Abstract
Background
Fabry disease is a rare X-linked inherited disorder caused by deficiency of α-Galactosidase A. Hundreds of mutations and non-coding haplotypes in the GLA gene have been described; however, many are variants of unknown significance, prompting doubts about the diagnosis and treatment. The α-Galactosidase A enzymatic activity in dried blood spot (DBS) samples are widely used for screening purposes; however, even when values below the normal are found, new tests are required to confirm the diagnosis. Here we describe an analysis of GLA variants and their correlation with DBS α-Galactosidase A enzymatic activity in a large Brazilian population with Fabry disease symptoms.
Results
We analyzed GLA variants by DNA sequencing of 803 male patients with suspected Fabry disease or belonging to high-risk populations; in 179 individuals, 58 different exonic variants were detected. From these, 50 are variants described as pathogenic and eight described as variants of unknown significance. The other individuals presented complex non-coding haplotypes or had no variants. Interestingly, the enzymatic activity in DBS was different among pathogenic variants and the other genotypes, including variants of unknown significance; the first presented mean of 12% of residual activity, while the others presented levels above 70% of the activity found in healthy controls.
Conclusion
The activity of α-Galactosidase A in DBS was markedly reduced in males with known pathogenic variants when compared with subjects presenting variants of unknown significance, non-coding haplotypes, or without variants, indicating a possible non-pathogenic potential of these latter genotypes. These findings bring a better understanding about the biochemical results of α-Galactosidase A in DBS samples, as well as the possible non-pathogenic potential of non-coding haplotypes and variants of unknown significance in GLA gene. These results certainly will help clinicians to decide about the treatment of patients carrying variants in the gene causing this rare but life-threatening disease.
Keywords: Fabry disease, GLA gene, Non-coding haplotypes, Molecular diagnosis, α-Galactosidase A activity, DBS enzymatic activity
Background
Fabry disease (FD - OMIM 301500) is a lysosomal storage disorder caused by pathogenic variants in the X-linked GLA gene (Xq22.1). GLA variants may produce α-Galactosidase A deficiency (α-Gal A; EC 3.2.1.22), which is required for degradation of glycosphingolipids. Deficiency in α-Gal A activity leads to storage of complex glycosphingolipids, mainly globotriaosylceramide (Gb3), inside of lysosomes in critical organs and tissues, impairing their functions and consequently resulting in a progressive multisystem disease, affecting people of all ethnic groups [1, 2].
FD presents a broad spectrum of heterogeneous clinical phenotypes, classified as classical and non-classical [3]. In the classical disease, the suspicion of FD begins with peculiar signs and symptoms such as angiokeratomas, acroparesthesia, abdominal pain, recurring headache, and progressive loss of renal function, cardiomyopathy and central nervous system microangiopathy. Non-classical phenotype frequently is associated with damage in a single organ system, principally kidney, heart and brain [4]. The clinical diagnosis of both phenotypes is challenging, since many of the main symptoms are common in other diseases [5]. Indeed, the time between the first symptoms and the diagnosis can take more than ten years.
The FD male phenotypes are directly linked to the residual α-Gal A activity. The exact threshold value of FD pathogenicity is unknown. However, it is estimated that the cutoff for diagnosing FD is 30–35% of mean normal α-Gal A. Some GLA mutations cause a reduction of enzyme activity to less than 10–15% of the wild type and are considered pathogenic [6]. However, others promoting a residual enzyme activity of at least 40% of the wild type protein can be considered as non-pathogenic [7].
The enzymatic activity measured in leukocytes or fibroblasts is considered a gold standard for the diagnosis of FD in male patients; however, the sample required for this analysis may be a limiting factor [8]. Thus, the analysis of α-Gal A activity in dried blood spot (DBS) samples has been shown to be a viable alternative, especially for screening in high-risk populations; however, confirmatory tests are required for the diagnosis [9].
The FD molecular analysis is important for family segregation studies, allowing the early diagnosis of family members with pathogenic mutations, enabling the monitoring before the first symptoms and therefore promoting a better management of the symptoms of the disease in these individuals. More than 960 mutations have been reported as causing FD disease in the Human Gene Mutation Database (HGMD) [10]; however, the pathogenicity of several exonic as well as, non-coding variants (NCV) is still controversial. We describe here an observational study based on biochemical analysis performed in DBS samples and genetic analysis in male individuals with suspicion of FD presenting characteristic clinical signs or belonging to high-risk populations, as patients with unexplained renal insufficiency, left ventricular hypertrophy or stroke without a known etiology. The results of the study show the profile of GLA variants in male Brazilian patients submitted to investigation of FD and the correlation between α-Gal A activity and genotype.
Methods
Patients and α-Gal A activity screening
This study included 803 male patients with suspicion of FD after clinical investigation, as well as individuals with symptoms reported as non-specific, observed in FD patients (high-risk populations). All patients were screened by α-Gal A enzymatic activity, determined by the hydrolysis of the substrate 4-methylumbelliferyl-α-D-galactopyranoside in DBS samples by a fluorometric assay as described by Muller and colleagues [11]. The cut-off value of enzymatic activity (compatible with FD diagnosis) used in this screening protocol was determined in healthy Brazilian volunteers [11] and a pilot screening protocol in DBS samples from Brazilian patients of hemodialysis centers [9].
DNA sequencing
DNA was extracted from the blood sample using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany), according to manufacturer’s instructions. Alternatively, FTA Classic Cards (Whatman™) were used to facilitate the collection; DNA extraction was performed using Chelex 100 resin (Bio-Rad Laboratories, Hercules, CA, USA) according to instructions. The amplification and sequencing of the GLA regions were performed according to Varela and colleagues [12]. We analyzed the seven exons, splicing sites and the regions flanking GLA exons. The introns in their entirety, as well as the promoter region, were not sequenced in this study.
Bioinformatic analysis
Data analysis was carried out using software Geneious® (Biomatters). Sequences were compared with the reference sequence (NCBI: NG_007119 (http://www.ncbi.nih.gov) and confirmed by sequencing the reverse strand. The variants were reviewed and annotated using dbSNP - Single-nucleotide polymorphism database and HGMD – The Human Gene Mutation Database [10]. Based on this analysis, the exonic mutations were divided into described pathogenic variants and variants of unknown significance (VUS). Mutations were correlated with the likely phenotype using the dbFGP - International Fabry Disease Genotype-Phenotype Database [13], previous publications and functional characterization. GnomAD - Genome Aggregation Database [14], 1000 Genomes Project Consortium [15] and ABraOM: Online Archive of Brazilian Mutations [16] were used to define the population frequency. The web-software Human Splicing finder [17] was used to identify significant splicing motif alterations. Non-coding variants were also analyzed by TRAP - Transcript-inferred pathogenicity score [18].
Non-coding variants analysis
Complex non-coding haplotypes (NCH) were assessed according to their frequency in The 1000 genomes database. Briefly, 2504 multisample (phased variant call format - vcf) of the X-chromosome were filtered to rule out variants with two alleles (female sample). The remaining files that comprise 1233 samples of healthy male individuals were used as the control group. A combinatorial analysis was performed to determine the haplotypes. Complex haplotypes found in the 1000 genomes were compared to the patients to determine their frequency.
Statistical analysis
Correlation between enzyme activity and GLA sequencing were analyzed by one-way analyses of variance (ANOVA) with Turkey as post-hoc, performed using IBM SPSS® software (version 18). The level of significance was set at p < 0.05.
Results
In this study, we analyzed patients with suspicion of FD with characteristic symptoms of the disease, as well as patients belonging to high-risk populations. Most of the patients presented renal disease and were screened in dialysis clinics (93%), while the other patients presented other symptoms that suggest FD. Details about these data are shown in Additional file 1: Table S1.
All individuals included were screened by α-Gal A enzymatic activity in DBS and presented low activity suggesting a possible FD diagnosis; however, other tests were requested to confirm. Of the total of males submitted to analysis, 783 were screened by enzymatic activity in DBS at the Laboratório de Erros Inatos do Metabolismo (LEIM – UNIFESP), and presented enzyme activity below the cutoff (2.2 μmol/L/h). Other 20 patients included were screened by other laboratories, and the results are reported as positive to FD. These 20 patients presented exonic alterations; therefore, they were included in this study; however, they were not included in the statistical analysis. We performed the GLA sequencing to confirm the diagnosis.
GLA sequencing revealed 179 patients (22.3%) with mutations in the coding regions (exons), 335 patients had no variants in the analyzed regions (41.7%), and 289 patients (36%) had only NCV. We found 58 previously described variants in the GLA exons; 98 patients (12.2%) presented 50 pathogenic mutations and 81 patients (10%) presented eight VUS. The most frequent VUS was D313Y found in 38 index cases, followed by R118C found in 30 individuals. The most frequent pathogenic mutations were R356W and M290I, found in 17 and 10 patients, respectively. A list of described variants, the enzymatic activity, functional tests, and likely phenotype is shown in Table 1.
Table 1.
Described mutations in the GLA gene found in suspected FD patients
| Aminoacid change (NM_000169.2) | Nucleotide change | Exon Location | Stop Codon Position | Type of alteration | Nr. of families | Enzymatic activity (μmol/L/h) | dbSNP | HGMD | Functional Characterization (%WT) | Likely Phenotype | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Range | ||||||||||
| Pathogenic Mutations | |||||||||||
| p.Q2* | c.4C>T | 1 | 2 | Nonsense | 1 | 0.12 | rs869312313 | DM | No FC | Classical (dbFGP) | |
| p.Gly11AlafsTer110 | c.32delG | 1 | 110 | Deletion | 2 | 0.09 | 0 - 0.19 | rs1057516967 | DM | 1.8±1.4 [19] | Classical (dbFGP; HGMD) |
| p.A15E | c.44C>A | 1 | Missense | 1 | 0.06 | rs869312304 | DM | 0±0 [42] | Classical (dbFGP) | ||
| p.A15G | c.44C>G | 1 | Missense | 1 | 0.14 | No dbSNP ID | DM | 19±0.7 [36] | Classical (dbFGP) | ||
| p.G35V | c.104G>A | 1 | Missense | 1 | 0.15 | No dbSNP ID | DM | No FC | Classical (HGMD) | ||
| p.M42I | c.126G>A | 1 | Missense | 1 | 0.17 | No dbSNP ID | DM | No FC | Likely Classical (dbFGP) / Classical (HGMD) | ||
| p.W47* | c.140G>A | 1 | 47 | Nonsense | 1 | + | No dbSNP ID | DM | No FC | Classical (dbFGP) | |
| p.R49C | c.145C>T | 1 | Missense | 1 | 0.31 | No dbSNP ID | DM | 0±0 [19] | Classical (dbFGP; HGMD) | ||
| p.R49G | c.145C>G | 1 | Missense | 1 | 0 | No dbSNP ID | DM | 0±0 [19] | Classical (dbFGP) | ||
| p.R49P | c.146G>C | 1 | Missense | 1 | 0.25 | rs398123205 | DM | No FC | Classical (dbFGP; HGMD) | ||
| p.C52* | c.156C>A | 1 | 52 | Nonsense | 1 | 0.41 | No dbSNP ID | DM | No FC | Classical (dbFGP; HGMD) | |
| p.F69L | c.207C>A | 2 | Missense | 1 | + | No dbSNP ID | DM | No FC | Mild/Late-onset - cardiac variant (dbFGP; HGMD) | ||
| p.W81* | c.242G>A | 2 | 81 | Nonsense | 1 | + | rs398123208 | DM | No FC | Classical (dbFGP; HGMD) | |
| p.C94Y | c.281G>A | 2 | Missense | 2 | 0 | rs113173389 | DM | 0±0 [19] | Classical (dbFGP; HGMD) | ||
| p.R100K | c.299G>A | 2 | Missense | 1 | + | rs869312273 | DM | 0±0 [20] | Classical (dbFGP; HGMD) | ||
| p.R112C | c.334C>T | 2 | Missense | 5 | 0.25 | 0 - 1.25 | rs104894834 | DM | 0±0 [20, 42] | Classical (dbFGP; HGMD) | |
| p.R112H | c.335G>A | 2 | Missense | 3 | 0.17 | 0.03 - 0.28 | rs869312273 | DM | 0±0 [20]; 1.6±0.6 [42] | Mild proteinuria -Later-onset (dbFGP; HGMD) | |
| p.F113L | c.337T>C | 2 | Missense | 1 | 0.27 | rs869312142 | DM | 17.3±3.6 [20]; 18.3±0.8 [36] | Later onset (dbFGP); Cardiac variant (HGMD) | ||
| p.G132E | c.395G>A | 3 | Missense | 1 | 0.01 | No dbSNP ID | DM | 0±0 [42] | Classical (dbFGP) | ||
| p.C142R | c.424T>C | 3 | Missense | 2 | 0 | 0 - 0.01 | No dbSNP ID | DM | 0±0 [20, 42] | Classical (dbFGP; HGMD) | |
| p.A156D | c.467C>A | 3 | Missense | 2 | 0.16 | 0 - 0.33 | rs869312307 | DM | 0±0 [42] | Classical (dbFGP) | |
| p.C172Y | c.515G>A | 3 | Missense | 1 | 0.03 | rs869312318 | DM | 0±0 [42] | Classical (dbFGP) | ||
| p.M187T | c.560T>C | 4 | Missense | 1 | + | rs869312342 | DM | 0±0 [42] | Classical (dbFGP; HGMD) | ||
| p.C202Y | c.605G>A | 4 | Missense | 1 | + | rs869312344 | DM | 0±0 [43] | Classical (dbFGP; HGMD) | ||
| p.I198T | c.593T>C | 4 | Missense | 1 | 0.15 | rs727503950 | DM | 38.7±3.1 [43] | Later onset (dbFGP) | ||
| p.W204* | c.611G>A | 4 | 204 | Nonsense | 1 | 0.7 | rs869312346 | DM | No FC | Classical (dbFGP) | |
| p.N215S | c.644A>G | 5 | Missense | 2 | 0.34 | 0.22 - 0.47 | rs28935197 | DM | 15.6±1.0 [36] /15.7±2.4 [20] / 39.5±1.5 [43] | Later onset (dbFGP) | |
| p.R220* | c.658C>T | 5 | 220 | Nonsense | 1 | 0.06 | rs727503949 | DM | 0±0 [43] | Classical (dbFGP) | |
| p.W226* | c.677G>A | 5 | 226 | Nonsense | 1 | 0.48 | rs398123219 | DM | No FC | Classical (dbFGP) | |
| p.R227* | c.679C>T | 5 | 227 | Nonsense | 3 | 0.08 | 0.06 - 0.11 | rs104894841 | DM | No FC | Classical (dbFGP; HGMD) |
| p.R227Q | c.680G>A | 5 | Missense | 1 | 0 | rs104894840 | DM | 0±0 [20, 42] | Classical (dbFGP; HGMD) | ||
| p.Lys240Glufs*9 | c.718_719delAA | 5 | 248 | Deletion | 1 | 0.01 | No dbSNP ID | DM | No FC | Classical (dbFGP) | |
| p.M267I | c.801G>A | 5 | Missense | 1 | 0.17 | rs869312408 | DM | No FC | Classical (dbFGP; HGMD) | ||
| p.V269M | c.805G>A | 6 | Missense | 1 | + | rs869312427 | DM | 0±0 [43] | Classical dbFGP; HGMD) | ||
| p.V269A | c.806T>C | 6 | Missense | 1 | 0.41 | rs28935488 | DM | 9.0±1.4 [42] | Classical (dbFGP; HGMD) | ||
| p.T282I | c.845C>T | 6 | Missense | 1 | 0.05 | No dbSNP ID | DM | 5.0±0.5 [42] / 5.2±0.2 [36] | Classical (dbFGP) | ||
| p.M290I | c.870G>A | 6 | Missense | 10 | 0.31 | 0 - 0.60 | rs869312438 | DM | 39±1.8 [42] | Later onset (dbFGP) - Classical (HGMD) | |
| p.A292V | c.875C>T | 6 | Missense | 1 | 0.54 | No dbSNP ID | DM | No FC | Classical (dbFGP) | ||
| p.P293S | c.877C>T | 6 | Missense | 1 | 0.10 | rs869312440 | DM | No FC | Classical (dbFGP) | ||
| p.R301G | c.901C>G | 6 | Missense | 1 | 0,34 | rs398123224 | DM | 19±4.1 [42] | VUS (dbFGP) | ||
| p.R301* | c.901C>T | 6 | 301 | Nonsense | 2 | 0.06 | 0.01 – 0.11 | rs398123224 | DM | No FC | Classical (dbFGP) - Kidney disease (HGMD) |
| p.Gln333Glufs*14 | c.996_999delACAG | 6 | 346 | Deletion | 2 | 1.84 | rs398123229 | DM | No FC | Classical (dbFGP; HGMD) | |
| p.R342Q | c.1025G>A | 7 | Missense | 4 | 0.07 | 0 -0.14 | rs28935493 | DM | 0±0 [20, 42] | Classical (dbFGP) | |
| p.Ser345Argfs*29 | c.1033_1034delTC | 7 | 373 | Deletion | 2 | 0.21 | rs398123198 | DM | No FC | Classical (dbFGP) | |
| p.W349* | c.1046G>A | 7 | 349 | Nonsense | 1 | 0,45 | No dbSNP ID | DM | No FC | Classical (dbFGP) | |
| p.R356W | c.1066C>T | 7 | Missense | 17 | 0.47 | 0.19 - 1.62 | rs104894827 | DM | 16.9±2.3 [42] | Later onset (dbFGP) | |
| p.R363H | c.1088G>A | 7 | Missense | 3 | 0.43 | 0.13 - 0.72 | rs111422676 | DM | 31.9±2.9 [42] | Later onset (dbFGP) - Renal presentation (HGMD) | |
| p.Y365* | c.1095T>A | 7 | 365 | Nonsense | 3 | 0.06 | 0 - 0.13 | rs104894849 | DM | No FC | Classical (dbFGP) |
| p.W399* | c.1196G>A | 7 | 399 | Nonsense | 1 | + | No dbSNP ID | DM | 2% [21] | Classical (dbFGP; HGMD) | |
| p.Thr412Serfs*? | c.1235_1236delCT | 7 | late termination codon | Deletion | 1 | 0 | rs797044777 | DM | No FC | Classical (dbFGP; HGMD) | |
| Variants of Unknown Significance | |||||||||||
| p.E66Q | c.196G>C | 2 | Missense | 1 | + | rs104894833 | DFP | 52.0±1.3 [36] | Benign (dbFGP) | ||
| p.R118C | c.352C>T | 2 | Missense | 30 | 1.68 | 1.15 - 2.10 | rs148158093 | DM | 24.0±1.3 [36] | Benign (dbFGP); Cardiac variant (HGMD) | |
| p.A143T | c.427G>A | 3 | Missense | 8 | 1.04 | 0.58 - 1.86 | rs104894845 | DM | 31.3±5.6 [42] | Benign (dbFGP) | |
| p.R220Q | c.659G>A | 5 | Missense | 1 | 1.17 | rs727503949 | DM? | 104±11.3 [42] | Likely bening (dbFGP) | ||
| p.N228S | c.683A>G | 5 | Missense | 1 | 2.03 | rs869312152 | DM | 59.5±9.8 [43] / 124.5±4.1 [36] | Likely benign (dbFGP) | ||
| p.D313Y | c.937G>T | 6 | Missense | 38 | 1.65 | 0.72 - 2.10 | rs28935490 | DM? | 83.9±21.1 [42] | Benign (dbFGP) | |
| p.R356Q | c.1067G>A | 7 | Missense | 1 | 0.9 | rs869312163 | DM | 89.1±5.0 [42] / 36.1±1.5 [36] | Later onset (dbFGP) | ||
| p.A368T | c.1102G>A | 7 | Missense | 1 | 1.69 | 1.69 | rs144994244 | DM? | 103.7±33.6 [42] | Benign (dbFGP) | |
F Female, M Male, No FC No functional characterization, NA Not Applicable, VUS Variant of Unknown Significance, DM Disease Causing Mutation, DM? Disease Causing Mutation?, DFP disease-associated polymorphism with supporting functional evidence, dbFGP (International Fabry Disease Genotype-Phenotype Database http://dbfgp.org/dbFgp/fabry/); DBS enzymatic activity applicable only for male: for samples screened by LEIM-UNIFESP the result is show as μmol/L/h; for samples screened by other laboratories, the result is show as + (positive)
Non-coding variants
Two hundred and eighty nine patients presented only NCV in the GLA. A list of all NCV, the population frequency and in silico predictor is shown in Additional file 2: Table S2. Seven NCV form nine NCH. To analyze the frequency of NCH, we used data from The 1000 Genomes (only male) as the control group. Except for the haplotype c.-10C > T / c.370-77_370-81delCAGCC / c.640-16A > G / c.802-67G > A / c.1000-22C > T, found only in one patient of this study, all other haplotypes were also found in the control group. The results are shown in Table 2.
Table 2.
Complex non-coding haplotypes found in male patients with suspicion of FD and population frequency in The 1000 Genomes Project
| DNA Region | Variant / Haplotype | Enzyme Activity (μmol/L/h) | Patients | The 1000 Genomes | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | Range | N = 624 | % | N = 1233 | % | |||
| Without GLA variants | Without intronic variants | 1.76 | 0.09–2.2 | 335 | 53.7 | 705 | 57.1 | |
| Variant 1 | 5’UTR | c.-30G > A | 1.60 | – | 1 | 0.16 | 2 | 0.16 |
| Variant 2 | 5’UTR | c.-10C > T | 1.72 | 1.13–2.10 | 6 | 0.74 | 1 | 0.08 |
| Variant 3 | Intron 1 | c.194 + 17A > G | 2.11 | – | 1 | 0.16 | 28 | 2.27 |
| Variant 4 | Intron 6 | c.1000-22C > T | 1.76 | 0.60–2.19 | 58 | 9.29 | 147 | 11,92 |
| Haplotype 1 | 5’UTR / intron 6 | c.-12G > A / c.1000-22C > T | 1.11 | 0.80–1.42 | 2 | 0.32 | 1 | 0.08 |
| Haplotype 2 | 5’UTR / intron4 / intron 6 | c.-12G > A / c.639 + 68 A > G / c.1000-22C > T | 1.64 | 0.90–2.20 | 23 | 3.68 | 127 | 10.3 |
| Haplotype 3 | 5’UTR / intron 2 / intron 4 | c.-12G > A / c.370-77_370-81delCAGCC / c.640-16A > G | – | – | 0 | 0 | 1 | 0.08 |
| Haplotype 4 | 5’UTR / intron 2 | c.-10C > T / c.370-77_370-81delCAGCC | – | – | 0 | 0 | 1 | 0.08 |
| Haplotype 5 | 5’UTR / intron 6 | c.-10C > T / c.1000-22C > T | 1.81 | 0.17–2.20 | 59 | 9.45 | 31 | 2.51 |
| Haplotype 6 | 5’UTR / intron 4 / intron 6 | c.-10C > T / c.640-16A > G / c.1000-22C > T | 2.07 | – | 1 | 0.16 | 4 | 0.32 |
| Haplotype 7 | 5’UTR / intron 2 / intron 4 / intron 6 | c.-10C > T / c.370-77_370-81delCAGCC / c.640-16A > G / c.1000-22C > T | 1.72 | 0.70–2.20 | 107 | 17.14 | 125 | 10.14 |
| Haplotype 8 | 5’UTR / intron 2 / intron 4 / intron 5 / intron 6 | c.-10C > T / c.370-77_370-81delCAGCC / c.640-16A > G / c.802-67G > A / c.1000-22C > T | 2.07 | – | 1 | 0.16 | 0 | 0 |
| Haplotype 9 | intron 2 / intron 4 / intron 6 | c.370-77_370-81delCAGCC / c.640-16A > G / c.1000-22C > T | 1.68 | 0.77–2.20 | 30 | 4.8 | 60 | 4.87 |
The most frequent haplotype is formed by the four variants c.-10C > T, c.370-77_370-81delCAGCC, c.640-16A > G and c.1000-22C > T. It was found in 107 (17.1%) patients and 125 individuals (10.1%) in the control group. The haplotype formed by c.-10C > T and c.1000-22C > T occurred with a frequency almost fourfold higher in patients with suspected FD than in the control group. The other haplotypes present similar frequency in patients and controls.
Enzymatic profile
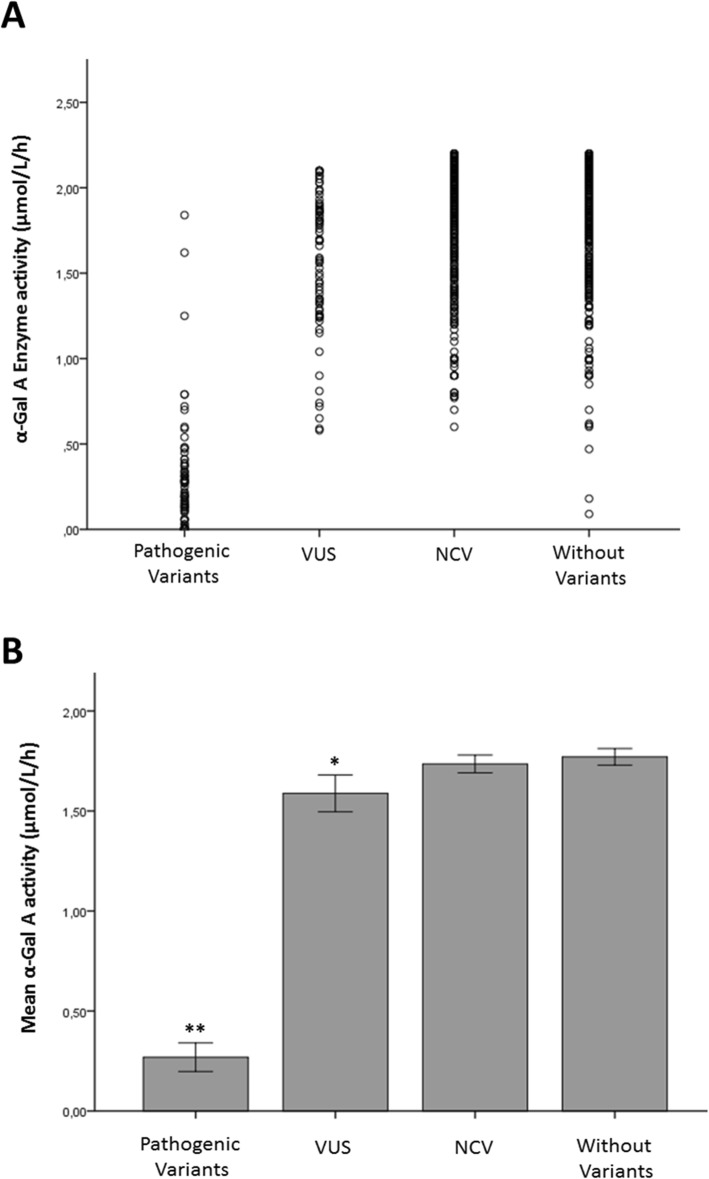
The correlation between the GLA variants and the α-Gal A activity levels were evaluated to estimate the impact of the variants in the enzyme in male patients screened by enzymatic activity in LEIM and presenting less than 2.2 μmol/L/h (N = 783). The patients were divided into groups according to the classification of their mutations. Figure 1a shows the distribution of enzymatic activity per patient in each group.
Fig. 1.
Enzymatic profile of GLA genotypes. (a) Scatter plot of the α-Gal A activity distribution in males with FD suspicion in different groups. The figure shows that most males with VUS, NCV and without variants present α-Gal A levels above 1 μmol/L/h, while patients with pathogenic variants presented α-Gal A levels lower than 1 μmol/L/h. Some outliers were found in each group. Three patients with pathogenic variants presented enzyme activity above 1 μmol/L/h, while twenty-four patients with only non-coding variants, twenty without variants and seven with VUS, being four with A143T, two with D313Y and one with R356Q, presented enzyme activity below 1 μmol/L/h. (b) Correlation analysis between α-Gal A level in DBS and GLA genotypes. The graphic shows the mean enzymatic activity detected in males in all the GLA variant groups. The data are expressed as mean ± S.E.M. **P < 0.001 known pathogenic mutation (0.27 μmol/L/h ± 0.03, N = 83) versus VUS (1.58 μmol/L/h ± 0.04, N = 76), non-coding variants (1.73 μmol/L/h ± 0.02, N = 289) and the group without variants in GLA (1.77 μmol/L/h ± 0.02, N = 335); *P = 0.013 VUS versus NCV and *P = 0.01 VUS versus patients without variants
Males with variants previously described as pathogenic had significantly lower enzymatic activity when compared with the other groups (mean 0.27 μmol/L/h, p < 0.001). The VUS showed a significant decrease of residual α-Gal A levels (mean 1.58 μmol/L/h) when compared with the group without variants (mean 1.73 μmol/L/h; p = 0.001) and the NCV group (mean 1.77 μmol/L/h; p = 0.013). There was no difference in α-Gal A levels between the group without mutation in GLA and the group with NCV (p = 0.64). The results are shown in Fig. 1b.
Discussion
GLA sequencing allows the identification of genetic mutations associated with FD and the detection of these variants is fundamental to support the diagnosis. Main FD symptoms are shared with other diseases, making the diagnosis based on such symptoms challenging. The FD clinical suspicion starts with characteristic signs and symptoms that appear over the years, promoting a delay of at least 10 years to diagnose a patient [5]. Therefore, in the last two decades, the number of screening studies in high-risk and in newborn population has increased.
The α-Gal A activity in DBS has been used for screening purposes and should be followed by enzymatic activity in leukocytes or DNA sequencing to confirm the diagnosis [9]. The efficacy of the applicability of enzymatic activity in DBS as an alternative screening test has been reported [8, 22–24]. Fuller and colleagues [25] tested DBS enzyme activity assay in FD hemizygous patients and found a clearly decrease in α-Gal A activity when compared with a control population. A comparison between the enzymatic activity assay in DBS versus leukocytes, conducted in male patients with known FD demonstrated that both assays were equally good [26]. Here, we analyzed by DNA sequencing 803 male individuals with low enzymatic activity in DBS. All patients presented suspicion of FD after clinical investigation or showed undefined symptoms as those observed in FD patients. However, a limitation of this study was the lack of detailed information on the patient’s clinic.
According to Van der Tol and coworkers [27], the prevalence of GLA variants in a high-risk population is 0.12%, when considered only pathogenic variants; when VUS are included, this frequency increases to 0.62%. FD is screened in dialysis centers as one possible cause of end-stage renal disease. Not surprisingly, nephrologists referred most patients included in this study, and they were predominantly followed in dialysis services. We performed the DNA sequencing only in individuals with low enzymatic activity screened by DBS assay. Interestingly, we found a high frequency of variants in our patients: 22.2% of individuals with enzymatic activity lower than 2.2 μmol/L/h presented GLA variants. Of these, 12.2% present pathogenic variants and 10% VUS. The numbers showed here do not reflect the Van der Tol data, which could be due to the fact that here we included only patients with low activity and not those with activity within the normal range.
In addition to exonic mutations, NCV were also detected. The comparison between patients and controls showed that seven NCV were observed in more than 1% of the control population, being considered as polymorphisms. Other two variants were extremely rare or were not found in any databank consulted. Despite rare, in silico pathogenic analysis did not consider any of the NCV found in this study as damaging.
Nine different NCH were found. Seven of them presented similar frequency in patients and control group. Our results are in agreement with the findings of Ferri and colleagues [28], who found seven different GLA haplotypes in control males, indicating that these NCH, per se, are not involved in the development of FD manifestations. However, the haplotypes 5 and 7 present higher frequency in patients when compared with controls. The haplotype 7 was already described in patients with FD suspicion [29, 30]. Both haplotypes contain the variant c.-10C > T, described as causing a decrease of approximately 25% of the α-Gal A activity [31]. As described by Oliveira and colleagues [31], we also found approximately 4-fold higher frequency of this variant in our patients when compared to general population. In our study, these haplotypes were found in males with enzymatic activity below the cut-off (~ 1.73 μmol/L/h), equivalent to a decrease of 21% of α-Gal A activity, indicating that c.-10C > T may cause this decrease. Residual enzyme activity of about 40% of the mean normal level can be considered enough to degrade the substrate, not promoting Gb3 accumulation [6, 7]. However, recent studies have demonstrated that, despite not altering the enzyme structure, patients with the haplotype 7 had significant levels of Gb3 accumulation when compared with controls [32, 33]. Gervas-Arruga and colleagues [32] suggest that in patients with this NCH, environmental factors, as a pro-inflammatory state, in addition to the accumulation of Gb3 may influence the symptoms.
An important finding of this study was the different levels of residual activity in DBS samples among the genotypes. By comparing the mean enzymatic activity, we have observed that described pathogenic variants showed significantly lower mean enzymatic activity, equivalent to 12% of the value found in healthy individuals. On the other hand, VUS including D313Y, R118C, and A143T, considered by many researchers as not causing FD [34–39] and by others as pathogenic [40, 41], presented higher enzymatic activity in comparison to individuals with pathogenic mutations. In contrast, patients carrying VUS present enzymatic levels statistically lower when compared to patients with NCV or patients without mutations in GLA. In fact, in in vitro experiments, the VUS found in this study showed a decreased α-Gal A activity, exception for R220Q and A368T, which have α-Gal A activity similar to that of the wild type [36, 42]. However, data from different groups showed that this decrease is not sufficient to promote glycosphingolipid accumulation, which would lead to disease [37, 42]. Although enzymatic activity of VUS were statistically different from NCH and patients without variants, these genotypes present values higher than 70% of the residual activity found in health population. It is already described that activity above 40% of the level found in health population is enough to degrade Gb3, therefore our results indicate that these genotypes are not compatible with FD. However, further studies are necessary to rule out FD in these patients.
In summary, in this study we sequenced a large group of male patients with suspicion of FD presenting enzymatic activity below the cutoff (2.2 μmol/L/h) and showed that pathogenic variants lead to a low residual enzymatic activity, while VUS, NCV and patients without GLA variants, lead to approximately 70% of the normal activity, indicating a possible non-pathogenicity. In addition, we showed, by bioinformatics correlation, that the frequency of most haplotypes formed by non-coding variants in the healthy population is similar to the frequency found in patients with suspicion of FD, and therefore, the haplotypes per se, do not correlate with FD. However, in the haplotypes most frequently observed in patients group, although presenting high levels of residual activity when compared with the pathogenic variants, other studies are necessary to discard the FD diagnosis. Moreover, the correlation between DBS enzyme activity and GLA variants revealed that this screening method is useful for diagnosing previously described mutations. However, when the patient presents VUS or NCH, although our study indicates a possible non-pathogenicity, the diagnosis may not be conclusive and other tools may be necessary to confirm or discard the disease. Indeed, new specific studies are necessary to correlate these genotypes with FD.
Conclusions
In this observational study, we identified 98 patients with described pathogenic variants in GLA gene, confirming FD diagnosis. In these patients, the enzymatic activity in DBS samples was below 0.3 μmol/L/h, equivalent to 12% of the residual activity of healthy individuals; significantly lower when compared with the other genotypes. On the other hand, 80 patients presented only VUS, and in these cases FD diagnosis was not confirmed, as well as in patients with NCV. Our study indicates a possible non-pathogenic potential of these latter genotypes by population frequency of haplotypes and correlation between the enzymatic phenotype in DBS samples and GLA variants. These findings highlight the importance of determining α-Gal A activity by DBS in the diagnosis of FD, considered as the only available tool for this purpose in many countries.
Supplementary information
Additional file 1: Table S1. Origin of samples for FD screening according to the main symptoms
Additional file 2: Table S2. Non-coding variants found in GLA gene in patients with suspicion of FD and the population frequency in 1000 genomes, GenomAD and ABraOM. Human Splicing Finder and TRAP were used to analyze potential pathogenicity.
Acknowledgements
We would like to thank all the patients and the physicians for providing the samples analyzed in this work and Julien Calais for reviewing the English idiom.
Disclosures
The authors report no disclosures.
Abbreviations
- ABraOM
Online Archive of Brazilian Mutations
- dbFGP
International Fabry Disease Genotype – Phenotype Database
- DBS
Dried Blood Spot
- FD
Fabry Disease
- Gb3
Globotriaosylceramide
- GnomAD
Genome Aggregation Database
- HGMD
Human Gene Mutation Database
- NCH
Non-coding haplotypes
- NCV
Non-coding variants
- TRAP
Transcript-inferred pathogenicity score
- VCF
Variant call format
- VUS
Variants of unknown significance
- α-GalA
α-Galalactosidase A
Authors’ contributions
PV conceptualized and designed the study, designed the data collection experiments, drafted the initial manuscript, carried out the analyses, reviewed and revised the manuscript and approved the final manuscript as submitted. JBP conceptualized and designed the study, drafted the initial manuscript, carried out the initial analyses, reviewed and revised the manuscript and approved the final manuscript as submitted. JBP is corresponding author and guarantor for the article. GMK, MCB, FLM, LTT, RPM, VD and AMM carried out the initial analyses, reviewed and revised the manuscript and approved the final manuscript as submitted. CPG, HLFF, PN, MMM and JGP technical support, reviewed and revised the manuscript, and approved the final manuscript as submitted.
Funding
This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2014/27198–8 and 2018/23367–0) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), finance code 001.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Informed consent was obtained from all individuals included in the study. The Research Ethics Committee of the Federal University of São Paulo, Brazil approved this protocol (0585/07 and 0354/18). All procedures were followed in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2013.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest and confirm independence from the sponsors. The sponsors have not influenced the content of the article.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13023-019-1274-3.
References
- 1.Auray-Blais Christiane, Ntwari Aimé, Clarke Joe T.R., Warnock David G., Oliveira João Paulo, Young Sarah P., Millington David S., Bichet Daniel G., Sirrs Sandra, West Michael L., Casey Robin, Hwu Wuh-Liang, Keutzer Joan M., Zhang X. Kate, Gagnon René. How well does urinary lyso-Gb3 function as a biomarker in Fabry disease? Clinica Chimica Acta. 2010;411(23-24):1906–1914. doi: 10.1016/j.cca.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 2.Boutin Michel, Gagnon René, Lavoie Pamela, Auray-Blais Christiane. LC–MS/MS analysis of plasma lyso-Gb3 in Fabry disease. Clinica Chimica Acta. 2012;414:273–280. doi: 10.1016/j.cca.2012.09.026. [DOI] [PubMed] [Google Scholar]
- 3.El-Abassi Rima, Singhal Divya, England John D. Fabry's disease. Journal of the Neurological Sciences. 2014;344(1-2):5–19. doi: 10.1016/j.jns.2014.06.029. [DOI] [PubMed] [Google Scholar]
- 4.Germain DP. Fabry disease. Orphanet J Rare Dis. 2010;22:5–30. doi: 10.1186/1750-1172-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terryn Wim, Cochat Pierre, Froissart Roseline, Ortiz Alberto, Pirson Yves, Poppe Bruce, Serra Andreas, Van Biesen Wim, Vanholder Raymond, Wanner Christoph. Fabry nephropathy: indications for screening and guidance for diagnosis and treatment by the European Renal Best Practice. Nephrology Dialysis Transplantation. 2012;28(3):505–517. doi: 10.1093/ndt/gfs526. [DOI] [PubMed] [Google Scholar]
- 6.Oliveira JP, Ferreira S. Multiple phenotypic domains of Fabry disease and their relevance for establishing genotype– phenotype correlations. Appl Clin Genet. 2019. https://doi.org/. 10.2147/TACG.S146022. [DOI] [PMC free article] [PubMed]
- 7.Schiffmann Raphael, Fuller Maria, Clarke Lorne A., Aerts Johannes M.F.G. Is it Fabry disease? Genetics in Medicine. 2016;18(12):1181–1185. doi: 10.1038/gim.2016.55. [DOI] [PubMed] [Google Scholar]
- 8.Caudron E, Prognon P, Germain DP. Enzymatic diagnosis of Fabry disease using a fluorometric assay on dried blood spots: An alternative methodology Eur J Med Genet 2015. https://doi: 10.1016/ej.ejmg.2015.10.014. [DOI] [PubMed]
- 9.Curiati Marco A., Aranda Carolina S., Kyosen Sandra O., Varela Patricia, Pereira Vanessa G., D’Almeida Vania, Pesquero João B., Martins Ana M. The Challenge of Diagnosis and Indication for Treatment in Fabry Disease. Journal of Inborn Errors of Metabolism and Screening. 2017;5:232640981668573. [Google Scholar]
- 10.Stenson Peter D, Mort Matthew, Ball Edward V, Howells Katy, Phillips Andrew D, Thomas Nick ST, Cooper David N. The Human Gene Mutation Database: 2008 update. Genome Medicine. 2009;1(1):13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller Karen B, Rodrigues Mayra DB, Pereira Vanessa G, Martins Ana M, D'Almeida Vânia. Reference values for lysosomal enzymes activities using dried blood spots samples - a Brazilian experience. Diagnostic Pathology. 2010;5(1):65. doi: 10.1186/1746-1596-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varela P, Mastroianni Kirsztajn, G, Ferrer H, Aranda C, Wallbach K, Mata GF et al. Functional characterization and pharmacological evaluation of a novel GLA missense mutation found in a severely affected Fabry disease family. Nephron. 2019. https://doi.org/10.1159/000503998. [DOI] [PubMed]
- 13.Desnick Robert J, Chen Rong, Srinivasan Ram, Doheny Dana O., Bishop DavidF. The Fabry disease genotype-phenotype database (dbFGP): an international expert consortium. Molecular Genetics and Metabolism. 2017;120(1-2):S41–S42. [Google Scholar]
- 14.Lek Monkol, Karczewski Konrad J., Minikel Eric V., Samocha Kaitlin E., Banks Eric, Fennell Timothy, O’Donnell-Luria Anne H., Ware James S., Hill Andrew J., Cummings Beryl B., Tukiainen Taru, Birnbaum Daniel P., Kosmicki Jack A., Duncan Laramie E., Estrada Karol, Zhao Fengmei, Zou James, Pierce-Hoffman Emma, Berghout Joanne, Cooper David N., Deflaux Nicole, DePristo Mark, Do Ron, Flannick Jason, Fromer Menachem, Gauthier Laura, Goldstein Jackie, Gupta Namrata, Howrigan Daniel, Kiezun Adam, Kurki Mitja I., Moonshine Ami Levy, Natarajan Pradeep, Orozco Lorena, Peloso Gina M., Poplin Ryan, Rivas Manuel A., Ruano-Rubio Valentin, Rose Samuel A., Ruderfer Douglas M., Shakir Khalid, Stenson Peter D., Stevens Christine, Thomas Brett P., Tiao Grace, Tusie-Luna Maria T., Weisburd Ben, Won Hong-Hee, Yu Dongmei, Altshuler David M., Ardissino Diego, Boehnke Michael, Danesh John, Donnelly Stacey, Elosua Roberto, Florez Jose C., Gabriel Stacey B., Getz Gad, Glatt Stephen J., Hultman Christina M., Kathiresan Sekar, Laakso Markku, McCarroll Steven, McCarthy Mark I., McGovern Dermot, McPherson Ruth, Neale Benjamin M., Palotie Aarno, Purcell Shaun M., Saleheen Danish, Scharf Jeremiah M., Sklar Pamela, Sullivan Patrick F., Tuomilehto Jaakko, Tsuang Ming T., Watkins Hugh C., Wilson James G., Daly Mark J., MacArthur Daniel G. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nat. 2015. https://doi.org/10.1038/nature15393. [DOI] [PMC free article] [PubMed]
- 16.Naslavsky Michel Satya, Yamamoto Guilherme Lopes, de Almeida Tatiana Ferreira, Ezquina Suzana A. M., Sunaga Daniele Yumi, Pho Nam, Bozoklian Daniel, Sandberg Tatiana Orli Milkewitz, Brito Luciano Abreu, Lazar Monize, Bernardo Danilo Vicensotto, Amaro Edson, Duarte Yeda A. O., Lebrão Maria Lúcia, Passos-Bueno Maria Rita, Zatz Mayana. Exomic variants of an elderly cohort of Brazilians in the ABraOM database. Human Mutation. 2017;38(7):751–763. doi: 10.1002/humu.23220. [DOI] [PubMed] [Google Scholar]
- 17.Desmet François-Olivier, Hamroun Dalil, Lalande Marine, Collod-Béroud Gwenaëlle, Claustres Mireille, Béroud Christophe. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Research. 2009;37(9):e67–e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelfman S, Wang Q, McSweeney KM, Ren Z, La Carpia F, Halvorsen M, Schoch K, Ratzon F, Heinzen EL, Boland MJ, Petrovski S, Goldstein DB. Annotating pathogenic non-coding variants in genic regions. Nat Commun 2017. https://doi.org/10.1038/s41467-017-00141-2. [DOI] [PMC free article] [PubMed]
- 19.Shin Sang-Hoon, Murray Gary J., Kluepfel-Stahl Stefanie, Cooney Adele M., Quirk Jane M., Schiffmann Raphael, Brady Roscoe O., Kaneski Christine R. Screening for pharmacological chaperones in Fabry disease. Biochemical and Biophysical Research Communications. 2007;359(1):168–173. doi: 10.1016/j.bbrc.2007.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Xiaoyang, Katz Evan, Valle Maria Cecilia Della, Mascioli Kirsten, Flanagan John J., Castelli Jeffrey P., Schiffmann Raphael, Boudes Pol, Lockhart David J., Valenzano Kenneth J., Benjamin Elfrida R. A pharmacogenetic approach to identify mutant forms of α-galactosidase a that respond to a pharmacological chaperone for Fabry disease. Human Mutation. 2011;32(8):965–977. doi: 10.1002/humu.21530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimotori Masaaki, Maruyama Hiroki, Nakamura Gen, Suyama Takayuki, Sakamoto Fumiko, Itoh Masaaki, Miyabayashi Shigeaki, Ohnishi Takahiro, Sakai Norio, Wataya-Kaneda Mari, Kubota Mitsuru, Takahashi Toshiyuki, Mori Tatsuhiko, Tamura Katsuhiko, Kageyama Shinji, Shio Nobuo, Maeba Teruhiko, Yahagi Hirokazu, Tanaka Motoko, Oka Masayo, Sugiyama Hitoshi, Sugawara Toshiyuki, Mori Noriko, Tsukamoto Hiroko, Tamagaki Keiichi, Tanda Shuuji, Suzuki Yuka, Shinonaga Chiya, Miyazaki Jun-ichi, Ishii Satoshi, Gejyo Fumitake. Novel mutations of the GLA gene in Japanese patients with Fabry disease and their functional characterization by active site specific chaperone. Human Mutation. 2008;29(2):331–331. doi: 10.1002/humu.9520. [DOI] [PubMed] [Google Scholar]
- 22.Chamoles NA, Blanco M. Gaggioli. Fabry disease: enzymatic diagnosis in dried blood spots on filter paper. Clin Chim Acta. 2001;308(1–2):195–196. doi: 10.1016/s0009-8981(01)00478-8. [DOI] [PubMed] [Google Scholar]
- 23.Okur Ilyas, Ezgu Fatih, Biberoglu Gursel, Tumer Leyla, Erten Yasemin, Isitman Muzeyyen, Eminoglu Fatma Tuba, Hasanoglu Alev. Screening for Fabry disease in patients undergoing dialysis for chronic renal failure in Turkey: Identification of new case with novel mutation. Gene. 2013;527(1):42–47. doi: 10.1016/j.gene.2013.05.050. [DOI] [PubMed] [Google Scholar]
- 24.Castilhos Cristina D., Mezzalira Jamila, Goldim Mariana P.S., Daitx Vanessa V., Garcia Cristina da S., Andrade Carla V., Breier Ana C., Cé Jaqueline, Mello Alexandre S., Coelho Janice C. Determination of the lysosomal hydrolase activity in blood collected on filter paper, an alternative to screen high risk populations. Gene. 2014;536(2):344–347. doi: 10.1016/j.gene.2013.11.101. [DOI] [PubMed] [Google Scholar]
- 25.Fuller Maria, Lovejoy Melanie, Brooks Doug A, Harkin Miriam L, Hopwood John J, Meikle Peter J. Immunoquantification of α-Galactosidase: Evaluation for the Diagnosis of Fabry Disease. Clinical Chemistry. 2004;50(11):1979–1985. doi: 10.1373/clinchem.2004.037937. [DOI] [PubMed] [Google Scholar]
- 26.Lukacs Z, Hartung R, Beck M, Mengel E. Direct comparison of enzyme measurements from dried blood and leukocytes from male and female Fabry disease patients. J. Inherit Metab Dis. 2007. https://doi::10.1007/s10545-007-0679-7. [DOI] [PubMed]
- 27.van der Tol L, Smid B E, Poorthuis B J H M, Biegstraaten M, Deprez R H Lekanne, Linthorst G E, Hollak C E M. A systematic review on screening for Fabry disease: prevalence of individuals with genetic variants of unknown significance. Journal of Medical Genetics. 2013;51(1):1–9. doi: 10.1136/jmedgenet-2013-101857. [DOI] [PubMed] [Google Scholar]
- 28.Ferri L, Guido C, la Marca G, Malvagia S, Cavicchi C, Fiumara A, Barone R, Parini R, Antuzzi D, Feliciani C, Zampetti A, Manna R, Giglio S, Della Valle CM, Wu X, Valenzano KJ, Benjamin ER, Donati MA, Guerrini R, Genuardi M, Morrone A. Fabry disease: polymorphic haplotypes and a novel missense mutation in the GLA gene. Clinical Genetics. 2011;81(3):224–233. doi: 10.1111/j.1399-0004.2011.01689.x. [DOI] [PubMed] [Google Scholar]
- 29.Tanislav C., Kaps M., Rolfs A., Böttcher T., Lackner K., Paschke E., Mascher H., Laue M., Blaes F. Frequency of Fabry disease in patients with small-fibre neuropathy of unknown aetiology: a pilot study. European Journal of Neurology. 2010;18(4):631–636. doi: 10.1111/j.1468-1331.2010.03227.x. [DOI] [PubMed] [Google Scholar]
- 30.Pisani A, Imbriaco M, Zizzo C, Albeggiani G, Colomba P, Alessandro R. A classical phenotype of Anderson-Fabry disease in a female patient with intronic mutations of the GLA gene: a case report. BMC Cardiovasc Disord 2012. https://doi.org/10.1186/1471-2261-12-39. [DOI] [PMC free article] [PubMed]
- 31.Oliveira J. P., Ferreira S., Barceló J., Gaspar P., Carvalho F., Sá Miranda M. C., Månsson J.-E. Effect of single-nucleotide polymorphisms of the 5′ untranslated region of the human α-galactosidase gene on enzyme activity, and their frequencies in Portuguese caucasians. Journal of Inherited Metabolic Disease. 2008;31(S2):247–253. doi: 10.1007/s10545-008-0818-9. [DOI] [PubMed] [Google Scholar]
- 32.Gervas-Arruga J, Cebolla J, Irun P, Perez-Lopez J, Plaza L, Roche J et al. Increased glycolipid storage produced by the inheritance of a complex intronic haplotype in the α-galactosidase a (GLA) gene. BMC Genet 2015. https://doi: 10.1186/s12863-015.0267-z. [DOI] [PMC free article] [PubMed]
- 33.Vieitez I, Souto-Rodriguez O, Fernandez-Mosquera L, San Millan B, Teijeira S, Fernandez-Martin J et al. Fabry disease in the Spanish population: observational study with detection of 77 patients. Orphanet of rare diseases 2018. https://doi:10.1186/s13023-018-0792-8. [DOI] [PMC free article] [PubMed]
- 34.Ferreira S, Ortiz A, Germain DP, Viana-Baptista M, Caldeira-Gomes A, Camprecios M, Fenollar-Cortés M, Gallegos-Villalobos Á, Garcia D, García-Robles JA, Egido J, Gutiérrez-Rivas E, Herrero JA, Mas S, Oancea R, Péres P, Salazar-Martín LM, Solera-Garcia J, Alves H, Garman SC, Oliveira JP. The alpha-galactosidase a p.Arg118Cys variant does not cause a Fabry disease phenotype: data from individual patients and family studies. Mol Genet Metab. 2015;114(2):248–258. doi: 10.1016/j.ymgme.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Froissart R, Guffon N, Vanier MT, Desnick RJ, Maire I. Fabry disease: D313Y is an alpha-galactosidase A sequence variant that causes pseudodeficient activity in plasma. Mol Genet Metab. 2003 Nov;80(3):307–314.Pubmed PMID:1468097. [DOI] [PubMed]
- 36.Benjamin Elfrida R., Della Valle Maria Cecilia, Wu Xiaoyang, Katz Evan, Pruthi Farhana, Bond Sarah, Bronfin Benjamin, Williams Hadis, Yu Julie, Bichet Daniel G., Germain Dominique P., Giugliani Roberto, Hughes Derralynn, Schiffmann Raphael, Wilcox William R., Desnick Robert J., Kirk John, Barth Jay, Barlow Carrolee, Valenzano Kenneth J., Castelli Jeff, Lockhart David J. The validation of pharmacogenetics for the identification of Fabry patients to be treated with migalastat. Genetics in Medicine. 2016;19(4):430–438. doi: 10.1038/gim.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yasuda Makiko, Shabbeer Junaid, Benson Stacy D., Maire Irene, Burnett Roger M., Desnick Robert J. Fabry disease: Characterization of ?-galactosidase A double mutations and the D313Y plasma enzyme pseudodeficiency allele. Human Mutation. 2003;22(6):486–492. doi: 10.1002/humu.10275. [DOI] [PubMed] [Google Scholar]
- 38.Hasholt Lis, Ballegaard Martin, Bundgaard Henning, Christiansen Michael, Law Ian, Lund Allan M., Norremolle Anne, Krogh Rasmussen Ase, Ravn Kirstine, Tumer Zeynep, Wibrand Flemming, Feldt-Rasmussen Ulla. The D313Y variant in theGLAgene – no evidence of a pathogenic role in Fabry disease. Scandinavian Journal of Clinical and Laboratory Investigation. 2017;77(8):617–621. doi: 10.1080/00365513.2017.1390782. [DOI] [PubMed] [Google Scholar]
- 39.Niemann Markus, Rolfs Arndt, Giese Anne, Mascher Hermann, Breunig Frank, Ertl Georg, Wanner Christoph, Weidemann Frank. JIMD Reports. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012. Lyso-Gb3 Indicates that the Alpha-Galactosidase A Mutation D313Y is not Clinically Relevant for Fabry Disease; pp. 99–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spada Marco, Pagliardini Severo, Yasuda Makiko, Tukel Turgut, Thiagarajan Geetha, Sakuraba Hitoshi, Ponzone Alberto, Desnick Robert J. High Incidence of Later-Onset Fabry Disease Revealed by Newborn Screening*. The American Journal of Human Genetics. 2006;79(1):31–40. doi: 10.1086/504601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eng CM, Resnick-Silverman LA, Niehaus DJ, Astrin KH, Desnick RJ. Nature and frequency of mutations in the alpha-galactosidase a gene that cause Fabry disease. Am J Hum Genet. 1993;53(6):1186–1197. [PMC free article] [PubMed] [Google Scholar]
- 42.Lukas Jan, Giese Anne-Katrin, Markoff Arseni, Grittner Ulrike, Kolodny Ed, Mascher Hermann, Lackner Karl J., Meyer Wolfgang, Wree Phillip, Saviouk Viatcheslav, Rolfs Arndt. Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease. PLoS Genetics. 2013;9(8):e1003632. doi: 10.1371/journal.pgen.1003632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukas Jan, Scalia Simone, Eichler Sabrina, Pockrandt Anne-Marie, Dehn Nicole, Cozma Claudia, Giese Anne-Katrin, Rolfs Arndt. Functional and Clinical Consequences of Novel α-Galactosidase A Mutations in Fabry Disease. Human Mutation. 2015;37(1):43–51. doi: 10.1002/humu.22910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Origin of samples for FD screening according to the main symptoms
Additional file 2: Table S2. Non-coding variants found in GLA gene in patients with suspicion of FD and the population frequency in 1000 genomes, GenomAD and ABraOM. Human Splicing Finder and TRAP were used to analyze potential pathogenicity.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.