This cohort study of patients with uveal melanomas with and without partial loss of chromosome 3 examines the association of partial chromosome 3 deletion in uveal melanomas with metastasis-free survival.
Key Points
Question
What is the association of partial chromosome 3 deletion in uveal melanomas with metastasis-free survival?
Findings
In this cohort study, partial deletions of chromosome 3 encompassing the BAP1 locus were associated with lower metastasis-free survival at 60 months compared with uveal melanomas without such deletion.
Meaning
These findings suggest that uveal melanomas that carry a partial deletion of chromosome 3 encompassing the BAP1 locus have a poor prognosis.
Abstract
Importance
Studies on uveal melanomas (UMs) have demonstrated the prognostic value of 8q gain and monosomy 3, but the prognosis of UMs with partial deletion of chromosome 3 remains to be defined.
Objective
To examine the association of partial chromosome 3 deletion in UMs with metastasis-free survival.
Design, Setting, and Participants
This retrospective cohort study of 1088 consecutive comparative genomic hybridization arrays performed from May 1, 2006, to July 31, 2015, assessed patients presenting with UMs with and without partial loss of chromosome 3 at a referral center. Data analysis was performed from September 1, 2017, to November 30, 2017.
Exposure
Uveal melanoma with or without partial loss of chromosome 3.
Main Outcomes and Measures
Metastasis-free survival and overall survival at 60 months.
Results
Of the 1088 consecutive comparative genomic hybridization arrays that were performed, 43 UMs (4.0%) in 43 patients (median age, 58 years [range, 12-79 years]; 22 [51%] female) carried partial deletions of chromosome 3. Median follow-up was 66 months (range, 1.2-126.2 months). Metastasis-free survival at 60 months was 33.6% (95% CI, 15.8%-71.4%) for UMs that carried a deletion of the BAP1 (BRCA1 associated protein 1) locus (BAP1del; 24 tumors) and 80.5% (95% CI, 64.8%-100%) for UMs without the loss of the BAP1 locus (BAP1 normal [BAP1nl]; 19 tumors) (log-rank P = .001). Overall survival at 60 months was 64.5% (95% CI, 43.5%-95.8%) in the BAP1del group vs 84.1% (95% CI, 69.0%-100%) in the BAP1nl group (log-rank P < .001). In these 43 cases, metastasis-free survival at 60 months was 100% for UMs without loss of the BAP1 locus or 8q gain, 70.0% (95% CI, 50.5%-96.9%) for UMs that carried 1 of these alterations, and 12.5% (95% CI, 2.1%-73.7%) for those that carried both (log-rank P < .001). Similarly, overall survival at 60 months was 100% for UMs without loss of the BAP1 locus or 8q gain, 80.8% (95% CI, 63.3%-100%) for UMs that carried 1 of these alterations, and 46.7% (95% CI, 23.3%-93.6%) for those that carried both (log-rank P < .001).
Conclusions and Relevance
These findings suggest that partial deletion of chromosome 3 encompassing the BAP1 locus is associated with poor prognosis. A cytogenetic classification of UMs could be proposed based on the status of the BAP1 locus instead of the chromosome 3 locus, while also taking chromosome 8q into account.
Introduction
Uveal melanoma (UM) is the most common primary malignant ocular tumor in adults of European ancestry.1 Despite efficient treatment, up to 50% of the patients will eventually develop metastases.2,3,4 Reliable prognostic assessment allows a closer monitoring of high-risk patients. Pathologic prognostic factors include large tumor basal diameter, thickness, ciliary body involvement, extraocular extension, epithelioid cell histologic findings, high mitotic rate, and lymphocytic infiltration.5 The gene expression profile (GEP) DecisionDx-UM (Castle Biosciences), based on the expression level of 12 genes, is frequently used in North America to complete the prognostic assessment.6,7
In the early 1990s, recurrent cytogenetic aberrations, including monosomy 3 (M3) and gain of 6p and 8q, were identified in UM samples.8 In 1996, M3 was empirically identified as a robust prognostic factor.9 Since then, genomic arrays have become routine tools to refine pathologic prognosis along with the GEP. We previously refined the prognostic value of M3 and gain of 8q by defining 3 groups: (1) high-risk patients whose tumors present an M3 and an 8q gain with a 2-year metastasis-free interval of 37%, (2) intermediate-risk patients with an M3 or an 8q gain (2-year metastasis-free interval of approximately 85%), and (3) low-risk patients without an M3 or an 8q gain (2-year metastasis-free interval of approximately 100%).10
The most common hypothesis to explain the poor prognosis of M3 tumors is the presence of 1 or more tumor suppressor genes on chromosome 3. BAP1 (BRCA1 associated protein 1) (OMIM 603089), a tumor suppressor gene located on the 3p21.1 cytoband, is now established as a main actor of UM malignant transformation because it is frequently mutated in M3 tumors, and germline mutations are associated with UM predisposition.11,12,13,14,15,16 However, all or most BAP1-mutated UMs intriguingly present a M3 (or a loss of heterozygosity of the whole chromosome 3 owing to an isodisomy), suggesting that the role of chromosome 3 loss in UM tumorigenesis may not be restricted to BAP1 inactivation. Therefore, prognostication of UM samples with partial deletions of chromosome 3, as sometimes observed in our daily practice and by other authors,17 is problematic. The goals of the present study were to explore these UMs with partial deletions of chromosome 3, as assessed by comparative genomic hybridization (array CGH), to assess their prognosis and to determine the minimal region of deletion associated with poor prognosis.
Methods
Patients
This cohort study was approved by our institutional ethics committee Institut Curie, PSL Research University. Written informed consent for the use of tissue samples and data for research was signed by each patient. The study complied with the principles of the Declaration of Helsinki.18 All patients were referred to Institut Curie, PSL Research University, Paris, France, and followed up by physicians at this institution from May 1, 2006, to September 30, 2017. Clinical diagnosis of UM was based on the presence of typical clinical findings as previously described.10 Local treatment consisted of proton beam radiotherapy, iodine 125 brachytherapy, or enucleation depending on the size and location of the tumors. Tumor samples were obtained by enucleation, endoresection, or fine-needle aspiration at the time of clip or plaque positioning. Liver ultrasonography, liver magnetic resonance imaging, or body computed tomography was performed at diagnosis and every 6 months thereafter. Diagnosis of metastasis was systematically confirmed by a biopsy.
Genomic Analysis
Tumor DNA was extracted and processed from May 1, 2006, to July 31, 2015, as previously described.10 Array CGH was performed on 3 different platforms according to the period when the test was performed: bacterial artificial chromosome arrays as previously described,19 NimbleGen 4 × 72 K arrays (Roche NimbleGen Inc), and Agilent 180K CGH/LOH custom chip (Agilent Technologies Inc). Array CGH results were interpreted by 3 of the authors (M.R., K.A.R., G.P.). Partial deletion of chromosome 3 was defined as the loss of at least 1 region of chromosome 3 but not the totality, whatever its size and location. Genomic positions in this article are defined in hg18 human genome assembly. Data analysis was performed from September 1, 2017, to November 30, 2017.
Statistical Analysis
Clinical, pathologic, and genomic data at diagnosis and follow-up events (local and distant recurrences, second cancers, or death from UM or from any other cause) were collected. The French Death Registry was consulted for patients who had not been followed up in consultation during the past 12 months. Metastasis-free survival (MFS) at 60 months was defined as the proportion of patients alive and free of metastasis at 60 months of follow-up after local treatment of primary UM. Overall survival (OS) at 60 months was defined as the proportion of patients alive at 60 months of follow-up after local treatment of primary UM, whatever the cause of death. Survival distributions were estimated by the Kaplan-Meier method and compared using the log-rank test. All tests were bilateral and performed with a 2-sided significance level of P < .05. To identify variables associated with MFS, a Cox proportional hazards regression analysis of candidate prognostic factors was performed using a forward stepwise selection procedure. The added value of each variable to the Cox proportional hazards regression model was determined using a likelihood ratio test with a significance level of P < .05. Statistical analysis was performed using R software, version 3.3 (R Foundation for Statistical Computing).
Results
We prospectively reanalyzed the array CGH profiles in 1088 UMs and detected 43 UMs (4.0%) that harbored a partial deletion of chromosome 3 in 43 patients (median age, 58 years [range, 12-79 years]; 22 [51%] female) (eTable in the Supplement). Median follow-up in these 43 cases was 66 months (range, 1.2-126.2 months). Median tumor diameter was 16 mm (range 10-22 mm), and median thickness was 10 mm (range, 5.3-18.2 mm). Ciliary body was involved in 14 of 43 cases (32.6%) and optic nerve in 4 of 43 cases (9.3%). Cell morphologic type was epithelioid or mixed in 13 cases (30.2%). Primary tumors were treated by enucleation in 18 cases (41.9%). The MFS at 60 months was 60.6% (95% CI, 46.0%-79.7%), and the OS at 60 months was 76.1% (95% CI, 62.8%-92.3%). A global overview of copy number profiles is provided in eFigure 1 in the Supplement. Size of deletions ranged from 1.36 to 110.88 Mb.
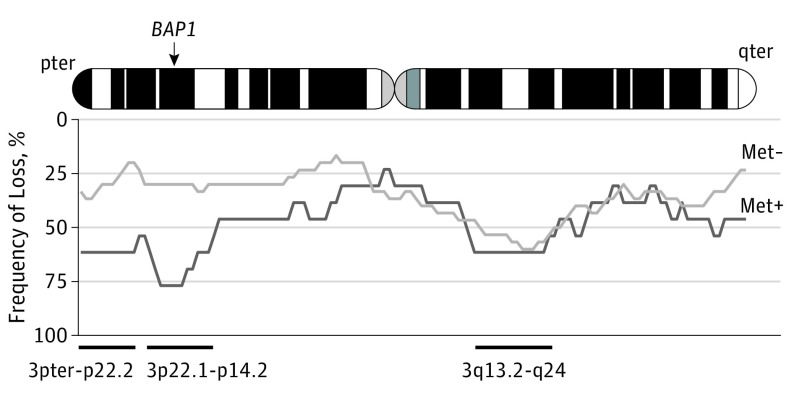
We initially explored survival data in an unsupervised manner and observed 3 recurrently lost regions in at least 8 metastatic samples: (1) from 3pter to p22.2, (2) from 3p22.1 to p14.2, and (3) from 3q13.2 to q24 (Figure 1). Of these, 2 regions were more frequently lost in metastatic cases than in nonmetastatic ones: the 3pter to p22.2 region (8 of 13 cases vs 6 of 30 cases; P = .01; odds ratio, 6.1; 95% CI, 1.2-34.1) and the 3p22.1 to p14.2 region, which encompasses BAP1 (10 of 13 cases vs 9 of 30 cases; P = .007; odds ratio, 7.4; 95% CI, 1.5-51.8). These 2 regions were close and highly correlated with each other because 8 of 10 metastatic cases presenting a 3p22.1 to p14.2 loss also presented a 3pter to p22.2 loss. The 3p22.1 to p14.2 region carries 290 other genes beside BAP1, but no recurrent mutations of these 290 genes were found in public and in-house databases.12,20,21
Figure 1. Copy Number Profiles in Metastatic (Met+) vs Nonmetastatic (Met−) Cases.
Frequencies of losses at a given position are shown at the bottom. Light gray indicates Met− cases (n = 30); dark gray, Met+ cases (n = 13).
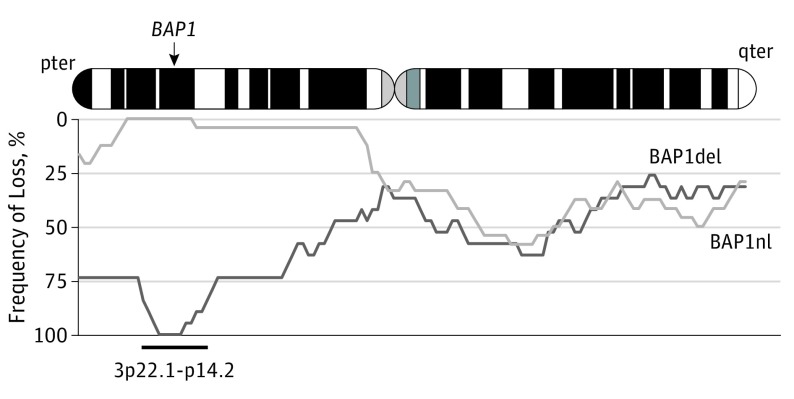
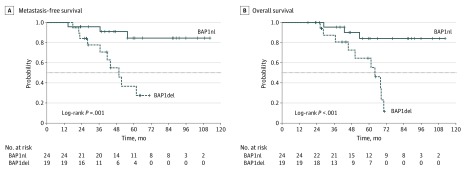
We then hypothesized that BAP1 loss was associated with poor prognosis in M3. To explore this hypothesis, we compared tumors with a chromosome 3 partial deletion that encompassed the BAP1 locus (BAP1del; 24 tumors) and tumors with a chromosome 3 partial deletion not encompassing the BAP1 locus (BAP1nl; 19 tumors). Tumors that carried a loss of the BAP1 locus frequently had large losses of the short arm of chromosome 3 (Figure 2). The MFS at 60 months was 80.5% (95% CI, 64.8%-100%) for the BAP1nl genomic group and 33.6% (95% CI, 15.8%-71.4%) for the BAP1del group (P = .001) (Figure 3). The OS at 60 months was 84.1% (95% CI, 69.0%-100%) for the BAP1nl genomic group and 64.5% (95% CI, 43.5%-95.8%) for the BAP1del group (P < .001). The only variables associated with MFS in univariate analysis were loss of the BAP1 locus and gain of 8q. These 2 variables were independently associated with MFS in multivariate analysis (Table).
Figure 2. Copy Number Profiles in Cases With Deletion of the BAP1 Locus (BAP1del) vs Cases Without the Loss of the BAP1 Locus (BAP1nl) .
Frequencies of deletion at a given position are shown at the bottom. Light gray indicates BAP1nl cases (n = 19); dark gray, BAP1del cases (n = 24).
Figure 3. Metastasis-Free Survival (MFS) and Overall Survival (OS) According to the Loss of the BAP1 Locus .
Survival curves in uveal melanomas with a partial loss of chromosome 3 encompassing the BAP1 locus or not. BAP1del indicates deletion of the BAP1 locus; BAP1nl, no loss of the BAP1 locus (normal).
Table. Univariate and Multivariate Analyses of Risk Factors for Metastasis.
| Characteristic | No. of Cases | HR (95% CI) | P Value |
|---|---|---|---|
| Univariate Analysis | |||
| Age, y | |||
| <60 | 23 | 1 [Reference] | .17 |
| ≥60 | 20 | 0.49 (0.17-1.4) | |
| Sex | |||
| Male | 21 | 1 [Reference] | .14 |
| Female | 22 | 0.46 (0.16-1.33) | |
| Diameter, mm | |||
| ≤15 | 17 | 1 [Reference] | .31 |
| >15 | 26 | 1.72 (0.6-4.95) | |
| Thickness, mm | |||
| ≤10 | 22 | 1 [Reference] | .43 |
| >10 | 21 | 1.49 (0.55-4) | |
| Tumor location | |||
| On the equator | 28 | 1 [Reference] | .07 |
| Anterior to the equator | 4 | 2.55 (0.69-9.36) | |
| Posterior to the equator | 10 | 0.36 (0.08-1.62) | |
| Retinal detachment | |||
| No | 4 | 1 [Reference] | .06 |
| Yes | 39 | 0.32 (0.09-1.11) | |
| Histologic type | |||
| Spindle cells | 10 | 1 [Reference] | >.99 |
| Epithelioid or mixed | 13 | 1 (0.28-3.55) | |
| BAP1 locus deletion | |||
| No | 24 | 1 [Reference] | .001 |
| Yes | 19 | 5.91 (1.89-18.54) | |
| 8q Gain | |||
| No | 16 | 1 [Reference] | .007 |
| Yes | 27 | 6.02 (1.36-26.61) | |
| Multivariate Analysis | |||
| BAP1 locus deletion | |||
| No | 24 | 1 [Reference] | .001 |
| Yes | 19 | 6.65 (2.09-21.18) | |
| 8q Gain | |||
| No | 16 | 1 [Reference] | .01 |
| Yes | 27 | 6.88 (1.53-30.86) | |
Abbreviation: HR, hazard ratio.
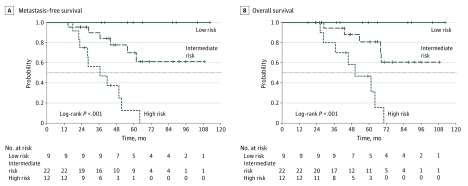
We defined 4 groups based on the BAP1 locus (lost or not lost) and 8q (gained or not gained) statuses. Prognoses of the BAP1 locus lost with 8q normal and BAP1 locus not lost with 8q gained were similar; thus, we merged these 2 groups, as in a previous classification (eFigure 2 in the Supplement).10 By analogy with previous work, we defined 3 prognosis groups as follows: (1) a group at low risk of metastasis without loss of the BAP1 locus or 8q gain (9 cases), (2) an intermediate risk group with tumors that carried loss of the BAP1 locus (7 cases) or 8q gain (15 cases), and (3) a high-risk group with loss of the BAP1 locus and 8q gain (12 cases). The MFS at 60 months was 100% for the low-risk group, 70.0% (95% CI, 50.5%-96.9%) for the intermediate-risk group, and 12.5% (95% CI, 2.1%-73.7%) for the high-risk group (P < .001; Figure 4). The OS at 60 months was 100% for the low-risk group, 80.8% (95% CI, 63.3%-100%) for the intermediate-risk group, and 46.7% (95% CI, 23.3%-93.6%) for the high-risk group (P < .001).
Figure 4. Metastasis-Free Survival (MFS) and Overall Survival (OS) According to the 3 Different Prognosis Groups .
Survival curves in uveal melanomas with a partial loss of chromosome 3 according to the 3 different prognosis groups.
Discussion
In this work, we explored a relatively large series of UMs with partial deletion of chromosome 3 and found that loss of the BAP1 locus may be associated with poor prognosis of M3 UM. This result was obtained by 2 different approaches investigating indirectly the prognostic value of the most frequently deleted regions of chromosome 3 and then directly assessing the prognostic value of the loss of the BAP1 locus in this series. The main consequence is to provide a potentially more accurate estimation of the prognosis of UMs that present with a partial deletion of chromosome 3. Our classification suggested efficiency in estimating metastatic outcome, identifying a group with a good MFS with no recurrence and a group with a high risk (87.5%) of recurrence with a median follow-up of more than 5 years. Survival rates were close to those observed in a previous series of UMs that presented with an M3 or disomy 3 associated or not associated with 8q gain.10 This hypothesis has yet to be verified in subsequent studies because direct comparison could not be done here.
Other teams are using different genomic technologies to assess UM prognosis. Fluorescence in situ hybridization (FISH) is widely used, but it may miss the loss of the BAP1 locus if the probe is not centered on this gene, as observed in several articles.2,22,23,24,25 Furthermore, FISH is often performed without chromosome 8q assessment, leading to suboptimal prognosis estimation. Multiplex ligation-dependent probe amplification assay that covers the BAP1 locus is a good alternative to characterize recurrent genomic imbalances in UM, but multiplex ligation-dependent probe amplification, as well as FISH and array CGH, only evaluate copy number and consequently do not identify isodisomic cases.26,27 Gene expression profile is a transcriptomic prognosis assay that is widely used in the United States.7 This assay distinguishes 2 subsets of UMs at low or high risk of metastasis by assessing the expression of 12 genes, including 4 that are located on the short arm of chromosome 3 (EIF1B [HGNC 30792], LMCD1 [OMIM 604859], ROBO1 [OMIM 602430], and SATB1 [OMIM 602075]) and 1 on the 3q (FXR1 [OMIM 600819]). Underexpression of these genes, possibly owing to M3, is associated with poor prognosis. A more accurate estimation by GEP is possible by adding the expression of PRAME (OMIM 606021), a gene located on an instable region of chromosome 22 exposed to duplication, which was correlated with the 8q status in a pivotal study.6 To our knowledge, GEP has never been specifically tested in a large series of UMs with partial chromosome 3 deletions. Furthermore, to our knowledge, GEP has never been compared with the combined M3/8q signature in a large cohort, impeding any conclusion on the superiority of one modality or the other. BAP1 immunohistochemistry is an alternative way to assess the prognosis of UMs.28,29 However, immunohistochemistry for BAP1 does not correlate in all cases with the BAP1 mutational status in UM and is therefore not a perfect surrogate.28
In the present series, partial deletions of chromosome 3 were found in 4.0% of cases, which is comparable with some previous series30,31,32 but lower than others.17,33,34 Recruitment bias may explain part of this discrepancy, but it is probably explained by the variety of technologies and the different classifications that were used. Comparison of all these studies is therefore limited. Similarly, the prognosis of these tumors was not clear because a discrepancy was observed, with some series associating partial loss with good prognosis17,30,33 and others associating it with intermediate or poor prognosis.26,34,35 These differences may be explained by the absence of distinction depending on the loss of the BAP1 locus compared with other losses.
One explanation for the low MFS associated with the loss of this locus may be that the loss of 1 BAP1 allele contributes to the inactivation of this gene and subsequent aggressiveness of the tumor. However, the minimal region of deletion that was associated with the lowest MFS in our series (3p22.1-p14.2) includes 291 genes. Even though this region encompasses BAP1, other important genes may be present there and haploinsufficiency of these genes may affect tumorigenesis. The 2 alleles of a tumor suppressor gene are commonly inactivated in the 2-hit model by a combination of different mechanisms, including total or partial loss of a chromosome, deleterious point mutations, short insertions and deletions, large-scale insertions and deletions, and promoter methylations.36 It is highly intriguing that BAP1 inactivation is frequently associated with monosomy 3 in UM, contrary to renal clear cell carcinomas and mesotheliomas, which carry losses of the short arm of chromosome 3 only or deleterious mutations of both alleles.16 Furthermore, haploinsufficiency of other genes on chromosome 3, possibly on its long arm, may play a role in UM tumorigenesis. This hypothesis may be of particular interest and should be put in perspective with the recent discovery of MBD4 (3q21.3) (OMIM 603574) recurrent, inactivating mutations in UM.37,38,39
To date, there is no standard treatment in the metastatic setting, but new drugs are being developed in UM.40 When an efficient treatment becomes available, the next step will consist of testing this treatment in the adjuvant setting in high-risk patients.41 Accurate prognosis evaluation is essential for such trials, and assays able to assess the status of the BAP1 locus and 8q status may then be required. Next-generation sequencing appears to be the best option in the near future because it not only assesses copy number, heterozygosity, and mutational statuses of UMs at low cost and with a lower amount of DNA but also allows circulating tumor DNA to be examined.42,43,44 Moving toward the implementation of such technologies in our daily practice would allow ocular oncology to enter the modern age of precision medicine while reducing costs and refining UM prognosis.
Limitations
The conclusions of this work are limited by its retrospective nature, but prospective series are unrealistic given the rarity of such tumors. Instead, the present work provides evidence to refine the current UM genomic classification, which may help ophthalmologists to better identify the metastatic evolution of their patients. Before generalization, other series from different centers are required. Furthermore, our series, composed of large tumors (median diameter of 16 mm and median thickness of 10 mm), is not reflective of the overall population of patients with UM, particularly because larger UMs are known to host a greater frequency of genomic alterations, including 8q gains.38,45 Other centers have reported genomic studies on biopsy samples of smaller UMs.46 However, this procedure is not consensual and must not be undertaken in inexperienced ocular oncology centers because of potential surgical complications. Multicenter collaborative studies of small UM genomics are required to address the question of partial chromosome 3 loss frequency at this stage of primary UM development. Another limitation of this study is that the array CGH technology is not adapted to detect chromosome 3 isodisomy, an infrequent alteration in UM, which may be associated with poor prognosis. A single-nucleotide polymorphism array can resolve this issue, but in the future, next-generation sequencing will probably be the privileged technology to circumvent this issue. Although the BAP1 locus hypothesis is a logical hypothesis, we cannot definitely affirm that BAP1 is the target of such deletions. Chromosome 3 is dense in cancer genes, and the BAP1 region, for instance, encompasses the tumor suppressor gene PBRM1 (OMIM 606083), which was recently found mutated in rare UMs. To confirm the BAP1 locus hypothesis and the classification, validation series are required, ideally together with further work sequencing BAP1 to confirm the presence of a second hit.
Conclusions
These findings suggest that partial deletion of chromosome 3 that encompasses the BAP1 locus is associated with poor prognosis. Consequently, a new cytogenetic classification of UMs was proposed based on the status of the BAP1 locus instead of chromosome 3. The frequent loss of the whole chromosome 3 in UMs raises the possibility of other genes associated with UM tumorigenesis on this chromosome.
eTable. Demographic and Disease Characteristics
eFigure 1. Copy Number Profile of Lost Regions on Chromosome 3 in the Whole Cohort
eFigure 2. Metastasis-Free Survival According to the Four Different Prognosis Groups
References
- 1.Mahendraraj K, Lau CS, Lee I, Chamberlain RS. Trends in incidence, survival, and management of uveal melanoma: a population-based study of 7,516 patients from the Surveillance, Epidemiology, and End Results database (1973-2012). Clin Ophthalmol. 2016;10:2113-2119. doi: 10.2147/OPTH.S113623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desjardins L, Levy-Gabriel C, Lumbroso-Lerouic L, et al. Prognostic factors for malignant uveal melanoma: retrospective study on 2,241 patients and recent contribution of monosomy-3 research [in French]. J Fr Ophtalmol. 2006;29(7):741-749. doi: 10.1016/S0181-5512(06)73843-8 [DOI] [PubMed] [Google Scholar]
- 3.Singh AD, Turell ME, Topham AK. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118(9):1881-1885. doi: 10.1016/j.ophtha.2011.01.040 [DOI] [PubMed] [Google Scholar]
- 4.Kujala E, Mäkitie T, Kivelä T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci. 2003;44(11):4651-4659. doi: 10.1167/iovs.03-0538 [DOI] [PubMed] [Google Scholar]
- 5.Brierley JD, Gospodarowicz MK, Wittekind C. The TNM Classification of Malignant Tumours. 8th ed New York, NY: Wiley-Blackwell; 2016. [Google Scholar]
- 6.Field MG, Decatur CL, Kurtenbach S, et al. PRAME as an independent biomarker for metastasis in uveal melanoma. Clin Cancer Res. 2016;22(5):1234-1242. doi: 10.1158/1078-0432.CCR-15-2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64(20):7205-7209. doi: 10.1158/0008-5472.CAN-04-1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prescher G, Bornfeld N, Becher R. Nonrandom chromosomal abnormalities in primary uveal melanoma. J Natl Cancer Inst. 1990;82(22):1765-1769. doi: 10.1093/jnci/82.22.1765 [DOI] [PubMed] [Google Scholar]
- 9.Prescher G, Bornfeld N, Hirche H, Horsthemke B, Jöckel KH, Becher R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347(9010):1222-1225. doi: 10.1016/S0140-6736(96)90736-9 [DOI] [PubMed] [Google Scholar]
- 10.Cassoux N, Rodrigues MJ, Plancher C, et al. Genome-wide profiling is a clinically relevant and affordable prognostic test in posterior uveal melanoma. Br J Ophthalmol. 2014;98(6):769-774. doi: 10.1136/bjophthalmol-2013-303867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48(12):856-859. doi: 10.1136/jmedgenet-2011-100156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson AG, Shih J, Yau C, et al. ; TCGA Research Network . Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2018;33(1):151. doi: 10.1016/j.ccell.2017.12.013 [DOI] [PubMed] [Google Scholar]
- 13.Field MG, Durante MA, Anbunathan H, et al. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat Commun. 2018;9(1):116. doi: 10.1038/s41467-017-02428-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rai K, Pilarski R, Boru G, et al. Germline BAP1 alterations in familial uveal melanoma. Genes Chromosomes Cancer. 2017;56(2):168-174. doi: 10.1002/gcc.22424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330(6009):1410-1413. doi: 10.1126/science.1194472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiesner T, Obenauf AC, Murali R, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43(10):1018-1021. doi: 10.1038/ng.910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdel-Rahman MH, Christopher BN, Faramawi MF, et al. Frequency, molecular pathology and potential clinical significance of partial chromosome 3 aberrations in uveal melanoma. Mod Pathol. 2011;24(7):954-962. doi: 10.1038/modpathol.2011.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-2194. doi: 10.1001/jama.2013.281053 [DOI] [PubMed] [Google Scholar]
- 19.Trolet J, Hupé P, Huon I, et al. Genomic profiling and identification of high-risk uveal melanoma by array CGH analysis of primary tumors and liver metastases. Invest Ophthalmol Vis Sci. 2009;50(6):2572-2580. doi: 10.1167/iovs.08-2296 [DOI] [PubMed] [Google Scholar]
- 20.Furney SJ, Pedersen M, Gentien D, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3(10):1122-1129. doi: 10.1158/2159-8290.CD-13-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson P, Aoude LG, Wadt K, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7(4):4624-4631. doi: 10.18632/oncotarget.6614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mensink HW, Vaarwater J, de Keizer RJ, et al. Chromosomal aberrations in iris melanomas. Br J Ophthalmol. 2011;95(3):424-428. doi: 10.1136/bjo.2010.181289 [DOI] [PubMed] [Google Scholar]
- 23.Worley LA, Onken MD, Person E, et al. Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clin Cancer Res. 2007;13(5):1466-1471. doi: 10.1158/1078-0432.CCR-06-2401 [DOI] [PubMed] [Google Scholar]
- 24.van Gils W, Lodder EM, Mensink HW, et al. Gene expression profiling in uveal melanoma: two regions on 3p related to prognosis. Invest Ophthalmol Vis Sci. 2008;49(10):4254-4262. doi: 10.1167/iovs.08-2033 [DOI] [PubMed] [Google Scholar]
- 25.Singh AD, Aronow ME, Sun Y, et al. Chromosome 3 status in uveal melanoma: a comparison of fluorescence in situ hybridization and single-nucleotide polymorphism array. Invest Ophthalmol Vis Sci. 2012;53(7):3331-3339. doi: 10.1167/iovs.11-9027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damato B, Dopierala J, Klaasen A, van Dijk M, Sibbring J, Coupland SE. Multiplex ligation-dependent probe amplification of uveal melanoma: correlation with metastatic death. Invest Ophthalmol Vis Sci. 2009;50(7):3048-3055. doi: 10.1167/iovs.08-3165 [DOI] [PubMed] [Google Scholar]
- 27.Larsen AC, Holst L, Kaczkowski B, et al. MicroRNA expression analysis and multiplex ligation-dependent probe amplification in metastatic and non-metastatic uveal melanoma. Acta Ophthalmol. 2014;92(6):541-549. doi: 10.1111/aos.12322 [DOI] [PubMed] [Google Scholar]
- 28.Kalirai H, Dodson A, Faqir S, Damato BE, Coupland SE. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br J Cancer. 2014;111(7):1373-1380. doi: 10.1038/bjc.2014.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Essen TH, van Pelt SI, Versluis M, et al. Prognostic parameters in uveal melanoma and their association with BAP1 expression. Br J Ophthalmol. 2014;98(12):1738-1743. doi: 10.1136/bjophthalmol-2014-305047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas S, Pütter C, Weber S, Bornfeld N, Lohmann DR, Zeschnigk M. Prognostic significance of chromosome 3 alterations determined by microsatellite analysis in uveal melanoma: a long-term follow-up study. Br J Cancer. 2012;106(6):1171-1176. doi: 10.1038/bjc.2012.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tschentscher F, Prescher G, Horsman DE, et al. Partial deletions of the long and short arm of chromosome 3 point to two tumor suppressor genes in uveal melanoma. Cancer Res. 2001;61(8):3439-3442. [PubMed] [Google Scholar]
- 32.Cross NA, Rennie IG, Murray AK, Sisley K. The identification of chromosome abnormalities associated with the invasive phenotype of uveal melanoma in vitro. Clin Exp Metastasis. 2005;22(2):107-113. doi: 10.1007/s10585-005-5142-2 [DOI] [PubMed] [Google Scholar]
- 33.Shields CL, Ganguly A, Bianciotto CG, Turaka K, Tavallali A, Shields JA. Prognosis of uveal melanoma in 500 cases using genetic testing of fine-needle aspiration biopsy specimens. Ophthalmology. 2011;118(2):396-401. doi: 10.1016/j.ophtha.2010.05.023 [DOI] [PubMed] [Google Scholar]
- 34.Damato B, Dopierala JA, Coupland SE. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin Cancer Res. 2010;16(24):6083-6092. doi: 10.1158/1078-0432.CCR-10-2076 [DOI] [PubMed] [Google Scholar]
- 35.Ewens KG, Kanetsky PA, Richards-Yutz J, et al. Genomic profile of 320 uveal melanoma cases: chromosome 8p-loss and metastatic outcome. Invest Ophthalmol Vis Sci. 2013;54(8):5721-5729. doi: 10.1167/iovs.13-12195 [DOI] [PubMed] [Google Scholar]
- 36.Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1(2):157-162. doi: 10.1038/35101031 [DOI] [PubMed] [Google Scholar]
- 37.Rodrigues M, Mobuchon L, Houy A, et al. Outlier response to anti-PD1 in uveal melanoma reveals germline MBD4 mutations in hypermutated tumors. Nat Commun. 2018;9(1):1866. doi: 10.1038/s41467-018-04322-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodrigues M, Mobuchon L, Houy A, et al. Evolutionary routes in metastatic uveal melanomas depend on MBD4 alterations. Clin Cancer Res. 2019;25(18):5513-5524. doi: 10.1158/1078-0432.CCR-19-1215 [DOI] [PubMed] [Google Scholar]
- 39.Johansson PA, Stark A, Palmer JM, et al. Prolonged stable disease in a uveal melanoma patient with germline MBD4 nonsense mutation treated with pembrolizumab and ipilimumab. Immunogenetics. 2019;71(5-6):433-436. doi: 10.1007/s00251-019-01108-x [DOI] [PubMed] [Google Scholar]
- 40.Carvajal RD, Schwartz GK, Tezel T, Marr B, Francis JH, Nathan PD. Metastatic disease from uveal melanoma: treatment options and future prospects. Br J Ophthalmol. 2017;101(1):38-44. doi: 10.1136/bjophthalmol-2016-309034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piperno-Neumann S, Rodrigues MJ, Servois V, et al. A randomized multicenter phase 3 trial of adjuvant fotemustine versus surveillance in high risk uveal melanoma (UM) patients (FOTEADJ). J Clin Oncol. 2017;35(15)(suppl):9502-9502. doi: 10.1200/JCO.2017.35.15_suppl.9502 [DOI] [Google Scholar]
- 42.Smit KN, van Poppelen NM, Vaarwater J, et al. Combined mutation and copy-number variation detection by targeted next-generation sequencing in uveal melanoma. Mod Pathol. 2018;31(5):763-771. doi: 10.1038/modpathol.2017.187 [DOI] [PubMed] [Google Scholar]
- 43.Afshar AR, Damato BE, Stewart JM, et al. Next-generation sequencing of uveal melanoma for detection of genetic alterations predicting metastasis. Transl Vis Sci Technol. 2019;8(2):18. doi: 10.1167/tvst.8.2.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matet A, Aït Raïs K, Malaise D, et al. Comparative cytogenetic abnormalities in paired choroidal melanoma samples obtained before and after proton beam irradiation by transscleral fine-needle aspiration biopsy and endoresection. Cancers (Basel). 2019;11(8):E1173. doi: 10.3390/cancers11081173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shain AH, Bagger MM, Yu R, et al. The genetic evolution of metastatic uveal melanoma. Nat Genet. 2019;51(7):1123-1130. doi: 10.1038/s41588-019-0440-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Angi M, Kalirai H, Taktak A, et al. Prognostic biopsy of choroidal melanoma: an optimised surgical and laboratory approach. Br J Ophthalmol. 2017;101(8):1143-1146. doi: 10.1136/bjophthalmol-2017-310361 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable. Demographic and Disease Characteristics
eFigure 1. Copy Number Profile of Lost Regions on Chromosome 3 in the Whole Cohort
eFigure 2. Metastasis-Free Survival According to the Four Different Prognosis Groups