Keywords: dorsal root ganglion, nerve regeneration, neural regeneration, peripheral nerve, Robo 1, Robo 2, sciatic nerve, Slit1, Slit2, Slit3
Abstract
The Slit family of axon guidance cues act as repulsive molecules for precise axon pathfinding and neuronal migration during nervous system development through interactions with specific Robo receptors. Although we previously reported that Slit1–3 and their receptors Robo1 and Robo2 are highly expressed in the adult mouse peripheral nervous system, how this expression changes after injury has not been well studied. Herein, we constructed a peripheral nerve injury mouse model by transecting the right sciatic nerve. At 14 days after injury, quantitative real-time polymerase chain reaction was used to detect mRNA expression of Slit1–3 and Robo1–2 in L4–5 spinal cord and dorsal root ganglia, as well as the sciatic nerve. Immunohistochemical analysis was performed to examine Slit1–3, Robo1–2, neurofilament heavy chain, F4/80, and vimentin in L4–5 spinal cord, L4 dorsal root ganglia, and the sciatic nerve. Co-expression of Slit1–3 and Robo1–2 in L4 dorsal root ganglia was detected by in situ hybridization. In addition, Slit1–3 and Robo1–2 protein expression in L4–5 spinal cord, L4 dorsal root ganglia, and sciatic nerve were detected by western blot assay. The results showed no significant changes of Slit1–3 or Robo1–2 mRNA expression in the spinal cord within 14 days after injury. In the dorsal root ganglion, Slit1–3 and Robo1–2 mRNA expression were initially downregulated within 4 days after injury; however, Robo1–2 mRNA expression returned to the control level, while Slit1–3 mRNA expression remained upregulated during regeneration from 4–14 days after injury. In the sciatic nerve, Slit1–3 and their receptors Robo1–2 were all expressed in the proximal nerve stump; however, Slit1, Slit2, and Robo2 were barely detectable in the nerve bridge and distal nerve stump within 14 days after injury. Slit3 was highly ex-pressed in macrophages surrounding the nerve bridge and slightly downregulated in the distal nerve stump within 14 days after injury. Robo1 was upregulated in vimentin-positive cells and migrating Schwann cells inside the nerve bridge. Robo1 was also upregulated in Schwann cells of the distal nerve stump within 14 days after injury. Our findings indicate that Slit3 is the major ligand expressed in the nerve bridge and distal nerve stump during peripheral nerve regeneration, and Slit3/Robo signaling could play a key role in peripheral nerve repair after injury. This study was approved by Plymouth University Animal Welfare Ethical Review Board (approval No. 30/3203) on April 12, 2014.
Chinese Library Classification No. R459.9; R741; R363
Introduction
The Slit family of secreted glycoproteins are members of four classic axon guidance protein families (Giger et al., 2010). The binding of Slits to their respective Roundabout (Robo) receptors activates one of the most crucial repulsive axon guidance signaling pathways to control precise axon pathfinding and neuronal migration during nervous system development (Ypsilanti et al., 2010; Blockus and Chédotal, 2016). To date, three Slits (Slit1–3) have been identified in mammals, with an expression pattern that varies spatiotemporally during nervous system development (Ypsilanti et al., 2010; Blockus and Chédotal, 2016). Numerous studies in worms, flies, zebrafish, chickens, and mammals have confirmed that Slit1–3 interact with Robo1 and Robo2 receptors with high affinity (Ypsilanti et al., 2010; Blockus and Chédotal, 2016). Notably, although Robo3 and Robo4 have also been named as Slit receptors based on their sequence homology with Robo1 and Robo2, recent studies have shown that Slit1–3 do not interact with Robo3 and Robo4 with high affinity in mammals (Koch et al., 2011; Zelina et al., 2014). Previously, we systematically examined the expression of Slit1–3, Robo1, and Robo2 in the adult mouse peripheral nervous system, and found that motor neurons and sensory neurons express Slit1–3 and Robo1–2, primarily in peripheral axons (Carr et al., 2017). However, Slit2, Slit3, and Robo1 were predominantly expressed by Schwann cells in peripheral nerves (Carr et al., 2017). Recently, we examined the expression of Slit1–3 and Robo1–2 in the nerve bridge after mouse sciatic nerve transection injury and found that repulsive Slit3-Robo1 signaling plays a key role in cell migration and axon pathfinding in the nerve bridge (Dun et al., 2019). However, a full description of Slit1–3 and Robo1–2 expression patterns in the adult mouse peripheral nervous system following injury was still missing.
To examine the expression pattern of Slit1–3 and Robo1–2 in the adult mouse peripheral nervous system following injury and reveal the function of Slit-Robo signaling in peripheral nerve repair, we performed mouse sciatic nerve transection injury and investigated the time course of Slit1–3 and Robo1–2 expression changes in spinal cord motor neurons, dorsal root ganglion (DRG) sensory neurons, the nerve bridge, and the distal nerve stump. We also identified cell types expressing Robo1–2 and/or Slit1–3 in the spinal cord, DRG, and different compartments of transected mouse sciatic nerve. Our results facilitate understanding of the function of Slit-Robo signaling in regulation of peripheral nerve regeneration.
Materials and Methods
Animals and peripheral nerve surgery
Mouse breeding and sciatic nerve transection injury were carried out according to Home Office regulations under the UK Animals (Scientific Procedures) Act of 1986. Ethical approval for the work in this paper was granted by Plymouth University Animal Welfare Ethical Review Board (approval No. 30/3203) on April 12, 2014. C57BL/6 mice were purchased from Charles River UK Ltd. (Harlow, UK), while myelin proteolipid protein (PLP)-green fluorescent protein (GFP) mice were obtained from Professor Thomas Misgeld (Technische Universität München, Munich, Germany) with permission from Professor Wendy Macklin (University of Colorado, Denver, CO, USA). Mice were housed in a 12-hour light/dark cycle with ad libitum access to food and water. A total of 15 male and 15 female C57BL/6 mice (20–25 g) and nine male PLP-GFP mice (20–25 g) were used for experiments. The PLP gene promoter in PLP-GFP mice drives cytoplasmic GFP expression in both myelinating and non-myelinating Schwann cells (Mallon et al., 2002). For sciatic nerve transection injury, 8-week-old C57BL/6 and PLP-GFP mice were anesthetized with isoflurane. The right sciatic nerve was exposed and transected at approximately 0.5 cm distal to the sciatic notch. Overlying muscle was sutured and the skin was closed with an autoclip applier. All animals undergoing surgery were given appropriate post-operative analgesia (0.025% bupivacaine solution, topically applied above the muscle suture before applying the surgical clip) and monitored daily (Dun and Parkinson, 2018). At the indicated time points post-surgery for each experiment described, animals were humanely euthanized by CO2.
Quantitative real-time polymerase chain reaction
Spinal cord (L4–5), DRG (L4 and L5), and sciatic nerve samples were dissected out at the stated time points post-surgery. Samples from three mice were pooled for each time point. Total mRNA was extracted using an miR-Neasy Mini Kit (#217004; Qiagen, Hilden, Germany). First-stand cDNA was synthesized using M-MLV reverse transcriptase (#M368; Promega, Southampton, UK) and random hexamer primers (#C1181, Promega). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed with a LightCycler480 Real-Time PCR Machine (Roche Applied Science, Burgess Hill, UK) using SYBR Green I Master Mix. Slit1–3, Robo1–2, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers used for qRT-PCR are shown in Table 1. Cross-point values were calculated using the software for the LightCycler480 instrument. Relative mRNA levels were calculated by the 2–ΔΔCt method (Livak and Schmittgen, 2001) using GAPDH as a reference gene for normalization. All reactions were carried out in triplicate (n = 3) for statistical analysis.
Table 1.
Primers used for quantitative real-time polymerase chain reaction
| Gene | Forward | Reverse |
|---|---|---|
| Slit1 | 5′-CTG CTC CCC GGA TAT GAA CC-3′ | 5′-TAG CAT GCA CTC ACA CCT GG-3′ |
| Slit2 | 5′-AAC TTG TAC TGC GAC TGC CA-3′ | 5′-TCC TCA TCA CTG CAG ACA AAC T-3′ |
| Slit3 | 5′-AGT TGT CTG CCT TCC GAC AG-3′ | 5′-TTT CCA TGG AGG GTC AGC AC-3′ |
| Robo1 | 5′-GCT GGC GAC ATG GGA TCA TA-3′ | 5′-AAT GTG GCG GCT CTT GAA CT-3′ |
| Robo2 | 5′-CGA GCT CCT CCA CAG TTT GT-3′ | 5′-GTA GGT TCT GGC TGC CTT CT-3′ |
| GAPDH | 5′-AAG GTC ATC CCA GAG CTG AA-3′ | 5′-CTG CTT CAC CAC CTT CTT GA-3′ |
GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; Robo: Roundabout.
Microarray data set analysis
Microarray data sets were searched in the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) using the key words ‘mouse’, ‘peripheral nerve’, and ‘injury’. Retrieved data sets were manually screened to identify data sets describing the time course of gene expression signatures in adult mouse sciatic nerve after injury. Two data sets were identified (GSE74087 and GSE22291) and further analyzed using the NCBI online tool GEO2R. Slit2, Slit3, and Robo1 fold-changes and P-values were obtained from analyzed data.
Immunohistochemistry
L4–5 spinal cord, L4 DRG, and sciatic nerve samples were dissected out and fixed overnight in 4% paraformaldehyde at 4°C. Samples were then washed in phosphate-buffered saline (PBS, 3 × 10 minutes) and dehydrated in 30% sucrose (in PBS) overnight at 4°C. Subsequently, samples were embedded in optimal cutting temperature medium and sectioned on a cryostat at a thickness of 12 µm. Sections were permeabilized with 0.25% Triton X-100 plus 1% bovine serum albumin (BSA) in PBS for 30 minutes, and then blocked with blocking buffer (3% BSA plus 0.05% Triton X-100 in PBS) for 45 minutes at room temperature. Sections were incubated with primary antibodies (diluted 1:100 in blocking buffer) overnight at 4°C. The following day, sections were washed with PBS (3 × 10 minutes) and then incubated with species-specific secondary antibodies plus Hoechst dye (1:300 diluted in blocking buffer) for 1 hour at room temperature. Finally, sections were washed with PBS (3 × 10 minutes) and mounted with Citiflour (#R1320; Agar Scientific, Stansted, UK) for imaging with a Leica SP8 confocal microscope (Wetzlar, Germany). For stained spinal cord and sciatic nerve whole sections, several images were captured covering the entire field of interest. Captured images were then combined into a whole image using Adobe Photoshop (Adobe Systems, San Jose, CA, USA). Primary and secondary antibodies for immunohistochemistry: Slit1 (#SAB1307048) and Slit3 (#SAB2104337) antibodies were purchased from Sigma (Gillingham, UK); Slit2 antibody was purchased from Chemicon (AB5701; Fisher Scientific, Loughborough, UK); Robo1 (#ab7279), neurofilament heavy chain (NF; #ab4680), F4/80 (#ab6640), GAPDH (#ab83485) and vimentin (#ab24525) antibodies were purchased from Abcam (Cambridge, UK); Robo2 was from purchased from Santa Cruz Biotechnology (#sc-25673, Dallas, TX, USA); NeuN was purchased from Merck Millipore (#ABN91; Watford, UK); Hoechst 33342 nuclear dye (#H3570) and Alexa Fluor 488 or 568 dye-conjugated donkey anti-rabbit and goat an-ti-chicken secondary antibodies were purchased from Thermo Scientific (Basingstoke, UK).
In situ hybridization
Slit1–3 and Robo1–2 in situ probes were described previously (Carr et al., 2017). Briefly, L4 DRG were fixed in 4% par-aformaldehyde at 4°C overnight and then embedded in optimal cutting temperature for cryostat sectioning. Subsequently, 15-µm thick sections were placed onto SuperFrost® Plus slides and then a Frame-Seal™ incubation chamber (#SLF1201; Bio-Rad, Hecules, CA, USA) was added onto each slide. Optimal cutting temperature compound was washed away with PBS and then 150 µL of hybridization buffer containing individual digoxigenin-labeled Slit1–3 or Robo1–2 probes was added into each incubation chamber, which were sealed with coverslips. Slides were placed into a hybridization box and kept at 65°C overnight for hybridization. The following day, coverslips were removed from the incubation chambers and sections were washed three times for 1 hour with a wash solution (50% formamide, 0.1% Tween-20, 150 mM NaCl, and 15 mM sodium citrate) at 65°C. Sections were then washed twice for 30 minutes with MABT solution (100 mM maleic acid, 150 mM NaCl, 0.1% Tween-20, pH 7.5) at room temperature. Next, sections were blocked with 500 µL blocking solution (10% BSA in MABT solution) for 1 hour at room temperature. Anti-digoxigenin-alkaline phosphatase antibody (1:1500 in blocking solution) was applied to the chambers, which were incubated overnight at 4°C. Sections were washed three times for 15 minutes each with MABT solution at room temperature, and signals were developed with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution (Roche Diagnostics GmbH; #1681451, Burgess Hill, UK) at 37°C overnight. Slides were then washed three times for 10 minutes each in water and mounted for imaging on a Nikon microscope (Eclipse 80i; Indigo Scientific, Baldock, UK). For stained spinal cord and sciatic nerve whole sections, several images were captured covering the entire field of interest. Captured images were then combined into a whole image using Adobe Photoshop (Adobe Systems, Version: 19.1.5).
Western blot assay
Sciatic nerve samples were directly sonicated into 1× sodium dodecyl sulfate loading buffer. Proteins were separated on 8% sodium dodecyl sulfate-polyacrylamide running gels and then transferred onto a polyvinylidene fluoride (Φ = 0.45 µm) transfer membrane using the wet transfer method. Membranes were blocked with 5% fat free milk in Tris-buffered saline plus 0.1% Tween-20 (TBST) for 1 hour at room temperature. Rabbit anti-Slit1 (#SAB1307048) and anti-Slit3 (#SAB2104337) antibodies were purchased from Sigma. Rabbit anti-Slit2 antibody was purchased from Chemicon (#AB5701, Fisher Scientific). Rabbit anti-Robo1 (#ab7279) and anti-GAPDH (#ab83485) antibodies were purchased from Abcam. Rabbit anti-Robo2 was from purchased from Santa Cruz (#sc-25673). Primary antibodies were diluted 1:500 in TBST containing 5% milk, and membranes were incubated in primary antibodies overnight at 4°C. The following day, membranes were washed with TBST three time for 10 minutes each and then incubated with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Sigma) for 1 hour at room temperature. Secondary antibodies were diluted 1:5000 in TBST containing 5% milk. After three 10-minute TBST washes, Pierce ECL western blotting substrate was added onto the membrane and incubated for 5 minutes to develop the chemilu-minescent signal. Amersham Hyperfilm™ ECL films (VWR, Leicestershire, UK) were used to capture the intensity of chemiluminescent signal. Exposed films were then developed in a Compact X4 automatic processor. The intensity of protein bands was quantified using ImageJ software (https://imagej.nih.gov/ij/).
Statistical analysis
Samples for qRT-PCR and western blot were grouped from three mice for each time point to create a single pooled sample. We used pooled biological replicates for three repetitions of these experiments. Statistical significance was analyzed using Student’s t-test and one-way analysis of variance in SPSS (IBM SPSS Statistics 24; Armonk, NY, USA) to determine whether there were any statistically significant differences between control samples and samples from each time point. Fold-changes are represented in figures as mean ± standard error of mean (SEM).
Results
Changes in Slit1–3 and Robo1–2 mRNA expression in the spinal cord and DRG in response to sciatic nerve transection injury
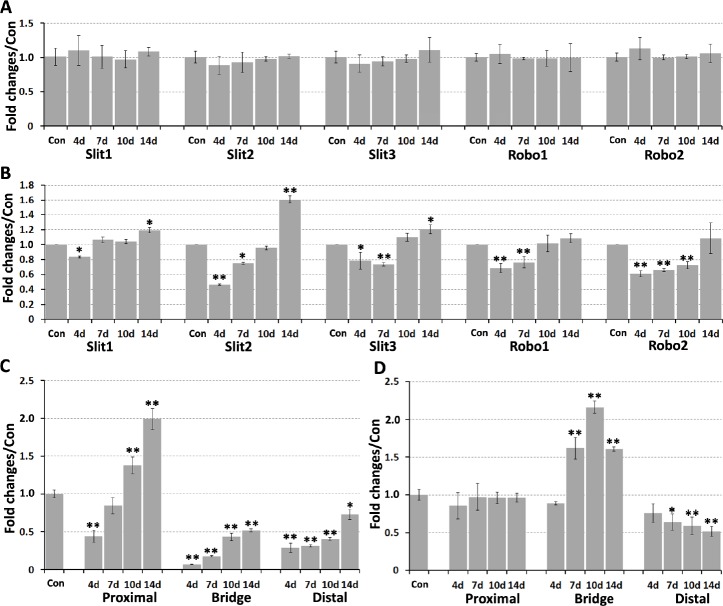
Recently, we showed that Slit1–3 and Robo1–2 mRNA are all expressed in the cell bodies of adult mouse motor and sensory neurons (Carr et al., 2017). As such, we first used qRT-PCR to compare mRNA expression changes in spinal cord (L4–5) and DRG (L4–5) after right-side sciatic nerve transection injury, using the left-side spinal cord and DRG as control samples. We previously showed four important time points (4, 7, 10, and 14 days) to visualize the pathfinding of regenerating axons crossing a mouse sciatic nerve gap after transection injury. Initial axon extension could be observed on day 4 and all regenerating axons had crossed the nerve bridge by day 14 (Dun and Parkinson, 2015). Therefore, we used these four time points for this study. We did not observe any significant changes in Slit1–3 or Robo1–2 mRNA levels at 4, 7, 10, or 14 days after sciatic nerve transection injury (Figure 1A). In DRG, Slit1–3 and Robo1–2 mRNA were all downregulated on day 4 (P < 0.05). Slit1 mRNA levels returned to control levels on day 7, but Slit2–3 and Robo1–2 mRNA remained downregulated (P < 0.05). Slit1–3 and Robo1 mRNA levels were similar to control side DRG on day 10, whereas Robo2 remained downregulated (P < 0.01). On day 14, Robo1–2 expression lev-els were similar to the control side DRG, however, Slit1–3 were upregulated (Figure 1B).
Figure 1.
Slit1–3 and Robo1–2 mRNA expression in mouse spinal cord, DRG, and sciatic nerve at 4, 7, 10, and 14 days after sciatic nerve transection injury detected by quantitative real-time polymerase chain reaction.
(A) No significant changes in Slit1–3 and Robo1–2 mRNA expression in mouse spinal cord from 4 days to 14 days after sciatic nerve transection injury. (B) Fold changes of Slit1–3 and Robo1–2 mRNA in DRG. (C) Fold changes of Slit2 mRNA in sciatic nerve. (D) Fold changes of Slit3 mRNA in sciatic nerve. Data are expressed as mean ± SEM (n = 3). *P < 0.05, **P < 0.01, vs. control group (one-way analysis of variance followed by Student’s t-test). Con: Control; DRG: dorsal root ganglion; Robo: Roundabout.
Changes in Slit2, Slit3, and Robo1 mRNA expression in the sciatic nerve after transection injury
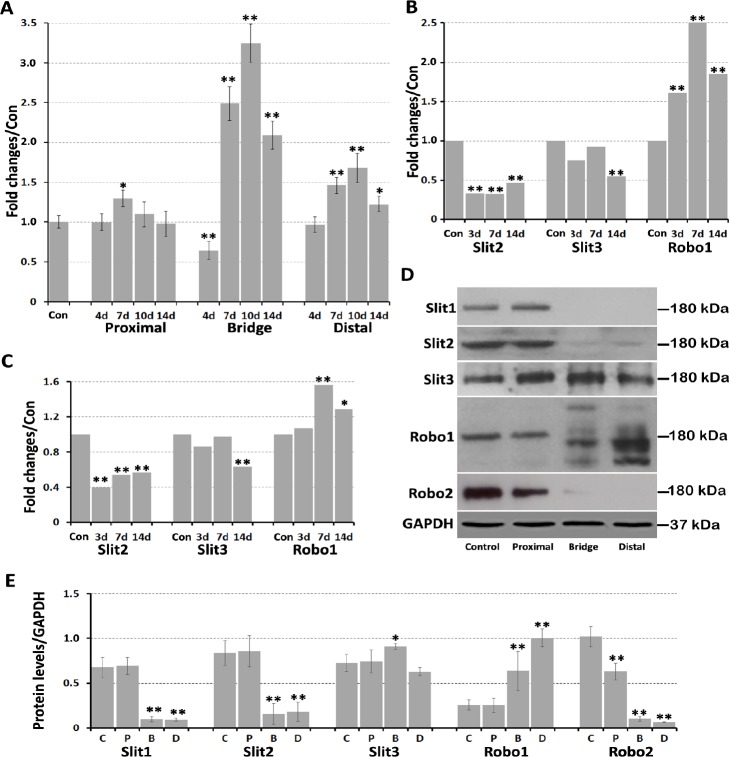
Previously, we showed that Slit1 and Robo2 mRNA are undetectable in intact mouse sciatic nerve (Carr et al., 2017) and remain undetectable after sciatic nerve transection injury. Therefore, Slit2, Slit3, and Robo1 mRNA expression changes were investigated by qRT-PCR in the proximal nerve stump, nerve bridge, and distal nerve stump using left-side sciatic nerves as control samples. In the proximal nerve stump, no significant changes in Slit3 mRNA occurred from day 4 to day 14 (Figure 1D), however Robo1 mRNA was upregulated on day 7 (P < 0.05; Figure 2A). Slit2 mRNA was downregulated on day 4 (P < 0.01) and remained lower than the control nerve on day 7 (Figure 1C). However, Slit2 mRNA levels were upregulated in the proximal nerve stump on days 10 and 14, exhibiting 1.38- and 2-fold upregulation, respectively (P < 0.01; Figure 1C).
Figure 2.
Fold-changes of Slit2, Slit3, and Robo1 mRNA in sciatic nerve, and Slit1–3 and Robo1–2 protein expression in sciatic nerve.
(A) Fold-changes of Robo1 mRNA in sciatic nerve detected by quantitative real-time polymerase chain reaction. (B) Slit2, Slit3, and Robo1 mRNA fold-changes in the mouse distal sciatic nerve from data set GSE74087. (C) Slit2, Slit3, and Robo1 mRNA fold-changes in the mouse distal sciatic nerve from data set GSE22291. (D) Western blot results for Slit1–3 and Robo1–2 protein expression in transected mouse sciatic nerve. (E) Quantification of Slit1–3 and Robo1–2 protein levels in intact sciatic nerve, proximal nerve stump, nerve bridge, and distal nerve stump. Data are expressed as mean ± SEM (n = 3). *P < 0.05, **P < 0.01, vs. control group (one-way analysis of variance followed by Student’s t-test). B: Bridge, C: control, D: distal; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; P: proximal. Robo: Roundabout.
In the nerve bridge, Slit2 mRNA levels were only upregulated 0.07-fold compared with control nerves on day 4, which slowly increased to 0.518-fold by day 14 (P < 0.01; Figure 1C). Slit3 mRNA levels were 0.9-fold higher than control nerves on day 4, but increased to 1.62-fold by day 7. Slit3 mRNA levels in the nerve bridge peaked on day 10 at 2.16-fold the control nerve value, but then dropped to 1.61-fold by day 14 (Figure 1D). Robo1 mRNA levels were 0.64-fold control nerve samples on day 4, but dramatically increased to 2.49-fold by day 7. Robo1 mRNA levels also peaked on day 10 at 3.24-fold the control nerve value, but then dropped to 2.09-fold by day 14 (Figure 2A).
In distal nerve stumps, Slit2 and Slit3 mRNA were downregulated from day 4 to day 14 (P < 0.05) (Figure 1C & D). There were no significant changes in Robo1 mRNA on day 4 in the distal nerve stumps, but Robo1 mRNA was upreg-ulated in the distal nerve stumps from day 7 to day 14 (P < 0.01; Figure 2A). To further validate our qRT-PCR results in the sciatic nerve, we also analyzed microarray data sets published in the GEO that described the time course of gene expression signatures in adult mouse sciatic nerve after injury. A search of the GEO database identified two data sets (GSE74087 and GSE22291) that measured gene expression profiles in the mouse distal sciatic nerve after 3, 7, and 14 days of injury (Barrette et al., 2010; Pan et al., 2017). In line with our qRT-PCR results, both data sets show Slit2 and Slit3 downregulation, and Robo1 upregulation in the distal sciatic nerve (Figure 2B & C).
Changes in Slit1–3 and Robo1–2 protein expression in the sciatic nerve after transection injury
We next used western blot to examine Slit1–3 and Robo1–2 protein expression in sciatic nerve 7 days after transec-tion injury, using left-side sciatic nerves as control samples. Our western blot results revealed that Slit1–3 and Robo1–2 proteins were all present in the intact sciatic nerve and proximal nerve stump. Slit2 and Robo2 were weakly expressed, but Slit1 was not present in the nerve bridge. In contrast, Slit3 and Robo1 proteins were upregulated in the nerve bridge. Slit1 and Robo2 were not present in the distal nerve stump. Slit2 was weakly expressed in the distal nerve stump, indicating downregulation of Slit2 after injury. Slit3 was slightly downregulated, whereas Robo1 was upregulated in the distal nerve stump (Figure 2D). Interestingly, Robo1 exhibited only one band around 180 kDa in the intact sciatic nerve and proximal nerve stump, but displayed bands of multiple sizes in the nerve bridge and distal nerve stump (Figure 2D). Moreover, major Robo1 bands in the nerve bridge and distal nerve stump were smaller than 180 kDa, indicating that Robo1 had undergone protein cleavage. Quantification of Slit1–3 and Robo1–2 protein levels from three independent western blots revealed no significant changes in Slit1–3 or Robo1 protein expression in the proximal nerve stump 7 days after injury, but Robo2 protein was downregulated. In contrast, Slit3 protein was upregulated in the nerve bridge, while Robo1 protein was upregulated in both the nerve bridge and distal nerve stump (Figure 2E).
Robo1 and Robo2 protein expression in motor and sensory neurons after sciatic nerve transection injury
Injury to the sciatic nerve damages two axonal populations: motor axons with cell bodies localized in the ventral horn of the spinal cord, and sensory axons with cell bodies localized in the DRG. Previously, we showed that Robo1 is highly expressed in both astrocytes and neurons of the spinal cord, whereas Robo2 is exclusively expressed by neurons (Carr et al., 2017). To understand the function of Slit/Robo signaling in peripheral axon regeneration, we examined Robo1 and Robo2 expression in the cell bodies of motor and sensory neurons 7 days after sciatic nerve transection injury using immunohistochemistry. We chose the 7-day timepoint because this is a key point for pioneer axons crossing the nerve bridge (Cattin et al., 2015; Dun and Parkinson, 2015). Immunostaining with Robo1 and Robo2 antibody in spinal cord sections (L4–5) revealed similar expression patterns of Robo1 and Robo2 on injured (right) and uninjured (left) sides (Figure 3A & B). Double staining with the neuronal marker NeuN confirmed that motor neurons in the ventral horn of the spinal cord express both Robo1 and Robo2 receptors after sciatic nerve transection injury (Figure 3C–H). Double staining of Robo1 and Robo2 with NeuN in DRG (L4–5) sections from the uninjured side revealed that Robo1 and Robo2 are expressed in sensory neurons before sciatic nerve transection injury (Figure 4A–F). Moreover, double staining Robo1 and Robo2 with NeuN in DRG (L4–5) sections from the injured side revealed that Robo1 and Robo2 are also expressed in sensory neurons after sciatic nerve transection injury (Figure 4G–L). Because the NeuN antibody showed both nucleus and cytoplasmic staining, so, we further used in situ hybridization of DRG sections to detect Robo1 and Robo2 mRNA expression. The results not only showed Robo1 and Robo2 expression in sensory neurons, but also confirmed the specificity of the Robo1 and Robo2 antibodies (Figure 4M & N). Thus, Robo1 and Robo2 receptors are expressed in both motor and sensory neurons during peripheral axon regeneration.
Figure 3.
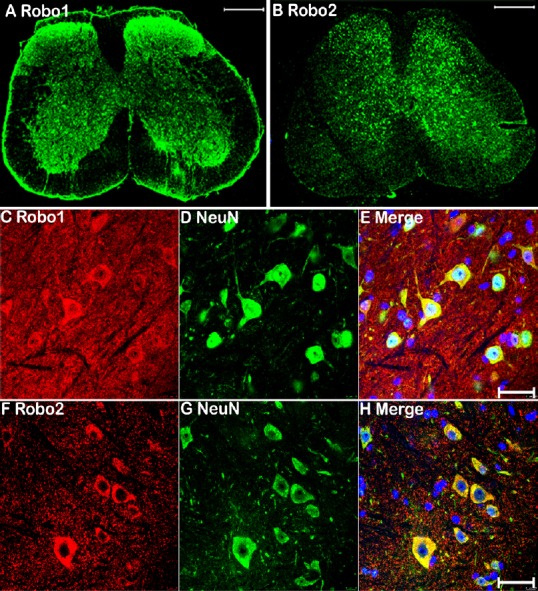
Immunohistochemical detection of Robo1 and Robo2 expression in motor neurons of the ventral spinal cord after sciatic nerve transection injury.
(A, B) Robo1 and Robo2 staining (green, Alexa Fluor 488) in L4–5 spinal cord sections 7 days after right side sciatic nerve transection injury. (C–E) Double staining Robo1 (red, Alexa Fluor 568) and NeuN (green, Alexa Fluor 488) showing Robo1 expression in motor neu-rons of the ventral spinal cord 7 days after sciatic nerve transection injury. (F–H) Double staining Robo2 (red, Alexa Fluor 568) and NeuN (green, Alexa Fluor 488) showing Robo2 expression in motor neurons of the ventral spinal cord 7 days after sciatic nerve transection injury. Scale bars: 400 μm in A, B; 40 μm in C–H. Robo: Roundabout.
Figure 4.
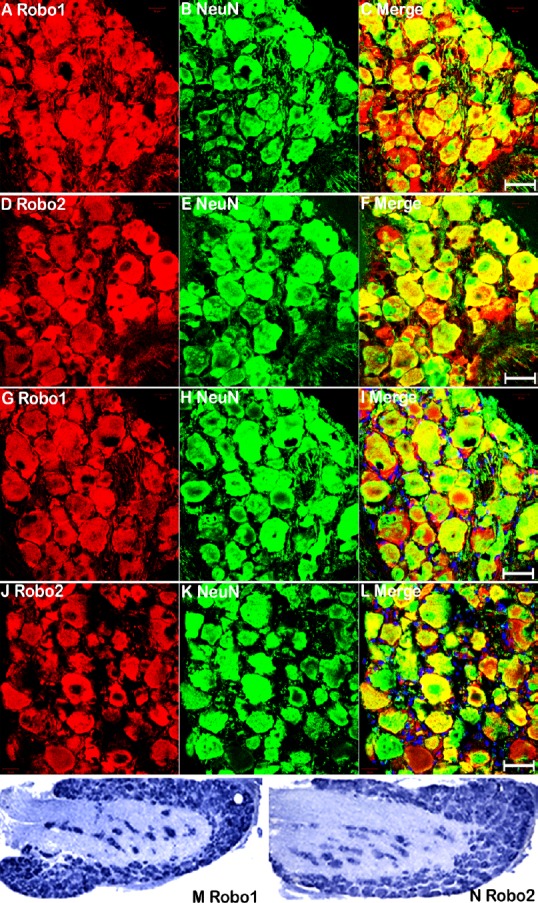
Sensory neurons in DRG express Robo1 and Robo2 after sciatic nerve transection injury, as detected by immunohistochemistry.
(A–C) Double staining for Robo1 (red, Alexa Fluor 568) and NeuN (green, Alexa Fluor 488) showing Robo1 immunopositivity in the cell bodies of adult sensory neurons. (D–F) Double staining Robo2 (red, Alexa Fluor 568) and NeuN (green, Alexa Fluor 488) showing Robo2 expression in the cell bodies of adult sensory neurons. (G–I) Double staining Robo1 (red, Alexa Fluor 568) and NeuN (green, Alexa Fluor 488) showing Robo1 immunopositivity in the cell bodies of sensory neurons in L4 DRG at 7 days after sciatic nerve transection injury. (J–L) Double staining Robo2 (red, Alexa Fluor 568) and NeuN (green, Alexa Fluor 488) showing Robo2 immunopositivity in the cell bodies of sensory neurons in L4 DRG at 7 days after sciatic nerve transection injury. (M–N) In situ hybridization indicated Robo1 and Robo2 mRNA expression in the cell bodies of sensory neurons in L4 DRG at 7 days after sciatic nerve transection injury. Scale bars: 40 μm. DRG: Dorsal root ganglion; Robo: Roundabout.
Slit1–3 and Robo1–2 protein expression pattern in the sciatic nerve after transection injury
Finally, we used immunohistochemistry to examine Slit1–3 and Robo1–2 protein expression in the sciatic nerve at 7 days after transection injury. Consistent with our previous findings that Slit1 and Robo2 are axon-specific proteins in peripheral nerves (Carr et al., 2017), double staining of Slit1 and Robo2 occurred with the axon marker neurofilament heavy chain (NF) on longitudinal sciatic nerve sections, indicating that Slit1 and Robo2 colocalize with NF in the proximal nerve stump. Slit1 and Robo2 were expressed in regenerating axons that had extended into the nerve bridge, but were undetectable further in the nerve bridge and in the distal nerve stump (Figures 5A–D and 6E–H). Slit1 and Robo2 immunostaining results were consistent with the western blot results described above, which indicated that Slit1 and Robo2 proteins are barely detectable in the nerve bridge and distal nerve stump (Figure 2D). Previously, we showed that Slit2, Slit3, and Robo1 are expressed in axons and Schwann cells of intact mouse sciatic nerve (Carr et al., 2017). Double staining Slit2, Slit3, or Robo1 with NF indicated that Slit2 is present in the proximal nerve stump, but barely detectable in the nerve bridge and distal nerve stump (Figure 5E–H). In contrast, both Slit3 and Robo1 were highly expressed in the nerve bridge. Slit3 and Robo1 were also expressed in the proximal and distal nerve (Figures 5I–L and 6A–D). Thus, our immunohistochemistry results were consistent with the qRT-PCR and western blot results described above, which indicated that Slit1–3 and Robo1–2 are all expressed in the proximal nerve stump. Slit1, Slit2, and Robo2 were barely detectable in the nerve bridge and distal nerve stump. In contrast, Slit3 and Robo1 not only exhibited distal nerve expression, but were also highly expressed in the nerve bridge (Figure 2D). Furthermore, the staining results showed that regenerating axons in the proximal nerve stump expressed both Robo1 and Robo2 (Figure 6I and J).
Figure 5.
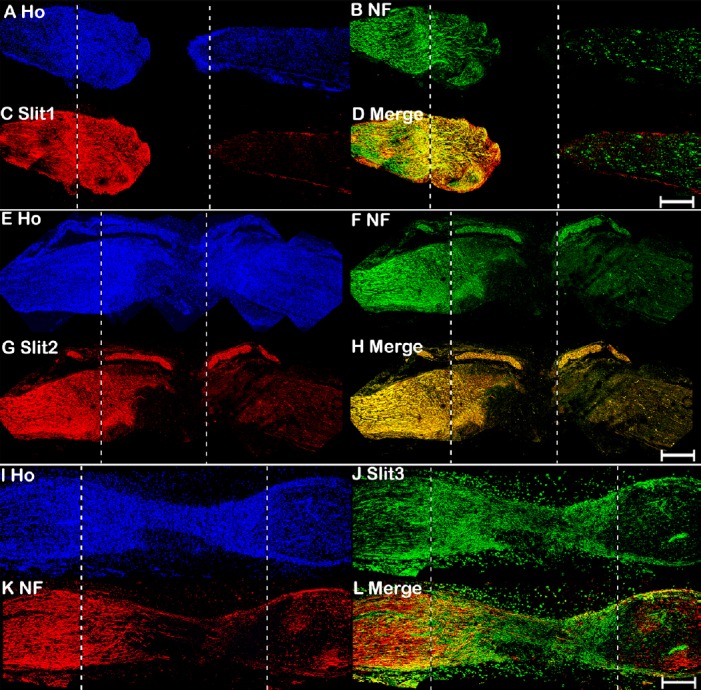
Double immunohistochemical staining of Slit1–3 and NF in mouse sciatic nerve at 7 days after transection injury.
(A–D) Double staining of Slit1 (red, Alexa Fluor 568) and NF (green, Alexa Fluor 488) in a 7-day post-transection mouse sciatic nerve showing Slit1 expression in regenerating axons of the proximal nerve stump. (E–H) Double staining of Slit2 (red, Alexa Fluor 568) and NF (green, Alexa Fluor 488) in a 7-day post-transection mouse sciatic nerve showing Slit2 expression in the proximal nerve stump. (I–L) Double staining of Slit3 (green, Alexa Fluor 488) and NF (red, Alexa Fluor 568) in a 7-day post-transection mouse sciatic nerve showing Slit3 expression in the proximal nerve stump, nerve bridge, and distal nerve stump. The proximal nerve stump is on the left, the distal nerve stump is on the right, and the nerve bridge is indicated between the two dashed lines. Scale bars: 400 μm. Ho: Hoechst 33342; NF: Neurofilament heavy chain; Robo: Roundabout.
Figure 6.
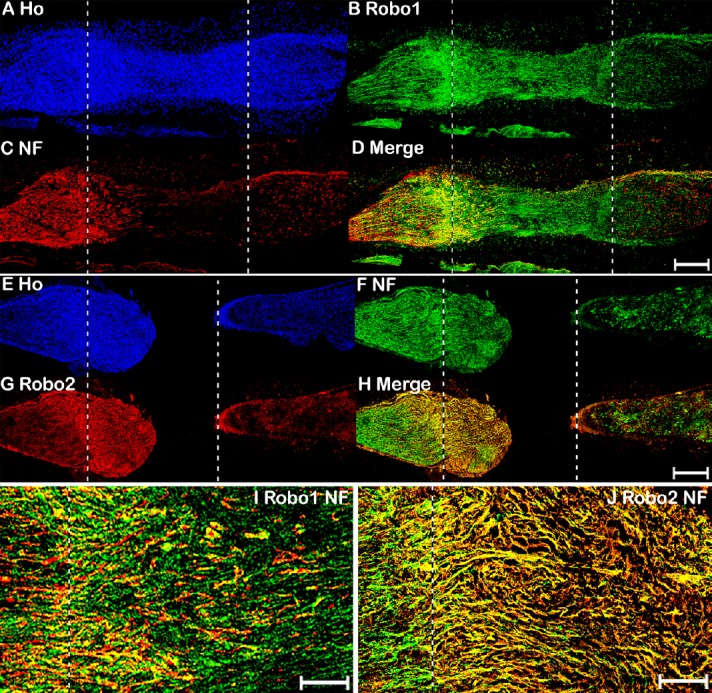
Double immunohistochemical staining of Robo1–2 and NF in mouse sciatic nerve 7 days after transection injury.
(A–D) Double staining of Robo1 (green, Alexa Fluor 488) and NF (red, Alexa Fluor 568) in a 7-day post-transection mouse sciatic nerve showing Robo1 expression in the proximal nerve stump, nerve bridge, and distal nerve stump. (E–H) Double staining of Robo2 (red, Alexa Fluor 568) and NF (green, Alexa Fluor 488) in a 7-day post-transection mouse sciatic nerve showing Robo2 expression in regenerating axons of the proximal nerve stump. (I–J) Higher magnification (40×) images of Robo1 and Robo2 expression in the tips of regenerating axons of the proximal nerve stump 7 days after transection injury. The proximal nerve stump is on the left, the distal nerve stump is on the right, and the nerve bridge is indicated between the two dashed lines. Scale bars: 400 μm in A–H, 20 μm in I, J. Ho: Hoechst 33342; NF: Neurofilament heavy chain; Robo: Roundabout.
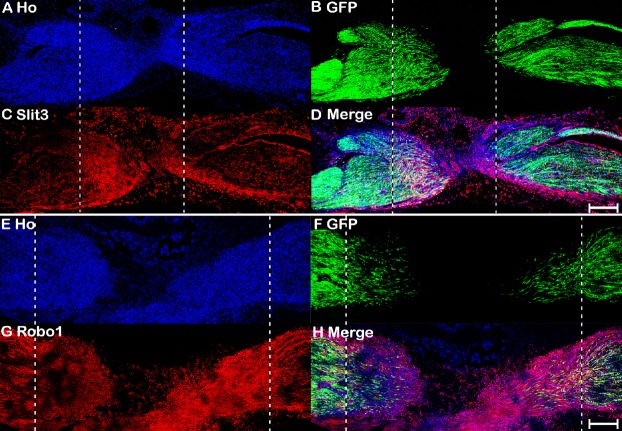
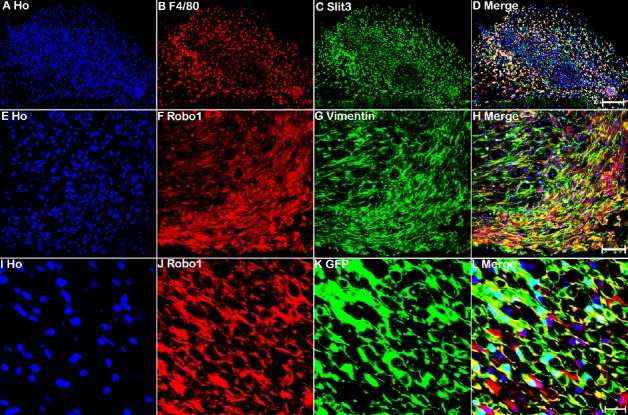
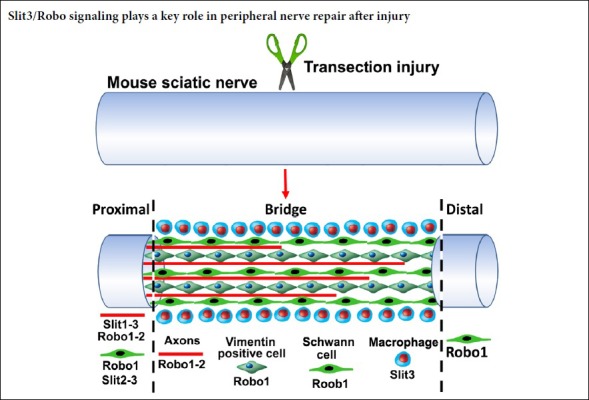
The nerve bridge, a newly formed tissue connecting the proximal and distal nerve stumps, consists of macrophages, endothelial cells, fibroblasts, Schwann cells, and perineurial cells (Williams et al., 1983; Schröder et al., 1993; Weis et al., 1994; Parrinello et al., 2010; Cattin et al., 2015). As Slit3 and Robo1 are highly expressed in the nerve bridge, we next used different cell markers to identify Slit3- and Robo1-positive cell types in the nerve bridge. First, we stained for Slit3 and Robo1 in 7-day post-transection longitudinal nerve bridge sections from PLP-GFP mice, which had GFP-labeled migrating Schwann cells in the nerve bridge (Mallon et al., 2002). Staining of Slit3 and Robo1 in nerve bridge sections from PLP-GFP mice showed strong Robo1 expression in migrating Schwann cells in the nerve bridge, whereas expression of Slit3 in migrating Schwann cells was weak (Figure 7). However, Slit3 and Robo1 staining of nerve bridge sections from PLP-GFP mice revealed that the majority Slit3- and Robo1-positive cells in the nerve bridge were not Schwann cells (Figure 7). Therefore, we further identified macrophages in the nerve bridge expressing high Slit3 levels using the macrophage marker F4/80 (Figure 8A–D). Notably, double staining of Robo1 with the mesenchymal cell marker vimentin showed that the majority of Robo1-positive cells in the nerve bridge were vimentin-positive cells (Figure 8E–H). Using qRT-PCR and western blot, we showed that Robo1 is upregulated in the distal nerve stump (Figure 2). Therefore, we further examined Robo1 immunostaining in sections from the distal nerve of PLP-GFP mice and found that Robo1 is highly expressed in Schwann cells of the distal nerve stump (Figure 8I–L). The dynamic expression patterns of Slit1–3 and Robo1–2 are summarized in Table 2.
Figure 7.
Slit3 and Robo1 immunopositivity of a longitudinal nerve bridge section from PLP-GFP mouse sciatic nerve 7 days after transection injury.
(A–D) Slit3 immunopositivity (red, Alexa Fluor 568) of a longitudinal nerve bridge section from PLP-GFP mice 7 days after transection injury showing Slit3-positive cells surrounding the nerve bridge. (E–H) Robo1 immunopositivity (red, Alexa Fluor 568) of a longitudinal nerve bridge section from PLP-GFP mice 7 days after transection injury showing that migrating cells inside the nerve bridge express high levels of Robo1. The proximal nerve stump is on the left, the distal nerve stump is on the right, and the nerve bridge is indicated between the two dashed lines. Scale bars: 300 μm. GFP: Green fluorescent protein; Ho: Hoechst 33342; PLP: myelin proteolipid protein; Robo: Roundabout.
Figure 8.
Macrophages in the nerve bridge expressed Slit3, while vimentin-positive cells in the nerve bridge and Schwann cells in the distal sciatic nerve expressed Robo1.
(A–D) Double staining of Slit3 (green, Alexa Fluor 488) with F4/80 (red, Alexa Fluor 568) in a transverse nerve bridge section showing that macrophages in the outermost layer of the nerve bridge express high levels of Slit3. (E–H) Double staining of Robo1 (red, Alexa Fluor 568) with the mesenchymal cell marker vimentin (green, Alexa Fluor 488) showing that vimentin-positive cells are the major cell type inside the nerve bridge expressing high levels of Robo1. (I–L) Immunopositivity of Robo1 (red, Alexa Fluor 568) in the distal nerve of PLP-GFP mice showing that Schwann cells in the distal nerve express high levels of Robo1. Ho: Hoechst. Scale bars: 120 μm in A–D, 50 μm in E–H, 20 μm in I–L. GFP: Green fluorescent protein; Ho: Hoechst 33342; PLP: myelin proteolipid protein; Robo: Roundabout.
Table 2.
Slit1–3 and Robo1–2 expression changes 7 days after mouse sciatic nerve transection injury
| Intact (Carr et al., 2017) | Proximal | Bridge | Distal | |
|---|---|---|---|---|
| Slit1 | Motor and sensory axons | Regenerating axons | Not expressed | Not expressed |
| Slit2 | Motor and sensory axons Schwann cells | Regenerating axons Schwann cells | Not expressed | Down-regulated in Schwann cells |
| Slit3 | Motor and sensory axons Schwann cells | Regenerating axons Schwann cells | Highly expressed in outermost layer of macrophages | Down-regulated in Schwann cells |
| Robo1 | Motor and sensory axons Schwann cells endothelial cells | Regenerating axons Schwann cells Endothelial cells | Up-regulated in migrating Schwann cells and Vimentin positive cells | Up-regulated in Schwann cells |
| Robo2 | Motor and sensory axons | Regenerating axons | Not expressed | Not expressed |
Robo: Roundabout.
Discussion
In this report, we examined the dynamic expression pattern of Slit1–3 and Robo1–2 in the adult mouse peripheral nervous system after sciatic nerve transection injury by qRT-PCR, western blot, and immunostaining. We showed that Slit1–3 and Robo1–2 are expressed in spinal cord motor neurons and DRG sensory neurons after mouse sciatic nerve injury. Slit1 and Slit2 were barely detectable in the nerve bridge and distal nerve stump. In contrast, Slit3 was upreg-ulated in macrophages surrounding the nerve bridge, but slightly downregulated in the distal nerve stump, indicating that Slit3 is the major ligand expressed in the nerve bridge and distal nerve stump. Robo1 and Robo2 were expressed in regenerating axons in the proximal nerve stump, while migrating Schwann cells and vimentin-positive cells in the nerve bridge expressed only Robo1, which was also upregulated in Schwann cells of the distal nerve stump. A major limitation for this study is that we were unable to identify the cell type that was Robo1 and vimentin double-positive in the nerve bridge because of a lack of cell markers to label this population of migrating cells. Moreover, the function of Slit/Robo signaling in peripheral nerve regeneration was not further investigated in this study.
Current clinical challenges for peripheral nerve repair come from transection injuries, whereby a gap is present be-tween two ends of a transected nerve that prevents precise axon retargeting to the distal nerve stump (Kou et al., 2019). In this study, we identified that macrophages surrounding the nerve bridge tissue secrete Slit3, which could interact with Robo1 and Robo2 on regenerating axons, as well as Robo1-expressing cells in the nerve bridge to regulate peripheral nerve regeneration. Nerve guidance conduits have been developed to repair peripheral nerve gaps. Thus, Slit3 could be delivered into the wall of nerve guidance conduits to control precise axon re-targeting for peripheral nerve repair.
Previously, we found that Slit1 and Robo2 mRNA are undetectable in adult mouse sciatic nerves, and showed by im-munostaining that they are axon-specific proteins in intact mouse sciatic nerve (Carr et al., 2017). In this study, we found that Slit1 and Robo2 are also expressed in regenerating axons of the proximal nerve stump by immunostaining and western blot assay. Indeed, Robo2 immunostaining results indicated that the weak Robo2 band in the nerve bridge observed by western blot comes from Robo2 expression in regenerating axons that have extended into the nerve bridge at day 7. Although we showed that Slit2 is downregulated in the distal nerve stump after peripheral nerve injury, our qRT-PCR results demonstrated that Slit2 is gradually upregulated in the proximal nerve stump upon axon regeneration. Slit2 is highly expressed in axons and Schwann cells of the adult mouse sciatic nerve (Carr et al., 2017). Thus, upregulation of Slit2 in the proximal nerve stump upon axon regeneration indicated that Slit2 is required to maintain the homeostasis of adult peripheral nerves, but may not play an important role during the early stages of peripheral nerve regeneration.
Slit1–3 have been identified as repellents for axonal guidance and cell migration through interactions with Robo1–2 receptors (Ypsilanti et al., 2010; Blockus and Chédotal, 2016). Previous studies suggested that shedding of the ecto-domain of Robo receptors is required for the initiation of Slit repulsive function (Coleman et al., 2010; Barak et al., 2014). Robo1 exhibited multiple-sized bands in the nerve bridge and distal sciatic nerve. Major bands in the nerve bridge and distal sciatic nerve were smaller than 180 kDa, indicating that Robo1 underwent proteolytic processing in injured peripheral nerves. Thus, Robo1 cleavage in the nerve bridge and distal nerve stump indicated that Robo1 mediates Slit3-repulsive signaling in the nerve bridge and distal sciatic nerve. In our western blots, Robo1 also exhib-ited a band larger than 180 kDa, indicating that Robo1 post-translational modification also occurred in the nerve bridge and distal sciatic nerve. In the future, it will be interesting to examine if Robo1 post-translational modification and proteolytic processing occur during nervous system development or only after central nervous system injury.
Using a zebrafish research model of motor axon regeneration, Isaacman-Beck et al. (2015) revealed that motor axons could regenerate into their original path with high accuracy. They demonstrated that Schwann cells in the zebrafish ventral motor nerve branch upregulate expression of collagen type IV, alpha 5 (col4a5) after motor axon transection injury. Upregulated col4a5 on the surface of Schwann cells binds Slit1a to repel dorsal branch motor axons regenerating into the ventral branch. They showed that differentiated Schwann cells in the distal part of the ventral motor nerve branch express Slit1a, the only Slit ligand in zebrafish, after transection injury (Isaacman-Beck et al., 2015). Their findings revealed that Slit1a/Robo signaling in zebrafish regulates the specificity of axon targeting during regeneration. In this report, we revealed that both Robo1 and Robo2 receptors are expressed in the cell bodies of motor and sensory neurons after mouse sciatic nerve transection injury. Robo1 and Robo2 pro-teins were also present in the tips of regenerating axons in the proximal sciatic nerve. We found that Slit3 is the major Robo ligand expressed in the nerve bridge and distal sciatic nerve. Thus, our results indicate that Slit3/Robo signaling could be an important axon guidance signaling pathway regulating the specificity of axon targeting during peripheral nerve regeneration.
Although binding of Slit to Robo receptors transduces repulsive signaling for axonal guidance and cell migration during nervous system development and tissue formation (Ypsilanti et al., 2010; Blockus and Chédotal, 2016), several reports have shown that Slit ligands and Robo receptors could interact with other families of guidance cues or receptors to transduce attractive signaling (Wayburn and Volk, 2009; Dascenco et al., 2015). Embryonic contractile tissue development in Drosophila requires muscle cells to migrate towards tendon cells. To understand the molecular mechanism regulating muscle cell migration towards their corresponding tendon cells, Wayburn and Volk (2009) discovered a novel tendon-specific transmembrane protein, which they named LRT for the leucine-rich repeat domain in its extracellular region. They found that LRT could initiate tendon-specific attractive signaling that effectively promoted muscle cell migration. Interestingly, they showed that LRT could interact with Robo receptors and this interaction activated an attractive signaling to promote muscle cell migration toward tendon cells (Wayburn and Volk, 2009). Other studies in sensory axons also found that Slit ligands could directly bind to Down syndrome cell adhesion molecule 1 (Dscam1) to induce an attractive signaling that stimulates sensory axon branching and arborization (Ma and Tessier-Lavigne, 2007; Dascenco et al., 2015). Therefore, it will be interesting to examine if LRT is expressed in the distal nerve stump while Dscam1 is expressed in sensory axons, and their potential functions in peripheral nerve regeneration.
In summary, we showed that there are dynamic changes of Slit1–3 and Robo1–2 expression in the mouse peripheral nervous system after sciatic nerve transection injury. Importantly, we found that Slit3 is the major ligand expressed in the nerve bridge and distal nerve stump. Both Robo1 and Robo2 were expressed in the cell bodies of motor and sensory neurons, as well as in regenerating axons in the proximal sciatic nerve stump. In the nerve bridge, Robo1 was expressed by migrating Schwann cells and vimentin-positive cells. In the distal nerve stump, Robo1 was highly expressed in Schwann cells. Collectively, our results indicate that the Slit3/Robo signaling pathway may play an important role in peripheral nerve regeneration.
Additional file: Open peer review reports 1 (76.4KB, pdf) and 2 (76.6KB, pdf) .
Footnotes
Conflicts of interest: The authors declare that there is no conflict of interests.
Financial support: This study was supported by the National Natural Science Foundation of China, No. 81371353 (to XPD). The funding body played no role in the study design, in the collection, analysis and interpretation of data, in the writing of the paper, or in the decision to submit the paper for publication.
Institutional review board statement: This study was approved by Plymouth University Animal Welfare and Ethical Review Board (approval No. 30/3203) on April 12, 2014.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewers: Christine A. Webber, University of Alberta, Canada; Sheng Yi, Nantong University, China.
Funding: This study was supported by the National Natural Science Foundation of China, No. 81371353 (to XPD).
P-Reviewers: Webber CA, Yi S; C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Van Deusen A, Yu J, Song LP; T-Editor: Jia Y
References
- 1.Barak R, Lahmi R, Gevorkyan-Airapetov L, Levy E, Tzur A, Opatowsky Y. Crystal structure of the extracellular juxtamembrane region of Robo1. J Struct Biol. 2014;186:283–291. doi: 10.1016/j.jsb.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 2.Barrette B, Calvo E, Vallieres N, Lacroix S. Transcriptional profiling of the injured sciatic nerve of mice carrying the Wld(S) mutant gene: identification of genes involved in neuroprotection, neuroinflammation, and nerve regeneration. Brain Behav Immun. 2010;24:1254–1267. doi: 10.1016/j.bbi.2010.07.249. [DOI] [PubMed] [Google Scholar]
- 3.Blockus H, Chédotal A. Slit-Robo signaling. Development. 2016;143:3037–3044. doi: 10.1242/dev.132829. [DOI] [PubMed] [Google Scholar]
- 4.Carr L, Parkinson DB, Dun XP. Expression patterns of Slit and Robo family members in adult mouse spinal cord and peripheral nervous system. PLoS One. 2017;12:e0172736. doi: 10.1371/journal.pone.0172736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattin AL, Burden JJ, Van Emmenis L, Mackenzie FE, Hoving JJ, Garcia Calavia N, Guo Y, McLaughlin M, Rosenberg LH, Quereda V, Jamecna D, Napoli I, Parrinello S, Enver T, Ruhrberg C, Lloyd AC. Macrophage-induced blood vessels guide Schwann cell-mediated regeneration of peripheral nerves. Cell. 2015;162:1127–1139. doi: 10.1016/j.cell.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman HA, Labrador JP, Chance RK, Bashaw GJ. The Adam family metalloprotease Kuzbanian regulates the cleavage of the roundabout receptor to control axon repulsion at the midline. Development. 2010;137:2417–2426. doi: 10.1242/dev.047993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dascenco D, Erfurth ML, Izadifar A, Song M, Sachse S, Bortnick R, Urwyler O, Petrovic M, Ayaz D, He H, Kise Y, Thomas F, Kidd T, Schmucker D. Slit and receptor tyrosine phosphatase 69D confer spatial specificity to axon branching via Dscam1. Cell. 2015;162:1140–1154. doi: 10.1016/j.cell.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dun XP, Parkinson DB. Visualizing peripheral nerve regeneration by whole mount staining. PLoS One. 2015;10:e0119168. doi: 10.1371/journal.pone.0119168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dun XP, Parkinson DB. Transection and crush models of nerve injury to measure repair and remyelination in peripheral nerve. Methods Mol Biol. 2018;1791:251–262. doi: 10.1007/978-1-4939-7862-5_20. [DOI] [PubMed] [Google Scholar]
- 10.Dun XP, Carr L, Woodley PK, Barry RW, Drake LK, Mindos T, Roberts SL, Lloyd AC, Parkinson DB. Macrophage-derived Slit3 controls cell migration and axon pathfinding in the peripheral nerve bridge. Cell Rep. 2019;26:1458–1472.e4. doi: 10.1016/j.celrep.2018.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Giger RJ, Hollis ER, 2nd, Tuszynski MH. Guidance molecules in axon regeneration. Cold Spring Harb Perspect Biol. 2010;2:a001867. doi: 10.1101/cshperspect.a001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isaacman-Beck J, Schneider V, Franzini-Armstrong C, Granato M. The lh3 glycosyltransferase directs target-selective peripheral nerve regeneration. Neuron. 2015;88:691–703. doi: 10.1016/j.neuron.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch AW, Mathivet T, Larrivee B, Tong RK, Kowalski J, Pibouin-Fragner L, Bouvree K, Stawicki S, Nicholes K, Rathore N, Scales SJ, Luis E, del Toro R, Freitas C, Breant C, Michaud A, Corvol P, Thomas JL, Wu Y, Peale F, et al. Robo4 maintains vessel integrity and inhibits angiogenesis by interacting with UNC5B. Dev Cell. 2011;20:33–46. doi: 10.1016/j.devcel.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Kou YH, Yu YL, Zhang YJ, Han N, Yin XF, Yuan YS, Yu F, Zhang DY, Zhang PX, Jiang BG. Repair of peripheral nerve defects by nerve transposition using small gap bio-sleeve suture with different inner diameters at both ends. Neural Regen Res. 2019;14:706–712. doi: 10.4103/1673-5374.247475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Ma L, Tessier-Lavigne M. Dual branch-promoting and branch-repelling actions of Slit/Robo signaling on peripheral and central branches of developing sensory axons. J Neurosci. 2007;27:6843–6851. doi: 10.1523/JNEUROSCI.1479-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mallon BS, Shick HE, Kidd GJ, Macklin WB. Proteolipid promoter activity distinguishes two populations of NG2-positive cells throughout neonatal cortical development. J Neurosci. 2002;22:876–885. doi: 10.1523/JNEUROSCI.22-03-00876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan B, Liu Y, Yan JY, Wang Y, Yao X, Zhou HX, Lu L, Kong XH, Feng SQ. Gene expression analysis at multiple time-points identifies key genes for nerve regeneration. Muscle Nerve. 2017;55:373–383. doi: 10.1002/mus.25225. [DOI] [PubMed] [Google Scholar]
- 19.Parrinello S, Napoli I, Ribeiro S, Wingfield Digby P, Fedorova M, Parkinson DB, Doddrell RD, Nakayama M, Adams RH, Lloyd AC. EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell sorting. Cell. 2010;143:145–155. doi: 10.1016/j.cell.2010.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schröder JM, May R, Weis J. Perineurial cells are the first to traverse gaps of peripheral nerves in silicone tubes. Clin Neurol Neurosurg. 1993;95(Suppl):S78–83. doi: 10.1016/0303-8467(93)90040-n. [DOI] [PubMed] [Google Scholar]
- 21.Wayburn B, Volk T. LRT, a tendon-specific leucine-rich repeat protein, promotes muscle-tendon targeting through its interaction with Robo. Development. 2009;136:3607–3615. doi: 10.1242/dev.040329. [DOI] [PubMed] [Google Scholar]
- 22.Weis J, May R, Schroder JM. Fine structural and immunohistochemical identification of perineurial cells connecting proximal and distal stumps of transected peripheral nerves at early stages of regeneration in silicone tubes. Acta Neuropathol. 1994;88:159–165. doi: 10.1007/BF00294509. [DOI] [PubMed] [Google Scholar]
- 23.Williams LR, Longo FM, Powell HC, Lundborg G, Varon S. Spatial-temporal progress of peripheral nerve regeneration within a silicone chamber: parameters for a bioassay. J Comp Neurol. 1983;218:460–470. doi: 10.1002/cne.902180409. [DOI] [PubMed] [Google Scholar]
- 24.Ypsilanti AR, Zagar Y, Chedotal A. Moving away from the midline: new developments for Slit and Robo. Development. 2010;137:1939–1952. doi: 10.1242/dev.044511. [DOI] [PubMed] [Google Scholar]
- 25.Zelina P, Blockus H, Zagar Y, Péres A, Friocourt F, Wu Z, Rama N, Fouquet C, Hohenester E, Tessier-Lavigne M, Schweitzer J, Roest Crollius H, Chédotal A. Signaling switch of the axon guidance receptor Robo3 during vertebrate evolution. Neuron. 2014;84:1258–1272. doi: 10.1016/j.neuron.2014.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.