Abstract
The 22q11.2 deletion syndrome (22q11DS), the most common survivable human genetic deletion disorder, is caused by a hemizygous deletion of 30–40 contiguous genes on chromosome 22, many of which have not been well characterized. Clinical features seen in patients with this deletion, including intellectual disability, are not completely penetrant and vary in severity between patients, suggesting the involvement of variants elsewhere in the genome in the manifestation of the phenotype. Given that it is a relatively rare disorder (1/2000–6000 in humans), limited research has shed light into the contribution of these second-site variants to the developmental pathogenesis that underlies 22q11DS. As CNVs throughout the genome might constitute such a genetic risk factor for variability in the 22q11DS phenotypes such as intellectual disability, we sought to determine if the overall burden of rare CNVs in the genetic background influenced the phenotypic variability. We analyzed CNV and clinical data from 66 individuals with 22q11DS, and found that 77% (51/66) of individuals with the 22q11DS also carry additional rare CNVs (< 0.1% frequency). We observed several trends between CNV burden and phenotype, including that the burden of large rare CNVs (> 200 Kb in size) was significantly higher in 22q11DS individuals with intellectual disability than with normal IQ. Our analysis shows that rare CNVs may contribute to intellectual disability 22q11DS, and further analysis on larger 22q11DS cohorts should be performed to confirm this correlation.
Keywords: 22q11DS, Copy number variations, Intellectual disabilities
1. Introduction
The 22q deletion syndrome (22q11DS), caused by a deletion of 1.5 or 3 Mb on chromosome 22q11.2, is the most common genomic disorder caused by a rare copy-number variant (CNV), with an estimated prevalence of 1 in 2000 to 1 in 6000 (Girirajan et al., 2011a,b). The syndrome is characterized by a common set of clinical features, including congenital heart defects, immune deficiencies, palatal abnormalities and hypocalcemia (McDonald-McGinn et al., 2013). Neuropsychiatric features, such as intellectual disability/developmental delay (ID/DD) (Swillen and McDonald-Mcginn, 2015), features of autism (Vorstman et al., 2006), a very high risk for schizophrenia in adulthood (Karayiorgou et al., 1995), and a range of other clinical features (Schneider et al., 2014) are also variably observed in patients with 22q11DS.
Despite a relatively clear clinical presentation, very little is known about which genes in the 22q11.2 region contribute towards specific clinical features, and whether combinatorial effects of reduced dosage of multiple genes act in concert to modulate phenotypic variations. Although expression levels of genes within the deleted region are expected to be reduced by half in 22q11DS, the remaining allele could be associated with different mRNA expression levels, as has been demonstrated for many genes. Indeed, it has been suggested that the 22q11 gene TBX1 (OMIM #602054) has a specific gene dosage threshold, and that very small reductions in the mRNA level of TBX1 result in a more severe cardiac phenotype (Baldini, 2006). More than a decade ago, TBX1 was identified as the major cause of conotruncal defects associated with the syndrome by using classical genetics and mouse models (Lindsay et al., 2001; Zweier et al., 2007). More recently, duplication of SLC2A3 (OMIM #138170) and deleterious single nucleotide variants within histone modifier genes were identified as second-site modifiers of cardiac phenotypes seen in 22q11DS (Guo et al., 2015; Mlynarski et al., 2015), and interactions between 22q11.2 genes and fibroblast growth factor genes were suggested to contribute towards palatal phenotypes (Widdershoven et al., 2013). Similarly, genetic and molecular dissection of schizophrenia phenotypes in 22q11DS has implicated variants both within and outside the deletion that may modulate the neurological phenotypes (Fénelon et al., 2011; Paylor et al., 2006), although one recent study showed that variants on the non-deleted allele do not carry a large effect towards schizophrenia phenotypes (Guipponi et al., 2017). However, which genetic variants contribute to ID features in 22q11DS is not entirely clear. In this study, we hypothesized that CNVs elsewhere in the genome contribute to the ID phenotypes observed in 22q11DS. We investigated CNV data from 66 individuals with 22q11DS for whom detailed phenotypic information was available to determine if large genomic variants in the genetic background modify the phenotypic expressivity of the observed ID features.
1.1. Patient data
Individuals with 22q11DS were recruited at the UC Davis Medical Investigation of Neurodevelopmental Disorders Institute, Sacramento, CA, USA. Participants provided informed consent according to protocols approved by the UC Davis Institutional Review Board. Participants included 66 individuals (41 males and 25 females, mean age = 11.67 years old, SD = 4.016 years) with a diagnosis of 22q11DS. Each subject with 22q11DS underwent a neuropsychological assessment, including full-scale IQ (FSIQ) using the Wechsler Intelligence Scale for Children (Weschler, 2008). Full-scale IQ ≤ 70 (2 SD below normed means) was used to define abnormally low FSIQ, in line with the accepted classifications for ID (Amer. Psy. Assoc, 2013). For other clinical features, patient medical history was collected from parents during visits with the developmental behavior pediatrician. Demographic, molecular and clinical information on all patients in the cohort are provided in Table 1, and clinical features of all patients are summarized in Supplementary Table 1.
Table 1.
Demographic, molecular, and full-scale intelligence quotient (FSIQ) data for study participants.
| Demographic Data | |||
|---|---|---|---|
| Participants | Male (n = 41) | Female (n = 25) | Total (n = 66) |
| Average age (SD) | 11.34 (4.526) | 12.2 (3.014) | 11.67 (4.016) |
| 22q11.2 Deletion Type | |||
| 3 Mb Deletion | 37 (90.24%) | 23 (92.00%) | 60 (90.91%) |
| 1.5 Mb Deletion | 3 (7.32%) | 2 (8.00%) | 5 (7.58%) |
| Atypical Deletion | 1 (2.44%) | 0 (0.00%) | 1 (1.52%) |
| Clinical Phenotypes | |||
| Average IQ (FSIQ > 100) | 10 (24.39%) | 3 (12.00%) | 13 (19.70%) |
| Below Average IQ (70 < FSIQ < 100) | 15 (36.59%) | 9 (36.00%) | 24 (36.36%) |
| ID (FSIQ < 70) | 16 (39.02%) | 13 (52.00%) | 29 (43.94%) |
2. Methods
Genomic DNA from each individual was isolated from 3 mL of peripheral blood leukocytes using standard procedures. The diagnosis of 22q11DS was first made by fluorescent in situ hybridization (FISH) using the TUPLE1 probe (McDonald-McGinn et al., 2013) and con-firmed using droplet digital PCR, as described previously (Hwang et al., 2014). Based on the FISH and ddPCR assays, we found that 60 subjects carried the 3 Mb deletion, five subjects carried the smaller 1.5 Mb deletion and one participant carried an atypical deletion.
Genome-wide CNV data were generated using HumanCytoSNP-12v2.1 beadchip single nucleotide polymorphism (SNP) arrays, according to manufacturer instructions (Illumina, San Diego, CA). CNV analysis was performed using CNV-WebStore as described previously (Vandeweyer et al., 2011) using human genome build hg19. We excluded CNVs spanning less than five SNPs on the microarray, and we filtered out common CNVs that were seen in greater than 0.1% of a control population of 8329 healthy individuals (Cooper et al., 2012). All statistical analysis was performed using R version 3.3.2 (R Foundation for Statistical Computing, Vienna, Austria).
3. Results
We used the genome-wide CNV data from our cohort of patients with 22q11DS to test our hypothesis that second-site CNVs may contribute to the phenotypic variability of IQ in 22q11DS. We identified 114 rare second-site CNVs greater than 50 kb, with an approximate 1.73 second-site CNVs per individual. Of these, 78 second-site CNVs included at least one RefSeq gene (an average of 1.20 CNVs per individual), seen in 51 out of 66 patients (77.2%). These CNVs are listed in Supplementary Table 2. Overall, each participant had an average second-site CNV burden of 476 kb, leading to the disruption of approximately six genes on average. The identified CNVs ranged up to 2.15 Mb in length, with 37 out of 66 individuals having at least 1 second-site CNV greater than 100 kb in length (56.1%) and 18 out of 66 individuals (27.3%) having at least 1 second-site CNV greater than 500 kb in length. We found that the total size of all CNVs in each individual (including the 22q11.2 deletion and all second-site CNVs) correlated with the number of affected genes within the CNV regions (Pearson correlation, r = 0.785, p = 5.927 × 10−15) (Suppl. Fig. 1A).
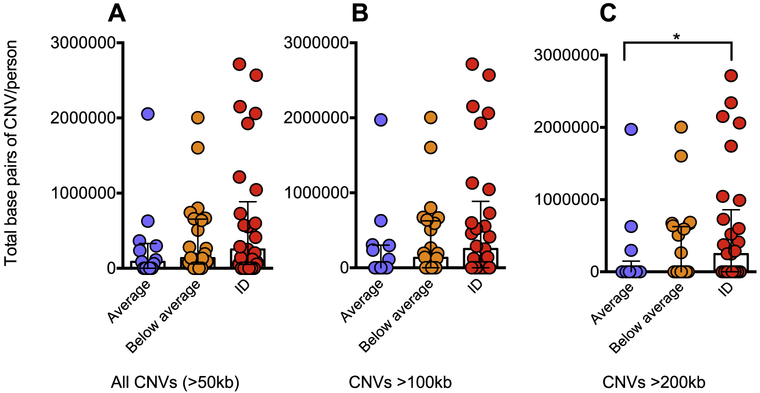
Based on the cognitive assessment, individuals were divided into three categories: average IQ (FSIQ > 100), below-average IQ (FSIQ between 70 and 100), and intellectual disability (FSIQ < 70). Using these definitions, we observed that 13 individuals (19.7%) had an average IQ, 24 individuals (36.4%) had a below-average IQ, and 29 individuals (43.9%) had intellectual disability. We first found that 22q11DS individuals with ID had a trend towards a higher CNV burden when compared to those with average IQ (Suppl. Fig. 1B). We then compared the burden of second-site rare CNVs with a minimum size of 50 kb in individuals across each IQ group, and found a trend towards a higher CNV burden in individuals with ID compared to those with average IQ (p = 0.0807, one-tailed Mann-Whitney test) (Fig. 1A). A higher CNV burden was not observed for individuals with below-average IQ (p = 0.1488, one-tailed Mann-Whitney test).
Fig. 1. Correlation of CNV burden with IQ in 22q11DS individuals.
Burden of (A) all rare CNVs, (B) rare CNVs > 100 kb, and (C) rare CNVs > 200 kb in individuals with average IQ (FSIQ > 100), below-average IQ (70 < FSIQ < 100), and intellectual disability (FSIQ < 70). * indicates p < 0.05 (one-tailed Mann-Whitney test).
When we examined the burden of large second-site CNVs at different size ranges, we found a higher burden of CNVs larger than 100 kb (p = 0.0938) and 200 kb (p = 0.0486, one-tailed Mann-Whitney test) in individuals with ID compared to those with average IQ (Fig. 1C). No trends were found for either size range in individuals with below-average IQ (p = 0.2714 for 100 kb and 0.1844 for 200 kb) (Fig. 1B), and we did not have a large enough sample size to examine burden of CNVs over 500 kb. The CNV burdens were also greater in individuals with intellectual disability when CNVs containing > 75% segmental duplications (Bailey et al., 2002) were removed from the analysis (Suppl. Fig. 2).
4. Discussion
CNV burden, defined as the total number of base pairs affected by CNV in an individual, has previously been linked to an increased risk in intellectual disability (Girirajan et al., 2012) and autism (Girirajan et al., 2013). Our data supports these previous findings, and suggests that a significant correlation between burden of large rare CNVs (> 200 kb) and intellectual disability also exists in individuals with 22q11DS. 22q11DS is a complex genetic disease characterized by a wide phenotypic variability, where phenotypes caused by the genetic variation are not seen in all individuals with the variation. In our cohort of 66 patients with 22q11DS, we observed a similar phenotypic penetrance for below-average intellect (36.2%) and intellectual disability (44.8%) as previously reported (Bassett et al., 2016). For many developmental disorders, second-site variations have been found to explain this phenotypic variability (Girirajan et al., 2010, 2012). We find that 51 of 66 patients in our 22q11DS cohort had at least one rare second-site CNV containing a RefSeq gene, indicating most individuals in our cohort have these potential sources of phenotypic variability.
IQ has previously been used to measure the correlation between second-site CNV burden and cognitive ability in neurodevelopmental disorders (Rees et al., 2016). For this study, we were only able to determine a significant association between the burden of large CNVs > 200 kb and ID, in addition to non-significant trends between CNV burden and IQ for other CNV size ranges. One of the limitations of our study is the size of the 22q11DS cohort; it is possible that a larger cohort of patients with 22q11DS would provide enough statistical power to identify a significant association between rare CNVs and IQ. However, it is also possible that second-site CNVs are not sufficient on their own to cause the observed phenotypic variability in 22q11DS. Second-site variants in single genes (loss-of-function or pathogenic missense mutations) related to neurodevelopment, or allelic variants in the genes within the CNV, could also contribute to the phenotypic variability. These variants should be considered along with CNVs when explaining the phenotypic variability in 22q11DS. Future whole-exome or whole-genome sequencing will help to more fully evaluate the contribution of second-site hits on phenotypic variability.
Supplementary Material
Acknowledgements
This work was supported by grants R01 HD042974 and R01 MH107108, by the NIH-funded MIND Institute Intellectual and Developmental Disabilities Research Center (U54 HD079125) and by UC Davis MIND funds to TJS and FT, NIH R01 GM121907 to SG, and NIH T32 GM102057 to MJ. This work is dedicated to the memory of Matteo.
Footnotes
Conflicts of interest
The authors have no conflicts of interest to declare.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ejmg.2017.11.016.
References
- Association A. P, 2013. Diagnostic and Statistical Manual of Mental Disorders.
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Eichler EE, 2002. Recent segmental duplications in the human genome. Science 297 (5583), 1003–1007. [DOI] [PubMed] [Google Scholar]
- Baldini A, 2006. The 22q11.2 deletion syndrome: a gene dosage perspective. Sci. World J 6, 1881–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Costain G, Marshall CR, 2016. Neuropsychiatric aspects of 22q11.2 deletion syndrome: considerations in the prenatal setting. Prenat. Diagn 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Baker C, Williams C, Gorski J, 2012. A copy number variation morbidity map of developmental delay. Nat. Genet 43 (9), 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fénelon K, Mukai J, Xu B, Hsu P-K, Drew LJ, Karayiorgou M, Gogos JA, 2011. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc. Natl. Acad. Sci. U. S. A 108 (11), 4447–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Brkanac Z, Coe BP, Baker C, Vives L, Vu TH, Eichler EE, 2011a. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 7 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Campbell CD, Eichler EE, 2011b. Human copy number variation and complex genetic disease. Annu. Rev. Genet 45 (1), 203–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Dennis MY, Baker C, Malig M, Coe BP, Campbell CD, Eichler EE, 2013a. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am. J. Hum. Genet 92 (2), 221–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Eichler EE, 2012. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N. Engl. J. Med 367 (14), 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, et al. , 2010. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet 42 (3), 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guipponi M, Santoni F, Schneider M, Gehrig C, Bustillo XB, Kates WR, Morrow B, Armando M, Vicari S, Sloan-Béna F, Gagnebin M, Shashi V, Hooper SR, Eliez S, Antonarakis SE, 2017. February 21 No evidence for the presence of genetic variants predisposing to psychotic disorders on the non-deleted 22q11.2 allele of VCFS patients. Transl. Psychiatry 7 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Chung JH, Wang T, McDonald-Mcginn DM, Kates WR, Hawuła W, Morrow BE, 2015. Histone modifier genes alter conotruncal heart phenotypes in 22q11.2 deletion syndrome. Am. J. Hum. Genet 97 (6), 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang VJ, Maar D, Regan J, Angkustsiri K, Simon TJ, Tassone F, 2014. Mapping the deletion endpoints in individuals with 22q11.2 deletion syndrome by droplet digital PCR. BMC Med. Genet 15, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, Gos A, Nestadt G, Wolyniec PS, Lasseter VK, Eisen H, Childs B, Kazazian HH, Kucherlapati R, Antonarakis SE, Pulver AE, Housman DE, 1995. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc. Natl. Acad. Sci. U. S. A 92 (17), 7612–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Baldini A, 2001. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410 (6824), 97–101. [DOI] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Emanuel B, Zackai E, 2013. 22q11.2 deletion syndrome In: Pagon R, Adam M, Ardinger H, Wallace S, Amemiya A, Bean L, Stephens K (Eds.), Gene Reviews. University of Wasington, Seattle, Seattle, WA. [Google Scholar]
- Mlynarski EE, Sheridan MB, Xie M, Guo T, Racedo SE, McDonald-Mcginn DM, Emanuel BS, 2015. Copy-number variation of the glucose transporter gene SLC2A3 and congenital heart defects in the 22q11.2 deletion syndrome. Am. J. Hum. Genet 96 (5), 753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paylor R, Glaser B, Mupo A, Ataliotis P, Spencer C, Sobotka A, Lindsay E, 2006. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc. Natl. Acad. Sci. U. S. A 103 (20), 7729–7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E, Kendall K, Pardiñas AF, Legge SE, Pocklington A, Escott-price V, Kirov G, 2016. Analysis of intellectual disability copy number variants for association with schizophrenia. JAMA Psychiatr. 13 (8), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, et al. , 2014. Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome. Results Int. Consortium Brain Behav. 22q11.2 Deletion Syndrome 171 (6), 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swillen A, McDonald-Mcginn DM, 2015. Developmental trajectories in 22q11.2 deletion. Am. J. Med. Genet. C Smin Med Genet 169 (2), 172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeweyer G, Reyniers E, Wuyts W, Rooms L, Kooy RF, 2011. CNV-WebStore: online CNV analysis, storage and interpretation. BMC Bioinforma. 12 (1), 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorstman JAS, Morcus MEJ, Duijff SN, Klaassen PWJ, Heineman-de Boer JA, Beemer FA, van Engeland H, 2006. The 22q11.2 deletion in children. J. Am. Acad. Child Adolesc. Psychiatry 45 (9), 1104–1113. [DOI] [PubMed] [Google Scholar]
- Weschler D, 2008. Wechsler Adult Intelligence Scale, fourth ed pp. 1–3. [Google Scholar]
- Widdershoven JC, Bowser M, Sheridan MB, McDonald-McGinn DM, Zackai EH, Solot CB, Kirschner RE, Beemer FA, Morrow BE, Devoto M, Emanuel BS, 2013. January A candidate gene approach to identify modifiers of the palatal phenotype in 22q11.2 deletion syndrome patients. Int. J. Pediatr. Otorhinolaryngol 77 (1), 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier C, Sticht H, Aydin-Yaylagul I, Campbell CE, Rauch A, 2007. Human TBX1 missense mutations cause gain of function resulting in the same phenotype as 22q11.2 deletions. Am. J. Hum. Genet 80 (3), 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.