| EF (%)b | 54 (49, 59) | 63 (60, 66) | < 0.001 | 59 (57, 60) | 63 (60, 66) | 65 (63, 68) | 61 (59, 64) | < 0.001 | < 0.001 | 0.004 | 0.023 |
| CO (l/min)b | 5.66 (4.74, 6.96) | 5.85 (4.90, 7.10) | 0.485 | 5.13 (4.53, 6.13) | 5.85 (4.91, 7.07) | 5.60 (4.96, 6.75) | 6.30 (4.89, 7.78) | 0.163 | 0.364 | 0.161 | 0.512 |
| LVM (g)b | 105 (82, 136) | 98 (84, 128) | 0.747 | 82 (67, 99) | 98 (83, 128) | 93 (81, 102) | 108 (84, 142) | 0.018 | 0.181 | 0.011 | 0.121 |
| LVMI (g/m2)b | 54.9 (48.9, 66.2) | 55.4 (50.1, 65.5) | 0.777 | 49.5 (43.7, 54.2) | 55.0 (50.0, 64.7) | 52.0 (48.9, 56.8) | 55.8 (50.1, 69.7) | 0.012 | 0.157 | 0.007 | 0.113 |
Data presented as n (%) or median (IQR)
LV left ventricle, LVNC left ventricular non-compaction cardiomyopathy, BSA body surface area, bpm beats per minute, EDV end-diastolic volume, EDVI end-diastolic volume indexed to BSA, ESV end-systolic volume, ESVI end-systolic volume indexed to BSA, SV stroke volume, SVi stroke volume indexed to BSA, EF ejection fraction, CO cardiac output, LVM left ventricular mass, LVMI LVM indexed to BSA
aFisher’s exact test
bWilcoxon rank sum test
cIndependent samples t-test
Subgroup analyses
Subgroups of subjects with normal LV volumetrics included 25 LVNC phenotype patients (subgroup A) and 34 healthy subjects (subgroup B); of the two healthy subjects excluded, both had borderline LV dilation and one had borderline impaired EF. Among the healthy subjects 16 met none of the CMR diagnostic criteria for LVNC (subgroup C) and 18 met one or more criteria (subgroup D).
Subgroup a vs. B
Comparison of LVNC phenotype patients (n = 25) and healthy suybjects (n = 34) with normal LV volumetrics revealed no statistically significant differences in sex (p = 0.598), age (p = 0.089), or BSA (p = 0.118).
Subgroup a vs. C
Comparison of LVNC phenotype patients with normal LV volumetrics (n = 25) and healthy subjects with normal LV volumetrics not meeting any LVNC criteria (n = 16) revealed a significant difference in age (p = 0.017) but no statistically significant differences in sex (p = 0.513), or BSA (p = 0.527).
Subgroup a vs. D
Comparison of LVNC phenotype patients with normal LV volumetrics (n = 25) and healthy subjects with normal LV volumetrics meeting one or more LVNC criteria (n = 18) revealed a significant difference in BSA (p = 0.045) but no statistically significant differences in sex (p = 0.130) or age (p = 0.588).
Subgroup C vs. D
Comparison between healthy subjects with normal LV volumetrics not meeting any of the LVNC criteria (n = 16) and those meeting one or more criteria (n = 18) revealed significant differences in sex (p = 0.045) and age (p = 0.012), but no statistically significant difference in BSA (p = 0.198).
LV Volumetrics
Results for LV volumetrics are detailed in Table 1. Statistically significant differences were observed between all LVNC phenotype patients and healthy subjects for EDVi (p = 0.036), ESV (p < 0.001), ESVi (end-systolic volume indexed to BSA) (p < 0.001) and EF (p < 0.001). The remaining LV volumetric parameters showed no statistically significant difference.
Subgroup a vs. B
Statistically significant differences were demonstrated in EF (p < 0.001), mass (p = 0.018), MASSi (p = 0.012) and SVi (stroke volume indexed to BSA) (p = 0.033).
Subgroup a vs. C
Statistically significant differences were demonstrated in EF (p < 0.001), ESV (p = 0.019) and ESVi (p = 0.002).
Subgroup a vs. D
Statistically significant differences were demonstrated in EF (p < 0.004), SV (p = 0.046), SVi (p = 0.008), mass (p = 0.011) and MASSi (p = 0.007).
Subgroup C vs. D
Statistically significant differences were demonstrated in EF (p < 0.023), EDVi (p = 0.032), ESV (p = 0.027) and ESVi (p = 0.019).
CMR diagnostic criteria for LVNC
Results for CMR diagnostic criteria for LVNC are detailed in Table 2. Although statistically significant differences were observed between healthy subjects and LVNC phenotype patients across all diagnostic criteria and measures of NC mass, a sizeable minority of the control group also met diagnostic criteria for LVNC. Seven healthy subjects (19.4%) demonstrated at least one segment of hypertrabeculation with an NC:C diameter ratio > 2.3, with 16 (44.4%) exceeding an NC mass of 20% (median 19.5%; IQR 17.2, 23.3%) and 5 (13.9%) exceeding 25% of the global LV myocardium, as well as 15 (41.7%) with an indexed NC mass > 15 g/m2 (median 13.6 g/m2; IQR 10.5, 17.2).
Table 2.
CMR Diagnostic Criteria for LVNC
| All patients and healthy subjects | Subgroup A | Subgroup B | Subgroup C | Subgroup D | A vs. B | A vs. C | A vs. D | C vs. D | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All LVNC phenotype patients | All healthy subjects | p-values | LVNC phenotype patients (normal LV volumetrics) | Healthy subjects (normal LV volumetrics) | Healthy subjects (normal LV volumetrics, not meeting any LVNC criteria) | Healthy subjects (normal LV volumetrics, meeting one or more LVNC criteria) | p-values | p-values | p-values | p-values | |
| (n = 59) | (n = 36) | (n = 25) | (n = 34) | (n = 16) | (n = 18) | ||||||
| Number of NC segmentsb | 8 (6, 9) | 0 (0, 0) | < 0.001 | 8 (6, 9) | 0 (0, 0) | 0 (0, 0) | 0 (0, 1) | < 0.001 | < 0.001 | < 0.001 | 0.013 |
| Maximum NC:C ratiob | 3.8 (3.0, 4.3) | 1.2 (0.9, 1.6) | < 0.001 | 3.5 (3.2, 4.3) | 1.1 (0.9, 1.3) | 1.0 (0.8, 1.2) | 1.2 (0.9, 2.5) | < 0.001 | < 0.001 | < 0.001 | 0.182 |
| NC:C > 2.3a | 59 (100%) | 7 (19.4%) | < 0.001 | 25 (100%) | 6 (17.7%) | 0 (0%) | 6 (33.3%) | < 0.001 | < 0.001 | < 0.001 | 0.020 |
| Percentage of NC mass (%)b | 40.8 (35.7, 45.9) | 19.5 (17.2, 23.3) | < 0.001 | 42.2 (38.7, 47.1) | 19.3 (17.1, 23.6) | 17.2 (14.8, 18.5) | 23.3 (20.1, 30.7) | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| NC mass > 20%a | 59 (100%) | 16 (44.4%) | < 0.001 | 25 (100%) | 14 (41.2%) | 0 (0%) | 14 (77.8%) | < 0.001 | < 0.001 | 0.025 | < 0.001 |
| NC mass > 25%a | 59 (100%) | 5 (13.9%) | < 0.001 | 25 (100%) | 5 (14.7%) | 0 (0%) | 5 (27.8%) | < 0.001 | < 0.001 | < 0.001 | 0.046 |
| NC mass/BSA (g/m2)b | 36.1 (32.3, 42.9) | 13.6 (10.5, 17.2) | < 0.001 | 35.1 (30.9, 39.2) | 12.7 (10.4, 17.0) | 10.0 (8.8, 11.6) | 17.0 (15.0, 23.7) | < 0.001 | < 0.001 | < 0.001 | < 0.001 |
| NC mass > 15 g/m2a | 59 (100%) | 15 (41.7%) | < 0.001 | 25 (100%) | 13 (38.2%) | 0 (0%) | 13 (72.2%) | < 0.001 | < 0.001 | 0.009 | < 0.001 |
Data presented as n (%) or median (IQR)
NC:C >2.3 = maximum end-diastolic non-compacted (NC): compacted (C) myocardium diameter ratio >2.3 in at least one segment.
CMR cardiovascular magnetic resonance, LVNC left ventricular non-compaction cardiomyopathy, LV left ventricle, NC non-compacted myocardium, C compacted myocardium, BSA body surface area
aFisher’s exact test
bWilcoxon rank sum test
Subgroup analyses
For subgroup comparisons A vs. B, A vs. C and A vs. D, statistically significant differences persisted across all LVNC diagnostic criteria and measures of NC myocardium. On comparison of healthy subjects with normal LV volumetrics meeting one or more LVNC criteria and those meeting none (subgroup C vs. D), only the maximum NC:C ratio did not show statistically significant difference. Of the diagnostic criteria for LVNC met by 18 (53%) healthy subjects with normal LV volumetrics, 5 healthy subjects (28%) met four criteria, 1 (6%) met three criteria, 6 (33%) met two criteria, and 8 (44%) met one of the four criteria.
CMR strain analysis
Results for strain analysis and torsion are detailed in Table 3.
Table 3.
Strain and Torsion
| All patients and healthy subjects | Subgroup A | Subgroup B | Subgroup C | Subgroup D | A vs. B | A vs. C | A vs. D | C vs. D | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All LVNC phenotype patients | All healthy volunteers | p-values | LVNC phenotype patients (normal LV volumetrics) | Healthy subjects (normal LV volumetrics) | Healthy subjects (normal LV volumetrics, not meeting any LVNC criteria) | Healthy subjects (normal LV volumetrics, meeting one or more LVNC criteria) | p-values | p-values | p-values | p-values | |
| (n = 59) | (n = 36) | (n = 25) | (n = 34) | (n = 16) | (n = 18) | ||||||
| GRS (%)a | 26.4 (21.8, 32.7) | 37.1 (33.4, 43.1) | < 0.001 | 31.2 (28.2, 34.7) | 37.5 (34.2, 43.5) | 40.2 (35.5, 44.3) | 35.8 (33.1, 40.8) | < 0.001 | < 0.001 | 0.006 | 0.103 |
| Basal RS (%)a | 26.8 (21.2, 30.7) | 34.6 (31.0, 39.1) | < 0.001 | 28.8 (27.4, 36.1) | 34.6 (31.4, 39.1) | 35.3 (33.9, 40.9) | 32.0 (30.3, 35.6) | 0.004 | 0.002 | 0.112 | 0.044 |
| Mid RS (%)a | 23.9 (19.7, 28.9) | 29.9 (24.9, 35.9) | < 0.001 | 28.5 (24.7, 31.8) | 30.5 (25.8, 36.2) | 32.4 (29.5, 37.1) | 26.6 (24.0, 35.5) | 0.065 | 0.008 | 0.516 | 0.087 |
| Apical RS (%)a | 29.4 (24.0, 37.3) | 46.4 (40.1, 56.4) | < 0.001 | 33.3 (29.4, 42.2) | 47.3 (41.8, 58.2) | 49.2 (42.8, 59.8) | 45.6 (40.3, 51.4) | < 0.001 | < 0.001 | 0.002 | 0.451 |
| GCS (%)b | −16.5 (−14.7, −18.6) | −20.5 (−19.1, −22.1) | < 0.001 | −18.5 (−16.8, −19.8) | −20.6 (−19.3, −22.1) | −21.5 (−20.1, −22.9) | −20.0 (−18.8, −21.7) | < 0.001 | < 0.001 | 0.005 | 0.073 |
| Basal CS (%)b | −16.3 (−14.2, −18.1) | −19.6 (−18.4, −21.1) | < 0.001 | −17.4 (−16.6, −20.0) | −19.6 (−18.4, −21.2) | −20.1 (−19.3, −21.7) | −18.5 (− 18.2, − 20.0) | 0.003 | 0.002 | 0.072 | 0.027 |
| Mid CS (%)b | −15.5 (−13.4, −17.8) | − 18.3 (− 16.1, − 20.3) | < 0.001 | −17.5 (− 16.1, − 18.9) | − 18.6 (− 16.5, − 20.4) | − 19.4 (− 18.1, − 21.0) | −17.2 (−15.9, − 19.9) | 0.061 | 0.005 | 0.658 | 0.079 |
| Apical CS (%)b | −17.8 (−15.4, − 19.9) | −23.4 (− 21.9, −25.9) | < 0.001 | − 19.4 (− 17.8, − 22.2) | −23.7 (− 22.3, − 26.3) | −24.4 (− 22.6, −26.6) | −23.0 (− 22.0, −24.5) | < 0.001 | < 0.001 | < 0.001 | 0.270 |
| GLS (%)b | −14.6 (−12.9, − 16.5) | − 17.1 (− 16.1, − 18.7) | < 0.001 | − 16.4 (− 15.3, − 17.3) | −17.1 (− 16.2, − 19.0) | −17.2 (− 16.7, − 19.6) | − 16.9 (− 16.1, − 18.3) | 0.011 | 0.009 | 0.099 | 0.143 |
| 4-chamber LS (%)b | −14.7 (− 12.9, − 16.9) | −18.1 (− 16.8, − 19.5) | < 0.001 | −16.0 (− 14.6, − 16.6) | −16.7 (− 15.4, − 18.4) | − 17.6 (− 16.2, − 20.4) | −16.2 (− 15.0, − 17.3) | 0.021 | 0.003 | 0.313 | 0.032 |
| 3-chamber LS (%)b | −14.7 (− 12.5, − 16.5) | −17.5 (− 15.9, − 19.1) | < 0.001 | − 16.4 (− 14.9, − 17.9) | − 17.6 (− 16.1, − 19.1) | −17.8 (− 16.4, − 19.3) | − 17.3 (− 16.0, − 18.6) | 0.046 | 0.048 | 0.168 | 0.427 |
| 2-chamber LS (%)b | −14.8 (−13.2, − 16.7) | − 17.6 (− 16.3, − 19.3) | < 0.001 | − 16.7 (− 14.8, − 18.5) | −17.6 (− 16.4, − 19.2) | −17.7 (− 16.3, − 18.7) | −17.5 (− 16.4, − 19.4) | 0.034 | 0.109 | 0.055 | 0.717 |
| Global Torsion (°/cm)b | 1.01 (0.68, 1.43) | 0.94 (0.74, 1.54) | 0.765 | 1.23 (0.99, 1.47) | 0.98 (0.78–1.55) | 1.01 (0.86, 1.75) | 0.93 (0.71, 1.42) | 0.470 | 0.852 | 0.184 | 0.178 |
Data presented as median (IQR)
LVNC left ventricular non-compaction cardiomyopathy, LV left ventricle, GRS global radial strain, RS radial strain, GCS global circumferential strain, CS circumferential strain, GLS global longitudinal strain, LS longitudinal strain
aIndependent samples t-test
bWilcoxon rank sum test
Radial, circumferential and longitudinal strain
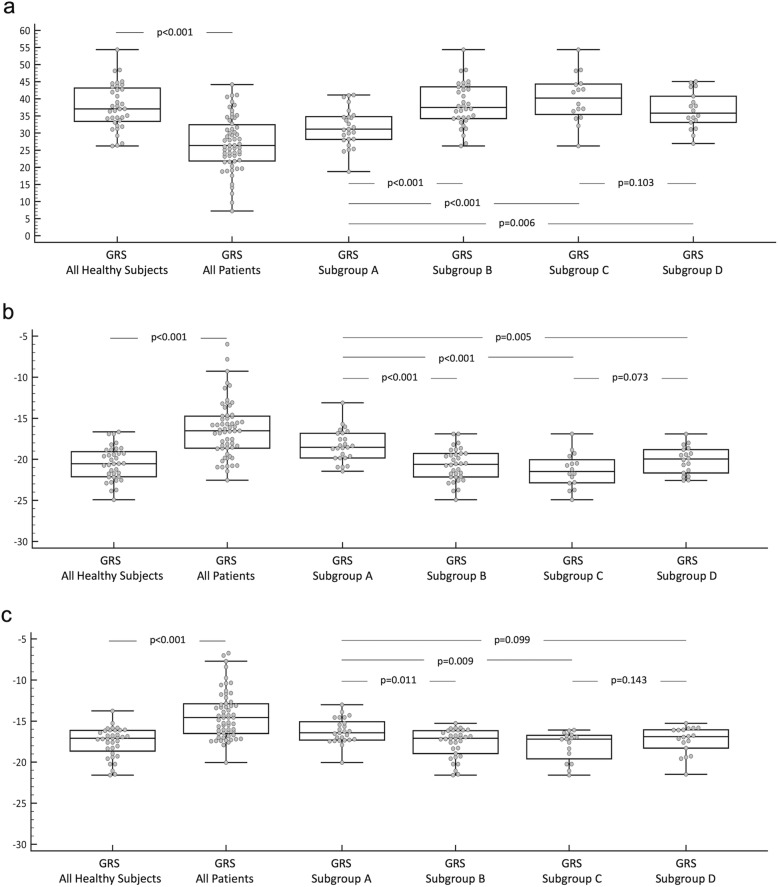
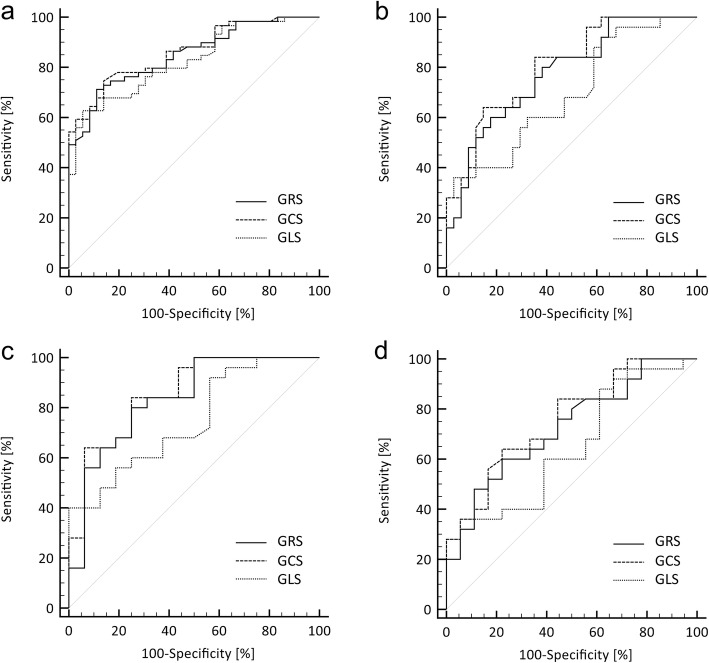
Reduced radial, circumferential and longitudinal strain in LVNC phenotype patients was observed compared to healthy subjects, with statistically significant differences at both global and slice specific levels (i.e. basal, mid and apical levels for radial and circumferential strain, and each LAX orientation for longitudinal strain). Comparisons between all LVNC phenotype patients and healthy subjects, as well as the subgroups, are illustrated with box plots in Fig. 3. There was no IQR overlap between patients and healthy controls in GRS (25th percentile of 33.4% for controls and 75th percentile of 32.7% for patients). IQR overlap was, however, observed between the groups for GCS and GLS. Results of the ROC analyses are detailed in Table 4. ROC curve analysis demonstrated an area under the curve (AUC) of 0.86 for GCS, 0.85 for GRS and 0.83 for GLS. ROC curves are illustrated in Fig. 4.
Fig. 3.
Strain: Box Plots. Box plots comparing a GRS, b GCS and c GLS between: all LVNC phenotype patients and healthy subjects; Subgroup A vs. B (LVNC phenotype patients with normal LV volumetrics and healthy subjects with normal LV volumetrics); Subgroup A vs. C (LVNC phenotype patients with normal LV volumetrics and healthy subjects with normal LV volumetrics not meeting any CMR diagnostic criteria for LVNC); Subgroup A vs. D (LVNC phenotype patients with normal LV volumetrics and healthy subjects with normal LV volumetrics meeting at least one of the CMR diagnostic criteria for LVNC); Subgroup C vs. D (healthy subjects with normal LV volumetrics meeting none of the CMR diagnostic criteria for LVNC and those meeting at least one criteria). GRS = global radial strain; GCS = global circumferential strain; GLS = global longitudinal strain; LVNC = left ventricular non-compaction cardiomyopathy; LV = left ventricle; CMR = cardiovascular magnetic resonance
Table 4.
ROC Analysis
| All LVNC phenotype patients and all healthy subjects | Subgroup A vs. B | Subgroup A vs. C | Subgroup A vs. D | Subgroup C vs. D | |
|---|---|---|---|---|---|
| LVNC phenotype patients (normal LV volumetrics) and healthy subjects (normal LV volumetrics) | LVNC phenotype patients (normal LV volumetrics) and healthy subjects (normal LV volumetrics, not meeting any LVNC criteria) | LVNC phenotype patients (normal LV volumetrics) and healthy subjects (normal LV volumetrics, meeting one or more LVNC criteria) | Healthy subjects (normal LV volumetrics, not meeting any LVNC criteria) and healthy subjects (normal LV volumetrics, meeting one or more LVNC criteria) | ||
| AUC | AUC | AUC | AUC | AUC | |
| GRS | 0.85 (0.78, 0.92) | 0.78 (0.66, 0.90) | 0.84 (0.70, 0.97) | 0.72 (0.57, 0.88) | 0.65 (0.46, 0.84) |
| GCS | 0.86 (0.79, 0.93) | 0.80 (0.69, 0.91) | 0.86 (0.74, 0.98) | 0.76 (0.61, 0.90) | 0.68 (0.49, 0.87) |
| GLS | 0.83 (0.75, 0.91) | 0.69 (0.56, 0.83) | 0.75 (0.59, 0.90) | 0.65 (0.48, 0.82) | 0.65 (0.46, 0.84) |
Data presented as AUC (95% CI)
LVNC left ventricular non-compaction cardiomyopathy, LV left ventricle, ROC receiver operating characteristic, AUC area under the curve, GRS global radial strain, GCS global circumferential strain, GLS global longitudinal strain, CI confidence interval
Fig. 4.
Strain: ROC Analysis. ROC curves for GRS, GCS and GLS comparing a All LVNC phenotype patients and healthy subjects ; b Subgroup A vs. B (LVNC phenotype patients with normal LV volumetrics and healthy subjects with normal LV volumetrics); c Subgroup A vs. C (LVNC phenotype patients with normal LV volumetrics and healthy subjects with normal LV volumetrics not meeting any CMR diagnostic criteria for LVNC); d Subgroup A vs. D (LVNC phenotype patients with normal LV volumetrics and healthy subjects with normal LV volumetrics meeting at least one of the CMR diagnostic criteria for LVNC). ROC = receiver operating characteristic; GRS = global radial strain; GCS = global circumferential strain; GLS = global longitudinal strain; LVNC = left ventricular non-compaction cardiomyopathy; LV = left ventricle; CMR = cardiovascular magnetic resonance
Comparing all healthy subjects and LVNC phenotype patients, GCS had independent and incremental diagnostic value beyond each of the CMR diagnostic criteria for LVNC: NC:C > 2.3 (OR 1.50, 95% CI: 1.04 to 2.18, p = 0.031, likelihood ration test χ2(df = 1) = 6.49, p = 0.011); NC mass > 20% (OR 1.69, 95% CI: 1.24 to 2.29, p = 0.001, likelihood ration test χ2(df = 1) = 18.15, p < 0.001); NC mass > 25% (OR 1.69, 95% CI: 1.04 to 2.73, p = 0.033, likelihood ration test χ2(df = 1) = 6.89, p = 0.009); NC mass > 15 g/m2 (OR 1.55, 95% CI: 1.17 to 2.07, p = 0.003, likelihood ratio test χ2(df = 1) = 13.39, p < 0.001).
Subgroup analyses
Statistically significant differences in GRS, GCS and GLS remained when LVNC phenotype patients were compared to healthy subjects with normal LV volumetrics (subgroup A vs. B) with AUCs of 0.80 for GCS, 0.78 for GRS and 0.69 for GLS, as well as when compared to healthy subjects not meeting any CMR diagnostic criteria for LVNC (subgroup A vs. C) with AUCs of 0.86 for GCS, 0.84 for GRS and 0.75 for GLS. Among subjects with normal LV volumetrics comparing LVNC phenotype patients with healthy subjects meeting at least one of the LVNC diagnostic criteria (subgroup A vs. D), statistically significant differences persisted for GCS (AUC 0.72) and GRS (AUC 0.69), but not GLS. However, only GCS remained significant in a bivariable logistic regression model after adjusting for EF (OR 1.59, 95% CI: 1.00 to 2.53, p = 0.049). In a nested logistic regression model with EF, model fit was significantly improved by the addition of GCS (χ2(df = 1) = 4.55, p = 0.033). No significant differences were demonstrated in global strain between healthy subjects with normal LV volumetrics meeting one or more LVNC criteria and those meeting none (subgroup C vs. D).
Torsion
Results for torsion are detailed in Table 3. No statistically significant differences were observed between all healthy subjects and LVNC phenotype patients or in the subgroup comparisons.
Discussion
The results demonstrate abnormal myocardial strain in LVNC phenotype patients compared to healthy subjects, also present on comparison of subjects with normal LV volumetrics, although the study was unable to demonstrate differences independent of LV volumetrics known to affect myocardial strain. The differences were accentuated among subjects with normal LV volumetrics when LVNC phenotype patients were compared to healthy subjects not meeting any CMR diagnostic criteria for LVNC, but still persisted for GRS and GCS (GCS only when corrected for EF) when compared to healthy subjects with ‘physiologic’ hypertrabeculation (meeting one or more diagnostic criteria). Importantly, there were no significant differences between healthy subjects with and without physiologic hypertrabeculation. The AUCs for global strain values in differentiating LVNC phenotype patients from healthy subjects with ‘physiologic’ hypertrabeculation are reasonable considering the important limitations of morphology-based diagnostic criteria and GCS provides significant incremental value in addition to EF. GCS also adds significant incremental value when added to any of the four morphologic diagnostic criteria for LVNC among all healthy subjects and LVNC phenotype patients. No significant differences in LV torsion, however, were demonstrated by CMR FT analysis.
The significant proportion of healthy subjects meeting CMR diagnostic criteria for LVNC has previously been observed by larger studies. In the TASCFORCE (Tayside Screening for Cardiovascular Events) study 14.8% of subjects met at least one of the diagnostic criteria assessed, including an NC:C diameter ≥ 2.3 measured on end-diastolic LAX images (12.6%), ≥3 on end-diastolic SAO images (7.2%), ≥2 on end-systolic SAO images (4.4%), and NC > 20% of global LV mass (4.1%) [12]. The MESA (Multi Ethnic Study of Atherosclerosis) population observed an even higher proportion of asymptomatic subjects meeting criteria, with 43% of subjects demonstrating at least one segment with an NC:C diameter ratio > 2.3 on end-diastolic LAX images [13]. This study observed a particularly high proportion of healthy subjects exceeding previously validated diagnostic thresholds of non-compacted mass, likely related to the inherent limitations of contouring the borders of finely trabeculated non-compacted myocardium on 6–8 mm thick cine bSSFP SAO slices and inevitable inclusion of intertrabecular blood pool. Variability of measurement may partly account for differences in prevalence of healthy subjects meeting LVNC criteria between studies, particularly measurement of the NC:C diameter ratio where some studies have found poor inter-observer agreement depending on the technique used [12]; however, the remaining CMR diagnostic criteria based on measurement of the NC mass have more consistently shown a high degree of inter-observer reproducibility [10–12]. In any event, the high proportion of healthy subjects meeting morphology-based CMR diagnostic criteria emphasizes the important potential complementary diagnostic value of strain in differentiating LVNC from physiologic hypertrabeculation.
Although there are multiple studies describing the strain characteristics of LVNC assessed by STE [15–18], CMR FT has some potential advantages that may provide a more accurate and reproducible assessment. The comparatively unrestricted access to multi-planar views and excellent contrast between the myocardium and blood pool allows reproducible imaging planes and clear boundary points for the measurement of myocardial strain [19]. However, the relatively poor temporal resolution of routine bSSFP CMR precludes reliable assessment of more sophisticated time-resolved measures of strain, including strain rate and velocity.
Among the healthy subjects GRS values were slightly higher, and GCS and GLS slightly lower, compared to a meta-analysis of CMR FT derived strain values in normal subjects [23]. Although a high degree of inter-observer reproducibility in the CMR assessment of strain has been previously demonstrated [24], recognized variations in strain on images obtained at different field strengths and significant variability across post-processing software vendors [25] may account for these differences.
To the best of the authors’ knowledge, this study represents the largest group of LVNC phenotype patients assessed by CMR FT strain analysis. The findings of impaired myocardial strain among LVNC phenotype patients are consistent with previous smaller studies [26, 27]. A study by Cai J et al. investigated the relationship between LV hypertrabeculation measured by fractal analysis and myocardial deformation by CMR FT among 180 healthy subjects and 10 LVNC patients, demonstrating an independent association between the degree of hypertrabeculation and reduced circumferential strain [26]. A retrospective study by Nucifora et al demonstrated impairment of radial, circumferential and longitudinal strain assessed by CMR FT in separate groups of children/adolescents and young adults with a total of 32 isolated LVNC patients compared to age-matched controls [27]. Impairment of myocardial deformation in the children/adolescent group with preserved LVEF but comparable degrees of non-compaction to young adult patients with reduced LVEF was observed, suggesting that abnormal strain may predict the development of clinically overt cardiomyopathy in LVNC.
The absence of a significant difference in LV twist and torsion is inconsistent with previous STE studies demonstrating significantly decreased twist in LVNC phenotype patients compared to healthy subjects [17, 18]. A disadvantage of CMR FT compared to STE that may partly account for these differences is the fairly uniform water content and thus homogenous signal within myocardium on bSSFP sequences. This may preclude accurate tracking of features within the myocardium required to measure rotational circumferential displacement and therefore LV twist, whereas other strain parameters dependent on tracking well-defined endo- and epicardial borders appear to be more robust [19]. Nevertheless, significantly greater degrees of LV torsion have been demonstrated amongst healthy subjects in previous STE [28] and CMR FT studies [29], with reported normal torsion values of 2.7°/cm (standard deviation ±1.5) at CMR FT compared to 0.94°/cm (IQR: 0.74, 1.54) observed in our study. These differences may again be related to differences in tissue tracking techniques employed by different post-processing software vendors.
Limitations
LVNC is a rare cardiomyopathy with a variety of clinical manifestations and growing number of genetic associations described in recent years that demonstrate phenotypic overlap with other cardiomyopathies [30]. Due to the limited number of LVNC phenotype patients, it was beyond the scope of this study to investigate potential associations between clinical manifestations and strain. Furthermore, it was not possible to limit the study to genetically confirmed cases as a minority of patients will have undergone testing for multiple associated genes. The retrospective nature of the study may have introduced an element of selection bias, as LVNC patients identified from clinic databases and CMR reports are more likely to have more severe disease. A further limitation is the use of a single post-processing software vendor for CMR FT strain analysis, as previous studies have demonstrated significant inter-vendor variability [25]; cut-points have therefore not been provided due to limited clinical applicability.
Conclusions
LVNC phenotype patients demonstrate impaired radial, circumferential and longitudinal strain by CMR FT, persistent on comparison of subjects with normal LV volumetrics meeting at least one diagnostic criteria for LVNC. The high proportion of healthy subjects meeting morphology-based CMR diagnostic criteria emphasizes the important potential complementary diagnostic value of strain in differentiating LVNC from physiologic hypertrabeculation.
Acknowledgements
Not applicable.
Abbreviations
- AUC
Area under the curve
- BSA
Body surface area
- bSSFP
Balanced steady-state free precession
- C
Compacted myocardium
- CMR
Cardiovascular magnetic resonance
- EDV
End-diastolic volume
- EDVI
End-diastolic volume indexed to body surface area
- EF
Ejection fraction
- ESV
End-systolic volume
- ESVI
End-systolic volume indexed to body surface area
- FT
Feature tracking
- GCS
Global circumferential strain
- GLS
Global longitudinal strain
- GRS
Global radial strain
- IQR
Interquartile range
- LAX
Long-axis
- LV
Left ventricle/left ventricular
- LVNC
Left ventricular non-compaction cardiomyopathy
- MASSi
Myocardial mass indexed to body surface area
- NC
Non-compacted myocardium
- ROC
Receiver operating characteristic
- SAO
Short-axis oblique
- STE
Speckle-tracking echocardiography
- SV
Stroke volume
- SVI
Stroke volume indexed to body surface area
Authors’ contributions
JGD and BJW were responsible for conception and design of the study. JGD and BJW were responsible for patient selection. JGD, SM and CH analysed the CMR studies and contributed to data acquisition. KH performed the statistical analysis and JGD, KH and BJW contributed to the data interpretation. JGD and BJW drafted the manuscript. All authors were involved in critical and intellectual review of the manuscript and made substantial contributions. All authors read and have approved the final manuscript version and agree to be accountable for all aspects of the work.
Authors’ information
Dr. Erwin Oechslin currently holds the Bitove Family Professorship of Adult Congenital Heart Disease.
Funding
Not applicable.
Availability of data and materials
The datasets used in the study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study protocol conformed to the Declaration of Helsinki and was approved by the Research Ethics Board of the University Health Network, Toronto (17–5741). Healthy Volunteer recruitment and scanning as part of the EMBRACE-MRI (13–6543-C) and MAFIO (13–5878) trials was also approved by the Research Ethics Board of the University Health Network, Toronto. Written informed consent was obtained from all volunteers and waived for the patient cohort.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
John G. Dreisbach, Email: john.dreisbach@nhs.net
Shobhit Mathur, Email: drshobhitmathur@yahoo.co.in.
Christian P. Houbois, Email: christian.houbois@uhn.ca
Erwin Oechslin, Email: erwin.oechslin@uhn.ca.
Heather Ross, Email: heather.ross@uhn.ca.
Kate Hanneman, Email: kate.hanneman@uhn.ca.
Bernd J. Wintersperger, Email: bernd.wintersperger@uhn.ca
References
- 1.Oechslin E, Jenni R. Left ventricular non-compaction revisited: a distinct phenotype with genetic heterogeneity? Eur Heart J. 2011;32(12):1446–1456. doi: 10.1093/eurheartj/ehq508. [DOI] [PubMed] [Google Scholar]
- 2.Jenni R, Oechslin E, Schneider J, Jost CA, Kaufmann PA. Echocardiographic and pathoanatomical characteristics of isolated left ventricular non-compaction: a step towards classification as a distinct cardiomyopathy. Heart. 2001;86(6):666 LP–666671. doi: 10.1136/heart.86.6.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oechslin EN, Attenhofer Jost CH, Rojas JR, Kaufmann PA, Jenni R. Long-term follow-up of 34 adults with isolated left ventricular noncompaction: a distinct cardiomyopathy with poor prognosis. J Am Coll Cardiol. 2000;36(2):493–500. doi: 10.1016/S0735-1097(00)00755-5. [DOI] [PubMed] [Google Scholar]
- 4.Anderson RH, Jensen B, Mohun TJ, Petersen SE, Aung N, Zemrak F, et al. Key Questions Relating to Left Ventricular Noncompaction Cardiomyopathy: Is the Emperor Still Wearing Any Clothes? Can J Cardiol. 2017;33(6):747–757. doi: 10.1016/j.cjca.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 5.Ichida F. Left ventricular noncompaction. Circ J. 2009;73(1):19–26. doi: 10.1253/circj.CJ-08-0995. [DOI] [PubMed] [Google Scholar]
- 6.Chin TK, Perloff JK, Williams RG, Jue K, Mohrmann R. Isolated noncompaction of left ventricular myocardium. A study of eight cases. Circulation. 1990;82(2):507–513. doi: 10.1161/01.CIR.82.2.507. [DOI] [PubMed] [Google Scholar]
- 7.Stöllberger C, Gerecke B, Finsterer J, Engberding R. Refinement of echocardiographic criteria for left ventricular noncompaction. Int J Cardiol. 2013;165(3):463–467. doi: 10.1016/j.ijcard.2011.08.845. [DOI] [PubMed] [Google Scholar]
- 8.Gebhard C, Stähli BE, Greutmann M, Biaggi P, Jenni R, Tanner FC. Reduced left ventricular compacta thickness: a novel echocardiographic criterion for non-compaction cardiomyopathy. J Am Soc Echocardiogr. 2012;25(10):1050–1057. doi: 10.1016/j.echo.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Petersen SE, Selvanayagam JB, Wiesmann F, Robson MD, Francis JM, Anderson RH, et al. Left ventricular non-compaction: insights from cardiovascular magnetic resonance imaging. J Am Coll Cardiol. 2005;46(1):101–105. doi: 10.1016/j.jacc.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 10.Jacquier A, Thuny F, Jop B, Giorgi R, Cohen F, Gaubert JY, et al. Measurement of trabeculated left ventricular mass using cardiac magnetic resonance imaging in the diagnosis of left ventricular non-compaction. Eur Heart J. 2010;31(9):1098–1104. doi: 10.1093/eurheartj/ehp595. [DOI] [PubMed] [Google Scholar]
- 11.Grothoff M, Pachowsky M, Hoffmann J, Posch M, Klaassen S, Lehmkuhl L, et al. Value of cardiovascular MR in diagnosing left ventricular non-compaction cardiomyopathy and in discriminating between other cardiomyopathies. Eur Radiol. 2012;22(12):2699–2709. doi: 10.1007/s00330-012-2554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weir-McCall JR, Yeap PM, Papagiorcopulo C, Fitzgerald K, Gandy SJ, Lambert M, et al. Left ventricular noncompaction: anatomical phenotype or distinct cardiomyopathy? J Am Coll Cardiol. 2016;68(20):2157–2165. doi: 10.1016/j.jacc.2016.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zemrak F, Ahlman MA, Captur G, Mohiddin SA, Kawel-Boehm N, Prince MR, et al. The relationship of left ventricular trabeculation to ventricular function and structure over a 9.5-year follow-up: the MESA study. J Am Coll Cardiol. 2014;64(19):1971–1980. doi: 10.1016/j.jacc.2014.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oechslin E, Jenni R. Left Ventricular Noncompaction. J Am Coll Cardiol. 2018;71(7):723 LP–723726. doi: 10.1016/j.jacc.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 15.Cortés M, Oliva MR, Orejas M, Navas MA, Rábago RM, Martínez ME, et al. Usefulness of speckle myocardial imaging modalities for differential diagnosis of left ventricular non-compaction of the myocardium. Int J Cardiol. 2016;223:813–818. doi: 10.1016/j.ijcard.2016.08.278. [DOI] [PubMed] [Google Scholar]
- 16.Kalapos A, Domsik P, Forster T, Nemes A. Left ventricular strain reduction is not confined to the noncompacted segments in noncompaction cardiomyopathy-insights from the three-dimensional speckle tracking echocardiographic MAGYAR-path study. Echocardiography. 2014;31(5):638–643. doi: 10.1111/echo.12447. [DOI] [PubMed] [Google Scholar]
- 17.Peters F, Khandheria BK, Libhaber E, Maharaj N, Dos Santos C, Matioda H, et al. Left ventricular twist in left ventricular noncompaction. Eur Heart J Cardiovasc Imaging. 2014;15(1):48–55. doi: 10.1093/ehjci/jet076. [DOI] [PubMed] [Google Scholar]
- 18.Rudolecká J, Veiser T, Plášek J, Homza M, Fürstová J. Ventricular twist in isolated left ventricular noncompaction. Cor Vasa. 2014;56(6):e471–e477. doi: 10.1016/j.crvasa.2014.05.004. [DOI] [Google Scholar]
- 19.Pedrizzetti G, Claus P, Kilner PJ, Nagel E. Principles of cardiovascular magnetic resonance feature tracking and echocardiographic speckle tracking for informed clinical use. J Cardiovasc Magn Reson. 2016;18(1):1–12. doi: 10.1186/s12968-016-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor RJ, Moody WE, Umar F, Edwards NC, Taylor TJ, Stegemann B, et al. Myocardial strain measurement with feature-tracking cardiovascular magnetic resonance: Normal values. Eur Heart J Cardiovasc Imaging. 2015;16(8):871–881. doi: 10.1093/ehjci/jev006. [DOI] [PubMed] [Google Scholar]
- 21.Hanneman K, Nguyen ET, Thavendiranathan P, Ward R, Greiser A, Jolly M-P, et al. Quantification of myocardial extracellular volume fraction with cardiac MR imaging in thalassemia major. Radiology. 2016;279(3):720–730. doi: 10.1148/radiol.2015150341. [DOI] [PubMed] [Google Scholar]
- 22.Alfakih K, Plein S, Thiele H, Jones T, Ridgway JP, Sivananthan MU. Normal human left and right ventricular dimensions for MRI as assessed by turbo gradient echo and steady-state free precession imaging sequences. J Magn Reson Imaging. 2003;17(3):323–329. doi: 10.1002/jmri.10262. [DOI] [PubMed] [Google Scholar]
- 23.Vo HQ, Marwick TH, Negishi K. MRI-Derived Myocardial Strain Measures in Normal Subjects. JACC Cardiovasc Imag. 2018;11(2P1):196–205. doi: 10.1016/j.jcmg.2016.12.025. [DOI] [PubMed] [Google Scholar]
- 24.Barreiro-pérez M, Curione D, Symons R, Claus P, Voigt J, Bogaert J. Left ventricular global myocardial strain assessment comparing the reproducibility of four commercially available CMR-feature tracking algorithms. 2018. pp. 5137–5147. [DOI] [PubMed] [Google Scholar]
- 25.Schuster A, Stahnke VC, Unterberg-Buchwald C, Kowallick JT, Lamata P, Steinmetz M, et al. Cardiovascular magnetic resonance feature-tracking assessment of myocardial mechanics: Intervendor agreement and considerations regarding reproducibility. Clin Radiol. 2015;70(9):989–998. doi: 10.1016/j.crad.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai J, Bryant JA, Le T-T, Su B, de Marvao A, O’Regan DP, et al. Fractal analysis of left ventricular trabeculations is associated with impaired myocardial deformation in healthy Chinese. J Cardiovasc Magn Reson. 2017;19(1):102. doi: 10.1186/s12968-017-0413-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nucifora G, Sree Raman K, Muser D, Shah R, Perry R, Awang Ramli KA, et al. Cardiac magnetic resonance evaluation of left ventricular functional, morphological, and structural features in children and adolescents vs. young adults with isolated left ventricular non-compaction. Int J Cardiol. 2017;246:68–73. doi: 10.1016/j.ijcard.2017.05.100. [DOI] [PubMed] [Google Scholar]
- 28.Abduch MCD, Alencar AM, Mathias W, Jr, de Vieira ML C. Cardiac Mechanics Evaluated by Speckle Tracking Echocardiography. Arq Bras Cardiol. 2014;102:403–412. doi: 10.5935/abc.20140041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowallick JT, Lamata P, Hussain ST, Kutty S, Steinmetz M, Sohns JM, et al. Quantification of left ventricular torsion and diastolic recoil using cardiovascular magnetic resonance myocardial feature tracking. PLoS One. 2014;9(10):e109164. doi: 10.1371/journal.pone.0109164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arbustini E, Favalli V, Narula N, Serio A, Grasso M. Left ventricular noncompaction: a distinct genetic cardiomyopathy? J Am Coll Cardiol. 2016;68(9):949–966. doi: 10.1016/j.jacc.2016.05.096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in the study are available from the corresponding author on reasonable request.