Abstract
Multidrug membrane transporters can extrude a wide range of substrates, which cause multidrug resistance and ineffective treatment of diseases. In this study, we used three different sized antibiotic drug nanocarriers to study their size-dependent inhibitory effects against Bacillus subtilis. We functionalized 2.4 ± 0.7, 13.0 ± 3.1, and 92.6 ± 4.4 nm silver nanoparticles (Ag NPs) with a monolayer of 11-amino-1-undecanethiol and covalently linked them with antibiotics (ofloxacin, Oflx). The labeling ratios of antibiotics with NPs are 8.6 × 102, 9.4 × 103, and 6.5 × 105 Oflx molecules per NP, respectively. We designed cell culture medium in which both BmrA and ΔBmrA cells grew and functioned normally while ensuring the stabilities of nanocarriers (nonaggregation). These approaches allow us to quantitatively study the dependence of their inhibitory effect against two isogenic strains of B. subtilis, WT (normal expression of BmrA) and ΔBmrA (deletion of bmrA), upon the NP size, antibiotic dose, and BmrA expression. Our results show that the inhibitory effects of nanocarriers highly depend on NP size and antibiotic dose. The same amount of Oflx on 2.4 ± 0.7, 13.0 ± 3.1, and 92.6 ± 4.4 nm nanocarriers shows the 3× lower, nearly the same, and 10× higher inhibitory effects than that of free Oflx, against both WT and ΔBmrA, respectively. Control experiments of the respective sized AgMUNH2 NPs (absence of Oflx) show insignificant inhibitory effects toward both strains. Taken together, the results show multiple factors, such as labeling ratios, multivalent effects, and pharmacodynamics (Oflx localization and distribution), which might play the roles in the size-dependent inhibitory effects on the growth of both WT and ΔBmrA strains. Interestingly, the inhibitory effects of nanocarriers are independent of the expression of BmrA, which could be attributed to the higher efflux of nanocarriers by other membrane transporters in both strains.
Introduction
ATP-binding cassette (ABC) membrane transporters are one of the largest membrane transport superfamilies, and they exist in all living organisms and play highly significant roles in biological functions.1−7 The ABC membrane transporters can selectively transport a wide variety of substrates across cellular membranes, even though they share a common modular architecture, two transmembrane domains (TMDs) and two nucleotide-binding domains (NBDs). TMDs exhibit variable sequence and topology and define substrate binding sites and transport passageway, while the NBDs possess conserved sequences and bind and hydrolyze ATP to provide the “power-stroke” for the transporters to translocate the specific substrates across the cellular membrane.7−17 The multidrug ABC membrane transporters can extrude chemotherapeutic agents out of bacteria or tumor cells, which causes multidrug resistance (MDR) and ineffective treatment of diseases, underscoring the importance of understanding their underlying mechanisms to design more effective therapy.3,18−22
Bacillus subtilis (a Gram-positive bacterium) is a widely used model organism to study multidrug ABC transporters. Current studies show that there are 78 ABC transporters in B. subtilis.23−25 BmrA (YvcC), the ABC transporter of B. subtilis, exhibits the highest homology to each half of P-gps (MDR1), HorA (40% identity), and LmrA (42% identity), which makes it an excellent choice to study multidrug ABC transporters.26 Fluoroquinolones (e.g., Oflx, norfloxacin, ciprofloxacin, levofloxacin, and gemifloxacin) are widely used antibiotics to treat a variety of respiratory and urinary tract infections, which include pneumonia, chronic bronchitis, and tuberculosis.27,28 The extensive use of conventional antibiotics has led to the development of multiantibiotics resistance and the creation of superbugs that cannot be effectively eradicated by traditional antibiotics; hence, there is an urgent need to design new types of antibiotics and unconventional drugs (e.g., nanomedicine) to treat the infections.
Noble metal nanoparticles (NPs) possess unique physicochemical properties. Their high surface-area-to-volume ratio allows them to serve as effective drug carriers that offer high payloads, high permeability, high local target concentrations, and high binding affinity due to multivalence effects.29−31 Furthermore, the high surface-area-to-volume ratio of NPs could also lead to high chemical reactivity, and bare NPs themselves could serve as unconventional drugs to eradicate bacteria.29,31−33 In our previous studies, we found that Ag and Au NPs at low concentrations do not inhibit cell growth and are biocompatible, while a higher concentration of NPs was unstable in the buffer or medium, which led to the aggregation of bare NPs on the surface of the cellular membrane that causes cell death.30,32−40
We have successfully imaged single Ag NPs in solution, in single live cells, and single embryos and used their size-dependent plasmonic optical properties to study size-dependent efflux functions of multidrug membrane transporters in single live cells (both Gram-positive and Gram-negative bacteria) in real time at nanometer resolution.33−36,38−51 We have also used their size-dependent plasmonic optical properties to characterize the mode of action of antibiotics, such as aztreonam and chloramphenicol, in single live Pseudomonas aeruginosa cells.37,40 Therefore, the plasmonic NP-based drug carriers can serve as size-dependent photostable-sized imaging probes to study size-dependent efflux kinetics of multidrug membrane transporters and size-dependent MDR in situ in real time, while nanocarriers are inhibiting the bacterial growth.
In our previous study, we have designed, synthesized, purified, and characterized three different sized antibiotic nanocarriers and developed cell culture medium, in which P. aeruginosa (Gram-negative bacteria) cells grew and functioned normally and the nanocarriers remained unchanged (stable, nonaggregated).30 We found that their inhibitory effect against two strains of P. aeruginosa, WT (normal expression of MexAB-OprM) and ΔABM (deletion of MexAB-OprM), highly depends upon the NP size, antibiotic dose, and multidrug membrane transporter (MexAB-OprM) expression. In this study, we used the same nanocarriers to study the dependence of inhibitory effect of the antibiotic nanocarriers against Gram-positive bacteria, two strains of B. subtilis (Gram-positive bacteria), WT (normal expression of BmrA) and ΔBmrA (deletion of bmrA), upon the size of nanocarriers, and dose of antibiotics (Oflx). Note that MexAB-OprM and BmrA are two completely different types of multidrug membrane transporters, energized by proton gradients across the cellular membrane and ATP hydrolysis, respectively. Further, P. aeruginosa are Gram-negative bacteria, while B. subtilis are Gram-positive bacteria, and they have very different cellular envelop structures. By comparing these two studies, we aim to understand the dependence of inhibitory effects of nanocarriers upon the types of multidrug membrane transporters and the types of bacteria.
Results and Discussion
Synthesis, Purification, and Characterization of Stable Drug Nanocarriers
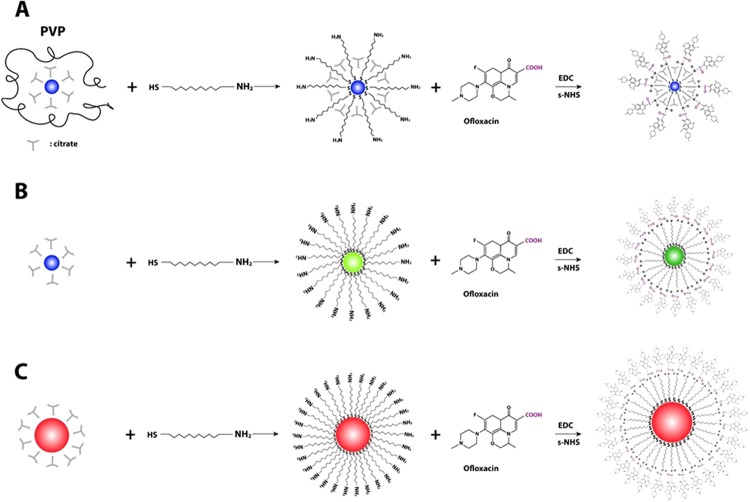
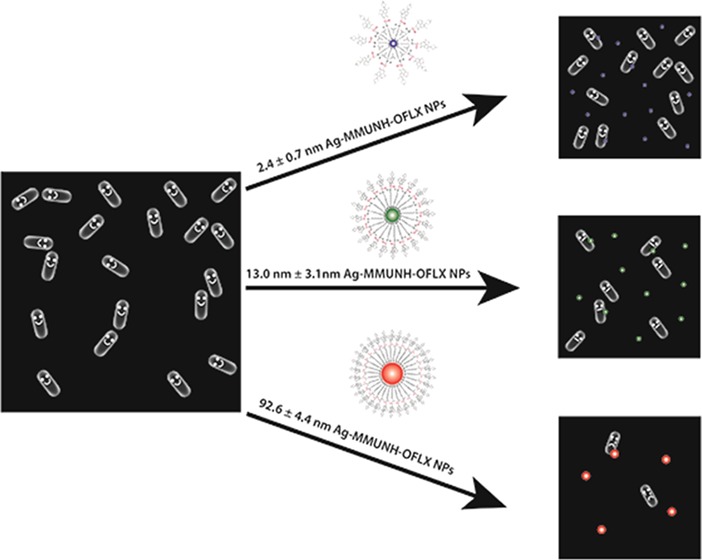
We synthesized, purified, and characterized 2.4 ± 0.7, 13.0 ± 3.1, and 92.6 ± 4.4 nm Ag NPs. We used the thiol group of 11-amino-1-undecanethiol hydrochloride (AUT) to attach a monolayer of AUT (MUNH2) onto surface of NPs to prepare AgMUNH2 NPs (Figure 1), as reported previously.30 We then used a peptide bond to covalently conjugate the carboxyl group of Oflx with the amine group of AgMUNH2 NPs to synthesize antibiotic drug nanocarriers (AgMUNH–Oflx) (Figure 1).30
Figure 1.
Chemical reactions for the preparation of antibiotic drug nanocarriers (AgMUNH–Oflx NPs): attaching monolayer of 11-amino-1-undecanethiol onto the surface of (A) 2.4 ± 0.7, (B) 13.0 ± 3.1, and (C) 92.6 ± 4.4 nm NPs to prepare AgMUNH2 NPs and then linking the amine group of 11-amino-1-undecanethiol with the carboxyl group of the Oflx via peptide bonds using 1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide hydrochloride (EDC) and sulfo-N-hydroxysulfosuccinimide (NHS) as mediators to generate AgMUNH–Oflx NPs (antibiotic drug nanocarriers).30
We purified the drug nanocarriers using centrifugation. We then characterized the sizes of the NPs, AUT (MUNH2) on the NPs, and conjugation ratios of Oflx molecules per NP for 2.4 ± 0.7, 13.0 ± 3.1, and 92.6 ± 4.4 nm NPs as 8.6 × 102, 9.4 × 103, and 6.5 × 105 using transmission electron microscopy and dynamic light scattering (DLS), NMR, and UV–vis spectroscopy, respectively.30
We used DLS and UV–vis spectroscopy to study the stability (nonaggregation) of antibiotic drug nanocarriers with desired concentrations in a commonly used standard Lysogeny broth (LB) medium (1% tryptone, 0.5% yeast extract, and 0.5% NaCl in deionized (DI) water, pH = 7.2) in the shaker (MaxQ 5000, 200 rpm, 37 °C) for 12 h. We found that they were aggregated.30 The aggregation of nanocarriers would alter their sizes and cause potential precipitation of nanocarriers from the medium that would reduce their doses and make the study of dose- and size-dependent unreliable. Therefore, we modified the cell culture medium by reducing the NaCl concentration from 0.5 to 0.1% and found that the drug nanocarriers with desired concentrations in the modified LB medium (1% tryptone, 0.5% yeast extract, and 0.1% NaCl in DI water, pH = 7.2) were stable (nonaggregated) under the cell culture condition and duration, in the shaker (MaxQ 5000, 200 rpm, 37 °C) for 12 h.
Characterization of Cellular Functions in the Modified Medium
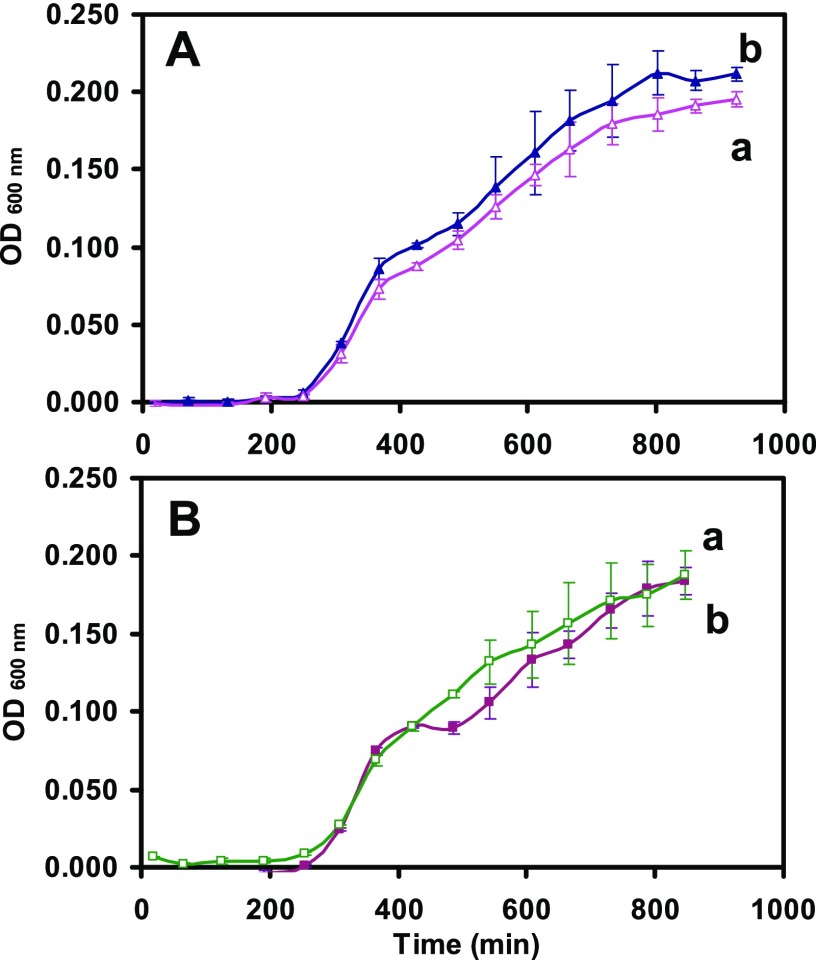
We characterized the viability and efflux function of the cells cultured in the modified medium to make certain that the modified cell culture medium can be used to culture healthy and well-functional cells as those cultured in the standard cell culture medium. We precultured the cells (WT-BmrA and ΔBmrA) in the standard medium for 12 h, then cultured the precultured cells in the standard medium and the modified medium, and followed the cell growth over time. The growth curves of the cells cultured in the standard medium (Figure 2a) and the modified medium (Figure 2b) for WT (Figure 2A) and ΔBmrA (Figure 2B) are the same, showing that the cells grow normally in the modified cell culture medium.
Figure 2.
Study of the suitability of the modified LB medium to culture (A) WT and ΔBmrA: the cellular growth curves of (A) WT and (B) ΔBmrA cultured in (a) standard and (b) modified LB media over time show that the growth rates of a given strain in either medium are nearly identical, which indicates that the modified LB medium can be used to culture both WT and ΔBmrA cells.
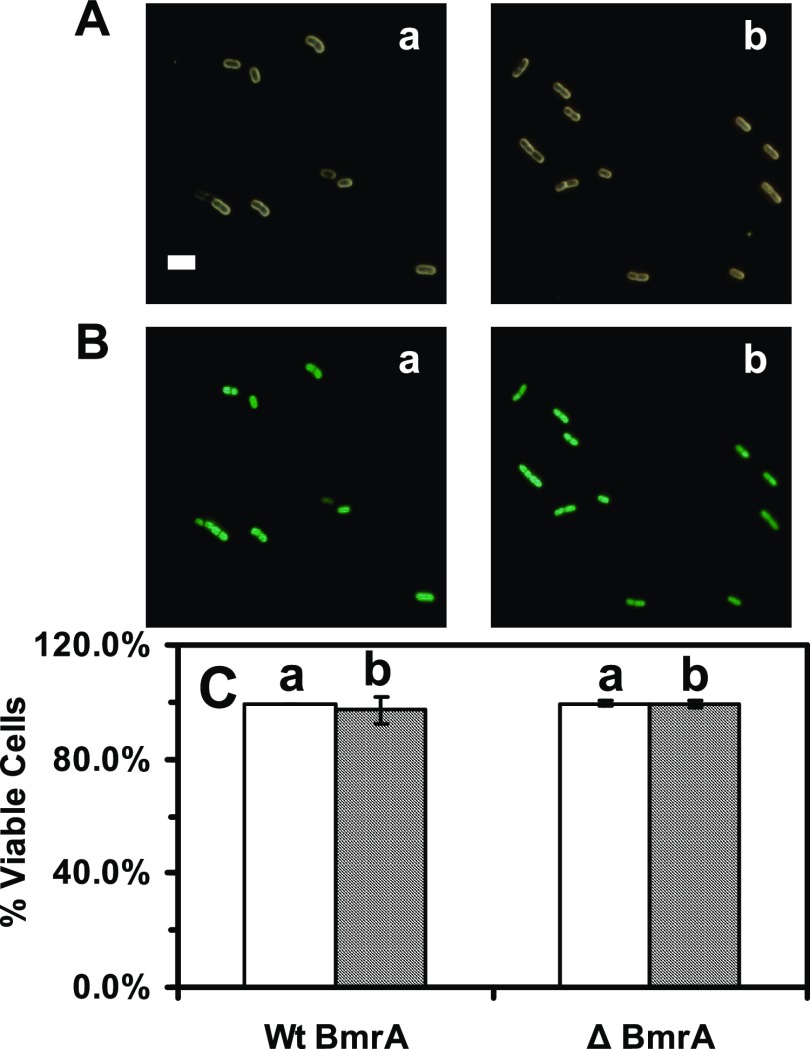
We then used live/dead BacLight assay to determine the viability of the cells, which had been cultured in the standard and modified media over 17 h. For live/dead BacLight assay,52 green fluorescent dye (SYTO9, λmax = 520 nm) stains only live cells, while a red fluorescent dye (propidium iodide, λmax = 610 nm) can only diffuse into the dead cells because of disintegrated cellular membranes of dead cells. Thus, the live cells exhibit green fluorescent, while dead cells display red fluorescent. The results in Figure 3 show that more than 97% of the cells cultured in both media over time exhibit SYTO9 green fluorescence, which demonstrates that 97% of the cells are viable. The results further demonstrate that the cells (BmrA and ΔBmrA) cultured in the modified medium grow normally and they are viable.
Figure 3.
Study of the viability of the cells (WT and ΔBmrA) cultured in (a) standard and (b) modified LB medium using live/dead BacLight assay. (A) Optical and (B) fluorescence images of the cells (e.g., WT) that were cultured over 12 h and suspended in the phosphate-buffered saline (PBS) buffer and assayed using live/dead BacLight assay. The cells exhibiting green fluorescence and red fluorescence are counted as live and dead cells, respectively. (C) Plot of the percent of the live cells (number of live cells divided by the total number of the cells) cultured in (a) standard and (b) modified LB media shows that more than 97% of the cells (WT and ΔBmrA) are viable, which further demonstrates that the modified LB medium can be used to culture the cells. The scale bar is 5 μm.
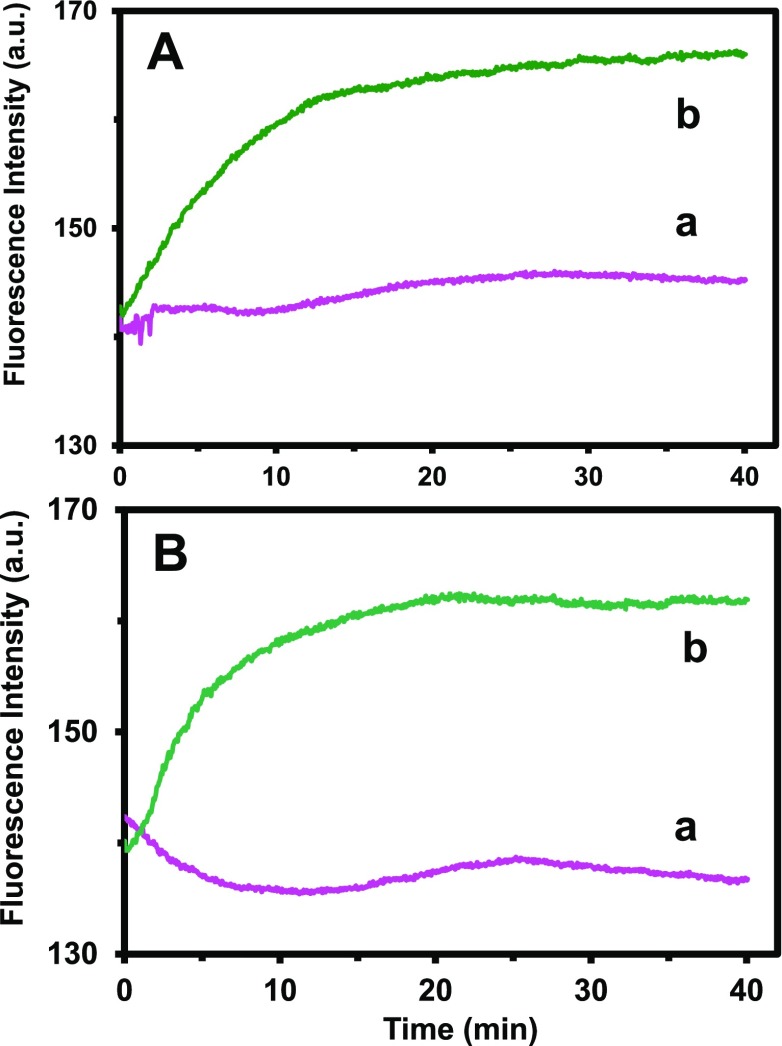
To characterize the efflux function of multidrug BmrA transporters of live cells, we studied the BmrA-dependent efflux kinetics of intracellular Hoechst dye (Hoechst 33342) in both WT and ΔBmrA live cells using fluorescence spectroscopy.39,51,53−57 Hoechst dye molecules emit weak fluorescent outside the cells, and their fluorescence intensity increases up to 10-folders as they enter the cells and intercalate with DNA.26,58 Thus, the time course fluorescence intensity of the dyes can be used to study their accumulation kinetics inside live cells in real time. The fluorescence intensity of the dyes incubated with ΔBmrA (absence of BmrA) increases with time more rapidly (Figure 4b) than those of WT (Figure 4a), showing that the efflux of the dye molecules out of the WT cells leads to a lower accumulation of intracellular dye molecules in the WT than in ΔBmrA cells. The results show the high BmrA-dependent efflux kinetics of intracellular dye in live cells that were cultured in both standard and modified cell culture media (Figure 4A,B), respectively. The efflux kinetics of cells cultured in the standard medium (Figure 4A) are similar to those cells cultured in the modified medium, further demonstrating that the modified cell culture medium is well suitable to culture WT and ΔBmrA cells that preserve the efflux function of multidrug BmrA membrane transporters.
Figure 4.
Characterization of the efflux function of membrane transporter (BmrA) in WT and ΔBmrA cells cultured in (A) standard and (B) modified LB medium. Time-dependent fluorescence intensity of 2 μM Hoechst dye incubated with the cells (OD600 nm = 0.1 in PBS buffer, pH 7.2): (a) WT and (b) ΔBmrA cultured in (A) standard and (B) modified LB media show nearly identical accumulation and efflux kinetics, demonstrating that the cells cultured in the modified LB medium possess well-functional efflux pump and the modified LB medium can be used to culture the cells for the study of accumulation and efflux kinetics of B. subtilis.
Study of Antibiotic Dose, NP Size, and BmrA Expression-Dependent Inhibitory Effects
We measured minimal inhibitory concentration (MIC) of Oflx attached onto the nanocarriers to determine whether inhibitory effects of antibiotic nanocarriers against the growth of WT-BmrA and ΔBmrA depend on the NP size, Oflx dose, and BmrA expression. The cells (WT or ΔBmrA) were cultured in the modified LB medium containing a dilution series of free Oflx alone, each type of the given sized drug nanocarriers (AgMUNH–Oflx NPs) and AgMUNH2 NPs (absence of Oflx, control experiment), and the cell growth was measured over time.
The dilution series contains 0, 0.055, 0.11, 0.22, 0.42, and 0.72 μM of free Oflx (Figure 5A,E) or Oflx carried by the NPs (AgMUNH–Oflx NPs) for WT-BmrA (Figure 5B–D) and ΔBmrA (Figure 5F–H), respectively. The corresponding concentrations of nanocarriers (NP concentration) are (Figure 5B: a–f) 0, 0.06, 0.13, 0.26, 0.49, and 0.83 nM for (2.4 ± 0.7) nm NPs with a conjugation ratio of 8.6 × 102 Oflx molecules per NP; (Figure 5C: a–f) 0, 5.8 × 10–3, 1.2 × 10–2, 2.3 × 10–2, 4.5 × 10–2, and 7.6 × 10–2 nM for (13.0 ± 3.1) nm NPs with a conjugation ratio of 9.4 × 103 Oflx molecules per NP; (Figure 5D: a–f) 0, 8.4 × 10–5, 1.7 × 10–4, 3.4 × 10–4, 6.4 × 10–4, and 1.1 × 10–3 nM for (92.6 ± 4.4) nm NPs with a conjugation ratio of 6.5 × 105 Oflx molecules per NP, respectively. The control experiments include the cells cultured with the LB medium alone (blank control, Figure 5A,E: a) or the medium containing 0.83, 7.6 × 10–2, or 1.1 × 10–3 nM AgMUNH2 NPs (in the absence of Oflx, Figure 5B–H: a) for the (2.4 ± 0.7), (13.0 ± 3.1), or (92.6 ± 4.4) nm NPs, respectively. The control experiments were conducted in parallel with the experiments with antibiotic nanocarriers and at the same conditions.
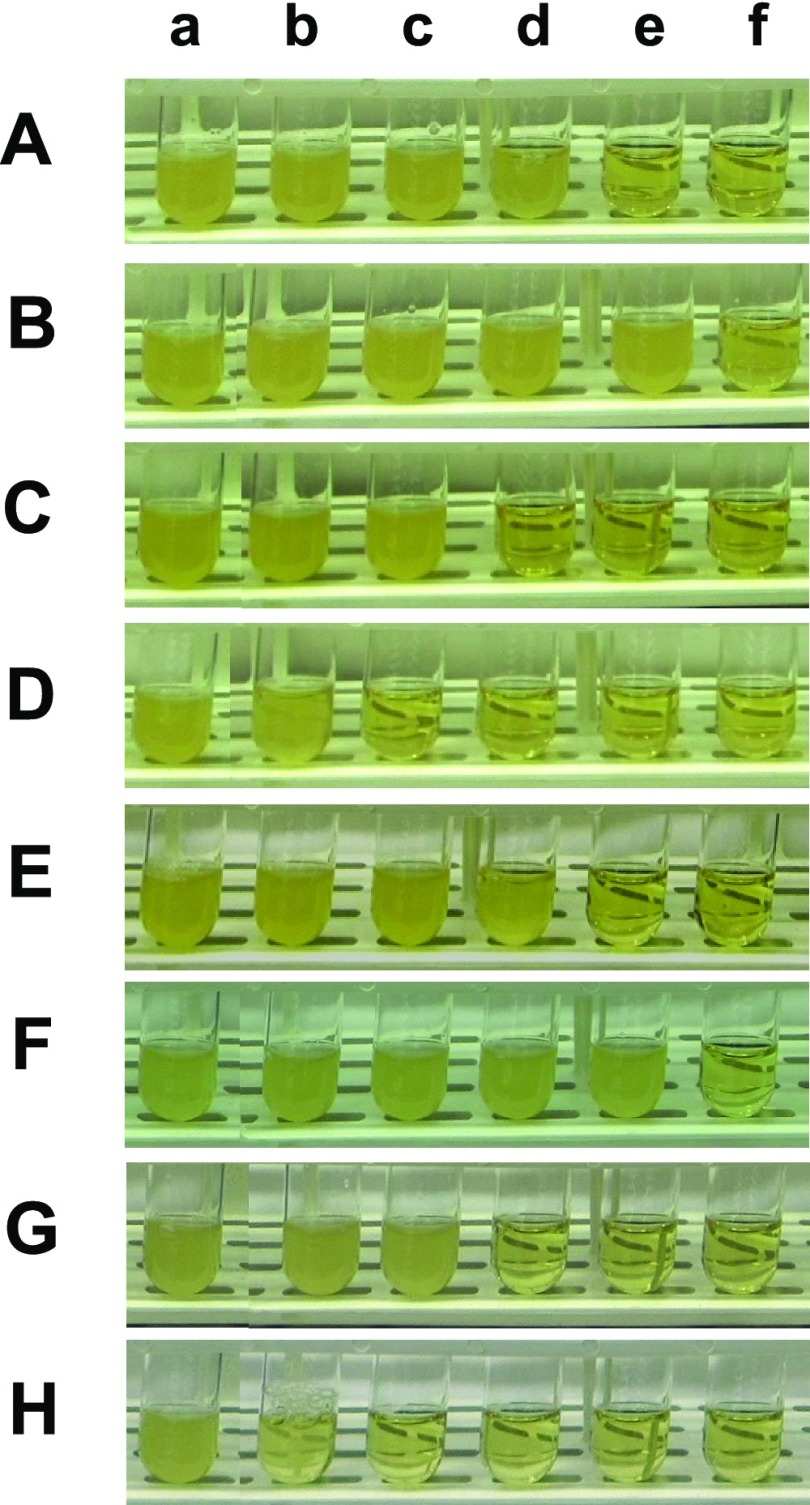
Figure 5.
Study of the dependence of inhibitory effects of antibiotic nanocarriers (AgMUNH–Oflx NPs) against the growth of (A–D) WT and (E–H) ΔBmrA cells upon antibiotic dose, NP size, and BmrA expression. Photographs of the LB medium cultured with (A–D) the WT cells and (E–H) ΔBmrA containing (a–f) 0, 0.055, 0.11, 0.22, 0.42, and 0.72 μM of (A and E) unconjugated free Oflx alone; (a–f) 0, 0.055, 0.11, 0.22, 0.42, and 0.72 μM of the Oflx conjugated with the Ag NPs of (B and F) 2.4 ± 0.7, (C and G) 13.0 ± 3.1, and (D and H) 92.6 ± 4.4 nm in diameters, respectively. The concentrations of Oflx conjugated onto the NPs are determined using their labeling ratios. The blank control experiments include the concentrations of AgMUNH2 NPs in (a) of (B–D and F–H) containing 0.83 nM, 7.6 × 10–2 nM, or 1.1 pM AgMUNH2 NPs (in the absence of Oflx) for 2.4 ± 0.7, 13.0 ± 3.1, or 92.6 ± 4.4 nm NPs, which are the same concentrations of the NPs of nanocarriers as those in (f) for each type of NPs (B–D and F–H), respectively.
We quantitatively measured the cell concentration over time by measuring their OD600 nm (optical density at 600 nm) at 5, 11, and 17 h, as shown in Figures S1 and S2 in the Electronic Online Supporting Information. Since nanocarriers and NPs possess plasmonic optical properties and molecular absorption at visible wavelengths, we subtracted the OD600 nm of the nanocarriers or AgMUNH2 NPs in the medium (in the absence of the cells) from the OD600 nm of the cell suspension with the nanocarriers or NPs to determine the cell concentration, respectively. The subtracted OD600 nm of the cell suspension was plotted over time to determine the duration (17 h) for the cell growth to reach equilibrium. The OD600 nm of the cell suspension at 17 h was then plotted versus the concentration of Oflx alone or Oflx covalently conjugated with a given sized antibiotic drug nanocarrier to determine the MIC of Oflx for each cell strain (Figure 6). The result shows that the inhibitory effects of Oflx highly depend on the dose of Oflx and size of the nanocarriers but not on the cellular expression of BmrA.
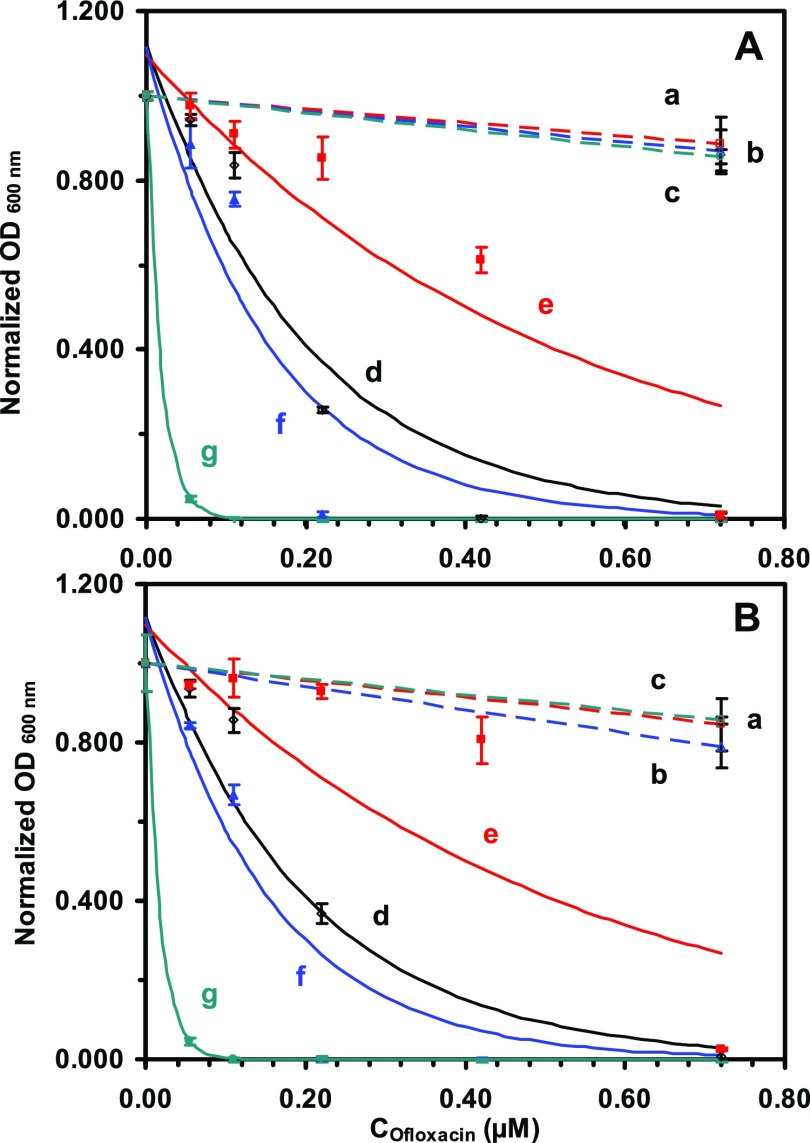
Figure 6.
Study of dose-, size-, and BmrA-dependent MICs of antibiotic nanocarriers (AgMUNH–Oflx NPs) against (A) WT and (B) ΔBmrA cells. Plots of normalized OD600 nm of the cells cultured for 17 h in the modified LB medium containing (a–c) AgMUNH2 NPs (absence of Oflx, control), (d) Oflx alone, and (e–g) Oflx linked with (e) 2.4 ± 0.7, (f) 13.0 ± 3.1, and (g) 92.6 ± 4.4 nm Ag NPs, respectively. The concentrations of AgMUNH2 NPs in (a–c) of (A) and (B) are the same as the given sized nanocarriers with the highest Oflx concentrations in (e–g) for each type of NPs in (A) and (B), but without Oflx (control experiments for the study of effects of NPs), respectively. The experimental data (points) are fitted with an equation y = a e–bx, solid line as follows: (A): (d) y = 1.12 e–5.02×, R2 = 0.918; (e) y = 1.10 e–1.964×, R2 = 0.841; (f) y = 1.11 e–6.53×, R2 = 0.881; (g) y = 1.00 e–55.48×, R2 = 1.000. (B): (d) y = 1.12 e–4.55×, R2 = 0.922; (e) y = 1.10 e–1.59×, R2 = 0.712; (f) y = 1.09 e–7.09×, R2 = 0.906; (g) y = 1.00 e–52.01×, R2 = 1.000. The MICs (IC50) of free Oflx and linked Oflx were determined at the half of the maximum of the normalized OD600 nm for each curve (solid line).
Control experiments (Figure 6: a–c) show that the OD600 nm of the cell suspension incubated with each of three different sized AgMUNH2 NPs (absence of Oflx) are nearly independent on the NP concentration and nearly the same as those cultured in the medium alone. The result indicates that the given concentration of AgMUNH2 NPs does not generate significant inhibitory effects against the growth of WT and ΔBmrA cells. Note that the NP concentration is the same as that of the highest concentration of the given sized nanocarriers, respectively. The studies have shown that the bare noble metal NPs (Ag NPs) themselves can inhibit the growth of bacteria in a concentration-dependent manner.32,59,60 Notably, the NPs that are functionalized with the surface molecules (e.g., peptides) exhibit biocompatibility of the surface molecules, instead of the bare NPs.41,45,61 Thus, the biocompatibility of AgMUNH2 NPs that we observed in the study is most likely attributed to the surface functional molecules (MUNH2) that are attached on the surface of the NPs.
On the contrary, the OD600 nm of the cell suspension incubated with Oflx alone or Oflx attached onto the given sized nanocarriers decrease as Oflx concentration increases, showing that their inhibitory effects against the growth of WT and ΔBmrA cells highly depend upon the dose of Oflx and size of nanocarriers (Figure 6A,B: d–f). We fitted the experimental data with an exponential decay equation (y = a e–bx), which represents the inhibitory effect upon the exponential cell growth. We define the concentration of Oflx needed to reduce the growth of the cells in the medium alone to the half as the MIC of Oflx. The results of MICs are summarized in Table 1, showing that the MICs of free Oflx and Oflx attached onto the nanocarriers highly depend upon the dose of Oflx and the size of nanocarriers but not the cellular expression of BmrA.
Table 1. Study of Dependence of MIC of Oflx upon the Size of Nanocarriers and Expression of BmrA of Two Strains of B. subtilis (WT and ΔBmrA).
| MIC50 of Oflx (μM)a |
||
|---|---|---|
| samples | WT | ΔBmrA |
| free Oflx alone | 0.16 ± 0.00 | 0.17 ± 0.02 |
| nanocarriers (2.4 ± 0.7 nm) | 0.40 ± 0.02 | 0.49 ± 0.03 |
| nanocarriers (13.0 ± 3.1 nm) | 0.12 ± 0.01 | 0.11 ± 0.01 |
| nanocarriers (92.6 ± 4.4 nm) | 0.012 ± 0.001 | 0.012 ± 0.001 |
For free Oflx alone (Figure 6A,B: d), the OD600 nm of the cell suspension decrease with the Oflx concentration, showing the MICs of 0.16 ± 0.00 and 0.17 ± 0.02 μM Oflx for the WT and ΔBmrA cells, respectively.
Interestingly, for 2.4 ± 0.7 nm drug nanocarriers with a conjugation ratio of 8.6 × 102 Oflx molecules/NP (Figure 6A,B: e), the OD600 nm of the cell suspension decrease with the Oflx concentration less rapidly than those of free Oflx and two other larger nanocarriers. Notably, for 13.0 ± 3.1 nm drug nanocarriers with a conjugation ratio of 9.4 × 103 Oflx molecules/NP (Figure 6A,B: f), the OD600 nm of the cell suspension decreases with the Oflx concentration are nearly the same as those of free Oflx, showing the nearly identical inhibitory and MICs of 0.12 ± 0.01 and 0.11 ± 0.01 μM Oflx for the WT and ΔBmrA cells, as those of free Oflx, respectively. For 92.6 ± 4.4 nm drug nanocarriers with a conjugation ratio of 6.5 × 105 Oflx molecules/NP (Figure 6A,B: g), the OD600 nm of the cell suspension decrease with the Oflx concentration the most rapidly, showing the highest inhibitory effects and the lowest MICs of 0.012 ± 0.001 and 0.012 ± 0.001 μM Oflx for the WT and ΔBmrA cells, respectively.
The MICs of either free Oflx or Oflx attached onto nanocarriers for WT-BmrA and ΔBmrA cells are the same, suggesting that either form of Oflx could be extruded out of the cells by other membrane transporters in both WT-BmrA and ΔBmrA. In other words, Oflx is not a specific substrate of BmrA, and other membrane transporters are primary forces to extrude the Oflx out of the cells, which leads to the insignificant (or masked) contribution of BmrA and the BmrA-independent MICs of Oflx.
Comparison of Both Studies and Distinctive Findings
By comparing the results in this study with our previous study,30 we found that Oflx attached onto the nanocarriers retains its efficacy (inhibitory effect) against both Gram-positive bacteria (B. subtilis) and Gram-negative bacteria (P. aeruginosa). Inhibitory effects of nanocarriers against both strains of bacteria highly depend upon the size of nanocarriers, further demonstrating that the closed-packed Oflx molecules on the NPs (multivalence) could increase local drug dose, enhance their binding affinity with the target, and offer higher potency against both B. subtilis and P. aeruginosa. Notably, the inhibitory effects of nanocarriers are not linearly proportional to their sizes. For example, the smallest antibiotic nanocarriers (2.4 ± 0.7 nm) show the lowest inhibitory effects and the highest MICs against both strains than free Oflx, while the mid-sized nanocarriers (13.0 ± 3.1 nm) display the nearly identical inhibitory effects and MICs as free Oflx, and the largest antibiotic nanocarriers (92.6 ± 4.4 nm) exhibit the highest inhibitory effects and the lowest MICs against both strains.
The primary difference between both studies is their inhibitory effects on two different types of the cells (B. subtilis and P. aeruginosa) and their dependence on the efflux pump. For the previous study,30 the inhibitory effects of nanocarriers highly depend upon the expression of MexAB-OprM (multidrug membrane transporter) of P. aeruginosa (Gram-negative bacterium). In this study, they are independent upon the expression of BmrA (multidrug membrane transporter) of B. subtilis (Gram-positive bacterium). Therefore, the inhibitory effects of nanocarriers against bacteria show the selectivity, suggesting the possibility of using them as medicines. Notably, all three sized nanocarriers can enter B. subtilis (Gram-positive bacteria) and P. aeruginosa(Gram-negative bacteria), even though B. subtilis (Gram-positive bacteria) possess a very different envelop structure from P. aeruginosa (Gram-negative bacteria), suggesting potential common applications against bacteria. Further, the results demonstrate that antibiotic drug nanocarriers show specificity toward the given membrane transporters and their potentially wide utility against both Gram-positive and Gram-negative bacteria.
Summary
In summary, we have successfully used three different sized antibiotic drug nanocarriers (AgMUNH–Oflx NPs) to study their size-dependent inhibitory effects against Gram-positive bacteria (Bacillus subtilis, WT-BmrA, and ΔBmrA). We have designed a modified cell culture medium, which enables the WT-BmrA and ΔBmrA cells to grow normally and the antibiotic drug nanocarriers to be stable (nonaggregated) in the medium over the entire duration of cell culture (17 h). Thus, the size and dose of nanocarriers remain unchanged during their incubation with the cells for 17 h, enabling us to study the size- and dose-dependent effects of the drug nanocarriers on the cell growth and efflux function of BmrA. Control experiments of three sized AgMUNH2 NPs (absence of Oflx) show insignificant inhibitory effects toward WT-BmrA and ΔBmrA, which are likely attributed to the surface functional molecules (MUNH2) attached onto the NPs. The significant findings include that (i) the largest nanocarriers create the highest inhibitory effects, while the smallest nanocarriers generate the lowest inhibitory effects, demonstrating that the same number of antibiotic molecules (Oflx) that are carried and delivered by the larger NPs can produce the higher inhibitory effects. These results indicate that the closed-packed Oflx molecules on the surface of NPs might strengthen their binding affinity with the target (multivalence) and provide larger payload to increase local targeting dose, leading to the higher potency. (ii) The inhibitory effects of Oflx depend upon the multivalent local targeting effects, as well as their intracellular distribution and concentrations. (iii) Their inhibitory effects do not significantly depend upon the cellular expression of multidrug BmrA membrane transporter, suggesting that other transporters in ΔBmrA could effectively extrude the antibiotic nanocarriers out of the ΔBmrA cells, leading to the same MICs as WT-BmrA. These findings show that the inhibitory effects of nanocarriers are not linearly proportional to their sizes, suggesting the possibility for one to design the optimal-sized nanocarriers to create the most potent effect of antibiotics against a specific given bacterial strain and to potentially evade a specific given multidrug membrane transporter. Efforts are being made to identify the specific membrane transporters that are responsible for the extrusion of drug nanocarriers out of WT-BmrA and ΔBmrA cells and to study their underlying molecular mechanisms.
Materials and Methods
Characterization of Cell Culture
We used two strains of Gram-positive bacterial cells (B. subtilis): WT (normal expression BmrA) and ΔBmrA previously named as ΔYvcC (a mutant strain that is devoid of the bmrA, also named as ΔyvcC or ΔbmrA) that were isogenic strains and provided by Jault.26 The cells were first precultured in a commonly used standard LB medium (1% tryptone, 0.5% yeast extract, and 0.5% NaCl in DI water, pH = 7.2) in a shaker (MaxQ 5000, 200 rpm, 37 °C) for 12 h. The precultured cells (20 μL) were then further cultured in the 2 mL standard LB medium or the modified LB medium (1% tryptone, 0.5% yeast extract, and 0.1% NaCl in DI water, pH = 7.2) in a shaker (MaxQ 5000, 200 rpm, 37 °C) for another 17 h. The cell growth curves were determined by measuring the OD600 nm of cell suspension in the medium over 17 h of cell culture. The cell suspension was diluted to the OD600 nm of cell suspension below 0.2 and measured.
By the end of cell culture, we used live/dead BacLight viability and counting assay (Invitrogen) to assay the viability of the cultured cells.52 We used dark-field optical microscopy and epi-fluorescence microscopy to image the cells in a microchamber containing the medium.33,37,39,40,53 We acquired the green and red fluorescence cell images and counted them as live and dead cells, respectively.
By the end of the cell culture, we used centrifugation to harvest the cells (Beckman JA-14, 7500 rpm) and rinsed the cells with the PBS buffer (0.5 mM phosphate buffer, 1.5 mM NaCl, pH 7.0) three times. We suspended the cells in the PBS buffer and adjusted to a desired cell suspension concentration (OD600 nm = 0.1) in the buffer. We continuously measured the fluorescence intensity of Hoechst 3342 dye (Invitrogen) of the cell suspension (OD600 nm = 0.1) containing 2 μM of the dye at a 3 s time interval for 2 h using a fluorescence spectrometer (PerkinElmer LS50B) with the excitation and emission wavelengths at 350 and 488 nm, respectively.
Synthesis, Purification, and Characterization of Drug Nanocarriers
We synthesized and characterized the antibiotic drug nanocarriers, as reported previously.30 Briefly, we synthesized, purified, and characterized (2.4 ± 0.7), (13.0 ± 3.1), and (92.6 ± 4.4) nm Ag NPs, as reported previously.39,42,62 We used centrifugation to thoroughly wash the NPs three times with the DI water immediately after the synthesis. We used UV–vis spectroscopy (Hitachi U-2010), dark-field optical microscopy and spectroscopy (DFOMS), high-resolution transmission electron microscopy (JEOL, JEM-2100F), and dynamic light scattering (DLS) (Nicomp 380ZLS particle sizing system) to measure the NP concentrations, the LSPR images, the spectra of single NPs, and the sizes of single NPs, respectively.33,36,37,39,42−45,47,49,55,62 We used the interaction of thiol groups of MUNH2 with the NPs to attach 11-amino-1-undecanethiol hydrochloride (MUNH2, AUT, 99%, Sigma-Aldrich) onto the surface of NPs to prepare functional AgMUNH2 NPs. We used centrifugation (Beckman Optima L90k, 4 °C) to wash the AgMUNH2 NPs thoroughly with DI water three times to remove excess MUNH2. We used a two-step method via 1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide hydrochloride (EDC) and N-hydroxysulfosuccinimide (s-NHS) as mediators to link the amine groups of each sized AgMUNH2 NPs with the carboxyl group of Oflx via peptide bonds (Figure 1). We used centrifugation to wash the nanocarriers with DI water to purify the drug nanocarriers (AgMUNH–Oflx NPs) and stored them at 4°C for future use. We used UV–vis spectroscopy, DFOMS, and DLS to characterize the concentrations, optical properties, and sizes of each sized AgMUNH2 NPs, respectively.
We used UV–vis absorbance spectra of Oflx at 288 nm and the plasmonic absorption spectra of the NPs to measure the molar concentration of Oflx and NPs, respectively. We then divided the molar concentrations of Oflx molecules on the surface of the nanocarriers by the molar concentration of the NPs to determine the molar labeling ratios of antibiotics (Oflx) to NPs for each sized drug nanocarriers.30
Study of Stability of Drug Nanocarriers in Cell Culture Medium
We used UV–vis absorption spectroscopy, DLS, and dark-field optical microscopy and spectroscopy to characterize the concentration, size, and optical properties of nanocarriers and studied their stability (nonaggregation) in the commonly used standard LB medium and the modified medium over 24 h, respectively. We found that the given concentrations of 2.4 ± 0.7, 13.0 ± 3.1, and 92.6 ± 4.4 nm nanocarriers (6.0 nM, 0.8 nM, and 7 pM) are stable (nonaggregated) in the modified medium over 24 h, but they are unstable (aggregated) in the standard medium.30
Characterization of Dose- and Size-Dependent Inhibitory Effects of Nanocarriers
The cells (WT or ΔBmrA) were precultured in the standard LB medium overnight and then cultured in the modified LB medium (2.5 mL) containing a dilution series of free Oflx alone, given sized drug nanocarriers (AgMUNH–Oflx NPs), and AgMUNH2 NPs (control experiments) by inoculating 104 precultured cells into the medium in a shaker (200 rpm, 37 °C) over 17 h.
The dilution series of 0, 0.055, 0.11, 0.22, 0.42, and 0.72 μM free Oflx or Oflx conjugated with the NPs (AgMUNH–Oflx NPs) were prepared in the modified cell culture medium to culture WT and ΔBmrA (Figure 5). They are correlated with the drug nanocarrier (NP) concentrations: (i) 0, 0.06, 0.13, 0.26, 0.49, and 0.83 nM for (2.4 ± 0.7) nm NPs with a conjugation ratio of 8.6 × 102 Oflx molecules per NP; (ii) 5.8 × 10–3, 1.2 × 10–2, 2.3 × 10–2, 4.6 × 10–2, and 7.6 × 10–2 nM for (13.0 ± 3.1) nm NPs with a conjugation ratio of 9.4 × 103 Oflx molecules per NP; and (iii) 8.4 × 10–2, 1.7 × 10–1, 3.4 × 10–1, 6.4 × 10–1, and 1.11 pM for (92.6 ± 4.4) nm NPs with a conjugation ratio of 6.5 × 105 Oflx molecules per NP. The control experiments include the modified LB medium only (blank control) and containing 0.83 nM, 7.6 × 10–2 nM, or 1.1 pM AgMUNH2 NPs (in the absence of Oflx) for 2.4 ± 0.7, 13.0 ± 3.1, or 92.6 ± 4.4 nm NPs, respectively.
We quantitatively determined the OD600 nm of the cell suspension in a 96-well plate using a plate reader (BioTek Synergy HT) equipped with a UV–vis absorption spectral detector at 5, 11, and 17 h. We plotted the OD600 nm of the cell suspension vs time to determine the duration (17 h) for the cells to reach the confluence. We then used the plots of the OD600 nm of each cell suspension at 17 h versus Oflx concentration (free Oflx or Oflx attached onto the nanocarrier) to determine their inhibitory effects (MIC). Specifically, we normalized the OD600 nm of each cell suspension with the maximum OD600 nm (the cells cultured in the medium alone, blank control), respectively. We plotted the normalized OD600 nm of the cell suspension versus the concentration of free Oflx (Oflx alone) or the concentration of Oflx attached onto a given sized drug nanocarrier. We used the exponential decay (y = a e–bx) to fit the experimental data and to determine the parameters (a, b) of the equation with the highest possible regression. The MIC50 (the concentration of Oflx at which the cell growth was inhibited to the half of the cell growth of the blank control experiment) was then determined. Unlike conventional semiquantitative methods to estimate the MICs using solid culture plates (a colony-forming unit), we developed this new approach to quantitatively determine the MIC50 using the dilution series of antibiotics or antibiotic nanocarriers in the liquid LB medium and mathematically fitting the experimental data. Further, the dilution series of antibiotics in the liquid LB medium enabled the nanocarriers to be well dispersed and avoided the aggregation of nanocarriers in the solid culture plates and allowed us to characterize the stability of nanocarriers in the liquid medium in real time at single NP resolution.
Acknowledgments
This work was supported, in part, by NSF (CBET 0507036 and 1450936) and NIH (R01 GM0764401, R21HL127580, and R15GM119116). All work was done in the Xu Lab. J.-M.J. provided ΔBmrA (also called as ΔyvcC or ΔbmrA) cells and discussion. The authors thank Tao Huang for participating in the synthesis and characterization of nanocarriers.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b03698.
Growth curve of WT-BmrA cells incubated with (A) free Oflx, (B) 2.4 ± 0.7, (C) 13.0 ± 3.1, and (D) 92.6 ± 4.4 nm Oflx nanocarriers, and AgMUNH2 NPs (in the absence of Oflx, control experiment) (Figure S1); and growth curve of ΔBmrA cells incubated with (A) free Oflx, (B) 2.4 ± 0.7, (C) 13.0 ± 3.1, and (D) 92.6 ± 4.4 nm Oflx nanocarriers, and AgMUNH2 NPs (in the absence of Oflx, control experiment) (Figure S2) (PDF)
Author Contributions
§ P.K.C., P.S., and F.D. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Horsey A. J.; Cox M. H.; Sarwat S.; Kerr I. D. The multidrug transporter ABCG2: still more questions than answers. Biochem. Soc. Trans. 2016, 44, 824–830. 10.1042/BST20160014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 1992, 8, 67–113. 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Robey R. W.; Pluchino K. M.; Hall M. D.; Fojo A. T.; Bates S. E.; Gottesman M. M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher T.; Benndorf R. A. ABC Transport Proteins in Cardiovascular Disease-A Brief Summary. Molecules 2017, 22, 589. 10.3390/molecules22040589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. P.; Bhardwaj G.; Gerlach J. H.; Mackie J. E.; Grant C. E.; Almquist K. C.; Stewart A. J.; Kurz E. U.; Duncan A. M.; Deeley R. G. Overxpression of a transporter gene in a mutidrug-resistant human lung cancer cell lin. Science 1992, 258, 1650–1654. 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- Grube M.; Hagen P.; Jedlitschky G. Neurosteroid Transport in the Brain: Role of ABC and SLC Transporters. Front Pharmacol. 2018, 9, 354. 10.3389/fphar.2018.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhalen B.; Dastvan R.; Thangapandian S.; Peskova Y.; Koteiche H. A.; Nakamoto R. K.; Tajkhorshid E.; Mchaourab H. S. Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein. Nature 2017, 543, 738–741. 10.1038/nature21414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M. M.; Ambudkar S. V.; Xia D. Structure of a multidrug transporter. Nat. Biotechnol. 2009, 27, 546–547. 10.1038/nbt0609-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F. ABC transporters: physiology, structure and mechanism--an overview. Res. Microbiol. 2001, 152, 205–210. 10.1016/S0923-2508(01)01193-7. [DOI] [PubMed] [Google Scholar]
- Hung L. W.; Wang I. X.; Nikaido K.; Liu P. Q.; Ames G. F.; Kim S. H. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature 1998, 396, 703–707. 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- Locher K. P.; Lee A. T.; Rees D. C. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 2002, 296, 1091–1098. 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- Davidson A. L.; Chen J. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 2004, 73, 241–268. 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- Holland I. B.; Blight M. A. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 1999, 293, 381–399. 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- Dawson R. J.; Locher K. P. Structure of a bacterial multidrug ABC transporter. Nature 2006, 443, 180–185. 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- Higgins L. K. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 2004, 11, 918–926. 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- Jones P. M.; George A. M. Mechanism of the ABC transporter ATPase domains: catalytic models and the biochemical and biophysical record. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 39–50. 10.3109/10409238.2012.735644. [DOI] [PubMed] [Google Scholar]
- Mishra S.; Verhalen B.; Stein R. A.; Wen P. C.; Tajkhorshid E.; Mchaourab H. S. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. elife 2014, 3, e0274. 10.7554/eLife.02740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. P. Multidrug resistance protein 1 (MRP1, ABCC1), a ″multitasking″ ATP-binding cassette (ABC) transporter. J. Biol. Chem. 2014, 289, 30880–30888. 10.1074/jbc.R114.609248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam S. K. What do drug transporters really do?. Nat. Rev. Drug Discovery 2015, 14, 29–44. 10.1038/nrd4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. I.; Williams R. T.; Henderson M. J.; Norris M. D.; Haber M. ABC transporters as mediators of drug resistance and contributors to cancer cell biology. Drug Resist. Updates 2016, 2, 1–9. 10.1016/j.drup.2016.03.001. [DOI] [PubMed] [Google Scholar]
- McIntosh K.; Balch C.; Tiwari A. K. Tackling multidrug resistance mediated by efflux transporters in tumor-initiating cells. Exp. Opin. Drug Metab. Toxicol. 2016, 12, 633–644. 10.1080/17425255.2016.1179280. [DOI] [PubMed] [Google Scholar]
- Trowitzsch S.; Tampé R. ABC Transporters in Dynamic Macromolecular Assemblies. J. Mol. Biol. 2018, 430, 4481–4495. 10.1016/j.jmb.2018.07.028. [DOI] [PubMed] [Google Scholar]
- Aguilar C.; Vlamakis H.; Losick R.; Kolter R. Thinking about Bacillus subtilis as a multicellular organism. Curr. Opin. Microbiol. 2007, 10, 638–643. 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y.; Fichant G.; Denizot F. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 1999, 287, 467–484. 10.1006/jmbi.1999.2624. [DOI] [PubMed] [Google Scholar]
- Völker U.; Hecker M. From genomics via proteomics to cellular physiology of the Gram-positive model organism Bacillus subtilis. Cell Microbiol. 2005, 7, 1077–1085. 10.1111/j.1462-5822.2005.00555.x. [DOI] [PubMed] [Google Scholar]
- Steinfels E.; Orelle C.; Fantino J. R.; Dalmas O.; Rigaud J. L.; Denizot F.; Di Pietro A.; Jault J. M. Characterization of YvcC (BmrA), a multidrug ABC transporter constitutively expressed in Bacillus subtilis. Biochemistry 2004, 43, 7491–7502. 10.1021/bi0362018. [DOI] [PubMed] [Google Scholar]
- Paton J. H.; Reeves D. S. Fluoroquinolone antibiotics. Microbiology, pharmacokinetics and clinical use. Drugs 1988, 36, 193–228. 10.2165/00003495-198836020-00004. [DOI] [PubMed] [Google Scholar]
- Marslin G.; Revina A. M.; Khandelwal V. K.; Balakumar K.; Sheeba C. J.; Franklin G. PEGylated ofloxacin nanoparticles render strong antibacterial activity against many clinically important human pathogens. Colloids Surf., B 2015, 132, 62–70. 10.1016/j.colsurfb.2015.04.050. [DOI] [PubMed] [Google Scholar]
- Baptista P. V.; McCusker M. P.; Carvalho A.; Ferreira D. A.; Mohan N. M.; Martins M.; Fernandes A. R. Nano-Strategies to Fight Multidrug Resistant Bacteria-″A Battle of the Titans″. Front Microbiol. 2018, 9, 1441. 10.3389/fmicb.2018.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F.; Songkiatisak P.; Cherukuri P. K.; Huang T.; Xu X. Size-Dependent Inhibitory Effects of Antibiotic Drug Nanocarriers against Pseudomonas aeruginosa. ACS Omega 2018, 3, 1231–1243. 10.1021/acsomega.7b01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natan M.; Banin E. From Nano to Micro: using nanotechnology to combat microorganisms and their multidrug resistance. FEMS Microbiol. Rev. 2017, 41, 302–322. 10.1093/femsre/fux003. [DOI] [PubMed] [Google Scholar]
- Kyriacou S. V.Real-time Study of Multidrug Resistance Mechanism in Pseudomonas aeruginosa Using Nanoparticle Optics and Single Live Cell Imaging; Old Dominion University: Norfolk, VA, 2003. [Google Scholar]
- Xu X.-H. N.; Chen J.; Jeffers R. B.; Kyriacou S. V. Direct measurement of sizes and dynamics of single living membrane transporters using nano-optics. Nano Lett. 2002, 2, 175–182. 10.1021/nl015682i. [DOI] [Google Scholar]
- Browning L. M.; Lee K. J.; Cherukuri P. K.; Huang T.; Songkiatisak P.; Warren S.; Xu X. Single gold nanoparticle plasmonic spectroscopy for study of chemical-dependent efflux function of single ABC transporters of single live Bacillus subtilis cells. Analyst 2018, 143, 1599–1608. 10.1039/C7AN01787A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning L. M.; Lee K. J.; Cherukuri P. K.; Huang T.; Warren S.; Xu X.-H. N. Single nanoparticle plasmonic spectroscopy for study of charge-dependent efflux function of multidrug ABC transporters of single live Bacillus subtilis cells. J. Phys. Chem. C 2016, 120, 21007–21016. 10.1021/acs.jpcc.6b03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning L. M.; Lee K. J.; Cherukuri P. K.; Nallathamby P. D.; Warren S.; Jault J.-M.; Xu X.-H. N. Single nanoparticle plasmonic spectroscopy for study of efflux function of multidrug ABC membrane transporters of single live cells. RSC Adv. 2016, 6, 36794–36802. 10.1039/C6RA05895G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriacou S. V.; Brownlow W. J.; Xu X. H. N. Using nanoparticle optics assay for direct observation of the function of antimicrobial agents in single live bacterial cells. Biochemistry 2004, 43, 140–147. 10.1021/bi0351110. [DOI] [PubMed] [Google Scholar]
- Lee K. J.; Browning L. M.; Huang T.; Ding F.; Nallathamby P. D.; Xu X.-H. N. Probing of multidrug ABC membrane transporters of single living cells using single plasmonic nanoparticle optical probes. Anal. Bioanal. Chem. 2010, 397, 3317–3328. 10.1007/s00216-010-3864-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallathamby P. D.; Lee K. J.; Desai T.; Xu X.-H. N. Study of multidrug membrane transporters of single living pseudomonas aeruginosa cells using size-dependent plasmonic nanoparticle optical probes. Biochemistry 2010, 49, 5942–5953. 10.1021/bi100268k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.-H. N.; Brownlow W. J.; Kyriacou S. V.; Wan Q.; Viola J. J. Real-time probing of membrane transport in living microbial cells using single nanoparticle optics and living cell imaging. Biochemistry 2004, 43, 10400–10413. 10.1021/bi036231a. [DOI] [PubMed] [Google Scholar]
- Huang T.; Browning L. M.; Xu X.-H. N. Far-field photostable optical nanoscopy (PHOTON) for real-time super-resolution single-molecular imaging of signaling pathways of single live cells. Nanoscale 2012, 4, 2797–2812. 10.1039/c2nr11739h. [DOI] [PubMed] [Google Scholar]
- Huang T.; Nallathamby P. D.; Gillet D.; Xu X.-H. N. Design and synthesis of single nanoparticle optical biosensors for imaging and characterization of single receptor molecules on single living cells. Anal. Chem. 2007, 79, 7708–7718. 10.1021/ac0709706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning L. M.; Lee K. J.; Huang T.; Nallathamby P. D.; Lowman J.; Xu X.-H. N. Random walk of single gold nanoparticles in zebrafish embryos leading to stochastic toxic effects on embryonic developments. Nanoscale 2009, 1, 138–152. 10.1039/b9nr00053d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. J.; Browning L. M.; Nallathamby P. D.; Desai T.; Cherukuri P.; Xu X.-H. N. In vivo quantitative study of size-dependent transport and toxicity of single silver nanoparticles using zebrafish embryos. Chem. Res. Toxicol. 2012, 25, 1029–1046. 10.1021/tx300021u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. J.; Nallathamby P. D.; Browning L. M.; Osgood C. J.; Xu X.-H. N. In vivo imaging of transport and biocompatibility of single silver nanoparticles in early development of zebrafish embryos. ACS Nano 2007, 1, 133–143. 10.1021/nn700048y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. J.; Nallathamby P. D.; Browning L. M.; Desai T.; Cherukuri P.; Xu X.-H. N. Single nanoparticle spectroscopy for real-time in vivo quantitative analysis of transport and toxicity of single nanoparticles in single embryos. Analyst 2012, 137, 2973–2986. 10.1039/c2an35293a. [DOI] [PubMed] [Google Scholar]
- Nallathamby P. D.; Lee K. J.; Xu X.-H. N. Design of stable and uniform single nanoparticle photonics for in vivo dynamics imaging of nanoenvironments of zebrafish embryonic fluids. ACS Nano 2008, 2, 1371–1380. 10.1021/nn800048x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallathamby P. D.; Xu X.-H. N. Study of cytotoxic and therapeutic effects of stable and purified silver nanoparticles on tumor cells. Nanoscale 2010, 2, 942–952. 10.1039/c0nr00080a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallathamby P. D.; Huang T.; Xu X.-H. N. Design and characterization of optical nano rulers of single nanoparticles using optical microscopy and spectroscopy. Nanoscale 2010, 2, 1715–1722. 10.1039/c0nr00303d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.-H. N.Far-Field Photostable Optical Nanoscopy (PHOTON). In Encyclopedia of Spectroscopy and Spectrometry, 3rd ed.; Lindon J., Tranter G. E., Koppenaal D., Eds.; Elsevier: Oxford, 2017; Vol. 1, pp 566–570. [Google Scholar]
- Xu X.-H. N.; Song Y.; Nallathamby P. D.. Probing Membrane Transport of Single Live Cells Using Single Molecule Detection and Single Nanoparticle Assay. In New Frontiers in Ultrasensitive Bioanalysis: Advanced Analytical Chemistry Applications in Nanobiotechnology, Single Molecule Detection, and Single Cell Analysis; Xu X.-H. N., Ed.; Wiley: NJ, 2007; pp 41–65. [Google Scholar]
- Berney M.; Hammes F.; Bosshard F.; Weilenmann H. U.; Egli T. Assessment and Interpretation of Bacterial Viability By Using The LIVE/DEAD Baclight Kit In Combination With Flow Cytometry. Appl. Environ. Microbiol. 2007, 73, 3283–3290. 10.1128/AEM.02750-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriacou S. V.; Nowak M. E.; Brownlow W. J.; Xu X.-H. N. Single live cell imaging for real-time monitoring of resistance mechanism in pseudomonas aeruginosa. J. Biomed. Opt. 2002, 7, 576–586. 10.1117/1.1506707. [DOI] [PubMed] [Google Scholar]
- Ocaktan A.; Yoneyama H.; Nakae T. Use of fluorescence probes to monitor function of the subunit proteins of the MexA-MexB-OprM drug extrusion machinery in Pseudomonas aeruginosa. J. Biol. Chem. 1997, 272, 21964–21969. 10.1074/jbc.272.35.21964. [DOI] [PubMed] [Google Scholar]
- Xu X.-H. N.; Brownlow W. J.; Huang S.; Chen J. Real-time measurements of single membrane pump efficiency of single living pseudomonas aeruginosa cells using fluorescence microscopy and spectroscopy. Biochem. Biophys. Res. Commun. 2003, 305, 79–86. 10.1016/S0006-291X(03)00886-6. [DOI] [PubMed] [Google Scholar]
- Xu X.-H. N.; Wan Q.; Kyriacou S. V.; Brownlow W. J.; Nowak M. E. Direct observation of substrate induction of resistance mechanism in Pseudomonas aeruginosa using single live cell imaging. Biochem. Biophys. Res. Commun. 2003, 305, 941–949. 10.1016/S0006-291X(03)00874-X. [DOI] [PubMed] [Google Scholar]
- Ding F.; Lee K. J.; Vahedi-Faridi A.; Yoneyama H.; Osgood C. J.; Xu X. H. N. Design and study of the efflux function of the EGFP fused MexAB-OprM membrane transporter in Pseudomonas aeruginosa using fluorescence spectroscopy. Analyst 2014, 139, 3088–3096. 10.1039/C4AN00108G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosa G.; Focsaneanu K.-S.; McLean J. R. N.; McNamee J. P.; Scaiano J. C. Photophysical properties of fluorescent DNA-dyes bound to single- and double-stranded DNA in aqueous buffered solution. Photochem. Photobiol. 2001, 73, 585–599. . [DOI] [PubMed] [Google Scholar]
- Nisar P.; Ali N.; Rahman L.; Ali M.; Shinwari Z. K. Antimicrobial activities of biologically synthesized metal nanoparticles: an insight into the mechanism of action. J. Biol. Inorg. Chem. 2019, 27, 927. 10.1007/s00775-019-01717-7. [DOI] [PubMed] [Google Scholar]
- Lv D.; Wang R.; Tang G.; Mou Z.; Lei J.; Han J.; De Smedt S.; Xiong R.; Huang C. Ecofriendly Electrospun Membranes Loaded with Visible-Light-Responding Nanoparticles for Multifunctional Usages: Highly Efficient Air Filtration, Dye Scavenging, and Bactericidal Activity. ACS Appl. Mater. Interfaces 2019, 11, 12880–12889. 10.1021/acsami.9b01508. [DOI] [PubMed] [Google Scholar]
- Lee K. J.; Browning L. M.; Nallathamby P. D.; Xu X. H. N. Study of charge-dependent transport and toxicity of peptide-functionalized silver nanoparticles using zebrafish embryos and single nanoparticle plasmonic spectroscopy. Chem. Res. Toxicol. 2013, 26, 904–917. 10.1021/tx400087d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.; Nallathamby P. D.; Xu X. H. N. Photostable Single-molecule Nanoparticle Optical Biosensors for Real-time Sensing of Single Cytokine Molecules and their Binding Reactions. J. Am. Chem. Soc. 2008, 130, 17095–17105. 10.1021/ja8068853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.