Abstract
Background
The current Brazilian population is the product of centuries of admixture between intercontinental founding groups. Although previous results have revealed a heterogeneous distribution of mitochondrial lineages in the Northeast region, the most targeted by foreign settlers during the sixteenth century, little is known about the paternal ancestry of this particular population. Considering historical records have documented a series of territorial invasions in the Northeast by various European populations, we aimed to characterize the male lineages found in Brazilian individuals in order to discover to what extent these migrations have influenced the present-day gene pool. Our approach consisted of employing four hierarchical multiplex assays for the investigation of 45 unique event polymorphisms in the non-recombining portion of the Y-chromosome of 280 unrelated men from several Northeast Brazilian states.
Results
Primary multiplex results allowed the identification of six major haplogroups, four of which were screened for downstream SNPs and enabled the observation of 19 additional lineages. Results reveal a majority of Western European haplogroups, among which R1b-S116* was the most common (63.9%), corroborating historical records of colonizations by Iberian populations. Nonetheless, FST genetic distances show similarities between Northeast Brazil and several other European populations, indicating multiple origins of settlers. Regarding Native American ancestry, our findings confirm a strong sexual bias against such haplogroups, which represented only 2.5% of individuals, highly contrasting previous results for maternal lineages. Furthermore, we document the presence of several Middle Eastern and African haplogroups, supporting a complex historical formation of this population and highlighting its uniqueness among other Brazilian regions.
Conclusions
We performed a comprehensive analysis of the major Y-chromosome lineages that form the most dynamic migratory region from the Brazilian colonial period. This evidence suggests that the ongoing entry of European, Middle Eastern, and African males in the Brazilian Northeast, since at least 500 years, was significantly responsible for the present-day genetic architecture of this population.
Keywords: Y-SNPs, Population genetics, Genetic ancestry, Asymmetric colonization, Admixed population
Background
Present-day Brazilians are the result of centuries of admixture between three main ethnic groups, namely Native Americans, Europeans and Africans (1). As the first colonizers of the American continent, Amerindians arrived via Bering strait and reached Brazilian territory during the late Pleistocene (2). Later, the European component of the Brazilian colonization had an important impact on the composition of the current day populations.
Besides Portuguese settlers, other European groups also invaded or populated the country, such as Spanish, Dutch, French, English, Italian, German, Middle-Eastern and Japanese populations. Most of these settlements initially took place in the Northeastern and Southern regions. In fact, the Northeast was the most targeted area for migratory events during the colonial period and received the greatest number of European and African individuals (1, 3–5). Therefore, admixture processes between distinct ethnic groups began much earlier and intensively in the Northeast than in the rest of the country, which makes this an important region for investigating Brazil’s demographic history.
Uniparental markers are useful for disentangling the complex processes that shaped the current population (6). Indeed, previous data from maternal and paternal lineages (using both slow and fast evolving markers) has shown a strong male biased colonization of the Brazilian territory, with the majority of mitochondrial DNA (mtDNA) haplogroups being of Native American and African origin, while Y-chromosome lineages are overwhelmingly dominated by European haplogroups (7–14). We have demonstrated heterogenous frequencies of mitochondrial Amerindian and African lineages in the Brazilian Northeast, which brought a new perspective to the understanding of the maternal ancestral contributions to this region (13). An insight into the paternal lineages of this same population is important to determine whether male contributions were also distinct from earlier reports and to what extent the numerous historical immigrations influenced the genetic architecture of this region (7, 9).
Given its non-recombining nature and low mutation rates, single nucleotide polymorphisms (SNPs) located in the Y-chromosome are a useful tool for investigating historical events and serve as a valuable counterpart for our mtDNA data (15–17). The geographic specificity enabled by this marker allows for an even greater characterization of the microevolutionary aspects of Brazil’s present-day population (6).
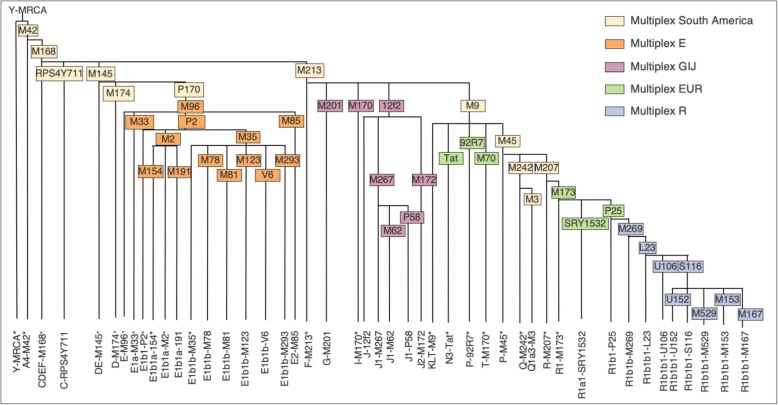
In this paper, a sample of 280 unrelated male individuals from several Northeastern states is analysed in a hierarchical assay for determining Y-chromosomal ancestral lineages (Fig. 1). Four subsequent high-resolution multiplex assays were also carried out to better characterize the haplogroups present in the Northeast region of Brazil (Fig. 2). Additionally, data was compared to previously obtained mitochondrial lineages from the same samples in order to provide a comprehensive description of the ancestral background and demographic history of this particular population.
Fig. 1.
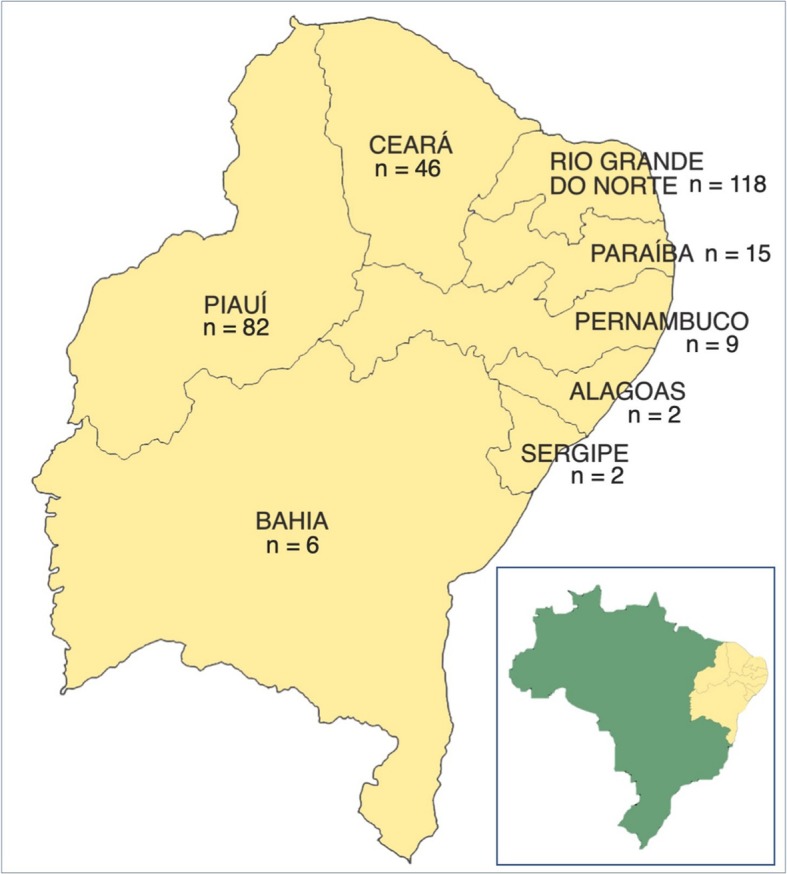
Map and sample sizes of the investigated regions in this study. MA – Maranhão, PI – Piauí, CE – Ceará, RN – Rio Grande do Norte, PB – Paraíba, PE – Pernambuco, AL – Alagoas, SE – Sergipe, BA – Bahia. Created with Adobe Photoshop CC 2019 (www.adobe.com/products/photoshop.html)
Fig. 2.
Y-Chromosome phylogeny of the SNPs investigated here through five Multiplex reactions
Results
Y-chromosome Haplogroups in the northeastern Brazilian population
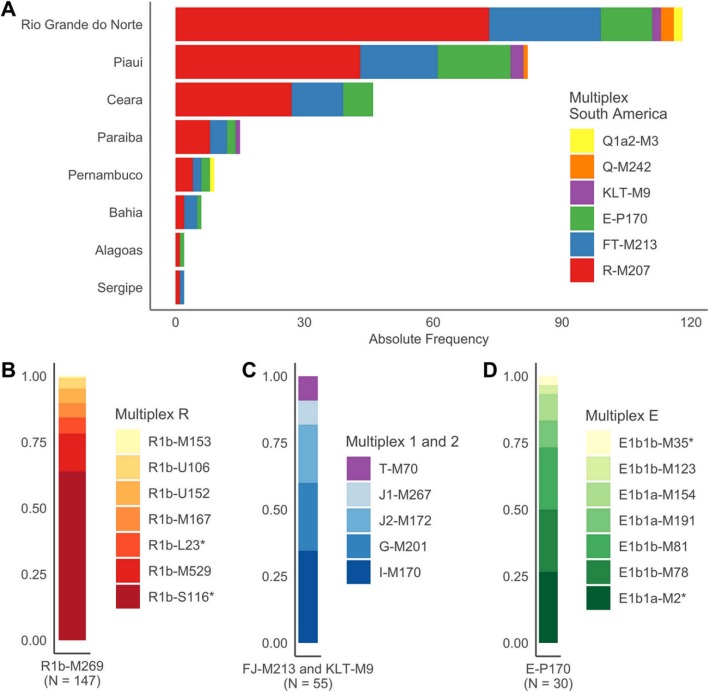
Data obtained from the Major South America Multiplex is shown in Table 1. All genotypes and relevant data regarding the SNPs and population chosen in this study are available in Additional file 1. In the Northeastern region, the majority of samples belonged to the R-M207 haplogroup (56.8%), followed by FJ-M213 (23.6%), E-P170 (15%), KLT-M9 (2.1%), Q-M242 (1.4%) and Q1a2-M3 (1.1%). This pattern is representative of all states surveyed in this study (with N ≥ 15), showing that there is a homogenous distribution of Y-chromosome haplogroups in the investigated locations (Pearson’s Chi-Square test, p = 0.6606).
Table 1.
Results obtained from Multiplex Major South America
| Population | N | Haplogroup frequencies | |||||
|---|---|---|---|---|---|---|---|
| E-P170 | FJ-M213 | KLT-M9 | Q-M242 | Q1a2-M3 | R-M207 | ||
| Northeast | 280 | 0.150 | 0.236 | 0.021 | 0.014 | 0.011 | 0.568 |
| Piauí | 82 | 0.207 | 0.220 | 0.037 | 0.012 | – | 0.524 |
| Ceará | 46 | 0.152 | 0.261 | – | – | – | 0.587 |
| Rio Grande do Norte | 118 | 0.102 | 0.220 | 0.017 | 0.025 | 0.017 | 0.619 |
| Paraíba | 15 | 0.133 | 0.267 | 0.067 | – | – | 0.533 |
| Pernambuco | 9 | 0.222 | 0.222 | – | – | 0.111 | 0.444 |
| Alagoas | 2 | 0.5 | – | – | – | – | 0.5 |
| Sergipe | 2 | – | 0.5 | – | – | – | 0.5 |
| Bahia | 6 | 0.167 | 0.500 | – | – | – | 0.333 |
n Sample size
Considering the globally widespread presence of certain Y-chromosome haplogroups, we performed the investigation of 33 downstream SNPs through multiple Multiplex reactions in order to provide higher phylogenetic resolution of the paternal ancestry composition of this population (Table 2, Fig. 3).
Table 2.
Diversity and relative frequencies of Y-SNP haplogroups found in eight Northeastern Brazilian states
| Haplogroups | Population | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Northeast | Piauí | Ceará | Rio Grande do Norte | Paraíba | Pernambuco | Alagoas | Bahia | Sergipe | |
| E1b1b-M123 | 0.004 | 0.017 | – | – | – | – | – | – | – |
| E1b1a-M154 | 0.013 | 0.017 | – | – | 0.067 | – | – | 0.167 | – |
| E-M2* | 0.033 | 0.068 | 0.024 | 0.019 | – | – | – | – | – |
| E-M35* | 0.004 | – | 0.024 | – | – | – | – | – | – |
| E1b1b-M78 | 0.029 | 0.017 | 0.073 | 0.019 | 0.067 | – | – | – | – |
| E1b1b-M81 | 0.029 | 0.034 | 0.024 | 0.028 | – | – | 0.500 | – | – |
| E1b1a-M191 | 0.013 | 0.034 | – | 0.009 | – | – | – | – | – |
| R-M153 | 0.004 | – | 0.024 | – | – | – | – | – | – |
| R-M167 | 0.033 | 0.017 | – | 0.047 | – | 0.125 | – | 0.167 | – |
| R1b-L23* | 0.038 | 0.068 | 0.024 | 0.028 | – | 0.125 | – | – | – |
| R1b-M529 | 0.088 | 0.051 | 0.024 | 0.132 | 0.133 | 0.125 | – | – | – |
| R1b-S116* | 0.393 | 0.492 | 0.366 | 0.377 | 0.400 | 0.125 | 0.500 | 0.167 | 0.500 |
| R1b-U106 | 0.025 | 0.034 | 0.049 | 0.019 | – | – | – | – | – |
| R1b-U152 | 0.033 | – | 0.122 | 0.028 | – | – | – | – | – |
| I-M170 | 0.079 | 0.051 | 0.049 | 0.104 | 0.133 | 0.125 | – | – | – |
| Q-M242 | 0.017 | 0.017 | – | 0.028 | – | – | – | – | – |
| Q-M3 | 0.013 | – | – | 0.019 | – | 0.125 | – | – | – |
| G-M201 | 0.059 | 0.051 | 0.098 | 0.047 | 0.067 | – | – | 0.167 | – |
| T-M70 | 0.021 | 0.034 | – | 0.019 | 0.067 | – | – | – | – |
| J1-M267 | 0.021 | – | 0.049 | 0.028 | – | – | – | – | – |
| J2-M172 | 0.050 | – | 0.049 | 0.047 | 0.067 | 0.125 | – | 0.333 | 0.500 |
| N | 239 | 59 | 41 | 106 | 15 | 8 | 2 | 6 | 2 |
| HD ± s.d. |
1.000 ± 0.0003 |
0.8440± 0.0322 |
0.8744 ± 0.0391 | 0.8559 ± 0.0256 | 0.8381 ± 0.0852 | 1.000 ± 0.0524 | 1.000 ± 0.5000 | 0.9333 ± 0.1217 | 1.000 ± 0.5000 |
N Number of subtyped samples; k Number of different haplogroups; HD Haplotype diversity; *Belongs to the clade but not a subclade
Fig. 3.
Frequency of Y-Chromosome Haplogroups in Northeastern Brazil
Sublineages of R1b-M269 haplogroup
To address the lack of data regarding European lineages in many states from Northeastern Brazil, we carried out the sub-typing of eight R1b-M269 downstream SNPs in 147 samples with the R-M207 derived allele (14). This methodology allowed the detection of lineages R1b-S116*, R1b-M529, R1b-U152, R1b-U106, R1b-L23*, R1b-M167, and R1b-M153. Sub-types derived from the M269 marker have been shown to be the most frequent in Western Europe, which make them useful for determining the origin of Brazilian patrilineages (18–21).
R1b-S116* was the most common haplogroup in this subset of samples, corresponding to 63.9% of individuals, while R1b-M529 showed a frequency of 14.3% as the second most frequent. Haplogroup R1b-L23* was observed in 6.1% of samples, R1b-M167 and R1b-U152 shared a frequency of 5.4%, R1b-U106 was assigned to 4.1% of samples, and lastly, R1b-M153 was the least frequent, accounting for 0.7% of the subjects.
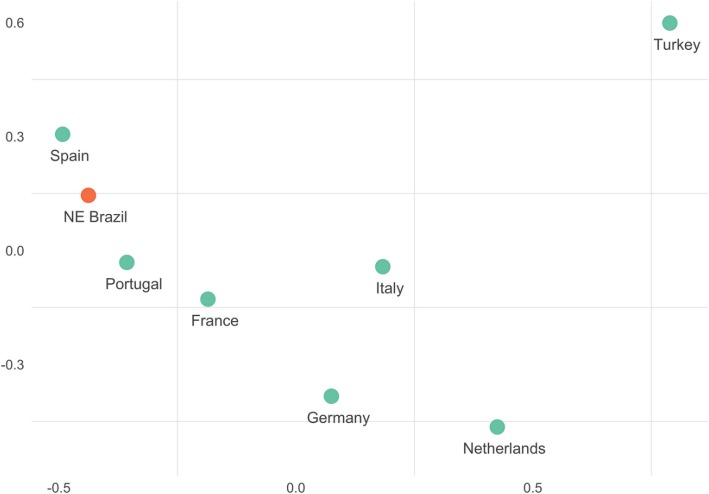
To test whether it was possible to observe diverse sources of European lineages in the Northeast, we computed FST genetic distances based on R1b-M269* sub-type frequencies. A multidimensional scaling plot demonstrates the distances between the Brazilian population investigated here and previous data obtained from potential European colonizer populations (19, 22) (Fig. 4). The Brazilian sample is closest to populations from the Iberian Peninsula, while still showing some proximity to other Western European populations.
Fig. 4.
Multidimensional scaling plot of the pairwise FST genetic distances based on the frequencies of R-L23*, R-U106, R-S116*, R-U152 and R-M529 haplogroups in Northeastern Brazil and in seven European populations that contributed to the formation of the Brazilian gene pool. Stress = 4.139478e-14
Sub-lineages of FJ-M213 and KLT-M9 haplogroups
In order to further investigate the European genepool of Brazil’s Northeastern population, it was necessary to consider sub-lineages derived from deep-rooting markers FJ-M213 and KLT-M9. These haplogroups contain lineages that are mostly present in current day Europeans, Middle Eastern and Near Eastern populations (23).
Accounting for 23.6% of individuals investigated in this study, 50 samples belonging to haplogroup FJ-M213 were genotyped for six downstream SNPs, which allowed the detection of four sub-lineages. Of these, sub-haplogroup I-M170 was the most frequent (38%), followed by G-M201 (28%), J2-M172 (24%), and J1-M267 (10%). Additionally, Multiplex 1 was performed on five samples assigned to haplogroup KLT-M9, all of which belonged to rare sub-haplogroup T-M70, typically attributed to Shepardic populations (24).
Sub-lineages of E-P170 haplogroup
A set of 13 downstream SNPs was chosen for increased resolution of 30 samples carrying the E-P170 marker. The importance of determining the ancestral origin of E haplogroup sub-lineages relies in the multiple colonization processes that took place in Northeast Brazil, which not only included the forced migration of many Sub-Saharan African groups, but also included subsequent North African and Middle Eastern immigrants (3).
Multiplex E results show that all samples belonged to subclades of the P2 marker, E-M35 and E-M2. The former, observed in 53.3% of these individuals, included E1b1b-M35*, E1b1b-M81, E1b1b-M78, and E1b1b-M123. From the latter, E1b1a-M2*, E1b1a-M191, and E1b1a-M154 represented 46.7% of samples.
Discussion
The homogenous distribution of European haplogroups in Northeastern Brazil showed by our results is expected in this region of the country due to historical reports and previous local data (7, 9). A greater European component of paternal lineages is also the case for the other geopolitical regions of Brazil, which is a demonstration of the colonization patterns in this territory (7, 10, 11, 14, 25–27). The investigation of subtypes of these deep-rooted haplogroups was important for elucidating the origin of foreign settlers that have historically contributed to the formation of the Brazilian gene pool, given that historical records describe intense migratory movements of diverse populations in this region since the colonial period (1).
The most frequent haplogroup in our sample, R-M207*, is the most commonly observed in Europe, indicating the origin of more than 50% of Northeast Brazilian lineages (19, 22, 28–30). Regarding sub-lineages derived from the M269 marker, S116* was the most common in our sample, which is in agreement with previous findings from the Northeastern states of Alagoas and Maranhão (9, 31). This haplogroups is also the most common in the Iberian peninsula, corroborating the historical occupation of Northeastern Brazil by men of Portuguese origin (32, 33). This haplogroup was followed by R1b-M529 as the second most common, which can be found at high frequencies in England and Ireland (34).
Findings for the state of Ceará (14) and Maranhão (31) show lineage R1b-M529 at 2.2 and 3% frequencies, respectively, in accordance with our findings for Ceará (2.4%). However, for the state of Rio Grande do Norte, this contribution is five times larger, making up 13.2% of the population. Interestingly, this same state has higher frequencies of haplogroup I-M170 (10.4%) when compared to most states investigated in this study. Considering both of these haplogroups are rarely found in western Europe, these results may reveal the continued presence of non-Iberian colonizers in Northeast Brazil, corroborating historical reports (1, 5).
Another example of differential haplogroup distribution in this region that may reflect historical occupations is the case of R1b-U152, which was observed four times more frequently in Ceará (12.2%) than in Rio Grande do Norte and Maranhão (31). This haplogroup is currently mostly found in Northern Italy, France and Germany (19).
Regarding the remaining M269 derived lineages found in frequencies ranging from 2.8–0.4% in our sample, R1b-U106 has been reported as most frequent in Northwestern Europe, R1b-M153 and M167 were reported in Iberian populations and their descendants, mostly in the Basques (19, 35–38), and R1b-L23* reaches its maximum frequency in the Balkans, Turkey, the Caucasus and the Circum-Uralic region (19). Our results are further substantiated by data from Carvalho-Silva (7), who described the interesting heterogeneity of European male haplogroups in the Northeastern region (which were also found to be common in the South) and brought attention to the fact that this region was largely inhabited by the Dutch during the seventeenth century. Therefore, the presence of multiple European lineages in this population is in agreement with historical records from the period of such settlements.
Indications of multiple colonizer sources are also observed through the presence of haplogroups G-M201, J-M267, and J-M172, common in the Middle East and Near Eastern regions (23, 39). These haplogroups have shown to be distributed in diverse frequencies throughout the Northeastern territory, with G-M201, for instance, varying from 11% in Bahia (40) to 3.6% in Alagoas (9). According to Resque et al. (14), the presence of these lineages is possibly a product of the immigration of Arab traders in the post-colonial period.
Such discussion may be further extended to the presence of other Middle Eastern and North African haplogroups E-M78 and E-M81 in our findings, both present in 2.9% of individuals. One should consider that similar frequencies are found in Iberian populations, meaning this contribution could be from a European colonizer source (33). However, it is worth noting that both haplogroups were found at a frequency of 8% in the state of Maranhão (31), suggesting they may indeed originate from North African and/or Middle Eastern groups. Data from that same study also shows that over 30% of Maranhão individuals have African haplogroups, potentially supporting a non-European origin for the aforementioned haplogroups in the Brazilian Northeast.
With regards to E haplogroups in our findings, the presence of M2 derived markers, which is restricted to Sub-Saharan Africa, seems to be a product of the transatlantic slave trade, responsible for the arrival of West African individuals mostly to the Northeast region of Brazil during the seventeenth century. The contribution of haplogroups E-M2*, E1b1a-M191, and E1b1a-M154 show that the ancestral background of these Brazilian men is derived from a signature of the Bantu expansion, which is consistent with prior studies (14, 41). Interestingly, a whole genome sequence investigation performed by Kehdy et al. (42) for the population of Salvador (capital of Bahia) yielded signatures of both Bantu and non-bantu genetic ancestry, indicating a greater complexity to the African background of Northeastern Brazilian men.
Finally, the least frequent haplogroups in our sample, accounting for a total of 1.4%, are derived from the Q-M242* polymorphism, which is confined to the American continent and Amerindian populations (43–45). Such small contributions of African and, ever more so, of Native American haplogroups are in accordance with Y-chromosome data from other European colonies in South America, despite them being the majority in mtDNA studies (46–49). In fact, previous mtDNA data obtained by Schaan et al. (13) for the same samples demonstrated a strong Amerindian (43.5%) and African (37.8%) female component in Northeast Brazil. These findings attest to the strong asymmetric colonization favoring the introgression of European Y lineages in this region, a pattern that has been reported for other Brazilian regions and is typical for South American countries as well (50).
Conclusions
In conclusion, our data brings biological evidence to historical records stating the importance of intercontinental arrivals to Northeast Brazil since the colonial period. Through the analyses of 45 Y-chromosome SNPs, we demonstrated that Iberian ancestry is represented in the majority of individuals. Still, the presence of other non-western European lineages is a strong indicator of the continued presence of multiple historically relevant occupations. Furthermore, the frequency of Middle Eastern haplogroups may suggest more recent immigrations, while common African Bantu lineages probably reflect the transatlantic slave trade. Overall, these results reveal the complex structure of the ancestral male genetic background of the Brazilian Northeast, and contribute to the knowledge of South American demographic history.
Methods
Population sample and DNA extraction
We tested a total of 280 unrelated male samples from the Northeastern region of Brazil, distributed in eight states as follows: i) 82 from Piauí, ii) 46 from Ceará; iii) 118 from Rio Grande do Norte; iv) 15 from Paraíba; v) nine from Pernambuco; vi) two from Alagoas; vii) two from Sergipe; viii) and six from Bahia, as shown in Fig. 1. These samples are a subset of those previously investigated for mtDNA data. See Schaan et al. (13) for biological material acquirement and DNA extraction methodology.
Genotyping
In total, 45 SNPs were analysed in this work (Fig. 2). SNP typing was performed through multiplex polymerase chain reactions (PCR) and Single Base Extension (SBE) analysis using the SNaPshot kit (Thermo Fisher Scientific, Waltham, MA.) The data obtained in this study is available in Additional file 1. For determining population substructure based on the main ethnic groups that compose the Brazilian gene pool, 12 SNPs were chosen based on the hierarchical Multiplex Major South America assay described by Geppert et al. (51). This initial screening allowed for the identification of five lineages, namely haplogroups from the E-P170, FJ-M213, KLT-M9, Q-M242, and Q1a2-M3 branches. Samples were subsequently genotyped according to obtained results, consisting in: i) Multiplex 1, for samples carrying derived allele M9 (Brion et al., 2004); ii) Multiplex GIJ, for samples with derived allele M213 (52); iii) Multiplex E, for samples with derived allele E-P170 (53); and iv) Multiplex R, for samples with derived allele R-M207 (14).
Statistical analysis
Haplogroup frequencies were determined by direct counting. Population genetics parameters such as comparisons, diversity values and population pairwise genetic distances (FST) were computed using the Arlequin software v.3.5.2.2 (54). FST values were visualized in a multi-dimensional scaling (MDS) analysis and haplogroup frequency distribution analysis (Pearson’s Chi-squared test) were performed on R software v.3.5.3 (55). Haplogroup frequencies of samples carrying the M269* derived allele were compared to those found in current European populations and data was extracted from Myres et al. and Busby et al. (19, 22). For this purpose, we also included the 41 R1b-M269 derived samples from Ceará tested by Resque et al. (14) into our analysis.
Availability of data and material
All data generated or analysed during this study are included in the supplementary information files.
Supplementary information
Additional file 1. Genotypic and SNP data. All genotypes found for the samples analysed in this study as well as chosen SNP information can be found in Additional file 1.
Acknowledgements
We thank the individuals who participated in this study by donating their time and DNA samples.
Abbreviations
- mtDNA
Mitochondrial DNA
- PCR
Polymerase chain reaction
- SNP
Single nucleotide polymorphism
Authors’ contributions
APS performed multiplex assays, analysed SNP data, and wrote the manuscript. LG and JJ performed multiplex assays and analysed SNP data. AM, LC, and MA performed multiplex assays. DM, SHR, RM, TL, FKY, GP, and VS performed sample collection and DNA extractions. SS and ARS conceived the study and participated in its design and writing of the manuscript. All authors have read and approved the final manuscript.
Funding
We thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Pró-Reitoria de Pesquisa (PROPESP) of Universidade Federal do Pará (UFPA), and Universidade do Estado do Rio de Janeiro (UERJ) for the received grants and/or support. This study is part of Rede de Pesquisa em Genômica Populacional Humana (Biocomputacional – Protocol no. 3381/2013/CAPES). Ândrea Ribeiro-dos-Santos is supported by CNPq/Produtividade (CNPQ 304413/2015–1). These funding sources had no role in the design of the study, collection, analysis, interpretation of data or in writing the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Ethics approval and consent to participate
Ethical consent was obtained according to the Helsinki Declaration. Ethical approval was obtained from the Research Ethics Committee of the Federal University of Rio Grande do Norte, Federal University of Piauí, and Federal University of Ceará, under protocol numbers 27,493,614.0.0000.5293, 0443.0.045.000–11, and 702/04, respectively. Before signing the Consent Form participants were informed regarding the nature of this research and use of their biological samples. All the analyses were performed preserving subjects anonymously throughout the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ana Paula Schaan and Leonor Gusmão contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12862-020-1579-9.
References
- 1.Ribeiro D. O Povo Brasileiro: A Formação e o Sentido do Brasil. 2. São Paulo: Companhia das Letras; 1995. [Google Scholar]
- 2.Moreno-Mayar JV, Potter BA, Vinner L, Steinrücken M, Rasmussen S, Terhorst J, et al. Terminal Pleistocene Alaskan genome reveals first founding population of native Americans. Nature. 2018;553:203. doi: 10.1038/nature25173. [DOI] [PubMed] [Google Scholar]
- 3.IBGE . Brasil, 500 Anos de Povoamento. Rio de Janeiro: Instituto Brasileiro de Geografia e Estatística; 2000. [Google Scholar]
- 4.Freyre G. Ingleses no Brasil. Rio de Janeiro: José Olympio; 1948. [Google Scholar]
- 5.Cascudo L. Os Holandeses no Rio Grande do Norte. Departamento de Educação: Natal; 1949. [Google Scholar]
- 6.Underhill PA, Kivisild T. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu Rev Genet. 2007;41(1):539–564. doi: 10.1146/annurev.genet.41.110306.130407. [DOI] [PubMed] [Google Scholar]
- 7.Carvalho-Silva DR, Santos FR, Rocha J, Pena SD. The phylogeography of Brazilian Y-chromosome lineages. Am J Hum Genet. 2001;68(1):281–286. doi: 10.1086/316931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho-Silva DR, Tarazona-Santos E, Rocha J, Pena SDJ, Santos FR. Y chromosome diversity in Brazilians: switching perspectives from slow to fast evolving markers. Genetica. 2006;126(1–2):251–260. doi: 10.1007/s10709-005-1454-z. [DOI] [PubMed] [Google Scholar]
- 9.de Azevedo DA, da Silva LAF, Gusmão L, de Carvalho EF. Analysis of Y chromosome SNPs in Alagoas, northeastern Brazil. Forensic Sci Int. 2009;2(1):421–422. [Google Scholar]
- 10.Carvalho M, Brito P, Lopes V, Andrade L, Anjos MJ, Real FC, et al. Analysis of paternal lineages in Brazilian and African populations. Genet Mol Biol. 2010;33(3):422–427. doi: 10.1590/S1415-47572010005000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palha T, Gusmão L, Ribeiro-Rodrigues E, Guerreiro JF, Ribeiro-dos-Santos Â, Santos S. Disclosing the genetic structure of Brazil through analysis of male lineages with highly discriminating haplotypes. PLoS One. 2012;7(7):1–8. doi: 10.1371/journal.pone.0040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batista Dos Santos SE, Rodrigues JD, Ribeiro-dos-Santos AKC, Zago MA. Differential contribution of indigenous men and women to the formation of an urban population in the amazon region as revealed by mtDNA and Y-DNA. Am J Phys Anthropol. 1999;109(2):175–180. doi: 10.1002/(SICI)1096-8644(199906)109:2<175::AID-AJPA3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 13.Schaan AP, Costa L, Santos D, Modesto A, Amador M, Lopes C, et al. mtDNA structure: the women who formed the Brazilian Northeast. BMC Evol Biol. 2017;17(1):185. doi: 10.1186/s12862-017-1027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Resque R, Gusmão L, Geppert M, Roewer L, Palha T, Alvarez L, et al. Male lineages in Brazil: intercontinental admixture and stratification of the European background. PLoS One. 2016;11(4):1–17. doi: 10.1371/journal.pone.0152573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oota H, Settheetham-Ishida W, Tiwawech D, Ishida T, Stoneking M. Human mtDNA and Y-chromosome variation is correlated with matrilocal versus patrilocal residence. Nat Genet. 2001;29(1):20–21. doi: 10.1038/ng711. [DOI] [PubMed] [Google Scholar]
- 16.Seielstad MT, Minch E, Cavalli-Sforza LL. Genetic evidence for a higher female migration rate in humans. Nat Genet. 1998;20(3):278–280. doi: 10.1038/3088. [DOI] [PubMed] [Google Scholar]
- 17.Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4(8):598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- 18.Balaresque P, Bowden GR, Adams SM, Leung HY, King TE, Rosser ZH, et al. A predominantly neolithic origin for European paternal lineages. PLoS Biol. 2010;8(1):e1000285. doi: 10.1371/journal.pbio.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myres NM, Rootsi S, Lin AA, Järve M, King RJ, Kutuev I, et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in central and Western Europe. Eur J Hum Genet. 2011;19(1):95–101. doi: 10.1038/ejhg.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiaroni J, Underhill PA, Cavalli-Sforza LL. Y chromosome diversity, human expansion, drift, and cultural evolution. Proc Natl Acad Sci. 2009;106(48):20174–20179. doi: 10.1073/pnas.0910803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alonso S, Flores C, Cabrera V, Alonso A, Martín P, Albarrán C, et al. The place of the Basques in the European Y-chromosome diversity landscape. Eur J Hum Genet. 2005;13:1293. doi: 10.1038/sj.ejhg.5201482. [DOI] [PubMed] [Google Scholar]
- 22.Busby GBJ, Brisighelli F, Sanchez-Diz P, Ramos-Luis E, Martinez-Cadenas C, Thomas MG, et al. The peopling of Europe and the cautionary tale of Y chromosome lineage R-M269. Proc R Soc B Biol Sci. 2012;279(1730):884–892. doi: 10.1098/rspb.2011.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semino O, Magri C, Benuzzi G, Lin AA, Al-Zahery N, Battaglia V, et al. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet. 2004;74(5):1023–1034. doi: 10.1086/386295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira AM, Domingues PM, Gomes V, Amorim A, Jannuzzi J, de Carvalho EF, et al. Male lineage strata of Brazilian population disclosed by the simultaneous analysis of STRs and SNPs. Forensic Sci Int Gen. 2014;13:264–268. doi: 10.1016/j.fsigen.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 26.de F Figueiredo R, Ambrosio IB, Braganholi DF, Chemale G, Martins JA, Gomes V, et al. Male-specific contributions to the Brazilian population of Espirito Santo. Int J Legal Med. 2016;130(3):679–681. doi: 10.1007/s00414-015-1214-2. [DOI] [PubMed] [Google Scholar]
- 27.Silva DA, Carvalho E, Costa G, Tavares L, Amorim A, Gusmão L. Y-chromosome genetic variation in Rio de Janeiro population. Am J Hum Biol. 2006;18(6):829–837. doi: 10.1002/ajhb.20567. [DOI] [PubMed] [Google Scholar]
- 28.Beleza S, Gusmão L, Lopes A, Alves C, Gomes I, Giouzeli M, et al. Micro-phylogeographic and demographic history of Portuguese male lineages. Ann Hum Genet. 2006;70(Pt 2):181–194. doi: 10.1111/j.1529-8817.2005.00221.x. [DOI] [PubMed] [Google Scholar]
- 29.Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, et al. The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci. 2001;98(18):10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batini C, Hallast P, Zadik D, Delser PM, Benazzo A, Ghirotto S, et al. Large-scale recent expansion of European patrilineages shown by population resequencing. Nat Commun. 2015;6:7152. doi: 10.1038/ncomms8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jannuzzi Juliana, Ribeiro Julyana, Alho Clarice, de Oliveira Lázaro e Arão Grasielly, Cicarelli Regina, Simões Dutra Corrêa Heitor, Ferreira Suelen, Fridman Cíntia, Gomes Verónica, Loiola Silvia, da Mota Mariana Flavia, Ribeiro-dos-Santos Ândrea, de Souza Carlos Antonio, de Sousa Azulay Rossana Santiago, Carvalho Elizeu F., Gusmão Leonor. Male lineages in Brazilian populations and performance of haplogroup prediction tools. Forensic Science International: Genetics. 2020;44:102163. doi: 10.1016/j.fsigen.2019.102163. [DOI] [PubMed] [Google Scholar]
- 32.Flores C, Maca-Meyer N, González AM, Oefner PJ, Shen P, Pérez JA, et al. Reduced genetic structure of the Iberian peninsula revealed by Y-chromosome analysis: implications for population demography. Eur J Hum Genet. 2004;12(10):855–863. doi: 10.1038/sj.ejhg.5201225. [DOI] [PubMed] [Google Scholar]
- 33.Gonçalves R, Freitas A, Branco M, Rosa A, Fernandes AT, Brehm A. Y-chromosome lineages from Portugal, Madeira and Açores record elements of Sephardim and Berber ancestry. Int Congr Ser. 2006;1288:237–239. doi: 10.1016/j.ics.2005.09.099. [DOI] [PubMed] [Google Scholar]
- 34.van Oven M, Van Geystelen A, Kayser M, Decorte R, Larmuseau MHD. Seeing the wood for the trees: a minimal reference phylogeny for the human Y chromosome. Hum Mutat. 2014;35(2):187–191. doi: 10.1002/humu.22468. [DOI] [PubMed] [Google Scholar]
- 35.Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, et al. Y chromosome sequence variation and the history of human populations. Nat Genet. 2000;26(3):358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 36.López-Parra AM, Gusmão L, Tavares L, Baeza C, Amorim A, Mesa MS, et al. In search of the pre- and post-Neolithic genetic substrates in Iberia: evidence from Y-chromosome in Pyrenean populations. Ann Hum Genet. 2009;73(1):42–53. doi: 10.1111/j.1469-1809.2008.00478.x. [DOI] [PubMed] [Google Scholar]
- 37.Hurles ME, Veitia R, Arroyo E, Armenteros M, Bertranpetit J, Pérez-Lezaun A, et al. Recent male-mediated gene flow over a linguistic barrier in Iberia, suggested by analysis of a Y-chromosomal DNA polymorphism. Am J Hum Genet. 1999;65(5):1437–1448. doi: 10.1086/302617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cruciani F, Trombetta B, Antonelli C, Pascone R, Valesini G, Scalzi V, et al. Strong intra- and inter-continental differentiation revealed by Y chromosome SNPs M269, U106 and U152. Forensic Sci Int Genet. 2011;5(3):e49–e52. doi: 10.1016/j.fsigen.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 39.Semino O. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000;290(5494):1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- 40.Nascimento E, Cerqueira E, Azevedo E, Freitas V, Azevedo D. The Africa male lineages of Bahia’s people—Northeast Brazil: a preliminary SNPs study. Forensic Sci Int Genet Suppl Series. 2009;2(1):349–350. doi: 10.1016/j.fsigss.2009.07.010. [DOI] [Google Scholar]
- 41.Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, et al. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet. 2002;70(5):1197–1214. doi: 10.1086/340257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kehdy FSG, Gouveia MH, Machado M, Magalhães WCS, Horimoto AR, Horta BL, et al. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. PNAS. 2015;112(28):8696–8701. doi: 10.1073/pnas.1504447112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roewer L, Nothnagel M, Gusmão L, Gomes V, González M, Corach D, et al. Continent-wide decoupling of Y-chromosomal genetic variation from language and geography in native south Americans. PLoS Genet. 2013;9(4):e1003460. doi: 10.1371/journal.pgen.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Battaglia V, Grugni V, Perego UA, Angerhofer N, Gomez-Palmieri JE, Woodward SR, et al. The first peopling of south america: new evidence from y-chromosome haplogroup q. PLoS ONE. 2013;8(8):e71390. doi: 10.1371/journal.pone.0071390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bisso-Machado R, Jota MS, Ramallo V, Paixão-Côrtes VR, Lacerda DR, Salzano FM, et al. Distribution of Y-chromosome Q lineages in native Americans. Am J Hum Biol. 2011;23(4):563–566. doi: 10.1002/ajhb.21173. [DOI] [PubMed] [Google Scholar]
- 46.Toscanini U, Gaviria A, Pardo-Seco J, Gómez-Carballa A, Moscoso F, Vela M, et al. The geographic mosaic of Ecuadorian Y-chromosome ancestry. Forensic Sci Int Genet. 2018;33(June):59–65. doi: 10.1016/j.fsigen.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 47.Corach D, Lao O, Bobillo C, van Der Gaag K, Zuniga S, Vermeulen M, et al. Inferring continental ancestry of argentineans from autosomal, Y-chromosomal and mitochondrial DNA. Ann Hum Genet. 2010;74(1):65–76. doi: 10.1111/j.1469-1809.2009.00556.x. [DOI] [PubMed] [Google Scholar]
- 48.Grugni V, Battaglia V, Perego UA, Raveane A, Lancioni H, Olivieri A, et al. Exploring the Y chromosomal ancestry of modern Panamanians. PLoS One. 2015;10(12):e0144223. doi: 10.1371/journal.pone.0144223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carvajal-Carmona LG, Soto ID, Pineda N, Ortíz-Barrientos D, Duque C, Ospina-Duque J, et al. Strong Amerind/white sex bias and a possible Sephardic contribution among the founders of a population in Northwest Colombia. Am J Hum Genet. 2000;67(5):1287–1295. doi: 10.1086/321216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salzano FM, Sans M. Interethnic admixture and the evolution of Latin American populations. Genet Mol Biol. 2014;37(1 Suppl):151–170. doi: 10.1590/S1415-47572014000200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geppert M, Baeta M, Núñez C, Martínez-Jarreta B, Zweynert S, Cruz OWV, et al. Hierarchical Y-SNP assay to study the hidden diversity and phylogenetic relationship of native populations in South America. Forensic Sci Int Genet. 2011;5(2):100–104. doi: 10.1016/j.fsigen.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 52.Geppert M, Roewer L. SNaPshot® minisequencing analysis of multiple ancestry-informative Y-SNPs using capillary electrophoresis. Methods Mol Biol (Clifton, NJ) 2012;830:127–140. doi: 10.1007/978-1-61779-461-2_9. [DOI] [PubMed] [Google Scholar]
- 53.Gomes V, Sánchez-Diz P, Amorim A, Carracedo A, Gusmão L. Digging deeper into east African human Y chromosome lineages. Hum Genet. 2010;127(5):603–613. doi: 10.1007/s00439-010-0808-5. [DOI] [PubMed] [Google Scholar]
- 54.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol Ecol Resour. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 55.Team RC. R: a language and environment for statistical computing. Vienna; 2016. https://www.R-project.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Genotypic and SNP data. All genotypes found for the samples analysed in this study as well as chosen SNP information can be found in Additional file 1.
Data Availability Statement
All data generated or analysed during this study are included in the supplementary information files.
All data generated or analysed during this study are included in this published article [and its supplementary information files].