Abstract
As self-assembling polyvalent nanoscale structures that can tolerate substantial genetic and chemical modification, virus-like particles are useful in a variety of fields. Here we describe the genetic modification and structural characterization of the Leviviridae PP7 capsid protein as a platform for the presentation of functional polypeptides. This particle was shown to tolerate the display of sequences from 1 kDa (a cell penetrating peptide) to 14 kDa (the Fc-binding double Z-domain) on its exterior surface as C-terminal genetic fusions to the coat protein. In addition, a dimeric construct allowed the presentation of exogenous loops between capsid monomers and the simultaneous presentation of two different peptides at different positions on the icosahedral structure. The PP7 particle is thereby significantly more tolerant of these types of polypeptide additions than Qβ and MS2, the other Leviviridae-derived VLPs in common use.
Keywords: protein nanoparticles, virus-like particles, peptide display, packaged enzymes, cryo-electron microscopy
Graphical Abstract

Virus-like particles (VLPs) are self-assembled protein nanostructures composed of multiple copies of one or more structural or envelope coat proteins (CPs). These particles resemble their corresponding natural viruses in structure but lack the genomic cargo necessary for replication.1,2 Similar to their natural precursors, VLPs typically are stable and biocompatible.3,4 Those derived from RNA bacteriophages are particularly amenable to high-yield expression and assembly in different systems, including bacteria, yeast, insect cells, and mammalian cells,5,6 and they have therefore been employed in a wide variety of biomedical applications.1,7–11 Many such uses derive from their polyvalency: the ability of VLPs to present multiple copies of functional peptides or protein domains to potential binding agents, cells, and tissues.
Both chemical and genetic approaches have been used to functionalize VLPs with foreign polypeptides.12 Chemical modification typically introduces biologically active moieties by making connections to side chains of natural amino acids as well as genetically introduced unnatural amino acid residues10,12 or by the introduction of binding and signal sequences for enzyme processing on the repeated coat protein units. The latter technology can provide an excellent means to fuse large functional moieties onto VLPs. Two examples the—SpyTag/SpyCatcher system13,14 and sortase A ligation15—have been most commonly used with VLPs, the latter for functionalization of P2216 and HBV17 particles. While powerful, these methods, as with any postassembly functionalization of polyvalent structures, do not ensure uniform loading except in rare cases in which reactions can be pushed to completion. This can be especially problematic when large domains are being attached.
Methods of controlled modification of VLPs that do not require chemical ligation include site-specific mutagenesis,18–20 the generation of peptide extensions at N- or C-termini, and insertions at exterior exposed CP loops.8 While the extension of CP sequences has been quite common (indeed, this is the basis of phage display on phagemid-assisted replicating particles), loop insertion has been more limited, with successful VLP examples reported for cowpea mosaic virus,21 flock house virus,22 PP7,23,24 and MS223,25 scaffolds. MS2 VLP loop display has also been used by Chackerian and colleagues as the basis of an epitope selection system;26–28 however, insertions are typically of short length to preserve particle assembly.
We have previously described a two-plasmid expression system for the production of hybrid Qβ VLPs composed of a mixture of truncated and extended coat proteins.29,30 While convenient and scalable, the Qβ particle will not assemble if every CP has even a small extension at either end of the truncated CP. Thus, the number of functional moieties presented on each VLP by this technique is a statistical average, and this average varies among different batches even if the expression is performed under the same conditions.
We report here an exploration of similar genetic manipulations of the related PP7 scaffold. We found this particle to be more tolerant of such changes, assembling into homogeneous icosahedral particles with extended peptides on every subunit and allowing for substantial loop insertions into a dimeric CP variant. We also describe the appearance of a T = 4 structure for this class of Leviviridae-derived capsids, adopted unexpectedly by particles self-assembled from CP dimers.
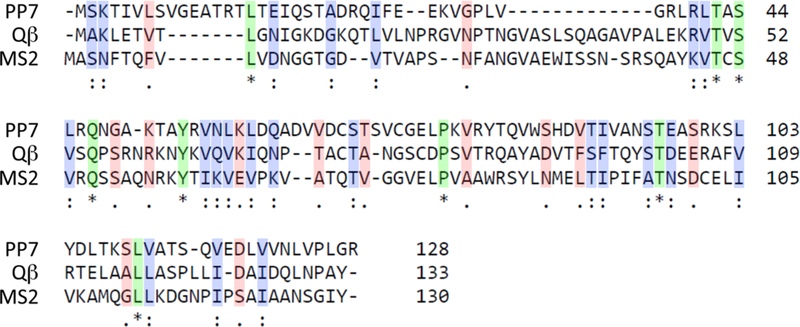
RESULTS AND DISCUSSION
ZZ-Domain Extensions on the PP7 Particle.
The “wild-type” (WT) PP7 VLP here corresponds to the 127 amino acid natural sequence of the virus reported by Olsthoorn et al.31 Expression of this protein in Escherichia coli from a standard T7 promoter plasmid gave rise to high yields (>50 mg per liter) of fully assembled virus-like particles.32 Its three-dimensional structure is nearly identical to other Leviviridae, featuring a noncovalent dimer in which interlocked α-helices dominate the particle exterior surface and β-sheet domains are contiguously arranged on the interior surface.33,34 However, the sequence homology of the PP7 coat protein with those of Qβ and MS2 are both less than 20% (Figure 1); as detailed below, the PP7 virus-like particle has proven to be the most physically and mutationally robust platform of the three.
Figure 1.
Sequence alignment of MS, PP7, and Qβ coat proteins. The coat protein sequences of MS2, Qβ, and PP7, aligned using the Clustal Omega35 multiple sequence alignment tool. Dashed lines mark alignment gaps, asterisks (green highlight) indicate identical residues, and single (red) and double (blue) dots indicate similar residues.
To explore the parameters of exterior surface display, we chose to use the Z-domain,36 a 58-amino acid analogue of the IgG-binding domain B in Staphylococcal Protein A,37,38 or a dimeric version (ZZ) joined by a four-amino acid linker.39 We previously fused these polypeptides to the Qβ CP as C-terminal extensions and were able to prepare hybrid particles bearing 24 Z-domains29 or more than 30 ZZ-domains40 per particle when expressed with the wild-type capsid protein. Other strategies, including amber stop codon suppression and ribosomal skip methods,41–43 have also been used to produce assembled hybrid Leviphage-derived particles. The largest structure appended to such capsids is human interleukin-1β (~150 amino acids) displayed on AP205 VLPs coassembled from the extended and wild-type CPs.44 A variety of other functional peptides have also been displayed in hybrid (mixed with WT capsid) form on VLPs from phage GA,44 fr,45 MS2,46,47 Qβ,41–43 and AP205.48–50 In all cases, no reliable way exists to predict the number of extensions that will be incorporated in such structures. The obvious solution to this problem—self-assembly of only the extended capsid protein—has been reported for Leviviridae particles only with short peptide appendages, with the notable exception of a C-terminal 55-amino acid extension derived from the Nef protein (amino acids 66–100 and 132–151 fusion) fused onto the AP205 CP.48
We tested the self-assembling properties of the PP7 capsid protein by appending Fc-binding domains in three different ways: ZZ-domains (14.2 kDa) added to the N-terminus (designated ZZ-PP7), single Z-domains at both the N- and C-termini of the PP7 CP (Z-PP7-Z), and ZZ-domains fused to the C-terminus (PP7-ZZ). When coexpressed with the wild-type CP, all three constructs were found to successfully self-assemble into icosahedral hybrid particles bearing significant numbers of polypeptide extensions, isolated in good yields (Table 1, entries 1–3). Characterization by transmission electron microscopy (TEM) of negatively stained particles, dynamic light scattering, and mass spectrometry of the denatured particles are given for each structure in Supporting Information. The hydrodynamic radii of all of these structures were found to be ~20–22 nm, significantly larger than the wild-type particle (17 nm). This is consistent with the presence of the added domains, and with the expectation derived from the PP7 virus atomic-resolution structure33,51 that both CP termini, and thus the extensions added to them, are located on the exterior surface of the particle. The addition of Z-domains to both N- and C-termini of the same CP is new in our experience, in that assembled particles are not found in large quantities in such cases with Qβ. Isolated yields were ~30–40 mg/L culture for hybrid particles and 20–30 mg/L for other particles described in this report.
Table 1.
Hybrid and Homogeneous PP7 Virus-like Particles
| entry | VLPa | incorporationb | Rhc (nm) |
|---|---|---|---|
| 1 | PP7 + Z-PP7-Z | 40 per capsid | 20.8 ± 0.1 |
| 2 | PP7 + ZZ-PP7 | 90 per capsid | 21.7 ± 1.1 |
| 3 | PP7 + PP7-ZZ | 40 per capsid | 21.0 ± 1.8 |
| 4 | PP7-ZZ | 180 per capsid | 21.5 ± 0.3 |
| 5 | ZZ-PP7 | 180 per capsid | 20.9 ± 0.4 |
| 6 | PP7-PP7-ZZ | 120 per capsidd | 22.6 ± 1.4 |
“Z“ = A M V D N K F N K E Q Q N A F Y E I L H L P N L N E E - QRNAFIQSLKDDPSQSANLLAEAKKLNDAQAPK. “ZZ” = Z-AYGG-Z.
Average number of extended coat proteins per VLP, determined for the hybrid particles (entries 1–3) by relative peak intensities in LC-MS spectra shown in Figure S1, assuming that both wild-type and extended polypeptides are detected with the same sensitivity.
Hydrodynamic radius measured by dynamic light scattering; see Figure S1 for representative measurement.
See text.
Interestingly, with an eight-amino acid linker to connect the ZZ-domain extension to the CP, we were able to express and isolate intact ZZ-PP7 and PP7-ZZ particles, each with 180 copies of the ZZ-domain and each exhibiting similar hydrodynamic radii as their hybrid versions (~22 nm, Table 1, entries 4 and 5; characterization data in Figure S2). The difference between PP7 and Qβ in this regard is striking: we found no extended Qβ capsid protein that is able to assemble into a discrete particle. A comparison of the X-ray crystal structures of these wild-type particles (PDB codes 1QBE and 1DWN) shows the N- and C-termini around the threefold axis in PP7 to be significantly less crowded, especially the C-termini, and therefore presumably better able to tolerate additional amino acids in those regions. The Z-PP7-Z protein, roughly similar in overall size, did not produce particles, even though plenty of the protein was expressed.
To decrease the number of extension incorporated onto the particle, we took the advantage of the well-characterized single-chain PP7-PP7 dimer construct, in which the N-terminus of one CP is linked to the C-terminus of another, in our case by a four-amino acid sequence (AYGG) instead of the dipeptide (YG) employed by Peabody and colleagues.32 The ZZ-domain was fused onto the C-terminus of the dimer with an eight-amino acid linker (GGPSESGA) to generate a protein that assembled in high yield into particles designated PP7-PP7-ZZ, which were isolated and characterized by the same procedures as used for the other particles described here (Table 1, entry 6; Figure S2).
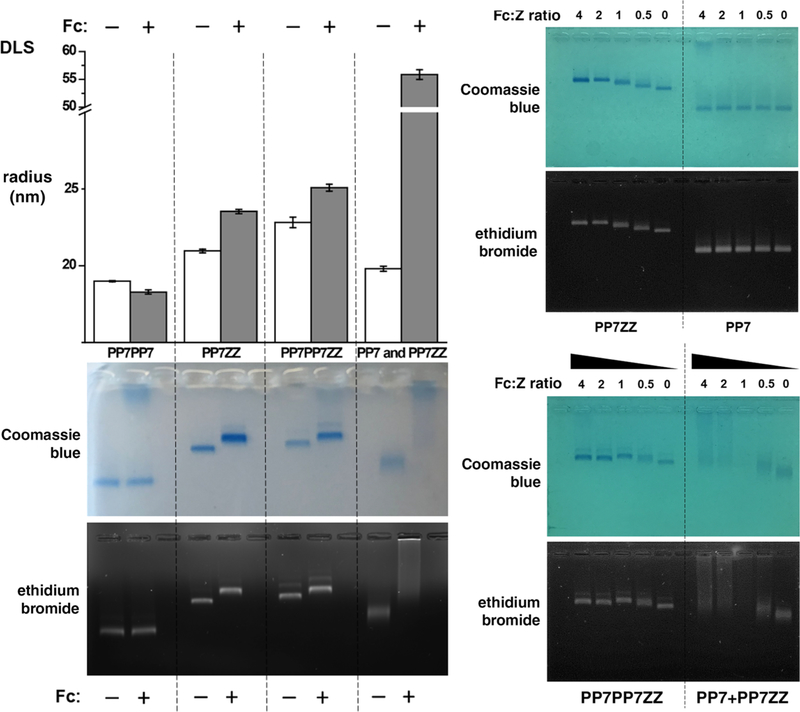
All of the Z-domain-bearing particles described above were found to bind to an antibody Fc domain, as shown in Figure 2. When Fc protein was used in excess to saturate the available Z-domains, native agarose gel electrophoresis and dynamic light scattering (DLS) both showed increased particle sizes and fairly narrow size distributions. No change was noted for the wild-type particle lacking Z-domain extensions. Titration of the amount of Fc added (Figure 2, right) showed some differences in dose–response behavior. Thus, a higher ratio of Fc to ZZ-domain was required for saturation of the PP7-ZZ particle than the PP7-PP7-ZZ particle, the former displaying ZZ-domains in higher density, suggesting perhaps a steric crowding effect. The mixture of hybrid PP7+PP7-ZZ particle with Fc protein appeared as a smear in native gel electrophoresis, presumably due to the distribution of different numbers of ZZ-domains presented on different particles and on the ability of Fc domains to aggregate particles when noncovalent cross-linking is sterically accessible.
Figure 2.
(top left) Hydrodynamic radii of the indicated particles in the presence and absence of excess Fc domain. (bottom left) Native agarose gel electrophoresis of the same mixtures. (right) Native agarose gel electrophoresis of the indicated particles (0.1 mg/mL) with decreasing concentrations of Fc domains, relative to the displayed Z- or ZZ-domains.
Other Functional Peptide Extensions on the PP7 Particle.
To further demonstrate the scope and limitations of the PP7 capsid terminal extension display technology, we appended several different functional peptides to the C-termini of the PP7 coat protein and its dimeric (PP7-PP7) analogue, in each case after a common eight-amino acid spacer sequence (GGASESGA). As indicated in Table 2, we were able to successfully produce intact PP7 constructs from both proteins alone (in other words, without the truncated capsid protein) bearing the following peptide extensions: a transferrin recognition peptide (TfR),52,53 an EGFR recognition peptide (GE7),54–57 and standard antigenic epitopes of the ovalbumin proteins (OVA1 and OVA2).58 In addition, two other functionally interesting peptides—a repeat of the NANP motif from Plasmodium falciparum circumsporozoite protein (CSP)59,60 and a Trypanosoma trans-sialidase (TS) epitope61,62—were successfully displayed on the PP7-PP7 particle. Longer extensions generally gave an increase in particle size as measured by DLS.
Table 2.
Successful C-Terminal Extension Modifications of the PP7 Virus-like Particle
| entry | VLPa | description or function | peptide sequences | Rhb (nm) |
|---|---|---|---|---|
| 1 | M, D | ZZ-domain (ZZ) | [see caption to Table 1] | 21.5 ± 0.3, 22.6 ± 1.4 |
| 2 | M, D | transferrin recognition (TfR) | HAIYPRH | 17.4 ± 0.8, 18.0 ± 0.2 |
| 3 | M, D | EGFR recognition (GE7) | NPVVGYIGERPQYRDL | 17.8 ± 0.3, 18.7 ± 1.2 |
| 4 | M, D | OVA1 antigen (OVA1) | SIINFEKL | 19.0 ± 1.5, 18.8 ± 1.2 |
| 5 | M, D | OVA2 antigen (OVA2) | ISQAVHAAHAEINEAGR | 18.8 ± 1.1, 18.5 ± 0.9 |
| 6 | D | Trypanosoma trans-sialidase epitope (TS) | ATIENRDVM | 18.0 ± 0.2 |
| 7 | D | P. falciparum circumsporozoite protein (NANP) | NANPNVDPNANPNANPNANPNANP | 20.0 ± 0.2 |
M = fusion to the PP7 monomeric coat protein; D = fusion to the PP7-PP7 dimeric coat protein.
Hydrodynamic radii measured by DLS (M, then D); see Figures S3 and S4 for representative measurements.
In contrast, fusions of the dimeric CP with several oligopeptides containing free thiols or with the epidermal growth factor (EGF) sequence (a protein known to be prone to aggregation63) did not provide intact particles from the standard expression and isolation protocol, suggesting that additional intersubunit interactions may interfere with particle assembly and/or isolation. This was previously observed for Qβ, although we managed in that case to isolate hybrid particles bearing a relatively small number of EGF extensions.64
PP7 Dimer VLP with Dual Displayed Zika Epitopes.
Although it is now common to display multiple copies of a single motif on VLPs, the simultaneous display of two or more different exogenous peptides on the same particle is quite rare.44 The only example of which we are aware in the Leviviridae family are reports of Peabody and colleagues of hybrid PP7 and MS2 particles displaying one human papillomavirus (HPV) minor capsid L2 epitope fused onto the N-terminus and another epitope inserted in the AB-loop of the capsid protein.47 Taking advantage of the ability of the PP7-PP7 dimeric CP to assemble when carrying a variety of extensions, we sought to incorporate both a C-terminal extension and a loop insertion, the latter placed in the linker sequence between the two copies of PP7 monomer. This may allow for the use of longer added sequences while maintaining self-assembly compared to loop insertions into the CP monomer.65
For preliminary exploration, we chose domain III of the Zika virus envelope protein, one of several regions on this and related Flaviviridae thought to be highly antigenic.66 We selected four loop epitope sequences—the so-called αB/I0 loop (a-loop),67 fusion loop (f-loop),68 the loop between the i and j strands (i-loop),67 and the 150-loop67—to be inserted into the linker region of the PP7-PP7 dimer to test the tolerance of this site for modification. Intact particles were isolated in good yield in three of these four cases; the f-loop insert failed perhaps because of aggregation caused by its unpaired cysteine residue. Particles bearing dimer loop insertions were similar to the wild-type PP7 particle in hydrodynamic radius (Table 3), in contrast to the larger values observed for particles bearing C-terminal extensions. To simultaneously present two epitopes, we added the 150-loop sequence to the C-termini of the PP7-a-loop-PP7 and PP7-i-loop-PP7 capsid proteins. The resulting proteins self-assembled as before, allowing for the routine isolation of good yields of particles bearing one Zika-related loop and one Zika-related linear oligopeptide per coat protein dimer.
Table 3.
Successful Loop Insertion Modifications of the PP7-PP7 Dimer Particle
| entry | VLP designation | added loop sequence | added C-term sequence | Rha (nm) |
|---|---|---|---|---|
| 1 | PP7-a-loop-PP7 | LEAEMDGAKGRL | 19.4 ± 0.9 | |
| 2 | PP7-i-loop-PP7 | VEFKDAHAKRQTVVV | 20.6 ± 2.6 | |
| 3 | PP7-a-loop-PP7–150 loop | LEAEMDGAKGRL | NDTGHETDEN | 19.3 ± 1.3 |
| 4 | PP7-i-loop-PP7–150 loop | VEFKDAHAKRQTVVV | NDTGHETDEN | 19.5 ± 0.9 |
| 8 | PP7-f-loop-PP7 | LVDRGWGNGCGLFGKG | no particles |
Hydrodynamic radii measured by DLS; see Figure S5 for representative measurements.
Thermal Stability.
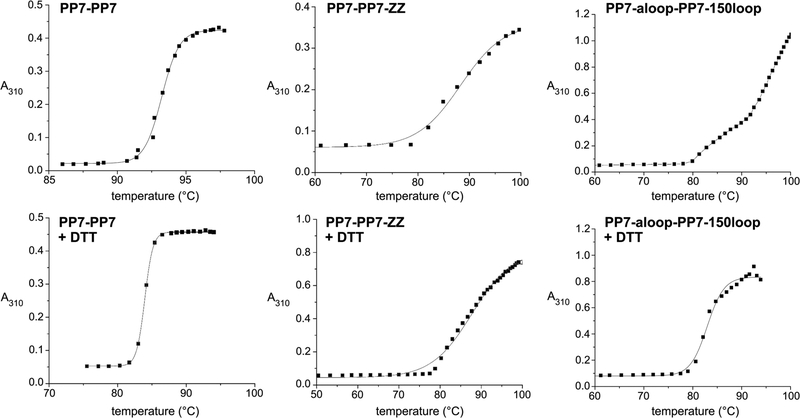
A gross measure of particle stability is its resistance to heating in standard buffer solution, conveniently monitored by UV–visible (UV–vis) absorbance, since denaturation and aggregation cause increased apparent absorption by light scattering. We previously measured Qβ VLP stability by circular dichroism as a function of temperature, finding an average decomposition temperature of 84 °C for the standard particle.69 The simpler UV–vis absorbance method gave the same result. Thus, this experiment was performed on each of the PP7 VLPs described above, with apparent melting temperature (Tm) defined as the temperature at which 50% of the maximum aggregation related absorbance at 310 nm was reached and the onset of aggregation (Tagg) defined as the temperature at which a significant increase in absorbance (10%) was observed. From a practical perspective, Tagg is the most important value, since it marks the upper temperature limit for chemical operations in which the capsid structure can be expected to remain intact. Representative data are shown in Figure 3, complete data in Supporting Information (Figure S6), and results collected in Table 4. It appears from these studies that the PP7 platform, regardless of how it is manipulated by dimerization, extension, or loop insertion, reliably maintains stability to ~80 °C if it forms particles at all. The PP7-PP7 dimer particle is very stable, essentially equivalent to that of the parent VLP made from CP monomers (Table 3; Tagg ≈ 90 °C, Tm ≈ 94 °C), both values ~10 °C higher than for Qβ.
Figure 3.
Representative analyses of thermal stability of the indicated PP7-PP7-based particles (0.1 mg/mL in PBS buffer, heated at 10 °C per minute) in the absence (top) and presence (bottom) of 1 mM DTT. Additional traces for other particles are provided in Supporting Information (Figure S1).
Table 4.
Melting (Tm) and Onset-of-Aggregation (Tagg) Temperatures of PP7 Particlesa
| Tm (°C) | Tagg (°C) | Tm (°C) | Tagg (°C) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| VLP | −DTT | +DTT | −DTT | +DTT | VLP | −DTT | +DTT | −DTT | +DTT |
| PP7 | 95.7 | 82.2 | 93.5 | 80.2 | PP7-PP7-OVA1 | 90.3 | 78.3 | 87.8 | 73.8 |
| PP7-PP7 | 93.2 | 84.0 | 91.8 | 82.3 | PP7-PP7-OVA2 | ND | 79.2 | ND | 77.4 |
| PP7-PP7-GE7 | 92.7 | 78.4 | 90.2 | 76.9 | |||||
| PP7-ZZ | 88.1 | 84.3 | 77.1 | 77.0 | PP7-PP7-TS | 97.4 | 82.2 | 94.4 | 80.7 |
| ZZ-PP7 | 87.9 | 83.2 | 79.3 | 78.6 | PP7-PP7-NANP | ND | 79.8 | ND | 77.7 |
| PP7-PP7-ZZ | 88.0 | 84.5 | 79.1 | 78.5 | PP7-PP7-TfR | 90.8 | 83.6 | 89.7 | 82.3 |
| PP7-a-loop-PP7 | 97.3 | 84.1 | 95.6 | 81.0 | PP7-a-loop-PP7–150-loop | ND | 82.9 | ND | 79.2 |
| PP7-i-loop-PP7 | 97.2 | 83.0 | 95.0 | 81.4 | PP7-i-loop-PP7–150-loop | ND | 83.0 | ND | 79.6 |
Measured as shown in Figure 3, grouped by type (parent particles, ZZ-domain extensions, smaller peptide C-terminal extensions, dimer loop insertions, and simultaneous dimer loop insertions and C-terminal extension). The standard deviations of the average Tm and Tagg were less than 1.0 °C for all experiments based upon repeat measurements of independently-prepared samples. ND means that a value could not be determined from the observed biphasic curve.
Reduction of the disulfide linkage between C67 and C72 (of the same or different CPs, depending on the symmetry position) gave a loss of ~10 °C in thermal stability for both PP7 and PP7-PP7 particles; in Qβ, this value is ~15 °C.69 In contrast, the MS2 VLP has no intersubunit disulfide linkages,23 and therefore dithiothreitol (DTT) treatment had no significant effect on the Tm of this particle (data not shown). Appending ZZ domains to either terminus of the PP7 CP monomer, or to the N-terminus of the PP7-PP7 CP dimer, gave equivalently stable particles, exhibiting a lower onset of aggregation (~80 °C) and a broader transition curve, resulting in Tm values of ~88 °C. All the other particles made and examined here showed very similar behavior, exhibiting flatter, and sometimes two-stage, temperature-aggregation curves (examples in Figure 3).
High-Resolution Structure Determination by Cryo-Electron Microscopy.
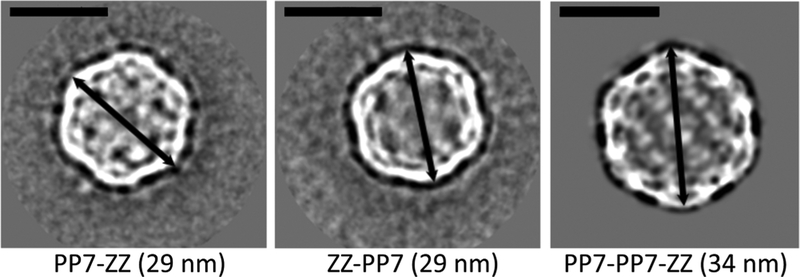
When we measured particle diameter by analysis of TEM images, we found that particles bearing the same extensions from the PP7-PP7 dimer platform were larger than the corresponding particles based on the monomeric PP7 capsid protein (Table 5). To better understand the three-dimensional display of the functional extensions on the dimer-based platform, we examined the PP7-PP7, PP7-PP7-ZZ, and PP7-a-loop-PP7 particles by cryo-electron microscopy (cryo-EM). Preliminary analysis in the form of two-dimensional (2D) class averages (Figure 4) confirmed the size difference between PP7 and PP7-PP7 extension particles.
Table 5.
Particle Sizes Measured from TEM Images
| monomeric version | diameter/nm | dimeric version | diameter/nm |
|---|---|---|---|
| PP7 | 26.4 ± 1.2 | PP7-PP7 | 32.8 ± 1.6 |
| ZZ-PP7 | 23.9 ± 1.7 | ||
| PP7-ZZ | 27.0 ± 1.6 | PP7-PP7-ZZ | 30.9 ± 2.2 |
| PP7-TfR | 28.5 ± 1.9 | PP7-PP7-TfR | 30.0 ± 3.4 |
| PP7-GE7 | 32.1 ± 1.8 | PP7-PP7-GE7 | 34.0 ± 2.0 |
| PP7-OVA1 | 27.8 ± 1.6 | PP7-PP7-OVA1 | 34.3 ± 2.3 |
| PP7-OVA2 | 28.2 ± 2.3 | PP7-PP7-OVA2 | 30.4 ± 2.0 |
| PP7-PP7-TS | 31.1 ± 2.0 | ||
| PP7-PP7-NANP | 32.2 ± 1.9 |
Figure 4.
Selected cryo-EM 2D class averages corresponding to the projection along the twofold axis of monomeric PP7-ZZ and ZZ-PP7 particles and dimeric PP7-PP7-ZZ particle. The apparent coronas around the first two particles are artifacts of sample preparation (scale bar = 20 nm).
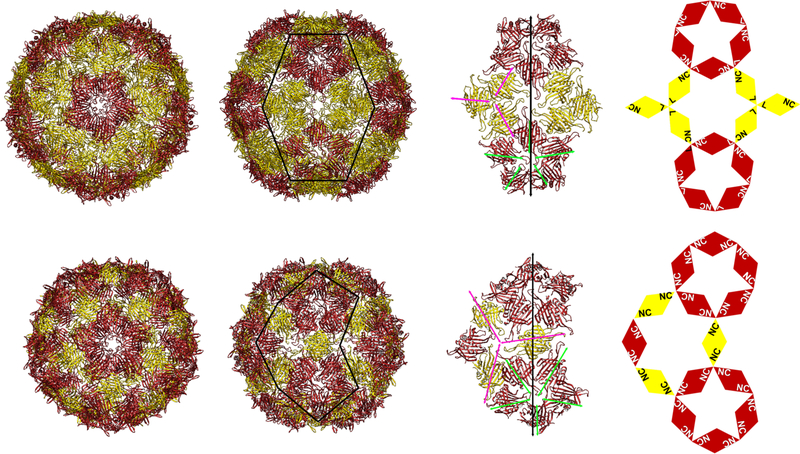
Structural resolution of ~3 Å was achieved for each, revealing the surprising finding that all of the dimer-based structures, including the simple PP7-PP7 dimer, formed T = 4 capsids.70 This stands in contrast to the T = 3 structure reported for the monomeric PP7 particle and confirmed here for ZZ-PP7 (Figure 5).33,34 Thus, the dimer-based particles are comprised of 120 copies of the PP7-PP7 CP and display 120 functional insertions/extensions. In contrast, the ZZ-PP7 reconstruction displayed 60 regions of low-resolution electron density protruding outward at the expected positions (Supporting Information Figure S8), each representing three N-terminal ZZ extensions and therefore a total of 180, not 240, ZZ-domains per particle.
Figure 5.
Cryo-EM structure of the PP7-PP7 virus-like particle [top, T = 4 (h = 2; k = 0) structure, PDB ID 6N4V] compared to the previously published structure of the PP7 particle [bottom, T = 3 (h = 1; k = 1) subunits colored the same way, PDB ID 1DWN]. (left) View down the fivefold symmetry axes; (middle) view down the twofold symmetry axis; (right) a cartoon representation showing the organization of the coat-protein dimers in the T = 4 structure. The linker loop is labeled “L”, and the termini are labeled “NC”.
While measurements of particle size in negative-stain images of the monomeric PP7 and dimeric PP7-PP7 particles (Table 2) showed the dimer-based particles to be larger than analogous structures based on the monomeric coat protein, results from dynamic light scattering were not consistent in this regard (Figures 2, 4, and 5). This is perhaps not surprising given the ability of flexible peptide or protein extensions to increase the apparent hydrodynamic radius of any particle.
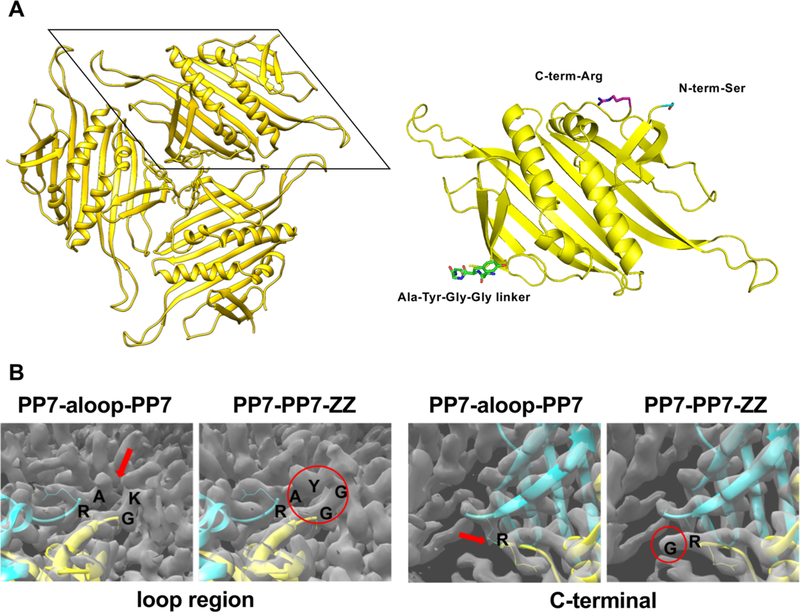
The added sequences and domains were not often well-resolved in the high-resolution cryo-EM structures, suggesting conformational flexibility in the appended regions. The dimer linker region was visible in the density map of the parent dimeric PP7-PP7 particle (Figure 6A), and the connecting tetrapeptide (AYGG) was clearly resolved in the PP7-PP7-ZZ structure. However, neither the C-terminal density corresponding to the ZZ polypeptide in PP7-PP7-ZZ nor the added loop residues in PP7-a-loop-PP7 could be resolved to high resolution (Figure 6B).
Figure 6.
Cryo-EM structural details. (A, left) PP7-PP7 dimer threefold organization, (A, right) expanded view of one single-chain dimer, with AYGG linker (green), N-terminal Ser (cyan), and C-terminal Arg (purple) residues shown in stick representation. (B) Comparison of particles bearing the a-loop insertion and C-terminal ZZ-domain extension. Arrows mark regions of poorly resolved amino acid density in the reconstruction of the PP7-a-loop-PP7 particle following the initial Ala residue of the a-loop (left) and following the last Arg residue at the C-terminus (Arg127) (right). Circles delineate well-resolved linker loop density for the PP7-PP7-ZZ particle (left) and the Gly residue that starts the linkage to the ZZ-domain but not the rest of that domain (right).
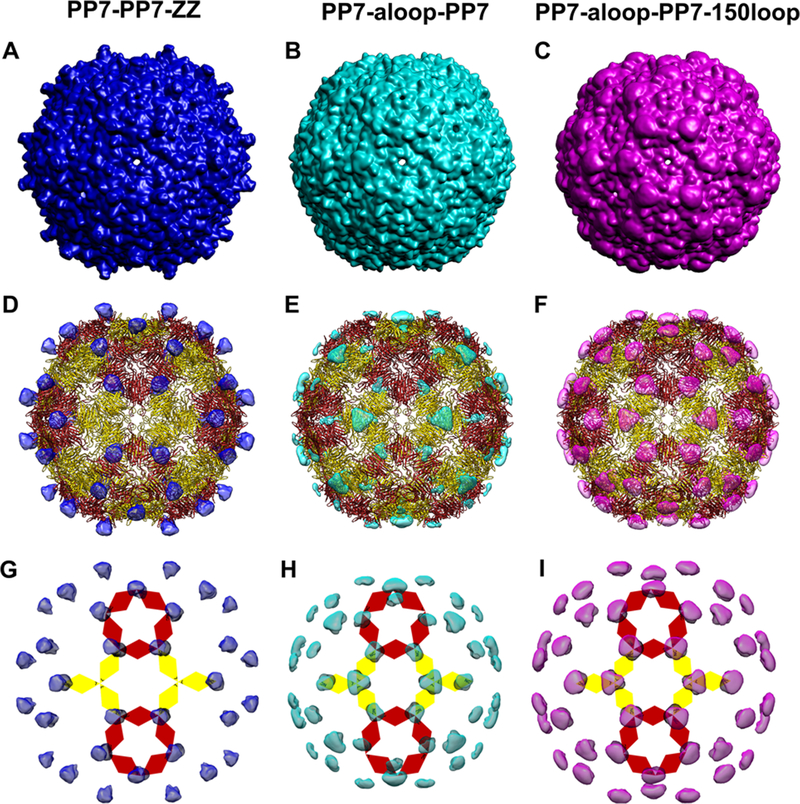
The extra peptides on the PP7-PP7-ZZ and PP7-a-loop-PP7 particles were best visualized using low-pass filtered maps displayed at low threshold (Figure 7). The PP7-PP7-ZZ particle, composed of 120 proteins, showed 60 doublet protrusions (Figure 7A,D,H), while the extra densities of the PP7-a-loop-PP7 particle had 20 trileaves (formed by the clustering of three loops at each of 20 threefold axis sites) and 60 monoleaves (formed by single loops; Figure 7B,E,I). These images provide insight as to how the covalent dimeric CP’s are organized and also suggest that the PP7-PP7-ZZ particles should be able to bind and display a maximum of 60 Fc ligands, since the paired ZZ-domains are too close together to be able to simultaneously bind Fc domains or full IgG molecules. Examination of the dual-epitope presenting PP7-a-loop-PP7–150-loop particle showed the two types of display to be compatible with each other (Figure 7C,F,J). Similar to the PP7-a-loop-PP7 particle, there were 20 trileaves (clustered at the 20 threefold axis sites) and 60 monoleaves (formed by single loop) of a-loop presentation. In addition, similar to the PP7-PP7-ZZ structure, the C-terminal 150-loop was visible as 60 doublet protrusions, which clustered with the single a-loop monoleaf.
Figure 7.
Mapping of extra density on the surface of PP7-a-loop-PP7, PP7-PP7-ZZ, and PP7-a-loop-PP7–150-loop particles. Overall surface density maps: (A) PP7-PP7-ZZ, (B) PP7-a-loop-PP7, and (C) PP7-a-loop-PP7–150-loop. Extra densities highlighted: (D) PP7-PP7-ZZ, (E) PP7-a-loop-PP7, and (F) PP7-a-loop-PP7–150-loop, comparing to PP7-PP7. (G) PP7-PP7-ZZ, (H) PP7-a-loop-PP7, and (I) PP7-a-loop-PP7–150-loop, showing the same loop insertions and C-terminal extensions as in (D–F). Subunit organization in (G–I) is indicated as in Figure 5.
While the core antigen of the Hepadnavirus hepatitis B is known to form both T = 3 (PDB ID 6BVN) and T = 4 (PDB ID 1QGT, 2G33, 2G34) structures, to our knowledge, Leviviridae capsids have previously been found only as T = 3 icosahedra Since the ZZ-PP7 particle is a T = 3 particle, it is apparently the covalent linkage of capsid proteins into dimers, and not the C-terminal appendage of a large polypeptide domain, that induces the structural change to T = 4.
Enzyme Encapsidation.
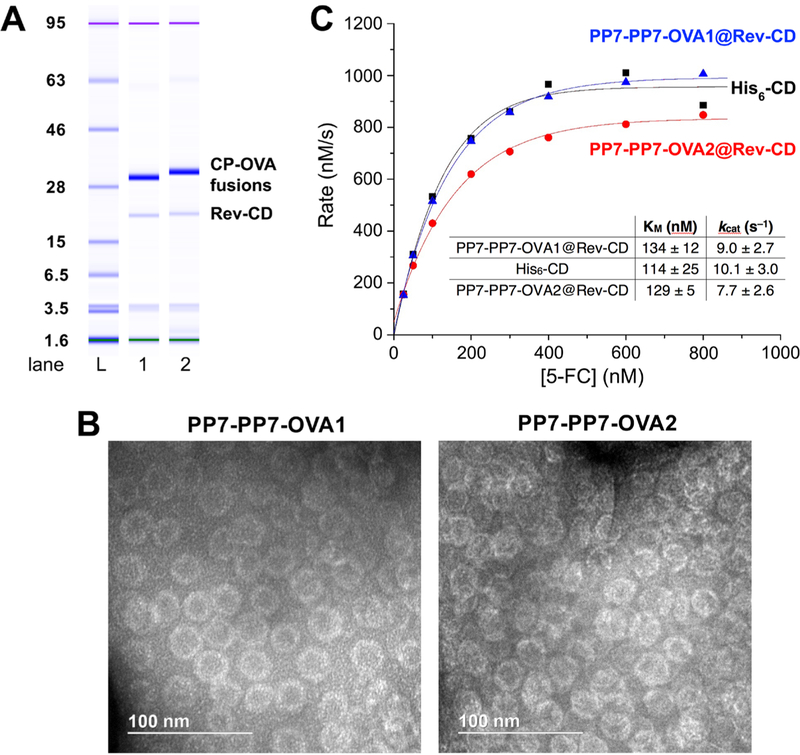
We have previously described the convenient packaging of enzymes bearing a positively charged Rev oligopeptide when coexpressed with the Qβ capsid protein and a bridging nontranslated RNA;71 we subsequently learned that the specific RNA is not necessary in some cases.40 To confirm or refute our expectation that the same technique should apply to PP7, and to demonstrate the simultaneous construction of a bifunctional particle with both targeting and catalytic function, we expressed the thermostable cytosine deaminase tagged with the Rev peptide at the N-terminus (Rev-CD)72 with each of the PP7-PP7-OVA1 and PP7-PP7-OVA2 VLP constructs. The resulting particles contained an average of 15–25 CD enzymes per particle, equivalent to packaging in the Qβ capsid, and were otherwise indistinguishable (DLS, TEM, size-exclusion chromatography (SEC)) from the PP7-PP7-OVA particles lacking the packaged enzyme (Figure 8A,B). Michaelis–Menten kinetics measurements of the conversion of 5-fluorocytosine (5-FC) to 5-fluorouracil (5-FU) showed the free and packaged enzymes to be essentially equivalent in activity (Figure 8C) and consistent with previously reported measurements.
Figure 8.
Characterization of Rev-CD encapsulated PP7-PP7-OVA particles. (A) Electrophoretic analysis: purified particles showing PP7-PP7-OVA1, PP7-PP7-OVA2, and Rev-CD bands. (B) TEM; images are indistinguishable from those of PP7-PP7-OVA1 and PP7-PP7-OVA2 without enzyme packaged. (C) Kinetic analysis, plotted on a per-enzyme basis, of the conversion of 5-FC to 5-FU catalyzed by the indicated forms of cytosine deaminase. Solid curves show the best fit using the Michaelis–Menten equation. The @ symbol designates the encapsidated enzyme.
CONCLUSIONS
Building on the work of Peabody and colleagues, we showed the robust capability of the PP7-derived virus-like particle for functional modification by polypeptide extension, insertion, and encapsulation. This system tolerates a wide variety of such changes, with the unexpected observation of expansion to a T = 4 structure for particles derived from the covalently linked coat protein dimer. In general, it appears that this dimer structure is somewhat less thermally stable than the T = 3 particle (decomposition onset at ~80 °C rather than 90 °C), but it is still a very robust platform. Unusually for the Leviviridae-based capsids, intact icosahedral particles can be made from identical copies of a coat protein fusion with a polypeptide (the 14.2 kDa ZZ-domain) of approximately the same size. This suggests that many other functional sequences will be amenable to the same type of presentation to give homogeneous polyvalent particles. In addition to this broad capability to tolerate the display of foreign oligo- and polypeptide sequences, PP7 allows dual display of both extended looped and linear peptides.
EXPERIMENTAL SECTION
Construction of PP7 with Extensions and Insertions.
The wild-type PP7 coding sequence was codon-optimized for expression in E. coli cells. The PP7-PP7 dimer construct was generated by Gibson Assembly protocol (NEB) with a four-amino acid linker AYGG that contained a built-in NdeI restriction site for downstream cloning. The PP7 C-terminal extensions were made by flanking polymerase chain reaction (PCR) in which the PCR primers contain a 3′-sequence base paring with the C-terminal sequence of PP7 and a 5′-flanking region coding for the extensions. The amplified PP7-extension products were purified, digested, and ligated into the expression vector. To construct the dimer loop insertions, similar flanking PCR strategy was used. The plasmid construct was shown in Figure S2. All the constructs were sequenced to verify the correct incorporation of the extension or insertion.
Protein Expression and Purification.
The PP7 VLPs were expressed and assembled in BL21(DE3) E. coli cells (Biogen) as previously described for Qβ.40 The transformed cells were grown in super optimal broth (SOB, Amresco) (Amresco) supplemented with kanamycin or streptomycin at 50 μg/mL. Starter cultures were grown overnight at 37 °C and used to inoculate larger cultures. Particle expression was induced with 1 mM isopropyl β-D-1-thiogalactopyr-anoside (IPTG) when the OD600 was between 0.8 and 1.0. After induction, expression cultures were kept at 37 °C for 4 h and then harvested by centrifugation at 6000 rpm. The resulting cell pellet was either immediately processed or stored at −80 °C.
For particle purification, the cell pellet was resuspended with an appropriate amount of potassium phosphate buffer (0.1 M, pH 7.0) and sonicated at 30–40 W for 10 min with 5 s bursts separated by 5 s intervals. The resulting clear suspension was centrifuged at 13 000 rpm for 10 min. The supernatant was carefully removed, and the protein precipitated with 0.265 g/mL ammonium sulfate, performed in a rotator at 4 °C for 2–3 h. The precipitate was collected by centrifugation at 13 000 rpm (Beckman JA-17 rotor) at 4 °C for 10 min, and the protein pellet was suspended with appropriate amount of phosphate-buffered saline (PBS; 0.1 M, pH 7.0). Lipids and membrane proteins carried through precipitation were removed by organic extraction using 1:1 n-butanol/chloroform. The aqueous layer containing VLPs was collected and further purified by sucrose density ultracentrifugation (10–40% w/v). The VLP fractions were collected and pelleted out by ultracentrifugation at 68 000 rpm (Beckman Type 70 Ti Rotor) for 2 h.
PP7 Particle Analysis.
The PP7 protein concentration was measured using the Coomasie Plus Protein Reagent (Pierce) with bovine serum albumin as standard. The purity and homogeneity of the assembled PP7 VLPs were assessed by SEC, DLS, and TEM. All the modified PP7 particles eluted between 10.8 and 12.0 mL from a Superose-6 SEC column, using 0.1 M potassium phosphate buffer (pH 7.0) with a 0.4 mL/min flow rate. All PP7 VLPs were assayed at0.1 mg/mL concentration with a Dynapro dynamic light scattering plate reader (Wyatt Technologies) in a 384-well plate with 10% laser power and 30% attenuation. In addition, the protein content of each sample was also analyzed with a Bioanalyzer 2100 Protein 80 microfluidics chip (Agilent). For accurate determination of the molecular weight, the PP7 VLPs were subjected to LC-MS analysis (Agilent).
UV–Vis Measurements of Aggregation.
All PP7-based VLPs (0.1 mg/mL) in PBS buffer (pH 7.0; 1 mL particle solution) were placed in 1 cm quartz cuvettes sealed with caps to avoid evaporation. Each sample was heated from 50 to 100 °C at a rate of 10 °C/min in a Thermo Evolution 201/220 UV–visible spectrophotometer equipped with a Peltier Control and Cooling Unit (PCCU1). Absorbance at 310 nm, a wavelength sensitive to light scattering by aggregates, was measured every 15 s. At the end of each temperature ramping experiment, white protein aggregates were observed inside the cuvette. Tm values were calculated by plotting the absorbance at 310 nm (A310) against temperature (T) using the Boltzmann sigmoid equation:
where Aini = initial A310, Aend = final A310, and S is the slope constant. Triplicate batches of PP7 VLP samples were independently analyzed, and the reported data are the average values ± standard deviation.
Tagg was defined as the temperature at which the absorbance value reaches 10% of the maximum observed increase:
Therefore, by equation transformation:
Functional Characterization of PP7 Z-Domain Particles.
The Z-domain fused PP7 VLPs (0.1 mg/mL) were mixed with human IgG Fc fragment (0.1 mg/mL) and incubated at 37 °C for 1 h. For IgG Fc fragment serial dilution experiment, the highest Fc fragment concentration was set to be 4 times the total molar concentration of the Z- or ZZ-domain and diluted by a factor of 2. The mixture was analyzed by DLS and/or native agarose gel electrophoresis.
Cryo-EM Sample Preparation, Data Collection and Data Processing.
Vitrified Sample Preparation.
300-mesh R1.2/1.3 ultrafoil holey grids (EMS) were plasma-cleaned using Solarus Gatan Plasma System for 7 s in an oxygen–argon environment at 15 W power. VLPs were diluted to 1.5 mg/mL, and an aliquot of 3 μL of sample was applied to the gold foil side of the grid. Grids were plunge-frozen into liquid ethane on a Leica EM GP automatic plunge freezer (Leica Microsystems) after a 1.5 s blotting time. Frozen grids were clipped in AutoGrid rings and stored in liquid nitrogen until data collection.
Data Collection.
Grids were imaged on Thermo Scientific Titan/Krios microscope (SEMC Krios 1) equipped with a Gatan K2 Summit direct electron detector camera. Movies were collected in counting mode with 8 e−/px/s dose rate on the camera and 5 s exposure fractionated into 25 frames—200 ms per frame. Images were acquired at a magnification of 22 500× corresponding to a calibrated pixel size of 1.072 Å and total dose on the specimen level of 35 e−/Å2. Automatic data acquisition was setup using Leginon/Appion.73,74 A total of 618 movies were collected for the PP7-PP7-WT construct, 776 movies for the PP7-a-loop-PP7, 181 movies for the PP7-PP7-ZZ construct, and 807 movies for ZZ-PP7.
Data Processing.
Raw movies were frame-aligned and dose-weighted using MotionCor2,75 and the contrast transfer function was estimated using CTFFIND4.76 An initial subset of ~1000 particles was picked using DoG picker77 and classified in 2D using XMIPP78 as implemented in Appion. Two 2D classes were selected as templates for automated particle picking using FindEM.79 Extracted particles were classified in 2D in RELION3,80 and selected classes were imported into CryoSPARC,81 where they were further classified in 2D. Selected classes were then used for ab initio model generation, which in turn was used as a starting model for the homogeneous refinement with icosahedral symmetry applied.
Model Building.
PP7-PP7 dimer was initially built in Coot using two copies of monomeric PP7 CP.82 The AYGG linker loop was built in manually on the side that corresponded to the linker density in PP7-PP7-ZZ structure (Figure 6). The resulting dimer was refined in PHENIX using real-space refine. The refined dimer was used to build the T4 cage, which was subjected to a final round of real-space refinement in PHENIX.83 The molecular model of the PP7-PP7-WT capsid was deposited to PDB with the accession code 6N4V. Cryo-EM maps were deposited to EMDB with the following accession codes:
PP7-PP7-WT: EMD-0344.
ZZ-PP7: EMD-0351.
PP7-PP7-ZZ: EMD-0352.
PP7-a-loop-PP7: EMD-0353.
PP7-a-loop-PP7–150-loop: EMD-0354.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a research partnership between Children’s Healthcare of Atlanta and the Georgia Institute of Technology, as well as the Shurl and Kay Curci Foundation. Structural analyses were performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247), NIH (GM103310, OD019994), and NYSTAR.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.8b09683.
Characterization data (TEM, mass spectrometry, dynamic light scattering) for all particles reported here, complete data for thermal stability analysis, plasmid designs, and representative images for the reconstruction of the ZZ-PP7 structure as an illustrative case (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Chroboczek J; Szurgot I; Szolajska E Virus-like Particles as Vaccine. Acta Biochim. Polym 2014, 61, 531–539. [PubMed] [Google Scholar]
- (2).Kushnir N; Streatfield SJ; Yusibov V Virus-like Particles as a Highly Efficient Vaccine Platform: Diversity of Targets and Production Systems and Advances in Clinical Development. Vaccine 2012, 31, 58–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ma Y; Nolte RJ; Cornelissen JJ Virus-based Nanocarriers for Drug Delivery. Adv. Drug Delivery Rev 2012, 64, 811–825. [DOI] [PubMed] [Google Scholar]
- (4).Cardinale D; Carette N; Michon T Virus Scaffolds as Enzyme Nano-carriers. Trends Biotechnol. 2012, 30, 369–376. [DOI] [PubMed] [Google Scholar]
- (5).Liu J; Dai S; Wang M; Hu Z; Wang H; Deng F Virus like Particle-based Vaccines Against Emerging Infectious Disease Viruses. Virol. Sin 2016, 31, 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Roldao A; Mellado MC; Castilho LR; Carrondo MJ; Alves PM Virus-like Particles in Vaccine Development. Expert Rev. Vaccines 2010, 9, 1149–1176. [DOI] [PubMed] [Google Scholar]
- (7).Naskalska A; Pyrc K Virus Like Particles as Immunogens and Universal Nanocarriers. Pol. J. Microbiol 2015, 64, 3–13. [PubMed] [Google Scholar]
- (8).Yan D; Wei YQ; Guo HC; Sun SQ The Application of Virus-like Particles as Vaccines and Biological Vehicles. Appl. Microbiol. Biotechnol 2015, 99, 10415–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Schwarz B; Douglas T Development of Virus-like Particles for Diagnostic and Prophylactic Biomedical Applications. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol 2015, 7, 722–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Mohsen MO; Zha L; Cabral-Miranda G; Bachmann MF Major Findings and Recent Advances in Virus-like Particle (VLP)-based Vaccines. Semin. Immunol 2017, 34, 123–132. [DOI] [PubMed] [Google Scholar]
- (11).Mohsen MO; Gomes AC; Vogel M; Bachmann MF Interaction of Viral Capsid-Derived Virus–Like Particles (VLPs) with the Innate Immune System. Vaccines 2018, 6, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Smith MT; Hawes AK; Bundy BC Reengineering Viruses and Virus-like Particles through Chemical Functionalization Strategies. Curr. Opin. Biotechnol 2013, 24, 620–626. [DOI] [PubMed] [Google Scholar]
- (13).Reddington SC; Howarth M Secrets of a Covalent Interaction for Biomaterials and Biotechnology: SpyTag and SpyCatcher. Curr. Opin. Chem. Biol 2015, 29, 94–99. [DOI] [PubMed] [Google Scholar]
- (14).Brune KD; Leneghan DB; Brian IJ; Ishizuka AS; Bachmann MF; Draper SJ; Biswas S; Howarth M Plug-and-Display: Decoration of Virus-Like Particles via Isopeptide Bonds for Modular Immunization. Sci. Rep 2016, 6, 19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ilangovan U; Ton-That H; Iwahara J; Schneewind O; Clubb RT Structure of Sortase, the Transpeptidase that Anchors Proteins to the Cell Wall of Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 6056–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Patterson D; Schwarz B; Avera J; Western B; Hicks M; Krugler P; Terra M; Uchida M; McCoy K; Douglas T Sortase-Mediated Ligation as a Modular Approach for the Covalent Attachment of Proteins to the Exterior of the Bacteriophage P22 Virus-like Particle. Bioconjugate Chem 2017, 28, 2114–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Tang S; Xuan B; Ye X; Huang Z; Qian Z A Modular Vaccine Development Platform Based on Sortase-Mediated Site-Specific Tagging of Antigens onto Virus-Like Particles. Sci. Rep 2016, 6, 25741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Strable E; Prasuhn DE Jr.; Udit AK; Brown S; Link AJ; Ngo JT; Lander G; Quispe J; Potter CS; Carragher B; Tirrell DA; Finn MG Unnatural Amino Acid Incorporation into Virus-like Particles. Bioconjugate Chem. 2008, 19, 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Patel KG; Swartz JR Surface Functionalization of Virus-like Particles by Direct Conjugation using Azide-Alkyne Click Chemistry. Bioconjugate Chem. 2011, 22, 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Stephanopoulos N; Tong GJ; Hsiao SC; Francis MB Dual-surface Modified Virus Capsids for Targeted Delivery of Photodynamic Agents to Cancer Cells. ACS Nano 2010, 4, 6014–6020. [DOI] [PubMed] [Google Scholar]
- (21).Hassani-Mehraban A; Creutzburg S; van Heereveld L; Kormelink R Feasibility of Cowpea Chlorotic Mottle Virus-like Particles as Scaffold for Epitope Presentations. BMC Biotechnol. 2015, 15, 80–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Barassi C; Soprana E; Pastori C; Longhi R; Buratti E; Lillo F; Marenzi C; Lazzarin A; Siccardi AG; Lopalco L Induction of Murine mMucosal CCR5-Reactive Antibodies as an Anti-human Immunodeficiency Virus Strategy. J. Virol 2005, 79, 6848–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Caldeira JC; Peabody DS Thermal Stability of RNA Phage Virus-like Particles Displaying Foreign Peptides. J. Nanobiotechnol 2011, 9, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Sun Y; Sun Y; Zhao R Establishment of MicroRNA Delivery System by PP7 Bacteriophage-like Particles Carrying Cell-Penetrating Peptide. J. Biosci. Bioeng 2017, 124, 242–249. [DOI] [PubMed] [Google Scholar]
- (25).Lino CA; Caldeira JC; Peabody DS Display of Single-Chain Variable Fragments on Bacteriophage MS2 Virus-like Particles. J. Nanobiotechnol 2017, 15, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Frietze KM; Roden RB; Lee JH; Shi Y; Peabody DS; Chackerian B Identification of Anti-CA125 Antibody Responses in Ovarian Cancer Patients by a Novel Deep Sequence-Coupled Biopanning Platform. Cancer Immunol. Res 2016, 4, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Crossey E; Frietze K; Narum DL; Peabody DS; Chackerian B Identification of an Immunogenic Mimic of a Conserved Epitope on the Plasmodium falciparum Blood Stage Antigen AMA1 Using Virus-Like Particle (VLP) Peptide Display. PLoS One 2015, 10, No. e0132560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Chackerian B; Caldeira J. d C.; Peabody J; Peabody DS Peptide Epitope Identification by Affinity Selection on Bacteriophage MS2 Virus-like Particles. J. Mol. Biol 2011, 409, 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Brown SD; Fiedler JD; Finn MG Assembly of Hybrid Bacteriophage Qβ Virus-Like Particles. Biochemistry 2009, 48, 11155–11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pokorski JK; Hovlid ML; Finn MG Cell Targeting with Hybrid Qβ Virus-Like Particles Displaying Epidermal Growth Factor. ChemBioChem 2011, 12, 2441–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Olsthoorn RC; Garde G; Dayhuff T; Atkins JF; Van Duin J Nucleotide Sequence of a Single-Stranded RNA Phage from Pseudomonas aeruginosa: Kinship to Coliphages and Conservation of Regulatory RNA Structures. Virology 1995, 206, 611–625. [DOI] [PubMed] [Google Scholar]
- (32).Caldeira JC; Peabody DS Stability and Assembly in vitro of Bacteriophage PP7 Virus-like Particles. J. Nanobiotechnol 2007, 5, 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Tars K; Fridborg K; Bundule M; Liljas L The Three-Dimensional Structure of Bacteriophage PP7 from Pseudomonas aeruginosa at 3.7-Å Resolution. Virology 2000, 272, 331–337. [DOI] [PubMed] [Google Scholar]
- (34).Tars K; Fridborg K; Bundule M; Liljas L Structure Determination of Bacteriophage PP7 from Pseudomonas aeruginosa: from Poor Data to a Good Map. Acta Crystallogr., Sect. D: Biol. Crystallogr 2000, 56, 398–405. [DOI] [PubMed] [Google Scholar]
- (35).Sievers F; Wilm A; Dineen D; Gibson TJ; Karplus K; Li W; Lopez R; McWilliam H; Remmert M; Soding J; Thompson JD; Higgins DG Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments using Clustal Omega. Mol. Syst. Biol 2011, 7, 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Mottershead DG; Alfthan K; Ojala K; Takkinen K; Oker-Blom C Baculoviral Display of Functional scFv and Synthetic IgG-Binding Domains. Biochem. Biophys. Res. Commun 2000, 275, 84–90. [DOI] [PubMed] [Google Scholar]
- (37).Nilsson B; Moks T; Jansson B; Abrahmsen L; Elmblad A; Holmgren E; Henrichson C; Jones TA; Uhlen M A Synthetic IgG-Binding Domain Based on Staphylococcal Protein A. Protein Eng., Des. Sel 1987, 1, 107–113. [DOI] [PubMed] [Google Scholar]
- (38).Braisted AC; Wells JA Minimizing a Binding Domain from Protein A. Proc. Natl. Acad. Sci. U. S. A 1996, 93, 5688–5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Chen C; Huang Q-L; Jiang S-H; Pan X; Hua Z-C Immobilized Protein ZZ, an Affinity Tool for Immunoglobulin Isolation and Immunological Experimentation. Biotechnol. Appl. Biochem 2006, 45, 87–92. [DOI] [PubMed] [Google Scholar]
- (40).Fiedler JL; Fishman MR; Brown SD; Lau J; Finn MG Multifunctional Enzyme Packaging and Catalysis in the Qβ Protein Nanoparticle. Biomacromolecules 2018, 19, 3945–3957. [DOI] [PubMed] [Google Scholar]
- (41).Skamel C; Aller SG; Bopda Waffo A In vitro Evolution and Affinity-Maturation with Coliphage Qβ Display. PLoS One 2014, 9, No. e113069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Kozlovska TM; Cielens I; Vasiljeva I; Strelnikova A; Kazaks A; Dislers A; Dreilina D; Ose V; Gusars I; Pumpens P RNA Phage Q beta Coat Protein as a Carrier for Foreign Epitopes. Intervirology 2004, 39, 9–15. [DOI] [PubMed] [Google Scholar]
- (43).Vasiljeva I; Kozlovska T; Cielens I; Strelnikova A; Kazaks A; Ose V; Pumpens P Mosaic Qbeta Coats as a New Presentation Model. FEBS Lett. 1998, 431, 7–11. [DOI] [PubMed] [Google Scholar]
- (44).Pumpens P; Renhofa R; Dishlers A; Kozlovska T; Ose V; Pushko P; Tars K; Grens E; Bachmann MF The True Story and Advantages of RNA Phage Capsids as Nanotools. Intervirology 2016, 59, 74–110. [DOI] [PubMed] [Google Scholar]
- (45).Voronkova T; Grosch A; Kazaks A; Ose V; Skrastina D; Sasnauskas K; Jandrig B; Arnold W; Scherneck S; Pumpens P; Ulrich R Chimeric Bacteriophage fr Virus-like Particles Harboring the Immunodominant C-terminal Region of Hamster Polyomavirus VP1 Induce a Strong VP1-Specific Antibody Response in Rabbits and Mice. Viral Immunol. 2002, 15, 627–643. [DOI] [PubMed] [Google Scholar]
- (46).Crossey E; Amar MJ; Sampson M; Peabody J; Schiller JT; Chackerian B; Remaley AT A Cholesterol-Lowering VLP Vaccine that Targets PCSK9. Vaccine 2015, 33, 5747–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Tyler M; Tumban E; Peabody DS; Chackerian B The Use of Hybrid Virus-like Particles to Enhance the Immunogenicity of a Broadly Protective HPV Vaccine. Biotechnol. Bioeng 2014, 111, 2398–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Tissot AC; Renhofa R; Schmitz N; Cielens I; Meijerink E; Ose V; Jennings GT; Saudan P; Pumpens P; Bachmann MF Versatile Virus-like Particle Carrier for Epitope Based Vaccines. PLoS One 2010, 5, No. e9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Cielens I; Jackevica L; Strods A; Kazaks A; Ose V; Bogans J; Pumpens P; Renhofa R Mosaic RNA phage VLPs Carrying Domain III of the West Nile Virus E Protein. Mol. Biotechnol 2014, 56, 459–469. [DOI] [PubMed] [Google Scholar]
- (50).Schmitz N; Beerli RR; Bauer M; Jegerlehner A; Dietmeier K; Maudrich M; Pumpens P; Saudan P; Bachmann MF Universal Vaccine Against Influenza Virus: Linking TLR Signaling to Anti-viral Protection. Eur. J. Immunol 2012, 42, 863–869. [DOI] [PubMed] [Google Scholar]
- (51).Van den Worm SHE; Koning RI; Warmenhoven HJ; Koerten HK; Van Duin J Cryo Electron Microscopy Reconstructions of the Leviviridae Unveil the Densest Icosahedral RNA Packing Possible. J. Mol. Biol 2006, 363, 858–865. [DOI] [PubMed] [Google Scholar]
- (52).Lee JH; Engler JA; Collawn JF; Moore BA Receptor Mediated Uptake of Peptides that Bind the Human Transferrin Receptor. Eur. J. Biochem 2001, 268, 2004–2012. [DOI] [PubMed] [Google Scholar]
- (53).Oh S; Kim BJ; Singh NP; Lai H; Sasaki T Synthesis and Anti-cancer Activity of Covalent CVonjugates of Artemisinin and a Transferrin-Receptor Targeting Peptide. Cancer Lett. 2009, 274, 33–39. [DOI] [PubMed] [Google Scholar]
- (54).Liu X; Tian P; Yu Y; Yao M; Cao X; Gu J Enhanced Antitumor Effect of EGF R-targeted p21WAF-1 and GM-CSF Gene Transfer in the Established Murine Hepatoma by Peritumoral Injection. Cancer Gene Ther. 2002, 9, 100–108. [DOI] [PubMed] [Google Scholar]
- (55).Tian P; Ren S; Ren C; Teng Q; Qu S; Yao M; Gu J A Novel Receptor-Targeted Gene Delivery System for Cancer Gene Therapy. Sci. China, Ser. C: Life Sci 1999, 42, 216–224. [DOI] [PubMed] [Google Scholar]
- (56).Liu X; Tian PK; Ju DW; Zhang MH; Yao M; Cao XT; Gu JR Systemic Genetic Transfer of p21WAF-1 and GM-CSF Utilizing of a Novel Oligopeptide-Based EGF Receptor Targeting Polyplex. Cancer Gene Ther. 2003, 10, 529–539. [DOI] [PubMed] [Google Scholar]
- (57).Studenovsky M; Pola R; Pechar M; Etrych T; Ulbrich K; Kovar L; Kabesova M; Rihova B Polymer Carriers for Anticancer Drugs Targeted to EGF Receptor. Macromol. Biosci 2012, 12, 1714–20. [DOI] [PubMed] [Google Scholar]
- (58).Mine Y; Zhang JW Comparative Studies on Antigenicity and Allergenicity of Native and Denatured Egg White Proteins. J. Agric. Food Chem 2002, 50, 2679–2683. [DOI] [PubMed] [Google Scholar]
- (59).Oyen D; Torres JL; Wille-Reece U; Ockenhouse CF; Emerling D; Glanville J; Volkmuth W; Flores-Garcia Y; Zavala F; Ward AB; King CR; Wilson IA Structural Basis for Antibody Recognition of the NANP Repeats in Plasmodium falciparum Circumsporozoite Protein. Proc. Natl. Acad. Sci. U. S. A 2017, 114, E10438–E10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Khan F; Porter M; Schwenk R; DeBot M; Saudan P; Dutta S Head-to-Head Comparison of Soluble vs. Qβ VLP Circumsporozoite Protein Vaccines Reveals Selective Enhancement of NANP Repeat Responses. PLoS One 2015, 10, No. e0142035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Rosenberg CS; Martin DL; Tarleton RL CD8+ T Cells Specific for Immunodominant Trans-sialidase Epitopes Contribute to Control of Trypanosoma cruzi Infection but Are Not Required for Resistance. J. Immunol 2010, 185, 560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Teixeira AA; de Vasconcelos V; Colli W; Alves MJ; Giordano RJ Trypanosoma cruzi Binds to Cytokeratin through Conserved Peptide Motifs Found in the Laminin-G-Like Domain of the gp85/Trans-sialidase Proteins. PLoS Neglected Trop. Dis 2015, 9, No. e0004099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Sharma K; Babu PV; Sasidhar P; Srinivas VK; Mohan VK; Krishna E Recombinant Human Epidermal Growth Factor Inclusion Body Solubilization and Refolding at Large Scale Using Expanded-bed Adsorption CVhromatography from Escherichia coli. Protein Expression Purif. 2008, 60, 7–14. [DOI] [PubMed] [Google Scholar]
- (64).Pokorski JK; Hovlid ML; Finn MG Cell Targeting with Hybrid Qβ Virus-like Particles Displaying Epidermal Growth Factor. ChemBioChem 2011, 12, 2441–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Caldeira JC; Medford A; Kines RC; Lino CA; Schiller JT; Chackerian B; Peabody DS Immunogenic Display of Diverse Peptides, Including a Broadly Cross-type Neutralizing Human Papillomavirus L2 Epitope, on Virus-like Particles of the RNA Bacteriophage PP7. Vaccine 2010, 28, 4384–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Jin J; Simmons G Inhibitory Antibodies Targeting Emerging Viruses: Advancements and Mechanisms. Clin. Vaccine Immunol 2016, 23, 535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Wang Q; Yang H; Liu X; Dai L; Ma T; Qi J; Wong G; Peng R; Liu S; Li J; Li S; Song J; Liu J; He J; Yuan H; Xiong Y; Liao Y; Li J; Yang J; Tong Z; Griffin BD; Bi Y; Liang M; Xu X; Qin C; Cheng G; Zhang X; Wang P; Qiu X; Kobinger G; Shi Y; Yan J; Gao GF Molecular Determinants of Human Neutralizing Antibodies Isolated from a Patient Infected with Zika Virus. Sci. Transl. Med 2016, 8, 369ra179. [DOI] [PubMed] [Google Scholar]
- (68).Dai L; Song J; Lu X; Deng YQ; Musyoki AM; Cheng H; Zhang Y; Yuan Y; Song H; Haywood J; Xiao H; Yan J; Shi Y; Qin CF; Qi J; Gao GF Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody. Cell Host Microbe 2016, 19, 696–704. [DOI] [PubMed] [Google Scholar]
- (69).Fiedler JD; Higginson C; Hovlid ML; Kislukhin AA; Castillejos A; Manzenrieder F; Campbell MG; Voss NR; Potter CS; Carragher B; Finn MG Engineered Mutations Change the Stability and Structure of a Virus-Like Particle. Biomacromolecules 2012, 13, 2339–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Caspar DL; Klug A Physical Principles in the Construction of Regular Viruses. Cold Spring Harbor Symp. Quant. Biol 1962, 27, 1–24. [DOI] [PubMed] [Google Scholar]
- (71).Fiedler JD; Brown SD; Lau J; Finn MG RNA-Directed Packaging of Enzymes within Virus-Like Particles. Angew. Chem., Int. Ed 2010, 49, 9648–9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Korkegian A; Black ME; Baker D; Stoddard BL Computational Thermostabilization of an Enzyme. Science 2005, 308, 857–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Suloway C; Pulokas J; Fellmann D; Cheng A; Guerra F; Quispe J; Stagg S; Potter CS; Carragher B Automated Molecular Microscopy: the New Leginon System. J. Struct. Biol 2005, 151, 41–60. [DOI] [PubMed] [Google Scholar]
- (74).Stagg SM; Lander GC; Quispe J; Voss NR; Cheng A; Bradlow H; Bradlow S; Carragher B; Potter CS A Test-bed for Optimizing High-Resolution Single Particle Reconstructions. J. Struct. Biol 2008, 163, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Zheng SQ; Palovcak E; Armache JP; Verba KA; Cheng Y; Agard DA MotionCor2: Anisotropic Correction of Beam-Induced Motion for Improved Cryo-Electron Microscopy. Nat. Methods 2017, 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Rohou A; Grigorieff N CTFFIND4: Fast and Accurate Defocus Estimation from Electron Micrographs. J. Struct. Biol 2015, 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Voss NR; Yoshioka CK; Radermacher M; Potter CS; Carragher B DoG Picker and TiltPicker: Software Tools to Facilitate Particle Selection in Single Particle Electron Microscopy. J. Struct. Biol 2009, 166, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Scheres SH; Valle M; Nunez R; Sorzano CO; Marabini R; Herman GT; Carazo JM Maximum-Likelihood Multi-Reference Refinement for Electron Microscopy Images. J. Mol. Biol 2005, 348, 139–149. [DOI] [PubMed] [Google Scholar]
- (79).Roseman AM FindEM–a Fast, Efficient Program for Automatic Selection of Particles from Electron Micrographs. J. Struct. Biol 2004, 145, 91–99. [DOI] [PubMed] [Google Scholar]
- (80).Zivanov J; Nakane T; Forsberg BO; Kimanius D; Hagen WJ; Lindahl E; Scheres SH New Tools for Automated High-Resolution Cryo-EM Structure Determination in RELION-3. eLife 2018, 7, No. e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Punjani A; Rubinstein JL; Fleet DJ; Brubaker MA cryoSPARC: Algorithms for Rapid Unsupervised Cryo-EM Structure Determination. Nat. Methods 2017, 14, 290–296. [DOI] [PubMed] [Google Scholar]
- (82).Emsley P; Lohkamp B; Scott WG; Cowtan K Features and Development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Adams PD; Afonine PV; Bunkoczi G; Chen VB; Davis IW; Echols N; Headd JJ; Hung LW; Kapral GJ; Grosse-Kunstleve RW; McCoy AJ; Moriarty NW; Oeffner R; Read RJ; Richardson DC; Richardson JS; Terwilliger TC; Zwart PH PHENIX: a Comprehensive Python-Based System for Macro-molecular Structure Solution. Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.