Key Points
Question
Which genetic loci are associated with keratoconus?
Findings
In this case-control genome-wide association study of a discovery cohort and 3 independent replication cohorts, a locus containing multiple variants across 6 protein-coding genes on chromosome 11 was associated with keratoconus. Several of these genes are likely involved in apoptotic pathways.
Meaning
This study of patients with keratoconus and control participants showed a potential role of genes involved in apoptotic pathways.
This case-control, genome-wide association study assesses genetic susceptibility regions for keratoconus via a genome-wide association study of a discovery cohort and 3 independent replication cohorts from the United States, Northern Ireland, and Australia.
Abstract
Importance
Keratoconus is a condition in which the cornea progressively thins and protrudes in a conical shape, severely affecting refraction and vision. It is a major indication for corneal transplant. To discover new genetic loci associated with keratoconus and better understand the causative mechanism of this disease, we performed a genome-wide association study on patients with keratoconus.
Objective
To identify genetic susceptibility regions for keratoconus in the human genome.
Design, Setting, and Participants
This study was conducted with data from eye clinics in Australia, the United States, and Northern Ireland. The discovery cohort of individuals with keratoconus and control participants from Australia was genotyped using the Illumina HumanCoreExome single-nucleotide polymorphism array. After quality control and data cleaning, genotypes were imputed against the 1000 Genomes Project reference panel (phase III; version 5), and association analyses were completed using PLINK version 1.90. Single-nucleotide polymorphisms with P < 1.00 × 10−6 were assessed for replication in 3 additional cohorts. Control participants were drawn from the cohorts of the Blue Mountains Eye Study and a previous study of glaucoma. Replication cohorts were from a previous keratoconus genome-wide association study data set from the United States, a cohort of affected and control participants from Australia and Northern Ireland, and a case-control cohort from Victoria, Australia. Data were collected from January 2006 to March 2019.
Main Outcomes and Measures
Associations between keratoconus and 6 252 612 genetic variants were estimated using logistic regression after adjusting for ancestry using the first 3 principal components.
Results
The discovery cohort included 522 affected individuals and 655 control participants, while the replication cohorts included 818 affected individuals (222 from the United States, 331 from Australia and Northern Ireland, and 265 from Victoria, Australia) and 3858 control participants (2927 from the United States, 229 from Australia and Northern Ireland, and 702 from Victoria, Australia). Two novel loci reached genome-wide significance (defined as P < 5.00 × 10−8), with a P value of 7.46 × 10−9 at rs61876744 in patatin-like phospholipase domain–containing 2 gene (PNPLA2) on chromosome 11 and a P value of 6.35 × 10−12 at rs138380, 2.2 kb upstream of casein kinase I isoform epsilon gene (CSNK1E) on chromosome 22. One additional locus was identified with a P value less than 1.00 × 10−6 in mastermind-like transcriptional coactivator 2 (MAML2) on chromosome 11 (P = 3.91 × 10−7). The novel locus in PNPLA2 reached genome-wide significance in an analysis of all 4 cohorts (P = 2.45 × 10−8).
Conclusions and Relevance
In this relatively large keratoconus genome-wide association study, we identified a genome-wide significant locus for keratoconus in the region of PNPLA2 on chromosome 11.
Introduction
Keratoconus is characterized by progressive thinning of the cornea, the clear tissue at the front of the eye. Asymmetrical bulging and conical protrusion of the cornea leads to extreme refractive error (myopia and irregular astigmatism), causing severe visual impairment.1 Keratoconus is relatively common, with a reported prevalence of around 55 per 100 000 individuals in white populations1 and up to 229 per 100 000 individuals in Asian populations.2 Because of recent advances in diagnostic imaging, it is now thought that the true incidence and prevalence of keratoconus may be 5 to 10 times higher than previously reported.3 The causative mechanism of keratoconus is poorly understood. Associations have been made with eye rubbing and atopy, but no direct connection has been established.4 Various biochemical pathways may be involved, including oxidative stress, apoptosis, and disruption to extracellular matrix turnover.5
Although many cases of keratoconus present as sporadic, there is a well-recognized genetic component to the disease. The estimated prevalence in relatives of patients with keratoconus is 3.34% (95% CI, 3.22%-3.46%), which is 15 to 67 times higher than in the general population.6 In addition, more than 20 syndromes are associated with keratoconus, including Down syndrome, Leber congenital amaurosis, and several connective tissue disorders.7 Linkage studies have identified at least 16 loci for keratoconus8,9; however, the causative genes and variants have remained elusive. Common variants in the dedicator of cytokinesis 9 (DOCK9)10 and lysyl oxidase (LOX)11 genes have been implicated, as well as rare mutations in microRNA 184 (MIR184),12,13 although these loci have not been broadly replicated. Genome-wide association studies (GWAS) have implicated several loci. Variation in the promoter region of the hepatocyte growth factor (HGF) gene14,15 and upstream of the Rab3 guanosine triphosphatase–activating protein catalytic subunit (RAB3GAP1) gene16,17 have both been associated in multiple independent studies. Furthermore, a GWAS for central corneal thickness (CCT) identified loci that are also associated with keratoconus, including retinoid X receptor alpha–collagen alpha-1 (RXRA–COL5A1), forkhead box protein O1 (FOXO1), and fibronectin type III domain containing 3B (FNDC3B),18 and more recently, a suggestive but nonsignificant association at the decorin (DCN) gene.19
We present findings from a GWAS of 522 patients with keratoconus, a relatively large sample for this complex disease, plus control participants. We also sought association and independent replication in additional cohorts.
Methods
Study Design
We report a study of 4 independent cohorts of white patients with keratoconus. The first case-control cohort used for the discovery phase included patients with keratoconus and control participants. All single-nucleotide polymorphisms (SNPs) with P values less than 1.00 × 10−6 were looked up in imputed genotypes from a previously published GWAS study of patients with keratoconus and control participants from the United States. In addition, SNPs were genotyped in an independent replication cohort of affected individuals and control participants from Australia and Northern Ireland and additional affected individuals and control participants from a Victoria, Australia, cohort.
Ethics
The protocol was approved by the Southern Adelaide Clinical Human Research Ethics Committee, and the Human Research Ethics Committee of the Royal Victorian Eye and Ear Hospital, and the Health and Medical Human Research Ethics Committee of the University of Tasmania. All participants gave written informed consent, and the study conformed to the tenets of the Declaration of Helsinki.
Discovery Cohort
Participants with keratoconus were ascertained through the eye clinic of Flinders Medical Centre, Adelaide, Australia; optometry and ophthalmology clinics in Adelaide and Melbourne, Australia; or an Australia-wide invitation to members of Keratoconus Australia, a community-based support group for patients. Clinical data were obtained from the participants’ eye care practitioners.
The diagnosis of keratoconus was based on both clinical examination and videokeratography pattern analysis, as described previously.20 Clinical examinations included slitlamp biomicroscopy, cycloplegic retinoscopy, and fundus evaluation. Slitlamp biomicroscopy was used to identify stromal corneal thinning, Vogts striae, or a Fleischer ring. Retinoscopy examinations were performed on a fully dilated pupil to determine the presence or absence of retroillumination signs of keratoconus, such as the oil-droplet sign and scissoring of the red reflex. Videokeratography evaluation was performed on each eye using the Orbscan (Orbtek [Bausch & Lomb]). Patients were classified as having keratoconus if they had at least 1 clinical sign of keratoconus and a confirmatory videokeratography.21 A history of penetrating keratoplasty performed because of keratoconus was also sufficient for inclusion as an affected individual. Patients with syndromic forms of keratoconus were excluded, and if multiple individuals from the same family presented, only 1 was included in the study.
Control participant data were obtained from the Australian cohort previously described in a GWAS for age-related macular degeneration (AMD) from the International AMD Genomics Consortium22; this cohort has been described in detail previously. For the current analysis, data from 676 Australian unaffected control participants (including 465 from the Blue Mountains Eye Study23 and healthy individuals previously recruited as control participants for a study of glaucoma24) were combined as control participants for keratoconus. Genetically related individuals and those who did not pass all sample quality-control procedures for the AMD GWAS were excluded. The DNA for affected individuals and control participants was extracted from whole blood using the QiaAMP DNA Maxi kit (Qiagen).
Genotyping and Data Quality Control
Individuals with keratoconus were genotyped for 551 839 variants with the HumanCoreExome array (HumanCoreExome-24v1-1_A [Illumina]). For the control participants, genotypes of 569 645 variants were generated with a customized Illumina HumanCoreExome array (called HumanCoreExome_Goncalo_15038949_A), as described previously.22
Quality-control procedures were carried out according to the protocol described by Anderson et al,25 modified as follows. Reverse and ambiguous-strand SNPs were detected using snpflip (https://github.com/biocore-ntnu/snpflip; accessed March 24, 2017) and flipped or excluded. Only SNPs common to both arrays were included. Individuals with discordant sex information, a missing genotype rate greater than 0.05, or heterozygosity more than 3 SDs from the mean were excluded. Genetically related individuals were detected by calculating pairwise identity by descent, and the individual with the lower genotyping rate in any pair with identity by descent greater than 0.185 was removed. Ancestry outliers were identified by principal component analysis using EIGENSTRAT26 and removed. Markers were excluded if they had missing genotype rate greater than 3%, had significantly different missing data rates between affected individuals and control participants, had minor allele frequency less than 0.01, or deviated significantly (P < 10−5) from Hardy-Weinberg equilibrium. Following all exclusions and quality control, affected individuals and control participants were genotyped for 264 115 common-platform SNPs.
Genomic Imputation and Association Analysis
We phased autosomal genotype data using Eagle (version 2.3.5 [Broad Institute])27 and then imputed genotypes on the basis of the European super population of the 1000 Genomes Project reference panel (phase III; version 5)28 using Minimac3 (version 2.0.1 [Center for Statistical Genetics]).29 We excluded indels, SNPs within 5 base pairs of an indel, rare variants (minor allele frequency <0.01), and variants with poor imputation quality (R2 <0.8). This filtering yielded a total of 6 252 612 quality-controlled variants, including 250 964 genotyped variants. Association analyses were performed on the most-likely genotypes under a logistic regression model using PLINK (version 1.90 [Christopher Chang/Grail Inc])30 using the first 3 principal components as covariates. Any P value less than 5.00 × 10−8 was considered significant.
Replication Cohorts and Analyses
The US cohort has been previously described.17 Briefly, white individuals who were clinically affected by keratoconus were enrolled into the GWAS17 as a part of the longitudinal videokeratography and genetic study at the Cornea Genetic Eye Institute.6 After removing samples with poor genotyping quality, samples were included in the analysis.
Control participants were obtained from the Cardiovascular Health Study (CHS), a population-based cohort study of risk factors for cardiovascular disease and stroke in adults 65 years or older, recruited at 4 field centers.31,32 The CHS was approved by the institutional review board at each recruitment site, and participants provided informed consent for the use of their genetic information. African American CHS participants were excluded from analysis because of an insufficient number of racially/ethnically matched affected individuals. The samples included in the analysis were therefore from self-reported white individuals. Participants did not have an eye examination to exclude keratoconus. Outliers detected by principal component analysis were excluded, and the analysis was adjusted for the top 3 principal components.
IMPUTE version 2.3.0 (Marchini Research Group) was used to perform imputation of the genotyping data from 370K BeadChip arrays (Illumina) in patients with keratoconus and control participants from the CHS using 1000 Genomes Phase I data as the reference panel. All SNPs with P values less than 1.00 × 10−6 identified in the discovery analysis were looked up and extracted, with the exception of 4 SNPs at the casein kinase I isoform epsilon gene (CSNK1E) locus, which were not imputed in the US study. The SNPs with P values less than 1.00 × 10−6 in the discovery cohort were selected for genotyping in additional replication cohorts using the MassARRAY System (Agena Bioscience) by the Australian Genome Research Facility. The SNPs were chosen from each locus that were compatible with the assay design, with preference given to SNPs with the smallest P value in the discovery cohort. Twenty-seven SNPs were genotyped in additional affected individuals recruited under the same protocol as the discovery cohort, as well as in patients from Northern Ireland, as described previously.14 These individuals were compared with unaffected examined control participants, consisting of individuals from the Blue Mountains Eye Study not included in the discovery cohort and older individuals recruited from nursing homes in the Launceston area of Tasmania, Australia. All control participants underwent a thorough ocular examination, and keratoconus was excluded. These SNPs were also genotyped in a replication cohort from Melbourne, Victoria, Australia, consisting of affected individuals and examined control participants, as described previously.15
Association was assessed in each cohort individually using logistic regression. Analysis of results from discovery and replication cohorts was performed using METAL version 2011-03-25 (University of Michigan Center for Statistical Genetics).33
Functional Annotation of Associated Variants
The lead SNP at the novel locus on chromosome 11, rs61876744, was queried in HaploReg version 4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php; accessed May 28, 2019),34 including data from the Genotype–Tissue Expression (GTex) pilot analysis.35 Genes in the associated region were assessed for ocular tissue expression using the Ocular Tissue Database (https://genome.uiowa.edu/otdb/)36 and differential expression between corneas from patients with keratoconus and myopia in a previously published study.37
Results
Cohort Characteristics
The first case-control cohort included 522 participants with keratoconus (290 men [55.6%]; mean [SD] age, 45 [15.2] years) and 655 control participants (307 men [46.9%]; mean [SD] age, 65 [10.6] years), after exclusions were completed. Candidate P values (<1.00 × 10−6) were looked up in a previous GWAS cohort of 222 US patients with keratoconus (123 men [55.4%]; mean [SD] age, 44 [13.3] years) and 3324 US control participants (1149 men [39.3%]; mean [SD] age, 72 [5.4] years). Another 27 SNPs were genotyped in the Australia and Northern Ireland cohort, which included 331 affected individuals (186 from Australia and 175 from Northern Ireland; 203 men [61.3%]; mean [SD] age, 41 [15.9] years) and 229 control participants (84 from the Blue Mountains Study in Australia who were not included in the discovery cohort and 145 nursing home residents in Launceston, Tasmania, Australia; 84 men [36.7%]; mean [SD] age, 75 [11.5] years), and the cohort from Melbourne, Victoria, Australia, which included 265 affected individuals (159 men [60.0%]; mean [SD] age, 35 [14.9] years) and 702 control participants (268 men [38.2%]; mean [SD] age, 52 [15.2] years). The demographic data for each cohort are given in Table 1.
Table 1. Demographics of the Australian Discovery Cohort and the 3 Replication Cohorts.
| Cohort | Cases | Controls | ||||
|---|---|---|---|---|---|---|
| Total, No. | Male, No. (%) | Age, Mean (SD), y | Total, No. | Male, No. (%) | Age, Mean (SD), y | |
| Discovery (Australia) | 522 | 290 (55.6) | 45 (15.2) | 655 | 307 (46.9) | 65 (10.6) |
| Replication | ||||||
| United States | 222 | 123 (55.4) | 44 (13.3) | 2927 | 1149 (39.3) | 72 (5.4) |
| Australia and Northern Ireland | 331 | 203 (61.3) | 41 (15.9) | 229 | 84 (36.7) | 75 (11.5) |
| Victoria, Australia | 265 | 159 (60.0) | 35 (14.9) | 702 | 268 (38.2) | 52 (15.2) |
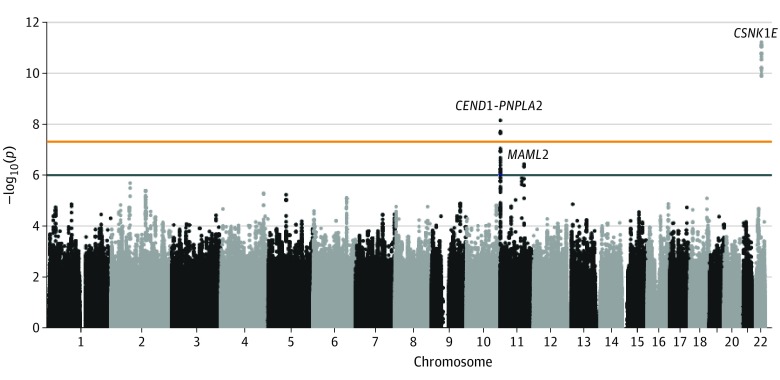
Genome-Wide Association Testing of the Discovery Cohort
Genome-wide association analysis was conducted in the discovery cohort (Figure; eFigure 1 in the Supplement). The genomic inflation factor was λ1000 = 1.023, and all included samples were from individuals of European ancestry (eFigure 2 in the Supplement). Two novel loci reached genome-wide significance (defined as P < 5.00 × 10−8), with a P value of 7.46 × 10−9 at rs61876744 in the patatin-like phospholipase domain–containing 2 gene (PNPLA2) on chromosome 11 and a P value of 6.35 × 10−12 at rs138380, 2.2 kb upstream of the CSNK1E gene, on chromosome 22. One additional locus was identified (P < 1.00 × 10−6) in the mastermind-like transcriptional coactivator 2 (MAML2) gene on chromosome 11 (rs10831500; P = 3.91 × 10−7). Association results for all SNPs with P values less than 1.00 × 10−6 are shown in eTable 1 in the Supplement. Locus-specific plots for all 3 loci are shown in eFigure 3 in the Supplement. The locus on chromosome 11 showing genome-wide significance included 25 SNPs with P values less than 1.00 × 10−6 spanning 6 protein-coding genes (cell cycle exit and neuronal differentiation 1 [CEND1], solute carrier family 25 member 22 [SLC25A22], proapoptotic nucleolar protein 1 [PANO1], P53-induced death domain protein 1 [PIDD1], 60S acidic ribosomal protein P2 [RPLP2], and PNPLA2) and multiple RNA genes.
Figure. Manhattan Plot of Association Results in the Discovery Cohort.
Results of logistic regression with the first 3 principal components as covariates (−log10 P values) are plotted for each chromosome. The orange and black lines represent the genome-wide significance threshold of a P value of 5.00 × 10−8 and the threshold for follow-up of a P value of 1.00 × 10−6, respectively.
Because previously reported keratoconus risk loci were initially identified in GWAS for CCT, we assessed each of the loci reaching suggestive significance in the current analysis in the previously reported analysis for CCT. As shown in Table 2, only the MAML2 locus showed nominal association with CCT (in this analysis: minor allele frequency: affected individuals, 0.422; control participants, 0.330; P = 3.91 × 10−7; in the CCT analysis: P = .01). We also looked up the lead SNPs from the CCT analysis in the results from the discovery cohort (eTable 2 in the Supplement). The SNPs at the FNDC3B gene (in this analysis: minor allele frequency: affected individuals, 0.240; control participants, 0.193; P = 1.82 × 10−3; in the CCT analysis: P = 7.22 × 10−14), the multiple PDZ domain crumbs cell polarity complex component (MPDZ) gene (in this analysis: minor allele frequency: affected individuals, 0.231; control participants, 0.170; P = 7.81 × 10−3; in the CCT analysis: P = 3.01 × 10−4), and the mothers against decapentaplegic homolog 3 (SMAD3) gene (in this analysis: minor allele frequency: affected individuals, 0.172; control participants, 0.211; P = 8.03 × 10−3; in the CCT analysis: P = 6.40 × 10−9) genes showed nominal associations in this analysis.
Table 2. Lead Single-Nucleotide Polymorphisms at All 3 Loci With Significant Results in the Discovery Cohort.
| Locusa | Chr | Lead Single-Nucleotide Polymorphism | Base Pairb | Allele 1/2 | Minor Allele Frequency in Affected Individuals | Minor Allele Frequency in Control Participants | Odds Ratio (95% CI)c | P Value | P Value in Central Corneal Thickness Studyd |
|---|---|---|---|---|---|---|---|---|---|
| PNPLA2 | 11 | rs61876744 | 820754 | T/C | 0.341 | 0.447 | 0.59 (0.49-0.71) | 7.46 × 10−9 | .55 |
| MAML2 | 11 | rs10831500 | 95982642 | G/T | 0.422 | 0.330 | 1.59 (1.33-1.91) | 3.91 × 10−7 | .01 |
| CSNK1E | 22 | rs138380 | 38796629 | G/A | 0.384 | 0.524 | 0.49 (0.40-0.60) | 6.35 × 10−12 | .79 |
Abbreviations: Chr, chromosome; CSNK1E, casein kinase I isoform epsilon; MAML2, mastermind-like transcriptional coactivator 2; PNPLA2, patatin-like phospholipase domain containing 2.
Locus assigned to the RefSeq protein-coding gene within or near the association signal interval.
Genomic positions are based on hg19.
Odds ratios are with respect to allele 1.
P values in the previously reported analysis for central corneal thickness.19
Association Testing of the Replication Cohorts
All SNPs with P values less than 1.00 × 10−6 identified in the discovery cohort were analyzed in the previously generated GWAS data for the US cohort. Twenty-seven SNPs compatible with a single-assay design were genotyped in the Australia and Northern Ireland and Victoria, Australia, replication cohorts (Table 3; eTable 3 in the Supplement).
Table 3. Replication and Analysis of Association Results at Lead SNPs.
| Locusa | Chr | Lead SNPb | Base Pairc | Allele 1/2 | Discovery Cohort | Replication Cohorts | Discovery and Replication Analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| United States | Australia and Northern Ireland | Victoria, Australia | ||||||||||||
| Odds Ratio (95% CI) | P Value | Odds Ratio (95% CI) | P Value | Odds Ratio (95% CI) | P Value | Odds Ratio (95% CI) | P Value | P Value | Direction | |||||
| PNPLA2 | 11 | rs61876744 | 820754 | T/C | 0.59 (0.49-0.71) | 7.46 × 10−9 | 0.77 (0.63-0.95) | 8.88 × 10−4 | 0.84 (0.65-1.08) | .18 | 0.97 (0.79-1.19) | .76 | 2.45 × 10−8 | −, −, −, − |
| MAML2 | 11 | rs10831500 | 95982642 | G/T | 1.59 (1.33-1.91) | 3.91 × 10−7 | 1.35 (1.11-1.65) | 5.69 × 10−3 | 1.09 (0.85-1.40) | .52 | 1.03 (0.84-1.26) | .79 | 3.83 × 10−6 | +, +, +, + |
| CSNK1E | 22 | rs138378 | 38796159 | A/G | 0.50 (0.41-0.61) | 1.77 × 10−11 | 0.95 (0.78-1.16) | .79 | 0.97 (0.76-1.24) | .82 | 0.93 (0.76-1.13) | .45 | 3.18 × 10−4 | −, −, −, − |
Abbreviations: Chr, chromosome; CSNK1E, casein kinase I isoform epsilon; MAML2, mastermind-like transcriptional coactivator 2; PNPLA2, patatin-like phospholipase domain containing 2; SNP, single-nucleotide polymorphism.
Locus assigned to the RefSeq protein-coding gene within or near the association signal interval.
The lead SNP at CSNK1E (rs138380) was not successfully genotyped in the Australia and Northern Ireland and Victoria, Australia, cohorts, and a proxy (rs138378, R2 = 0.938) was used for replication instead.
Genomic positions are based on hg19.
Multiple SNPs in both novel loci at PNPLA2 and MAML2 showed association with P values less than .05 in the US cohort. None of the SNPs in the CSNK1E locus reached significance, although all showed the same direction of negative association as in the discovery cohort. None of the 3 loci reached significance in either the Australia and Northern Ireland or the Victoria, Australia, replication cohort, but most SNPs showed the same (varying) directions of association as in the discovery cohort. Minor allele frequencies for affected individuals and control participants in each cohort are given in eTable 3 in the Supplement.
Analysis of Combined Data
Analysis of the combined data from the discovery and all 3 replication cohorts found 12 SNPs at the novel PNPLA2 locus to be associated with keratoconus at genome-wide significance (P < 5.00 × 10−8). The MAML2 locus on chromosome 11 showed suggestive but nonsignificant association, with a P value of 3.83 × 10−6 at rs10831500. The CSNK1E locus on chromosome 22 reached a P value of 3.18 × 10−4 at rs138378. For all 3 loci, there were other SNPs with smaller P values in the combined analysis, but these did not include data for all replication cohorts (eTable 3 in the Supplement).
Functional Annotations of Novel Associated Loci
At the significant locus on chromosome 11, the lead SNP rs61876744 is located in the second intron of the PNPLA2 gene (NM_020376.3). The associated region extends for around 40 kbp, encompassing multiple transcripts. The PNPLA2 gene is highly expressed in all eye tissues assessed in the ocular tissue database, including the cornea, as are other protein-coding genes at this locus (eTable 4 in the Supplement). A recent study37 compared gene expression in corneal epithelium from patients with keratoconus and myopia and found that PNPLA2 and PIDD1 were differentially expressed, with a false-discovery rate (FDR) of less than 0.05 (PNPLA2: log2 fold change, −1.277; P = 1.38 × 10−4; FDR, 0.028; PIDD1: log2 fold change, −1.429; P = 1.19 × 10−3; FDR, 0.046; eTable 4 in the Supplement), while RPLP2 and CSNK1E are significant at an FDR less than 0.10 (RPLP2: log2 fold change, −0.659; P = 5.37 × 10−3; FDR, 0.078; CSNK1E: log2 fold change, −0.436; P = 5.30 × 10−3; FDR, 0.078).
HaploReg identified 7 SNPs in strong linkage disequilibrium with the lead SNP (rs61876744), and all report an expression quantitative trait locus (eQTL) for an antisense RNA transcript AP006621 (multiple transcripts 1 through 8) in multiple tissues assessed in GTex (eTable 5 in the Supplement), in which the more common allele, C, is associated with increased transcript levels (P = 4.6 × 10−18 for AP006621.5 in sun-exposed skin; eFigure 4 in the Supplement). This RNA gene is not represented in the ocular tissue database. A similar finding is seen in GTex for the PIDD1 gene in sun-exposed skin (P = 1.5 × 10−16; eFigure 4 in the Supplement).
Discussion
This study has identified a candidate locus for keratoconus on chromosome 11 that shows replication in the US data and consistent direction of association in the other cohorts. The lead SNP is located in an intron of PNPLA2. This gene encodes patatin-like phospholipase domain-containing protein 2, which catalyzes the initial step in triglyceride hydrolysis. The relevance of this pathway to keratoconus is not obvious, but it is well known that the closest gene to an association signal is not necessarily the causative gene. At least 4 other protein-coding genes at this locus are also expressed in the cornea, and RNA-coding genes are also annotated in the region. There is a strong eQTL signal of the lead SNP rs61876744 for an antisense RNA gene, AP006621.8, which is located on the opposite strand to the protein-coding PNPLA2 gene. The antisense RNA AP006621 transcripts may have a role in regulating PNPLA2 or other genes at this locus and elsewhere. The minor allele at rs61876744, T, is associated with reduced risk of keratoconus and reduced expression of AP006621 in many tissues. This suggests that overexpression of AP006621 may destabilize corneal structures. Oxidative stress and apoptosis have been suggested as part of the pathogenesis of keratoconus5 and sun (or ultraviolet light) exposure is known to trigger oxidative stress and DNA-damage pathways.38 Several genes at this locus likely play a role in apoptotic pathways, including PPID and PANO1.
The chromosome 11 locus overlaps with a previously reported (although not genome-wide significant) association signal for Fuchs endothelial corneal dystrophy (FECD).39 The lead SNP in the FECD GWAS is rs12223324 in the Parkinson disease 7 domain-containing protein 1 (PDDC1) gene, which is upstream of PNPLA2. This SNP does not reach significance in this keratoconus GWAS. It is unknown how this locus might lead to FECD, but the overlap of genetic association with keratoconus is intriguing, given that both diseases affect the cornea. Although rare, there are reports in the literature of patients with both FECD and keratoconus.40,41 The participants in the current study do not have FECD, and thus this disease does not account for the association observed here.
Although we observed the strongest association in the discovery cohort at the CSNK1E locus, this result was not replicated. The signal appears to be driven by a single genotyped SNP that has influenced the imputation of a surrounding linkage disequilibrium block (eTable 1 and eFigure 3 in the Supplement). The signal at the MAML2 locus is supported by the US replication cohort. Further replication of these loci is required before any firm conclusions can be drawn.
Previous GWAS for keratoconus (using a subset of cases involved in the current study) reported RAB3GAP117 and a region upstream of the HGF gene,14 although neither study reached genome-wide significance. The lead SNPs at these loci reached nonsignificant P values in the current discovery cohort, suggesting these loci are not major contributors in this better-powered study. Previously reported keratoconus-associated loci with genome-wide significance (RXRA–COL5A1, FOXO1, and FNDC3B) are also associated with CCT. The current findings suggest that mechanisms other than susceptibility to a thinner cornea may also be at play in the genetic risk of keratoconus.
Limitations
All control cohorts used in this study had older mean ages than the case cohorts; thus, it will be important to assess these loci for age-associated effects in future studies. Batch effects are a potential problem in an analysis in which cases and controls are genotyped separately; however, the low inflation factor seen in this analysis offers reassurance that batch effects are unlikely to have a major influence.
Conclusions
In summary, this study has identified a locus for keratoconus on chromosome 11. The lead SNP is in an intron of the PNPLA2 gene and an eQTL for a long noncoding RNA, AP006621.8. We have also assessed loci near MAML2 and CSNK1E that require further replication. It is very likely that additional risk loci exist for keratoconus, and larger studies will be needed to identify them.
eTable 1. Association results for all SNPs with P < 10-6 in the discovery cohort
eTable 2. Association results for lead SNPs at CCT-mediated keratoconus loci
eTable 3. Replication and meta-analysis of top ranked SNPs
eTable 4. Expression of genes in associated regions
eTable 5. Functional annotations of the lead SNP at the PNPLA2 locus, rs61876744, and SNPs in high linkage disequilibrium
eFigure 1. Q-Q plot of p-values from logistic regression analysis of discovery cohort with first three principal components as covariates
eFigure 2. Ancestry clustering based on genome-wide association data
eFigure 3. Locus specific plots for the three loci reaching suggestive or genome-wide significance in the discovery cohort
eFigure 4. eQTL data from GTex project for SNP rs61876744
eReferences.
References
- 1.Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42(4):297-319. doi: 10.1016/S0039-6257(97)00119-7 [DOI] [PubMed] [Google Scholar]
- 2.Pearson AR, Soneji B, Sarvananthan N, Sandford-Smith JH. Does ethnic origin influence the incidence or severity of keratoconus? Eye (Lond). 2000;14(Pt 4):625-628. doi: 10.1038/eye.2000.154 [DOI] [PubMed] [Google Scholar]
- 3.Godefrooij DA, de Wit GA, Uiterwaal CS, Imhof SM, Wisse RP. Age-specific incidence and prevalence of keratoconus: a nationwide registration study. Am J Ophthalmol. 2017;175:169-172. doi: 10.1016/j.ajo.2016.12.015 [DOI] [PubMed] [Google Scholar]
- 4.Zadnik K, Barr JT, Edrington TB, et al. Baseline findings in the Collaborative Longitudinal Evaluation of Keratoconus (CLEK) Study. Invest Ophthalmol Vis Sci. 1998;39(13):2537-2546. [PubMed] [Google Scholar]
- 5.Wojcik KA, Blasiak J, Szaflik J, Szaflik JP. Role of biochemical factors in the pathogenesis of keratoconus. Acta Biochim Pol. 2014;61(1):55-62. doi: 10.18388/abp.2014_1923 [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Rabinowitz YS, Rotter JI, Yang H. Genetic epidemiological study of keratoconus: evidence for major gene determination. Am J Med Genet. 2000;93(5):403-409. doi: [DOI] [PubMed] [Google Scholar]
- 7.Nowak DM, Gajecka M. The genetics of keratoconus. Middle East Afr J Ophthalmol. 2011;18(1):2-6. doi: 10.4103/0974-9233.75876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abu-Amero KK, Al-Muammar AM, Kondkar AA. Genetics of keratoconus: where do we stand? J Ophthalmol. 2014;2014:641708. doi: 10.1155/2014/641708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burdon KP, Vincent AL. Insights into keratoconus from a genetic perspective. Clin Exp Optom. 2013;96(2):146-154. doi: 10.1111/cxo.12024 [DOI] [PubMed] [Google Scholar]
- 10.Czugala M, Karolak JA, Nowak DM, et al. Novel mutation and three other sequence variants segregating with phenotype at keratoconus 13q32 susceptibility locus. Eur J Hum Genet. 2012;20(4):389-397. doi: 10.1038/ejhg.2011.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bykhovskaya Y, Li X, Epifantseva I, et al. Variation in the lysyl oxidase (LOX) gene is associated with keratoconus in family-based and case-control studies. Invest Ophthalmol Vis Sci. 2012;53(7):4152-4157. doi: 10.1167/iovs.11-9268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes AE, Bradley DT, Campbell M, et al. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am J Hum Genet. 2011;89(5):628-633. doi: 10.1016/j.ajhg.2011.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lechner J, Bae HA, Guduric-Fuchs J, et al. Mutational analysis of MIR184 in sporadic keratoconus and myopia. Invest Ophthalmol Vis Sci. 2013;54(8):5266-5272. doi: 10.1167/iovs.13-12035 [DOI] [PubMed] [Google Scholar]
- 14.Burdon KP, Macgregor S, Bykhovskaya Y, et al. Association of polymorphisms in the hepatocyte growth factor gene promoter with keratoconus. Invest Ophthalmol Vis Sci. 2011;52(11):8514-8519. doi: 10.1167/iovs.11-8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahebjada S, Schache M, Richardson AJ, Snibson G, Daniell M, Baird PN. Association of the hepatocyte growth factor gene with keratoconus in an Australian population. PLoS One. 2014;9(1):e84067. doi: 10.1371/journal.pone.0084067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae HA, Mills RA, Lindsay RG, et al. Replication and meta-analysis of candidate loci identified variation at RAB3GAP1 associated with keratoconus. Invest Ophthalmol Vis Sci. 2013;54(7):5132-5135. doi: 10.1167/iovs.13-12377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Bykhovskaya Y, Haritunians T, et al. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum Mol Genet. 2012;21(2):421-429. doi: 10.1093/hmg/ddr460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y, Vitart V, Burdon KP, et al. ; NEIGHBOR Consortium . Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet. 2013;45(2):155-163. doi: 10.1038/ng.2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iglesias AI, Mishra A, Vitart V, et al. ; Blue Mountains Eye Study—GWAS group; NEIGHBORHOOD Consortium; Wellcome Trust Case Control Consortium 2 (WTCCC2) . Cross-ancestry genome-wide association analysis of corneal thickness strengthens link between complex and mendelian eye diseases. Nat Commun. 2018;9(1):1864. doi: 10.1038/s41467-018-03646-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burdon KP, Coster DJ, Charlesworth JC, et al. Apparent autosomal dominant keratoconus in a large Australian pedigree accounted for by digenic inheritance of two novel loci. Hum Genet. 2008;124(4):379-386. doi: 10.1007/s00439-008-0555-z [DOI] [PubMed] [Google Scholar]
- 21.Rabinowitz YS. Videokeratographic indices to aid in screening for keratoconus. J Refract Surg. 1995;11(5):371-379. [DOI] [PubMed] [Google Scholar]
- 22.Fritsche LG, Igl W, Bailey JN, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48(2):134-143. doi: 10.1038/ng.3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burdon KP, Mitchell P, Lee A, et al. Association of open-angle glaucoma loci with incident glaucoma in the Blue Mountains Eye Study. Am J Ophthalmol. 2015;159(1):31-6.e1. doi: 10.1016/j.ajo.2014.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimasi DP, Burdon KP, Hewitt AW, et al. Genetic investigation into the endophenotypic status of central corneal thickness and optic disc parameters in relation to open-angle glaucoma. Am J Ophthalmol. 2012;154(5):833-842.e2. doi: 10.1016/j.ajo.2012.04.023 [DOI] [PubMed] [Google Scholar]
- 25.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5(9):1564-1573. doi: 10.1038/nprot.2010.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904-909. doi: 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 27.Loh P-R, Danecek P, Palamara PF, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48(11):1443-1448. doi: 10.1038/ng.3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium . A global reference for human genetic variation. Nature. 2015;526(7571):68-74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284-1287. doi: 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fried LP, Borhani NO, Enright P, et al. The cardiovascular health study: design and rationale. Ann Epidemiol. 1991;1(3):263-276. doi: 10.1016/1047-2797(91)90005-W [DOI] [PubMed] [Google Scholar]
- 32.Psaty BM, O’Donnell CJ, Gudnason V, et al. ; CHARGE Consortium . Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73-80. doi: 10.1161/CIRCGENETICS.108.829747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190-2191. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930-D934. doi: 10.1093/nar/gkr917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.GTEx Consortium Human genomics, The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648-660. doi: 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner AH, Anand VN, Wang WH, et al. Exon-level expression profiling of ocular tissues. Exp Eye Res. 2013;111:105-111. doi: 10.1016/j.exer.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You J, Corley SM, Wen L, et al. RNA-Seq analysis and comparison of corneal epithelium in keratoconus and myopia patients. Sci Rep. 2018;8(1):389. doi: 10.1038/s41598-017-18480-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birch-Machin MA, Russell EV, Latimer JA. Mitochondrial DNA damage as a biomarker for ultraviolet radiation exposure and oxidative stress. Br J Dermatol. 2013;169(suppl 2):9-14. doi: 10.1111/bjd.12207 [DOI] [PubMed] [Google Scholar]
- 39.Afshari NA, Igo RP Jr, Morris NJ, et al. Genome-wide association study identifies three novel loci in Fuchs endothelial corneal dystrophy. Nat Commun. 2017;8:14898. doi: 10.1038/ncomms14898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jurkunas U, Azar DT. Potential complications of ocular surgery in patients with coexistent keratoconus and Fuchs’ endothelial dystrophy. Ophthalmology. 2006;113(12):2187-2197. doi: 10.1016/j.ophtha.2006.06.036 [DOI] [PubMed] [Google Scholar]
- 41.Vira S, Abugo U, Shih CY, et al. Descemet stripping endothelial keratoplasty for the treatment of combined fuchs corneal endothelial dystrophy and keratoconus. Cornea. 2014;33(1):1-5. doi: 10.1097/ICO.0b013e3182a7389c [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Association results for all SNPs with P < 10-6 in the discovery cohort
eTable 2. Association results for lead SNPs at CCT-mediated keratoconus loci
eTable 3. Replication and meta-analysis of top ranked SNPs
eTable 4. Expression of genes in associated regions
eTable 5. Functional annotations of the lead SNP at the PNPLA2 locus, rs61876744, and SNPs in high linkage disequilibrium
eFigure 1. Q-Q plot of p-values from logistic regression analysis of discovery cohort with first three principal components as covariates
eFigure 2. Ancestry clustering based on genome-wide association data
eFigure 3. Locus specific plots for the three loci reaching suggestive or genome-wide significance in the discovery cohort
eFigure 4. eQTL data from GTex project for SNP rs61876744
eReferences.