Abstract
Background:
Standards-based clinical data normalization has become a key component of effective data integration and accurate phenotyping for secondary use of electronic healthcare records (EHR) data. HL7 Fast Healthcare Interoperability Resources (FHIR) is an emerging clinical data standard for exchanging electronic healthcare data and has been used in modeling and integrating both structured and unstructured EHR data for a variety of clinical research applications. The overall objective of this study is to develop and evaluate a FHIR-based EHR phenotyping framework for identification of patients with obesity and its multiple comorbidities from semi-structured discharge summaries leveraging a FHIR-based clinical data normalization pipeline (known as NLP2FHIR).
Methods:
We implemented a multi-class and multi-label classification system based on the i2b2 Obesity Challenge task to evaluate the FHIR-based EHR phenotyping framework. Two core parts of the framework are: (a) the conversion of discharge summaries into corresponding FHIR resources – Composition, Condition, MedicationStatement, Procedure and FamilyMemberHistory using the NLP2FHIR pipeline, and (b) the implementation of four machine learning algorithms (logistic regression, support vector machine, decision tree, and random forest) to train classifiers to predict disease state of obesity and 15 comorbidities using features extracted from standard FHIR resources and terminology expansions. We used the macro- and micro-averaged precision (P), recall (R), and F1 score (F1) measures to evaluate the classifier performance. We validated the framework using a second obesity dataset extracted from the MIMIC-III database.
Results:
Using the NLP2FHIR pipeline, 1237 clinical discharge summaries from the 2008 i2b2 obesity challenge dataset were represented as the instances of the FHIR Composition resource consisting of 5677 records with 16 unique section types. After the NLP processing and FHIR modeling, a set of 244,438 FHIR clinical resource instances were generated. As the results of the four machine learning classifiers, the random forest algorithm performed the best with F1-micro(0.9466)/F1-macro(0.7887) and F1-micro(0.9536)/F1-macro(0.6524) for intuitive classification (reflecting medical professionals’ judgments) and textual classification (reflecting the judgments based on explicitly reported information of diseases), respectively. The MIMIC-III obesity dataset was successfully integrated for prediction with minimal configuration of the NLP2FHIR pipeline and machine learning models.
Conclusions:
The study demonstrated that the FHIR-based EHR phenotyping approach could effectively identify the state of obesity and multiple comorbidities using semi-structured discharge summaries. Our FHIR-based phenotyping approach is a first concrete step towards improving the data aspect of phenotyping portability across EHR systems and enhancing interpretability of the machine learning-based phenotyping algorithms.
Keywords: Clinical phenotyping, HL7 Fast Healthcare Interoperability Resources (FHIR), Electronic Health Records (EHRs), Natural language processing, Algorithm portability
1. Introduction
Electronic health records (EHR) offer many opportunities for their secondary use in support of clinical and translational research. EHR data may exist as structured data (eg, patient demographics and laboratory test results), or unstructured clinical notes (eg, history of present illness and family history), or in some cases, as unstructured data embedded within semi-structured EHR data (eg, discharge summaries and clinical interviews). These essential data sources, containing considerable valuable information, are increasingly being utilized for information extraction-based applications to support patient care and clinical research. Due to the heterogeneity of data sources from different EHR systems, EHR data for the current phenotyping process (eg, transforming the raw EHR data into clinically relevant features) carries many challenges in terms of completeness, accuracy, complexity and systematic errors, which leads to barriers in large-scale EHR data integration and analytics [1]. The next generation phenotyping of EHRs requires identification of true patient states in an accurate and high-throughput manner, leveraging a rigorous healthcare process model [1]. To realize the vision of next generation phenotyping, there is an urgent need to improve the portability and interpretability of the phenotyping algorithms through a standards-based framework.
As a number of clinical data standards and common data models (CDMs) have been developed to provide a normalized representation of EHR data, it becomes feasible to establish a standards-based EHR phenotyping framework. These clinical data standards and CDMs include, but are not limited to, the Health Level Seven International (HL7) Fast Healthcare Interoperability Resource (FHIR) specification [2], the HL7 Clinical Document Architecture (CDA) [3], the National Quality Forum (NQF) Quality Data Model (QDM) [4], the Observational Health Data Sciences and Informatics (OHDSI) Common Data Model (CDM) [5], and the National Patient-Centered Clinical Research Network (PCORnet) CDM [6]. Among these data standards and models, the FHIR standard was developed by HL7 as the next generation clinical data standards framework to meet clinical interoperability needs, and the FHIR specification puts a strong emphasis on implementation of the standard using APIs and supporting tools.
FHIR has increasingly been used in modeling and integrating both structured and unstructured EHR data for a variety of clinical research applications. In ongoing work, we developed a clinical data normalization pipeline – referred to as NLP2FHIR [7,8]. The work was motivated by unmet needs on standardizing unstructured clinical data, as exemplified by the recent proposal from the Office of the National Coordinator for Health Information Technology (ONC) and the Centers for Medicare & Medicaid Services (CMS) that FHIR APIs be required for certified EHR systems [9]. Particularly, using natural language processing (NLP) to gain access to the narrative content in EHRs via FHIR will be of great value to data analytics, quality improvement, and advanced decision support as a significant portion of clinically relevant information remains locked away in unstructured data. However, the current HL7 Argonaut project [10] has not yet provided a solution to standardize unstructured data. The NLP2FHIR pipeline utilizes a FHIR-based Unstructured Information Management Architecture (UIMA) type system as a core target model to integrate and normalize the outputs from a number of UIMA-based clinical NLP tools such as cTAKES [11], MedXN [12] and MedTime [13] with the goal of automatically generating standardized FHIR resource instances from unstructured text. Note that cTAKES [11] is an open-source NLP system for extraction of information from clinical narratives; MedXN [12] system is designed to extract comprehensive medication information and normalize it to the most appropriate RxNorm concept unique identifier (RxCUI) as specifically as possible; and MedTime [13] is an automated system for EVENT/YIMEX3 extraction and temporal link identification from clinical text.
Machine learning-based computable phenotypes have increasingly become a common practice to identify patients’ true disease states from raw EHR data [1,14]. Once true disease status is known, additional models can be derived to predict future onset of diseases, quantify severity of diseases and/or identify sub-populations within diseases. A portable machine learning system has many advantages, including lowering development costs when it runs on multiple targets (eg, multiple datasets); helping ensure compatibility and functional correctness across different platforms; and encouraging collaborations among researchers [15]. In a previous study, we proposed a framework for improving the portability of EHR-based phenotyping algorithms [16], in which we demonstrated that the concept of portability is multi-faceted with many steps and potential impact points. Specifically, portability can be achieved by tradeoffs across three domains: data, authoring and implementation. For data preparation, the following factors would have an impact on the portability of phenotype algorithms: (1) efforts to transform the shape of the data into a CDM; (2) efforts to convert data from one modality to another (eg, NLP to obtain structured results); and (3) efforts to map local terms to a standard vocabulary with handling information loss or semantic drift. In this study, we place a stronger emphasis on the data aspect of the portability as we argue that with standard input using FHIR-based EHR data, the machine learning algorithms that represent computable phenotypes become more portable across heterogeneous data systems and different institutions. This argument will become stronger as more and more major EHR vendors adopt the FHIR standard for the exchange of EHR data [10]. In addition, features selected from standard FHIR data instances would potentially increase the interpretability of the machine learning models used in generating computable phenotypes as interpretability of a machine learning algorithm is a huge barrier for its adoption in many clinical applications [17].
Our overall objective of this study is to develop and evaluate a portable FHIR-based EHR phenotyping framework leveraging the NLP2FHIR clinical data normalization pipeline. To assess feasibility, we conducted a case study based on the “2008 i2b2 NLP Obesity Challenge” [18], which is a well-defined phenotyping problem to identify patients with obesity and their comorbidities from semi-structured discharge summaries. We validated the framework using a second obesity dataset extracted from a freely accessible critical care database known as MIMIC-III [19].
2. Related work
According to the results of the 2008 i2b2 NLP Obesity Challenge, the top 10 solutions were primarily rule-based systems [20,21], while machine learning-based studies and combined methods also demonstrated promising results [22,23]. From the portability perspective, the tradeoff between phenotyping accuracy and portability should be considered for implementing any NLP-driven phenotyping system across different institutions and data systems. The proof-of-concept of this notion has been demonstrated in a number of recent studies. Sharma and Mao, et al. presented a portable NLP-based phenotyping system that leverages the OHDSI CDM and standard vocabularies [24]. Specifically, textual discharge summaries were parsed using MetaMap, and all of the extracted features were mapped to UMLS’s Concept Unique Identifiers (CUIs). The MetaMap’s output was stored in a MySQL database using the schemas defined in the OHDSI CDM to standardize necessary data elements. Once data from multiple sources get harmonized into the CDM, researchers can conduct systematic analysis at a larger scale to perfect new secondary research techniques in biomedical data mining. The eMERGE [25] and PhEMA [50] projects aim at the implementation of an open-sourced, standard-based system to improve the portability of EHR-driven phentotype algorithms. For example, the Quality Data Model (QDM) [4] has been leveraged in the PhEMA project to standardize phenoype algorithm definitions, which can be further translated and executed against different clinical data repositories (eg, local data warehouse, or i2b2) [27]. The DeepPhe project [28] developed an NLP system for extracting cancer phenotypes from clinical records leveraging a conceptual model (known as the DeepPhe Ontology) and the FHIR standard, with the assumption that current information models of clinical and genomic data are not sufficiently expressive to represent individual observations and to aggregate those observations into longitudinal summaries over the course of cancer care. In addition, Google actively investigated the FHIR integration with its deep learning framework using electronic health records [29], in which one of main contributions is to provide a generic data processing pipeline to produce FHIR outputs without manual feature harmonization. This makes it relatively easy to deploy their system to a new hospital. However, such a data processing pipeline is not publicly available as open source and it is not clear whether the pipeline provides the functionality to represent unstructured EHR data in FHIR.
3. Materials and methods
3.1. Materials
3.1.1. The i2b2 obesity Challenge dataset
The dataset contains a total of 1237 annotated discharge summaries, each corresponding to one patient [18,21]. For each discharge summary, the status of patients’ obesity and their fifteen comorbidities are annotated using a simple classification scheme: present (Y), absent (N), questionable (Q), or unmentioned (U). These fifteen obesity comorbidities are: asthma, atherosclerotic cardiovascular disease (CAD), congestive heart failure (CHF), depression, diabetes mellitus (DM), gallstones/cholecystectomy, gastroesophageal reflux disease (GERD), gout, hypercholesterolemia, hypertension (HTN), hypertriglyceridemia, obstructive sleep apnea (OSA), osteoarthritis (OA), peripheral vascular disease (PVD), and venous insufficiency [21]. For each patient, two types of judgments, ie, textual and intuitive, are provided by the original obesity challenge dataset. Textual judgments are those based on explicit mentions in the text recognized by experts without reasoning (eg, the statement “the patient is obese”), and Y/N/Q/U were labeled for each condition. Intuitive judgments are those made by experts by applying their intuition and judgment to infer the presence of unmentioned conditions based on what is mentioned in the text (eg, the statement “the patient weighs 230 lbs and is 5 ft 2 in.”), and as such only the status categories Y/N/Q were labeled for each condition. The dataset is divided into training and testing corpus, and all records have been fully de-identified and publicly available for study purposes.
3.1.2. MIMIC-III database
MIMIC-III (Medical Information Mart for Intensive Care) is a publicly and freely available database developed by the Massachusetts Institution of Technology (MIT), comprising deidentified health information data related to patients admitted to critical care units [19]. Data includes vital signs, medications, laboratory measurements, observations and notes charted by care providers, and more. It is disseminated in a relational database consisting of 26 tables. In this study, we identified a cohort of obesity patients as a case group (using the criteria: Age at admission > =18 years old and BMI > = 30 kg/m2) and a control group (using the criteria: Age at admission > =18 years old and BMI > = 18.5 kg/m2 and < 25 kg/m2). Note that BMI stands for Body Mass Index (BMI) which is calculated as a patient’s weight (kg) divided by their height (m) squared and is a measure used for screening for obesity [30]. For adult men and women, a BMI value of 18.5–24.9 indicates normal weight and a BMI value of 30 or greater indicates obesity. We extracted a collection of discharge summaries for both obesity case and control groups and used them as a second dataset for validating the NLP2FHIR pipeline and prediction models.
3.1.3. FHIR specification
FHIR is an emerging HL7 standard specification for exchanging electronic healthcare information. It provides a consistent, easy to implement, and rigorous mechanism for exchanging data between healthcare applications [2]. A FHIR resource is the basic building block that can be maintained independently and is the smallest unit of exchange. The core FHIR resources satisfy the majority of common use cases, and represent a wide range of healthcare related concepts and elements, both clinical and administrative. FHIR supports three standard data exchange formats: XML (eXtensible Markup Language), JSON (JavaScript Object Notation) and RDF (Resource Description Framework). As of Feb 21, 2019, the version FHIR R4 has been released officially while we used an earlier version, the Standards for Trial Use Version 3 (STU3) (released on April 19, 2017) in this study. Fig. 1 shows the FHIR Condition resource definition and an example of its instance data in JSON. The elements and datatypes that are not covered by the core FHIR resource definitions, but are required in an implementation scenario, can be handled with its built-in extension mechanism. In this study, we used FHIR resources and extensions to represent discharge summaries in a structured and standardized way.
Fig. 1.
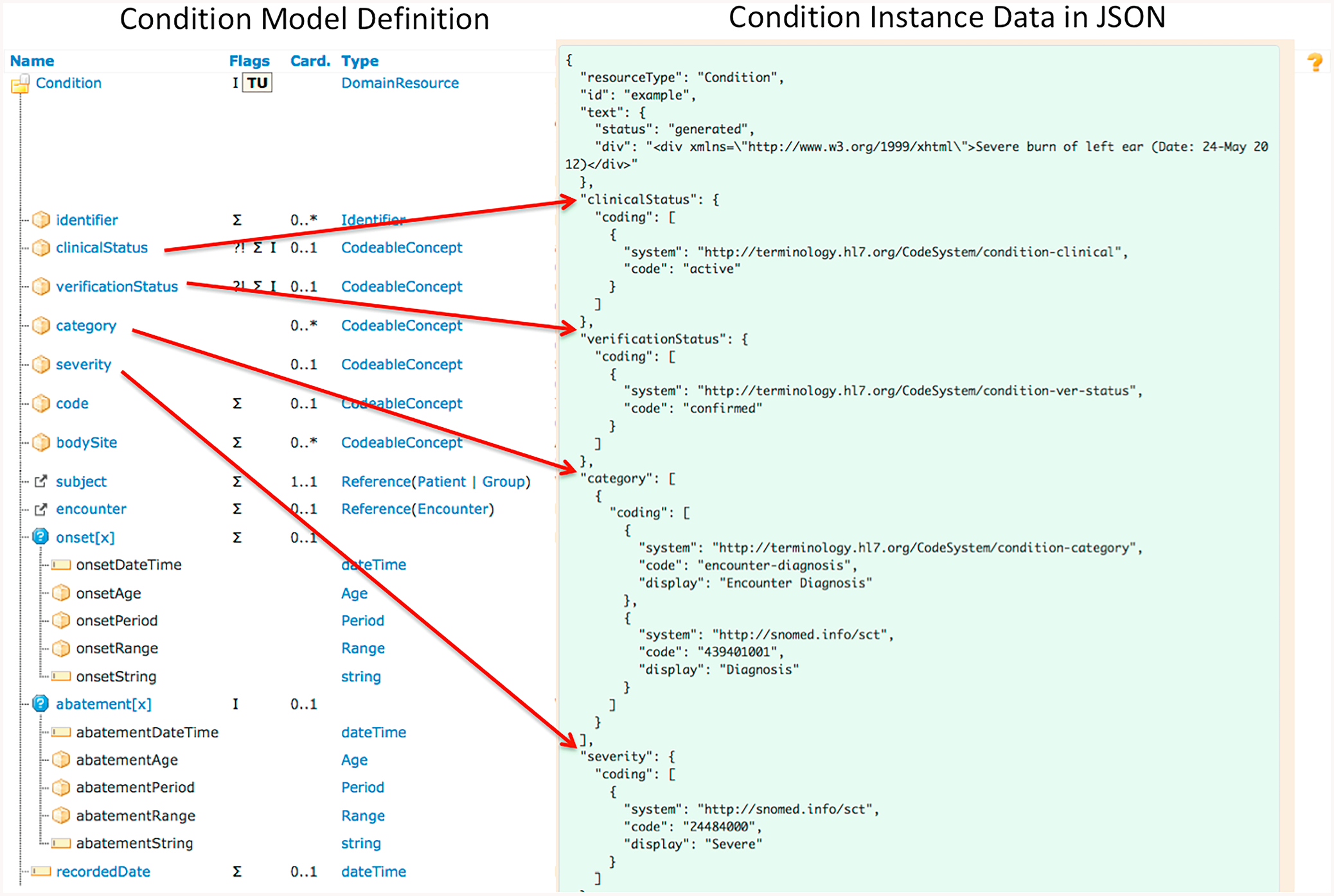
The FHIR model of Condition Resource (partial) and an example of its instance data in JSON.
In the current NLP2FHIR implementation, clinical FHIR resources Condition, Procedure, MedicationStatement (including Medication), and FamilyMemberHistory are used to capture core clinical data. Note that the MedicationStatement resource represents a record of a medication that is being consumed by a patient whereas the Medication resource is primarily used for the identification and definition of a medication. In other words, the MedicationStatement resource covers more clinical context information about how a medication is used for a patient. In addition, the FHIR document resources [31] Composition and Bundle are used to build documents to represent unstructured discharge summaries used in this study. The FHIR Composition resource is the foundation of a clinical document that provides identity, context, and purpose of the document, and divides the documents into a series of sections, each with their own narrative. The resource also supports references to other clinical resources that are used as semantic annotations of the clinical documents (we call these references as semantic links). The FHIR Bundle resource serves as a container for a collection of resources, which is used to contain both the instances of the FHIR Composition resource and its referenced clinical resources in this study.
3.2. Methods
Fig. 2 shows the system architecture of our FHIR-based EHR phenotyping framework which is comprised of the following two core components: (1) NLP2FHIR: a FHIR-based data normalization pipeline component, where the semi-structured discharge summaries were processed and represented in the standard FHIR resource instances; and (2) a FHIR-based EHR phenotyping component, where machine learning algorithms were implemented to predict obesity and its 15 aforementioned comorbidities.
Fig. 2.

The framework for FHIR-based phenotyping using electronic health records data.
3.2.1. NLP2FHIR: A FHIR-based data normalization pipeline
We used the NLP2FHIR pipeline [32] to support automatic fine-grained semantic modeling and enhanced its functionalities based on the requirements for this obesity use case under study. The enhanced functionalities include section detection and concept identifier detection. The sub-components of the NLP2FHIR pipeline are described as follows:
Section detection. Use of section headings in unstructured clinical narratives are not standardized across EHR systems and institutions, or even across practitioners within a single institution. Therefore, section detection is one of NLP tasks that can help detect and standardize major sections and downstream data integration. Through a dictionary lookup at the beginning of each section, we separated each unstructured document with a collection of different sections leveraging the SecTag section dictionary [33,34], and used the matched words as a section name to label each section. Although each section of the document was labeled with section names, these section names do not have a normalized expression. For example, the section name “History of Present Illness” may exist as different expressions, such as “patient hpi”, “summary of present illness”, “present illness”, etc. We reused and extended the SecTag section index with section names identified from both i2b2 Challenge and MIMIC-III datasets, and normalized the section names using LOINC codes. Note that the updated section index file is available in the project GitHub website (see link in the Conclusion section). The recall and precision of our section detection algorithm were 92.6% and 93.8% for major sections in a preliminary evaluation of 100 documents (with 911 identified sections), which is less optimal than the performance of original SecTag algorithm but is satisfactory for this study. With the normalized section names, our pipeline supports the representation of a variety of documents using the FHIR Composition.
A core NLP engine. We integrated a number of existing clinical NLP tools, including cTAKES [11], MedXN [12], and MedTime [13], to extract clinical entities from corresponding document sections. In the current implementation, the NLP2FHIR pipeline invokes cTAKES and MedTime for extracting entities and temporal relationships using the FHIR resources Condition, Procedure, and FamilyMemberHistory, and utilizes MedXN and MedTime for extracting and standardizing drug names using the FHIR resources MedicationStatement and Medication. Furthermore, to allow for rapid integration of NLP outputs or particularly extracted FHIR elements, we developed a FHIR-based type system [7] based on the FHIR STU 3 v1.8.0 specification, which is used as a core target model to integrate and normalize the outputs from the three UIMA-based NLP pipelines (as mentioned above) in the current NLP2FHIR implementation. This FHIR-based type system (rendered in the UIMA-compliant format) defines core clinical data models (eg, Condition, Procedure, Medication, Observation) based on the FHIR specification for capturing and standardizing entities extracted from unstructured clinical narratives.
Rules for detecting concept modifiers. “Uncertainty” and “Negation” are two common linguistic phenomena existing in clinical documents, eg, “The patient is suspicious for breast cancer” or “Patient denies headache”. Detecting these modifiers around clinical concepts helps to judge if an event is uncertain or negated. We define the rules and a set of trigger words, and used the ConText algorithm [35] to capture them. The ConText algorithm was applied for determining entity contexts such as negation and certainty. The algorithm works by detecting defined start triggers (a span of text, the context (negation, assertion, etc.) it represents, and a lookup direction) and extends a span to either the left or the right of the said trigger, as defined in the trigger definition, until such a point that a terminal trigger (a different span of text and the context to terminate) is found. Any extracted entities contained within these detected spans are annotated with the contexts relevant to the span in question; for instance, if the entity is contained within a span that was created by a negation trigger word, the said entity would be negated. The results from this contextual annotation were represented in generated FHIR resources through the use of FHIR NLP extensions [36]. If the words/phrases around each concept matched one of pre-defined rules and triggers from the original ConText algorithm, we treated it as negative or uncertain. The ConText algorithm has a well-defined performance on clinical narratives (including discharge summaries) [35] so we did not evaluate its performance in this study.
Normalized clinical elements representation in FHIR clinical resources. Our NLP2FHIR pipeline generates a set of the FHIR clinical resource instances, including those instances defined in the resources Condition, Procedure, MedicationStatement and FamilyMemberHistory. FHIR supports gathering a collection of resource instances into a single instance of FHIR Bundle to support flexible contents representation and exchange. Accordingly, we combined the FHIR Composition documents and the generated clinical resource instances into exchangeable resource bundles through their references. Fig. 3 shows an example of a FHIR Composition instance that includes a standard section of ‘Problem List – Reported (LOINC: 11450–4)’ and its referred FHIR resources.
Fig. 3.

An example of an FHIR Composition instance that includes a standard section of ‘Problem List – Reported (LOINC: 11450–4)’ and its referred FHIR resources.
3.2.2. FHIR-based EHR phenotyping module
Fig. 4 shows the system architecture for the module of FHIR-based EHR phenotyping. In this module, four types of machine learning algorithms were used to classify patients with obesity and its 15 comorbidities: logistic regression (LR) [37], support vector machine (SVM) [38], decision tree (DT) [39], and random forest (RF) [40]. The packages are implemented using the package scikit-learn [41]. LR is a statistical model that uses a logistic function to model a binary dependent variable. It can be treated as a special case of the generalized linear model. It has extensions such as multinomial LR or ordinal LR to model more than two levels of the dependent variables. SVM is a supervised learning method that maps training examples as points in a feature space and identifies a hyperplane that can separate examples into their corresponding categories. DT is a decision support method that splits a dataset into separate branches and each split labeled with an attribute produces the most homogeneous groups. The resulting tree structure allows for easy visualization and interpretation. RF is an ensemble learning algorithm that uses multiple decision trees to obtain better prediction performance. RF is robust in correcting the overfitting of decision trees. All four machine learning models are widely used in the classification tasks in healthcare domains; in this study, these models were mainly used for cross-validation purposes.
Fig. 4.

The module of FHIR-based EHR phenotyping to classify patients with obesity and its comorbidities.
-
1Feature extraction. We extracted features for classifier training from the instances of standard FHIR Bundle resource, which contains instance data of the following FHIR resources: Composition, Condition, Procedure, FamilyMemberHistory, and MedicationStatement. The key features extracted for classifier training include:
- Concept codes and attributes extracted from FHIR resources
- Negation modifiers extracted from FHIR extensions
- Section codes extracted from the FHIR Composition to discriminate the source of concept.
To enhance the classification performance, we extended the above FHIR-based feature sets by using external terminologies and incorporating additional semantic information. The extended features include the following two aspects. (1) Medication ingredients from RxNorm [42], their corresponding drug classes in the Anatomical Therapeutic Chemical Classification (ATC) [43], and the parent/ancestor concepts (medication classes) from the ATC. The same drug ingredient can be assigned multiple different RxNorm codes due to differences in form and dosage in addition to branding. To associate the same drug ingredients with different RxNorm codes, the ATC terminology is applied to provide corresponding drug class codes. Note that the ATC Classification System is structured at five levels to allow for the classification of drugs based on anatomical, pharmacological, and chemical subgroups. For example, for an ATC code of C10AA01 identified as the chemical substance for simvastatin in RxNorm, the corresponding levels of classification C, C10, C10A, C10AA are added to the extended feature set. (2) Conditions and procedures defined in SNOMED CT [26] and the parent/ancestor concepts tracing back two levels of ancestry. For example, for the SNOMED CT code “73430006|Sleep apnea (disorder)” identified, its parent codes for Apnea (finding), Sleep disorder (disorder) and Disorder of respiratory system (disorder) are added to the extended feature set.
-
2
Classifiers that consume the FHIR-based data. According to two classification tasks of the Obesity Challenge, ie, textual and intuitive classification, we trained each classifier separately for each of the 16 conditions outlined in the Challenge. A random search approach was performed to find a single set of optimal hyperparameters (ie, parameters of a prior distribution) for each classifier based on performance on all 16 conditions. Note that there are many other approaches for the hyperparameter optimization [44], and the argument about whether truly “optimal” hyperparamters can be found or not is beyond the scope of the study. Recursive feature elimination with 3-fold cross-validation was employed to reduce overfitting using the ranking of the coefficients calculated by scikit-learn classifiers. Based on the cross-validation performance, the top 1% of features was used for the final results.
3.2.3. Evaluation methods
We used macro- and micro-averaged precision (P), recall (R), and F1 score (F1) evaluation measures to evaluate the performance. Equations used for the calculation are shown below.
| (1) |
| (2) |
| (3) |
Eqs. (1)–(3): Micro averaged measures, where TP = True Positives, FP = False Positives, FN = False Negatives.
| (4) |
| (5) |
| (6) |
Eqs. (4)–(6): Macro-averaged measures. For each class (Y/N/Q/U), the measures are calculated. These measures are averaged to yield macro-averaged measures, where M = number of classes (eg, M = 4 in this study).
The macro-averaged precision, recall, and F-measure of the system are obtained from the precision, recall, and F-measure on the classes observed in the ground truth for all diseases [21]. F1 macro scores are based on the average performance for each class (Y/N/Q/U), so that a small class is weighted the same as a large class. In contrast, F1 micro measures give all samples equal weight such that results are predominantly influenced by performance in large classes. As many of the comorbidities in the i2b2 dataset have highly unbalanced classes, especially for those with a judgment of Questionable or Absent, F1 macro scores are more useful for reflecting class-imbalance than F1 micro scores. This is the reason that in the original i2b2 Obesity Challenge, F1 macro scores were used as the primary metric for performance.
The Obesity Challenge task provided testing data and ground truth for the analysis. We evaluated the performances of classification tasks in two ways: (1) using features from FHIR resource instances; (2) adding additional features by terminology expansion as described above.
The Shapley Additive Explanations (SHAP) tool [45] was used to aid in the interpretation of the performance. SHAP is a unified approach [46] to explain the output of any machine learning model and produce graphs for visualization. The resulting SHAP graphs display features that have a large contribution to the classification results.
We validated our NLP2FHIR pipeline and prediction models using the MIMIC-III obesity dataset as a second dataset. First, we created SQL queries to extract a collection of discharge summaries for both obesity case and control groups. Fig. 5 shows a flow chart diagram illustrating the algorithms we used to create SQL queries. We also identified 15 comorbidities (as used in the i2b2 obesity dataset) using the ICD9 codes from these two groups (cases vs. controls), meaning that if an ICD9 code for a specific comorbidity condition is present, then the patient had the condition. For validation purposes, we randomly selected a total of 2000 discharge summary notes with their comorbidity annotations − 1000 notes from each of the case and control groups. Second, we invoked our pipeline and produced the FHIR-based representation for the discharge summary notes in each ICU stay. Third, we invoked the same four classifiers that were trained by the i2b2 obesity dataset to predict obesity and its 15 comorbidities: logistic regression (LR)[37], support vector machine (SVM) [38], decision tree (DT) [39], and random forest (RF) [40]. We conducted three experiments: (1) using the training data in the i2b2 obesity dataset and treating the MIMIC-III obesity dataset as the testing data); (2) using 70% of discharge summary notes (ie, 70% * 2000 = 1400) in the MIMIC-III obesity dataset as training data and treating the rest of 30% discharge summary notes in the MIMIC-III obesity dataset as the testing data; and (3) using the i2b2 training data plus 70% of discharge summary notes (ie, 70% * 2000 = 1400) in the MIMIC-III obesity dataset as the training data and treating the rest of 30% discharge summary notes in the MIMIC-III obesity dataset as the testing data.
Fig. 5.

A flow chart diagram illustrating the algorithms used to create SQL queries for identifying the case and control groups.
4. Results
We detected 16 unique types of sections with a total of 5677 records from the Obesity Challenge discharge summary data, and assigned LOINC document section codes to each section type detected. Table 1 shows our detected sections with normalized LOINC names and their distributions.
Table 1.
Detected sections with normalized LOINC codes and names and their occur-rences.
| LOINC codes | LOINC Section Name | Counts |
|---|---|---|
| 29545–1 | Physical findings Narrative | 1021 |
| 8648–8 | Hospital course Narrative | 1020 |
| 10155–0 | History of allergies | 910 |
| 10164–2 | History of Present illness Narrative | 827 |
| 29548–5 | Diagnosis Narrative | 789 |
| 11322–5 | History of General health Narrative | 738 |
| 10160–0 | History of Medication use Narrative | 687 |
| 10167–5 | History of Surgical procedures Narrative | 677 |
| 18776–5 | Plan of care note | 512 |
| 10157–6 | History of family member diseases Narrative | 253 |
| 29546–9 | History of disorders Narrative | 158 |
| 29299–5 | Reason for visit | 128 |
| 29762–2 | Social history Narrative | 110 |
| 10188–1 | Review of systems overview Narrative | 59 |
| 61144–2 | Diet and nutrition Narrative | 52 |
| 61133–5 | Clinical impression [TIMP] | 48 |
| 11450–4 | Reported Problem List | 45 |
| 11338–1 | History of Major illnesses and injuries Narrative | 12 |
| 10186–5 | Identifying information Narrative | 11 |
| 11451–2 | Psychiatric findings Narrative | 5 |
| 11366–2 | History of Tobacco use Narrative | 4 |
| 11330–8 | History of Alcohol use Narrative | 3 |
| Others (No LOINC code matched) | 1187 |
In total, 204,855 records of resource instances consisting of 73,977 Conditions (36.1%), 39,130 Procedures (19.1%), 90,941 MedicationStatements (44.4%), and 807 FamilyMemberHistory resources (0.3%) were generated and assembled as the instances of the FHIR Bundle resource after the discharge summaries were processed by the NLP2FHIR pipeline. The high frequency concepts of each FHIR resource type can be found in Appendix A.
Using the features extracted from FHIR resources, we obtained the classification results of both intuitive and textual judgments for the four classifiers as shown in Table 2. Adding the extended features derived from using the external terminologies slightly improved the classification results (Table 3) with RF performing the best on most measures. Table 4 shows a comparison of our performance to top results of original rule-based approach (ie, the state-of-the-art) from Obesity Challenge. In general, it indicated that our results are comparable with top results that were based on rule-based approach. Interestingly, we found that the macro-averaged precision, recall, and F-measures of our FHIR-based approach outperformed the baseline performance for the intuitive classification task, whereas the performance of our approach is suboptimal in comparison with the baseline performance for the textual classification task (Table 4). Textual judgments based on whether or not there is a mention in text, can be usually retrieved with high performance by pure rule-based approaches. In other words, the rule-based approaches as used in baseline outperformed in representing textual judgments that would require no reasoning. On the other hand, because of the data standardization defined by FHIR models resulting in a reduced input feature dimensionality, we consider that statistical machine learning-based approaches benefited in the intuitive classification task, where rule-based approaches are not as strong. In other words, the machine learning-based approaches as used in this study outperformed in representing intuitive judgments that would allow inference of diseases from information including mentions of examination and test results (eg, blood sugar), physical characteristics (eg, body mass index), and the medications and diseases discussed in the discharge summaries.
Table 2.
Overall Classification Results of Intuitive and Textual Judgments (FHIR Features Only).
| P-micro | P-macro | R-micro | R-macro | F1-micro | F1-macro | ||
|---|---|---|---|---|---|---|---|
| DT | Intuitive | 0.9388 | 0.7975 | 0.9388 | 0.7743 | 0.9388 | 0.7837 |
| Textual | 0.9508 | 0.6893 | 0.9508 | 0.6519 | 0.9508 | 0.6578 | |
| LR | Intuitive | 0.9146 | 0.7209 | 0.9146 | 0.7099 | 0.9146 | 0.7145 |
| Textual | 0.9207 | 0.6148 | 0.9207 | 0.5993 | 0.9207 | 0.6058 | |
| RF | Intuitive | 0.9430 | 0.8220 | 0.9430 | 0.7701 | 0.9430 | 0.7814 |
| Textual | 0.9516 | 0.6885 | 0.9516 | 0.6505 | 0.9516 | 0.6566 | |
| SVM | Intuitive | 0.9105 | 0.6905 | 0.9105 | 0.6870 | 0.9105 | 0.6883 |
| Textual | 0.9155 | 0.6069 | 0.9155 | 0.6100 | 0.9155 | 0.6039 |
Table 3.
Overall Classification Results of Intuitive and Textual Judgments (FHIR Features + Extended Features).
| P-micro | P-macro | R-micro | R-macro | F1-micro | F1-macro | ||
|---|---|---|---|---|---|---|---|
| DT | Intuitive | 0.9417 | 0.8146 | 0.9417 | 0.7988 | 0.9417 | 0.8044 |
| Textual | 0.9497 | 0.6679 | 0.9497 | 0.6593 | 0.9497 | 0.6620 | |
| LR | Intuitive | 0.9021 | 0.7145 | 0.9021 | 0.7016 | 0.9021 | 0.7067 |
| Textual | 0.9029 | 0.6030 | 0.9029 | 0.5918 | 0.9029 | 0.5954 | |
| RF | Intuitive | 0.9466 | 0.8080 | 0.9466 | 0.7780 | 0.9466 | 0.7887 |
| Textual | 0.9536 | 0.6574 | 0.9536 | 0.6498 | 0.9536 | 0.6524 | |
| SVM | Intuitive | 0.9002 | 0.7132 | 0.9002 | 0.6997 | 0.9002 | 0.7049 |
| Textual | 0.8937 | 0.5911 | 0.8937 | 0.5950 | 0.8937 | 0.5887 |
Table 4.
Comparison of our results to top results of original rule-based approach from i2b2 Obesity Challenge.
| P-micro | P-macro | R-micro | R-macro | F1-micro | F1-macro | ||
|---|---|---|---|---|---|---|---|
| Baseline | Intuitive | 0.9590 | 0.7485 | 0.9590 | 0.6571 | 0.9590 | 0.6745 |
| Textual | 0.9756 | 0.8318 | 0.9756 | 0.7776 | 0.9756 | 0.8000 | |
| FHIR (RF) | Intuitive | 0.9466 | 0.8080 | 0.9466 | 0.7780 | 0.9466 | 0.7887 |
| Textual | 0.9536 | 0.6574 | 0.9536 | 0.6498 | 0.9536 | 0.6524 |
To compare the classification performance for the 16 specific conditions (ie, obesity and its 15 comorbidities) separately, charts for each condition performance were displayed in Fig. 6 on the basis of the method of Table 3, where F1-micro scores are shown in bars and F1-macro scores are shown with an asterisk. Our evaluation work provides evidence on the value of our solutions in the real-world use cases.
Fig. 6.

The classification performance of 4 classifiers for 16 specific conditions: (A) intuitive classification; (B) textual classification. (F1-micro scores are shown in bars and F1-macro scores are shown as asterisk).
To better explain which features are most important for each machine learning model, we used the SHAP graph to visualize the SHAP values of every feature for all patients in each classification task. Fig. 7 shows the SHAP graphs that sort features by the sum of SHAP value magnitudes over all patients in the classification tasks (intuitive vs. textual) of the comorbidity CHF (Congestive Heart Failure) as an example. The results indicated that standard vocabulary codes and their expansions played an important role on the classifications tasks (unsurprisingly). We also noticed that the negation of some SNOMED CT codes contributed towards classification as top features, which reflected the importance of the negation detection algorithm as an integral component of the NLP2FHIR pipeline. The SHAP graphs for other comorbidities are available in the project GitHub website (see link in the Conclusion section).
Fig. 7.

The SHAP graphs to visualize the SHAP values of every feature for all patients in the classification tasks of the comorbidity CHF (Congestive Heart Failure): (A) intuitive classification; (B) textual classification.
For validation using the MIMIC-III obesity dataset, we successfully ran the NLP2FHIR pipeline without additional configuration, which produced the instance data of the FHIR Composition for representing semi-structured discharge summaries and also generated instance data of clinical FHIR resources (ie, Condition, Procedure, MedicationStatement, and FamilyMemberHistory) that is linked as references of the FHIR Composition. We also invoked the four machine learning models with minimal code changes (eg, adjusting labels for the MIMIC-III dataset as it only has two labels - present and absent for all conditions). Table 5 shows that all classifiers achieved satisfactory results on performance in classifying patients with obesity and its comorbidities in the MIMIC-III dataset although the performance is suboptimal in comparison with that for the i2b2 dataset. As expected, the performance of all 4 classifiers improved universally when including some MIMIC-III training data, but the best isn’t always the combination of both i2b2 and MIMIC-III training data. We also compared the resource instance occurrences in the generated bundles for both i2b2 and MIMIC-III datasets as shown in Fig. 8. We observed that the resource instance occurrences in many of the sections between two datasets are different, which may partially explain why the performance in classifying the MIMIC-III dataset is suboptimal when using combined datasets.
Table 5.
Overall Classification Results of MIMIC Obesity Dataset (FHIR features + Extended features).
| P-micro | P-macro | R-micro | R-macro | F1-micro | F1-macro | ||
|---|---|---|---|---|---|---|---|
| DT | i2b2* | 0.8468 | 0.7102 | 0.8468 | 0.7813 | 0.8468 | 0.7126 |
| mimic** | 0.8913 | 0.7356 | 0.8913 | 0.7128 | 0.8913 | 0.7169 | |
| i2b2 + mimic*** | 0.8808 | 0.7265 | 0.8808 | 0.7417 | 0.8808 | 0.7207 | |
| LR | i2b2 | 0.8342 | 0.6854 | 0.8342 | 0.7321 | 0.8342 | 0.6823 |
| mimic | 0.8587 | 0.6811 | 0.8587 | 0.6810 | 0.8587 | 0.6799 | |
| i2b2 + mimic | 0.8748 | 0.7239 | 0.8748 | 0.7341 | 0.8748 | 0.7216 | |
| RF | i2b2 | 0.8491 | 0.7121 | 0.8491 | 0.7739 | 0.8491 | 0.7106 |
| mimic | 0.9315 | 0.8151 | 0.9006 | 0.7056 | 0.9006 | 0.7252 | |
| i2b2 + mimic | 0.8908 | 0.7257 | 0.8908 | 0.6928 | 0.8908 | 0.6987 | |
| SVM | i2b2 | 0.8120 | 0.6695 | 0.8120 | 0.7357 | 0.8120 | 0.6658 |
| mimic | 0.8424 | 0.6315 | 0.8424 | 0.6026 | 0.8424 | 0.6074 | |
| i2b2 + mimic | 0.8760 | 0.7230 | 0.8760 | 0.7402 | 0.8760 | 0.7199 |
indicates using i2b2 training data only;
indicates using mimic training data only;
indicates using a combination of both i2b2 and mimic training data.
Fig. 8.

Comparison of the resource instance occurrences in the generated FHIR resource bundles between the i2b2 and MIMIC-III obesity datasets. (A) i2b2 obesity dataset; (B) MIMIC-III obesity dataset.
5. Discussions
We developed a FHIR-based EHR phenotyping framework for identifying the conditions that a patient may have from EHR data. Our study is innovative in the following aspects. First, we adopted the FHIR document resource Composition and associated clinical resources to represent unstructured EHR documents. As illustrated in Fig. 8, with such a standard representation, we were able to look into the semantic distributions based on different clinical resources. This semantic distribution can allow researchers to integrate additional datasets in a transparent manner and compare the performance of machine learning algorithms against different datasets. We demonstrated this point using a second obesity dataset extracted from the MIMIC-III database. The MIMIC-III obesity dataset was successfully integrated for prediction with minimal configuration of the NLP2FHIR pipeline and machine learning models. While the performance of classifiers for combined datasets is suboptimal in prediction, we argue that it is more generalizable for the prediction of obesity and its comorbidities.
Second, we implemented a section detection algorithm to standardize clinical note section headers. One of the main reasons to choose the SecTag section dictionary is because it defines the section names with standard LOINC codes and LOINC is the preferred vocabulary for standardizing the section names in FHIR. Although the implementations of section headers in different EHR systems and institutions vary, we consider that the impact of section detection on portability of our phenotyping approach is mild because of the following reasons: First, the section detection is a data preprocessing option to users. Users can choose whether or not to use the section detection option based on whether users would like to use sections as contextual features for their downstream machine learning models. Second, more and more EHR systems (eg, Epic) are adopting standardized section headers, which would make preprocessing of the section headers much easier. Third, the sections represent different clinical contexts that may contribute differently to different machine learning targets. A previous study on the Obesity classifications found that incorporating the features from the family history section had a negative impact on the performance of the machine learning algorithms [24]. We were able to reproduce the finding, although the negative impact is mild in our experiments (data not shown) and it indicated that our machine learning model is more robust to a certain degree. In the future, we will systematically investigate the effect of different sections on the machine learning performance.
Third, we built a FHIR-based data access mechanism between normalized EHR data and the machine learning modules to increase the portability of our machine learning modules to different data sources. This kind of data access mechanism would allow a researcher to design machine learning experiments with flexible feature selections to understand which semantic data types have contributions to the overall performance of the machine learning algorithms, thus increase the interpretability of the machine learning model. We have leveraged the SHAP visualization tool [45] to help explain the output of the machine learning models (Fig. 7). Note that we only touched a limited aspect of a post hoc interpretability method (ie, applying methods that analyze the model after training) [47] that returns feature importance and summary visualization. There are many other interpretability methods (either model-specific or model-agnostic) that can be applied to enhance the interpretability, which may be explored in the future. In addition, this data access mechanism enables a future extension of our FHIR-based EHR phenotyping framework to support advanced deep learning. The viability of such a mechanism has been illustrated in Google’s deep learning framework with EHRs [29], where a FHIR-based interface was used to generate predictors (eg, the length of stay in hospital) and to standardize the feature selections in their Tensorflow implementation [48].
Finally, we have leveraged external terminologies linked to the FHIR resource instances to improve the performance. RxNorm [42] consists of generic and branded drugs and those drugs may have different codes based on their ingredients, dosage, and dose form. As mentioned previously, to associate the same drug ingredients with different RxNorm codes, the ATC terminology is applied to provide corresponding drug class codes. Similarly, we expanded the SNOMED CT [26] codes to their parent codes tracing back two levels of ancestry. According to the visualization results using the SHAP graphs, the terminology expansions are an important component that contributed to the performance improvement of the classification tasks. In the future, we will conduct a systematic analysis on determining an appropriate granularity (ie, correct level for terminology expansions) that results in the best performance of the machine learning model.
There are a number of challenges in this study. The first challenge is the model element mapping. We created mappings from different NLP outputs to their corresponding FHIR resources as reflected in the FHIR-based type system. Generally we did not apply section-filtering rules for clinical concepts identified as Condition, Procedure, and Medication. For example, medication mentions were detected in most sections, and we currently mapped MedicationStatement/Medication resources from multiple sections. But when we want to identify the instances of the FamilyMemberHistory resource, we decided to only use the text under the sections detected as the History of Family Member Diseases Narrative (LOINC code: 10157–6) because that the NLP2FHIR pipeline needs a restricted context to perform well for this specific resource. The second challenge is the number of NLP-specific elements not covered by the FHIR specification. To preserve this useful information, we utilized the FHIR extension mechanism. The third challenge is related to the terminology binding. Many elements in the FHIR resources have a coded value; some in the form of a fixed string (a sequence of characters) as defined in the FHIR specification; some in the form of “concept” codes as defined in the external terminologies or ontologies (eg, LOINC, RxNorm or SNOMED CT). Normalizing non-standard data into the coded FHIR elements thus poses a challenge. In this study, we handle the terminology binding using the UMLS lookup for vocabulary mappings. In the future, FHIR-based terminology services should be established and leveraged to tackle the challenge although this is a nontrivial task.
This study on the FHIR-based EHR phenotyping framework also has a number of limitations. First, this study only used two datasets (ie, the i2b2 Obesity Challenge dataset and the MIMIC-III obesity dataset) to demonstrate the portability of our FHIR-based EHR phenotyping framework as a proof-of-concept. A rigorous assessment of the framework involving more datasets and multiple institutions will be needed in a future study. Second, we compared our results with other previous results on the same Obesity Challenge task. Our results are comparable with other previous results but with performance limitations, compared with the top 10 rule-based methods’ results reported previously [21] and the hybrid machine learning methods [22]. As previously mentioned, the tradeoff between phenotyping accuracy and portability should be considered for implementing any NLP-driven phenotyping system across different institutions and data systems. Rule-based methods with higher performances may be tailored for this specific i2b2 Obesity Challenge task, whereas our FHIR-based EHR phenotyping methods in this study are more focused on issues around generalizability, portability and interpretability while maintaining comparable performance. Third, some information loss is inevitable when modeling unstructured data due to different information representation granularity between NLP outputs and FHIR models.
6. Conclusions
We demonstrated the value of a FHIR-based clinical data normalization pipeline in enabling precise clinical data capturing and EHR-based phenotyping. Our FHIR-based phenotyping approach is a first concrete step towards improving portability of phenotyping across EHR systems, enhancing the reproducibility and interpretability of the machine learning based phenotyping algorithms, and ultimately enabling large-scale phenotyping practice on semi-structured, unstructured, as well as structured EHR data. The source code of the study and all SHAP visualization results are publicly available at the project GitHub website at: https://github.com/BD2KOnFHIR/nlp2fhir-obesity-phenotyping.
Supplementary Material
Acknowledgement
This paper is an extension of a podium abstract presented at AMIA Informatics Summit 2019. This study is supported in part by the United States National Institutes of Health (NIH) grants U01 HG009450 and R01 GM105688.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbi.2019.103310.
References
- [1].Hripcsak G, Albers DJ, Next-generation phenotyping of electronic health records, J. Am. Med. Inform. Assoc 20 (1) (2013) 117–121, 10.1136/amiajnl-2012-001145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].FHIR R4 2019, Available from: https://www.hl7.org/fhir/.
- [3].Dolin RH, Alschuler L, Boyer S, Beebe C, Behlen FM, Biron PV, Shvo A Shabo, HL7 clinical document architecture, release 2, J. Am. Med. Inform. Assoc 13 (1) (2006) 30–39, 10.1197/jamia.M1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Quality Data Model 2019. [June 10, 2019]. Available from: http://www.qualityforum.org/QualityDataModel.aspx.
- [5].Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, Suchard MA, Park RW, Wong IC, Rijnbeek PR, van der Lei J, Pratt N, Noren GN, Li YC, Stang PE, Madigan D, Ryan PB, Observational health data sciences and informatics (OHDSI): opportunities for observational researchers, Stud. Health Technol. Inform 216 (2015) 574–578 [PMC free article] [PubMed] [Google Scholar]
- [6].PCORI, PCORnet Common Data Model (CDM) [cited 2017 Mar.]. Available from: http://www.pcornet.org/pcornet-common-data-model/.
- [7].Hong N, Wen A, Shen F, Sohn S, Liu S, Liu H, Jiang G, Integrating structured and unstructured EHR Data using an FHIR-based type system: A case study with medication data, AMIA Summits on Translat. Sci. Proc 2017 (2018) 74. [PMC free article] [PubMed] [Google Scholar]
- [8].Hong N, Wen A, Mojarad RM, Sohn S, Liu H, Jiang G, Standardizing heterogeneous annotation corpora using HL7 FHIR for facilitating their reuse and integration in clinical NLP, AMIA Annu. Symp. Proc (2018). [PMC free article] [PubMed] [Google Scholar]
- [9].21ST Century cures act: interoperability, information blocking, and the ONC HEALTH IT certification program proposed Rule 2019 2019. [July 16, 2019]. Available from: https://www.healthit.gov/sites/default/files/nprm/ONCCuresNPRMAPICertification.pdf.
- [10].HL7 Argonaut Project 2019. [March 29, 2019]. Available from: http://argonautwiki.hl7.org/index.php?title=Main_Page.
- [11].Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG, Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications, J. Am. Med. Inform. Assoc 17 (5) (2010) 507–513, 10.1136/jamia.2009.001560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sohn S, Clark C, Halgrim SR, Murphy SP, Chute CG, Liu H, MedXN: an open source medication extraction and normalization tool for clinical text, J. Am. Med. Inform. Assoc 21 (5) (2014) 858–865, 10.1136/amiajnl-2013-002190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sohn S, Wagholikar KB, Li D, Jonnalagadda SR, Tao C, Komandur Elayavilli R, Liu H, Comprehensive temporal information detection from clinical text: medical events, time, and TLINK identification, J. Am. Med. Inform. Assoc 20 (5) (2013) 836–842, 10.1136/amiajnl-2013-001622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kohane IS, Using electronic health records to drive discovery in disease genomics, Nat. Rev. Genet 12 (6) (2011) 417–428, 10.1038/nrg2999 [DOI] [PubMed] [Google Scholar]
- [15].Moskewicz MW, Implementing Efficient Portable Computations for Machine Learning, UC; Berkeley, 2017. [Google Scholar]
- [16].Rasmussen LV, Brandt PS, Jiang G, Kiefer RC, Pacheco JA, Adekkanattu P, Ancker JS, Wang F, Xu Z, Pathak J, Luo Y, Considerations for improving the portability of electronic health record-based phenotype algorithms, AMIA Annual Symposium 2019, (2019) (in press). [PMC free article] [PubMed] [Google Scholar]
- [17].Adkins DE, Machine learning and electronic health records: A paradigm shift, Am. J. Psychiatry 174 (2) (2017) 93–94, 10.1176/appi.ajp.2016.16101169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].i2b2, The i2b2 NLP Obesity Challenge [cited 2018 Aug.]. Available from: https://www.i2b2.org/NLP/Obesity/.
- [19].Johnson AE, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG, MIMIC-III, a freely accessible critical care database, Sci. Data 3 (2016) 160035, 10.1038/sdata.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Solt I, Tikk D, Gál V, Kardkovács ZT, Semantic classification of diseases in discharge summaries using a context-aware rule-based classifier, J. Am. Med. Inform. Assoc 16 (4) (2009) 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Uzuner O, Recognizing obesity and comorbidities in sparse data, J. Am. Med. Inform. Assoc 16 (4) (2009) 561–570, 10.1197/jamia.M3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Savova G, Clark C, Zheng J, Cohen KB, Murphy S, Wellner B, Harris D, Lazo M, Aberdeen J, Hu Q (Eds.), The Mayo/MITRE system for discovery of obesity and its comorbidities, in: Proceedings of the i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data, 2008. [Google Scholar]
- [23].Ambert KH, Cohen AM, A system for classifying disease comorbidity status from medical discharge summaries using automated hotspot and negated concept detection, J. Am. Med. Inform. Assoc 16 (4) (2009) 590–595, 10.1197/jamia.M3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sharma H, Mao C, Zhang Y, Vatani H, Yao L, Zhong Y, Rasmussen L, Jiang G, Pathak J, Luo Y, Developing a portable natural language processing based phenotyping system, BMC Med. Inf. Decis. Making 19 (Suppl) (2019) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, Sanderson SC, Kannry J, Zinberg R, Basford MA, Brilliant M, Carey DJ, Chisholm RL, Chute CG, Connolly JJ, Crosslin D, Denny JC, Gallego CJ, Haines JL, Hakonarson H, Harley J, Jarvik GP, Kohane I, Kullo IJ, Larson EB, McCarty C, Ritchie MD, Roden DM, Smith ME, Bottinger EP, Williams MS, e MN, The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future, Genet Med. 15 (10) (2013) 761–771, 10.1038/gim.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].SNOMED CT 2019. [June 10, 2019]. Available from: http://www.snomed.org/snomed-ct/.
- [27].Pacheco JA, Rasmussen LV, Kiefer RC, Campion TR, Speltz P, Carroll RJ, Stallings SC, Mo H, Ahuja M, Jiang G, LaRose ER, Peissig PL, Shang N, Benoit B, Gainer VS, Borthwick K, Jackson KL, Sharma A, Wu AY, Kho AN, Roden DM, Pathak J, Denny JC, Thompson WK, A case study evaluating the portability of an executable computable phenotype algorithm across multiple institutions and electronic health record environments, J Am Med Inform Assoc. 25 (11) (2018) 1540–1546, 10.1093/jamia/ocy101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Savova GK, Tseytlin E, Finan S, Castine M, Miller T, Medvedeva O, Harris D, Hochheiser H, Lin C, Chavan G, Jacobson RS, DeepPhe: A natural language processing system for extracting cancer phenotypes from clinical records, Cancer Res. 77 (21) (2017) e115–e118, 10.1158/0008-5472.CAN-17-0615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, Liu PJ, Liu XB, Marcus J, Sun MM, Sundberg P, Yee H, Zhang K, Zhang Y, Flores G, Duggan GE, Irvine J, Le Q, Litsch K, Mossin A, Tansuwan J, Wang D, Wexler J, Wilson J, Ludwig D, Volchenboum SL, Chou K, Pearson M, Madabushi S, Shah NH, Butte AJ, Howell MD, Cui C, Corrado GS, Dean J, Scalable and accurate deep learning with electronic health records, Npj Digit. Med (2018) 1, 10.1038/s41746-018-0029-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Body Mass Index (BMI) 2019. [June 10, 2019]. Available from: https://www.nhlbi.nih.gov/health/educational/lose_wt/BMI/bmicalc.htm.
- [31].FHIR Document Resource 2019. [Feburary 21, 2019]. Available from: https://www.hl7.org/fhir/documents.html.
- [32].NLP2FHIR GitHub Website. 2019. Available from: https://github.com/BD2KOnFHIR/NLP2FHIR.
- [33].SecTag – Tagging Clinical Note Section Headers 2019. [June 10, 2019]. Available from: https://www.vumc.org/cpm/sectag-tagging-clinical-note-section-headers.
- [34].Denny JC, Spickard A 3rd, Johnson KB, Peterson NB, Peterson JF, Miller RA, Evaluation of a method to identify and categorize section headers in clinical documents, J. Am. Med. Inform. Assoc 16 (6) (2009) 806–815, 10.1197/jamia.M3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Harkema H, Dowling JN, Thornblade T, Chapman WW, ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports, J. Biomed. Inform 42 (5) (2009) 839–851, 10.1016/j.jbi.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].FHIR Extensibility 2019. [June 10, 2019]. Available from: https://www.hl7.org/fhir/extensibility.html.
- [37].LogisticRegression 2019. [June 10, 2019]. Available from: http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html.
- [38].SVM.svc 2019. [June 10, 2019]. Available from: http://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html-sklearn.svm.SVC.
- [39].DecisionTreeClassifier 2019. [June 10, 2019]. Available from: http://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeClassifier.html.
- [40].RandomForestClassifier 2019. [June 10, 2019]. Available from: http://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html.
- [41].scikit-learn 2019. [June 10, 2019]. Available from: http://scikit-learn.org/stable/.
- [42].RxNorm 2019. [June 10, 2019]. Available from: https://www.nlm.nih.gov/research/umls/rxnorm/.
- [43].Anatomical Therapeutic Chemical Classification (ATC) 2019. [June 10, 2019]. Available from: http://bioportal.bioontology.org/ontologies/ATC.
- [44].Hyperparameter optimization 2019. [September 4, 2019]. Available from: https://en.wikipedia.org/wiki/Hyperparameter_optimization
- [45].SHAP (SHapley Additive exPlanations) tool 2019. [March 22, 2019]. Available from: https://github.com/slundberg/shap.
- [46].Lundberg SM, Lee S (Eds.), A Unified Approach to Interpreting Model Predictions, Advances in Neural Information Processing Systems 30 (NIPS 2017), 2017. [Google Scholar]
- [47].Molnar C, Interpretable Machine Learning: A guide for making black box models explainable 2019. [September 4, 2019]. Available from: https://christophm.github.io/interpretable-ml-book/.
- [48].Google’s FHIR Protocol Buffers 2019. [June 6, 2019]. Available from: https://github.com/google/fhir/.
- [49].PhEMA Project. [accessed 14 October 2019]. Available from: https://github.com/phema.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


