Abstract
Arsenic, in the trivalent form (AsIII), is a human co-carcinogen reported to enhance mutagenesis effects of other carcinogens such as UV radiation by inhibiting DNA repair. The zinc finger DNA repair protein Poly (ADP-ribose) polymerase 1 (PARP-1) is a sensitive target of AsIII and both reactive oxygen and nitrogen species (ROS/RNS) generated by AsIII contribute to PARP-1 inhibition. However, the mechanisms of ROS/RNS-mediated PARP inhibition and how AsIII-generated ROS/RNS may be interconnected are still unclear. In this study, we found AsIII exposure of normal human keratinocyte (HEKn) cells generated peroxynitrite through superoxide and nitric oxide production in an AsIII concentration dependent manner. Peroxynitrite inhibited PARP-1 activity and caused zinc loss from PARP-1 protein while scavenging peroxynitrite was protective of the impacts on PARP-1. We identified peroxynitrite was responsible for S-nitrosation on cysteine residues resulting in PARP-1 zinc finger conformational changes. Taken together, the evidence indicates AsIII generates peroxynitrite through superoxide and nitric oxide production, induces S-nitrosation on PARP-1, leading to zinc loss and activity inhibition of PARP-1, thus enhancing DNA damage caused by UV radiation. These findings highlight a role for peroxynitrite as a key molecule of ROS/RNS mediated DNA repair inhibition by AsIII which should inform the development of prevention and intervention strategies against AsIII co-carcinogenesis.
Keywords: arsenic, peroxynitrite, PARP-1, reactive oxygen species, reactive nitrogen species
Introduction
Arsenic is a well-known environmental hazard to promote lung, bladder, and skin cancer by acting as a co-carcinogen. Environmental arsenic exposure at low and non-cytotoxic concentrations enhances the mutagenic and carcinogenic effects of ultraviolet radiation (UVR) (1–3). The impact on DNA repair system of arsenic is suggested to be an important molecular mechanism (4,5). The DNA repair system is highly sensitive to arsenic exposure (2,6) largely because certain zinc finger DNA repair proteins, such as Poly (ADP-ribose) polymerase-1 (PARP-1), are molecular targets of arsenic (2,6,7). Trivalent arsenite (AsIII) selectively binds with C3H1 (3 × Cys and 1 × His) and C4 (4 × Cys) zinc fingers (7), causing the loss of zinc from the zinc finger domain of the protein. C3H1 and C4 configurations are a minority in the zinc finger protein family, and PARP-1 is a C3H1 zinc finger protein (8). Moreover, our research revealed AsIII interacts with the PARP-1 zinc finger by directly binding to 3 Cys residues (7). Conversely, AsIII exposure has been reported to generate reactive oxygen/nitrogen species (ROS/RNS) (9–11), which contribute to PARP-1 inhibition (11). Cys residues of PARP-1 zinc finger are highly sensitive to redox-related post-translational modifications (PTMs) (12,13). Binding of AsIII to PARP-1 zinc finger domain Cys residues leads to conformational change, zinc loss and dysfunction of PARP-1. Arsenic-induced PAPR-1 modifications can be a direct (arsenic binding) or indirect (redox modifications) function of arsenic exposure (11,12). These two mechanisms work together in that selective AsIII binding leads to selective PARP-1 zinc finger oxidation (12). Therefore, clarifying the mechanism of redox PTMs on PARP-1 by AsIII is critical for improved understanding of arsenic co-carcinogenesis.
Redox-dependent PTMs of PARP-1 are on redox-sensitive Cys residues of zinc finger motifs, due to AsIII-generated ROS/RNS. AsIII generates ROS and RNS (1,11), both of which contribute to zinc loss and protein dysfunction of PARP-1 (11). PARP-1 can be oxidized by AsIII-generated ROS (12). Furthermore, AsIII-generated nitric oxide (NO) leads to S-nitrosation on the PARP-1 zinc finger motif, which contributes to PARP-1 inhibition (13) and may lead to permanent disruption of zinc finger Cys residues (14,15). However, it is still unclear whether ROS and RNS mechanisms are interrelated. Moreover, the relationship between ROS/RNS with regard to PARP-1 inhibition and modification remain unclear, and the molecules involved in integrating ROS/RNS mechanisms are unknown. Addressing these questions is an essential step to gaining a more complete understanding of the redox mechanisms involved in DNA repair inhibition by AsIII exposure.
In this study, we provide evidence that AsIII-generated superoxide and nitric oxide produce peroxynitrite. Moreover, we demonstrate peroxynitrite contributes to activity inhibition, zinc loss, S-nitrosation, and conformational change of PARP-1. These observations not only demonstrate AsIII-induced ROS/RNS work together in disrupting PARP-1, but they also reveal ROS and RNS are both required for AsIII inhibition of PARP-1. These findings shed light on the molecular mechanisms underlying arsenic-mediated DNA repair inhibition and identify a key molecule, peroxynitrite, as a possible target for prevention and intervention of AsIII co-carcinogenesis.
Materials and Methods
Cell culture and chemicals
Normal human neonatal epidermal keratinocytes (HEKn) were purchased from Lifeline Cell Technologies (Oceanside, CA). HEKn cells were cultured in DermaLife K media (Lifeline Cell Tech) and maintained according to manufacturer’s instructions. Peroxynitrite fluorescent label HKGreen-4 (16) was a kind gift from Dr. Dan Yang and Dr. Jiangang Shen (University of Hong Kong). Sodium arsenite (AsIII, purity > 99%) was purchased from Fluka Chemie. SIN-1 and carboxy-PTIO (c-PTIO) were from Cayman Chemical. Other chemicals such as MnTMPyP and Uric Acid (UA)were from Sigma-Aldrich unless otherwise indicated.
Peroxynitrite measurement
HEKn cells were treated as described in the results section and corresponding figure legends. HKGreen-4 was added to cell culture medium at 1:1000 and incubated at 37 °C for 30 min. After washing twice with Phosphate-buffered saline (PBS, Sigma-Aldrich), cells were imaged in the colorless DermaLife K media using fluorescence microscope (Zeiss) for green fluorescence. The peroxynitrite production was represented by mean intensity of green fluorescence using Image-Pro Plus Software. 6 independent wells in each group were analyzed to generate statistical summary.
PARP activity assay
HEKn cells were treated as described in figure legends and the results section. PARP activity assay was performed as described (11,13). Briefly, HEKn cells were collected and lysed using ice-cold RIPA buffer (Thermo Fisher Pierce). Protein concentrations in cell extract were determined using BCA protein detection kit (Thermo Fisher Pierce). PARP activity was then measured using the HT Colorimetric PARP/Apoptosis Assay kit (Trevigen, Gaithersburg, MD) following the manufacturer’s instructions. The relative PARP activation value was presented using the absorbance at 450 nm on a SpectraMax 450 plate reader.
PARP-1 zinc content measurement
After treatments of HEKn cells as described in the results section, PARP-1 protein was isolated by immunoprecipitation and zinc content was measured with the 4-(2-pyridylazo) resorcinol (PAR) colorimetric assay as previously described (7,13). Briefly, PARP-1 protein was purified using Dynabeads Protein A Immunoprecipitation Kit (Thermo Fisher Scientific) from cell extracts according to manufacturer’s instructions. PARP-1 was eluted from the beads, and pH was adjusted to greater than 7 using the neutralizing buffer provided in the immunoprecipitation kit. Purified PARP-1 was then incubated with 10 mM hydrogen peroxide at 4 °C for 4 h to release zinc. After adding 100 μM PAR, absorbance at 493 nm was recorded and used to determine relative zinc content in PARP-1 protein.
Protein S-nitrosation analysis
S-nitrosated proteome from cell extract was purified using a modified “biotin-switch” assay (13,17,18). After corresponding treatments on HEKn cells, the following steps were performed in the dark. Briefly, cells were harvested in RIPA cell lysis buffer (Thermo Scientific), sonicated, and centrifuged at 13,500 rpm for 15 min at 4 °C to remove cellular debris. 10 mM N-ethylmaleimide (NEM) was added and incubated for 1 h at room temperature to block all free Cys residues. Excess NEM in samples was removed by ice-cold acetone precipitation repeated three times for 20 min each. 1 mM ascorbic acid (Vc, Sigma-Aldrich) and 500 μM biotin-NEM (Thermo Scientific Pierce) were then added and incubated at room temperature for 1 h in order to reduce and label S-nitrosated Cys residues. Labeled S-nitrosated proteome was purified using streptavidin agarose beads (Thermo Scientific Pierce). Then the samples were resolved by electrophoresis through 10% SDS-polyacrylamide gels. Proteins were transferred to nitrocellulose membranes (Bio-Rad) and probed with PARP antibody (Cell Signaling Inc. #9532). The membranes were developed using the SuperSignal chemiluminescent detection system (Thermo Scientific Pierce). Quantification of immunoblotting results was performed using software ImageJ. Statistical summaries were obtained from 3 independent samples.
Intrinsic Fluorescence Measurement
The peptide representing human PARP-1 zinc finger 1 was custom-synthesized (Genemed Synthesis Inc. San Antonio TX). The sequence of this 29-AA (amino acid) peptide is GRASCKKCSESIPKDKVPHWYHFSCFWKV. The purity was above 95% as assessed by HPLC. SIN-1 at corresponding concentrations was mixed with 100 μM peptide in 10 mM tris, pH 7.4. After 30 min incubation at room temperature, fluorescent spectra were collected using a SpectraMax M2 Multi-Mode Microplate Reader (Molecular Devices, Sunnyvale, CA) at room temperature. Emission were scanned between 300 and 400 nm with the excitation wavelength at 280 nm.
UV radiation and 8-OHdG measurement
HEKn cells were treated as described in results and figure legends for 24 h. Cells were then exposed to 1 kJ/m2 solar-simulated UV radiation (ss-UVR) using an Oriel 300 W Solar Ultraviolet Simulator (Newport Corporation, Irvine, CA). Immunocytochemistry detection of 8-OHdG was then performed as described previously (19). After fixing, blocking, labeling, washing and drying, cells on coverslips were imaged using an Olympus BH2-RFCA fluorescence microscope (Melville, NY) and Omegafire digital camera with MagnaFire software. The 8-OHdG production was represented by relative intensity of staining, which was quantified by measuring mean intensity of fluorescence using the Image-Pro Plus software (Media Cybernetics, Inc., Silver Spring, MD). 20 to 30 randomly selected cells on each slide were measured and 6 independent slides in each group were analyzed.
Statistical analysis
At least three independent experiments were conducted. Differences between two groups were evaluated by Student’s t-test. Differences across three or more groups were evaluated using a one-way analysis of variance (ANOVA) followed by a Tukey’s multiple comparison post hoc test. p < 0.05 was considered significantly different. All data were analyzed by GraphPad Prism 6.
Results
AsIII generates peroxynitrite in HEKn cells
Our previous work revealed ROS/RNS generated by AsIII contributes to DNA repair inhibition (11–13). To test whether peroxynitrite, which is produced by the reaction of superoxide with nitric oxide, is generated by AsIII treatment, HEKn cells were treated with sodium arsenite (AsIII) for 24 h. Previously, we’ve shown AsIII significantly induces iNOS expression and nitric oxide production in HaCat cells (1) and HEKn cells 24 hours following treatment (13). To visualize AsIII-induced peroxynitrite production, HEKn cells were treated with 0.1 to 2 μM AsIII and peroxynitrite was fluorescently labeled using a specific probe HKGreen-4 (16). The green fluorescence in microscopic images indicated peroxynitrite production (Fig. 1A). The statistical analysis of the fluorescent intensities showed peroxynitrite was generated in an AsIII concentration dependent manner (Fig. 1B). Peroxynitrite levels were significantly elevated at or above 0.5 μM of AsIII.
Figure 1. Peroxynitrite induced by AsIII in HEKn cells.
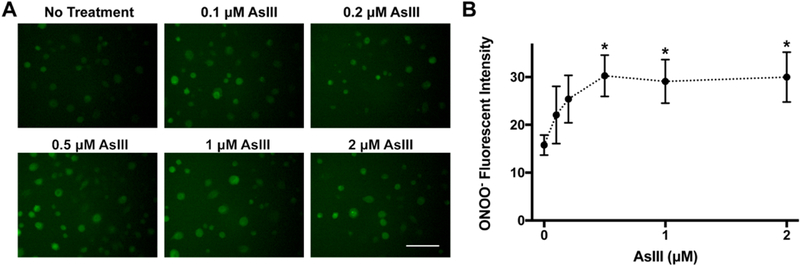
A) HEKn cells were treated with various concentrations of AsIII, as labeled on images, for 24 h. Peroxynitrite was detected by probe HKGreen-4 under fluorescent microscopy. Scale bar = 100 μm. B) Statistical summary of green fluorescence indicating AsIII concentration dependence of peroxynitrite production in HEKn cells. At concentrations at or above 0.5 μM AsIII, peroxynitrite level was significantly higher than no-treatment group. Bar chart shows mean ± SD. *, p < 0.05, n = 6.
We previously reported AsIII generates both superoxide (12) and nitric oxide (NO) species (1,13). To test whether AsIII-generated peroxynitrite production is dependent on superoxide and NO, we treated HEKn cells with superoxide or nitric oxide scavengers and quantified the amount of peroxynitrite generated using the HKGreen-4 fluorescent labeling protocol. Cells were treated with 1 μM AsIII, and co-treated with either 100 μM superoxide scavenger MnTMPyP (manganese(III) tetrakis(1-methyl-4-pyridyl)porphyrin), 100 μM nitric oxide scavenger c-PTIO, or 100 μM peroxynitrite scavenger Uric Acid (UA) for 24 h. Statistical analysis of fluorescent microscopy images revealed a decrease in peroxynitrite generation following co-treatment with superoxide or nitric oxide scavengers compared to treatment with AsIII alone (Fig. 2A&B). Moreover, peroxynitrite production was reduced in the presence of UA (Fig. 2B). In summary, these results illustrate, in HEKn cells, AsIII induces peroxynitrite production in a dose-dependent manner and peroxynitrite production is a downstream effect of superoxide and nitric oxide generation.
Figure 2. Peroxynitrite production by AsIII in a superoxide and nitric oxide dependent manner in HEKn cells.
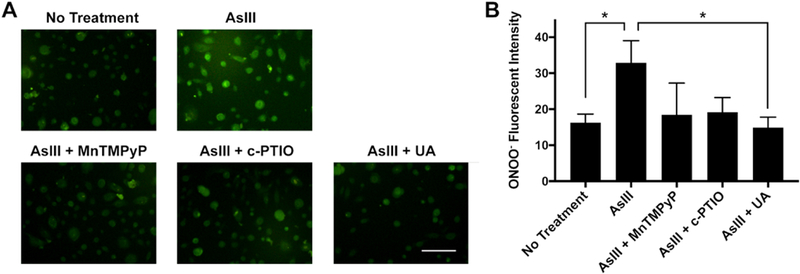
A) Fluorescent microscopic images of peroxynitrite by HKGreen-4 labeling. HEKn cells were treated with 1 μM AsIII, or co-treated with 100 μM MnTMPyP (SOD mimic), 100 μM c-PTIO (nitric oxide scavenger), 100 μM Uric Acid (UA, peroxynitrite scavenger) for 24 h. Scale bar = 100 μm. B) Statistical summary of fluorescent intensity. AsIII significantly induced peroxynitrite production, which was reduced by scavenging superoxide, nitric oxide, or peroxynitrite. Bar chart shows mean ± SD. *, p < 0.05, n = 6.
Peroxynitrite contributes to PARP-1 inhibition and zinc loss by AsIII
We’ve previously reported AsIII inhibits DNA repair by targeting zinc finger DNA repair proteins such as PARP-1 (6) through ROS/RNS generation (11). In the present study, we demonstrate ROS/RNS generated by AsIII upregulates peroxynitrite production. We then asked whether AsIII-generated peroxynitrite contributes to PARP-1 activity inhibition. To this end, we treated HEKn cells with 10 μM SIN-1 (3-Morpholino-sydnonimine, peroxynitrite generator), 1 μM AsIII, or co-treatment of AsIII with 100 μM Uric Acid (UA, peroxynitrite scavenger) for 24 h and measured PARP-1 activity using a colorimetric PARP-1 activity assay (Fig. 3A). SIN-1 treatment inhibited PARP-1 activity, and peroxynitrite scavenging by UA significantly restored PARP-1 inhibition by AsIII (Fig. 3A). Previously, we’ve reported zinc loss from PARP-1 depends on ROS/RNS inhibition of PARP-1 (11). Therefore, we tested whether interruption of PARP-1 activity by peroxynitrite is an effect of zinc finger disruption and zinc loss. After treating HEKn cells with 10 μM SIN-1, 1 μM AsIII, or co-treatment of AsIII with 100 μM UA for 24 h, we immunoprecipitated PARP-1 and analyzed zinc content using a 4-(2-pyridylazo) resorcinol (PAR) colorimetric assay. Colorimetric analysis revealed the peroxynitrite generator SIN-1 induces PARP-1 zinc loss and treatment with the peroxynitrite scavenger UA restored PARP-1 zinc content (Fig. 3B). These results confirmed AsIII-generated peroxynitrite results in zinc loss and functional inhibition of PARP-1.
Figure 3. Peroxynitrite contributes to AsIII-induced dysfunction and zinc loss from PARP-1.
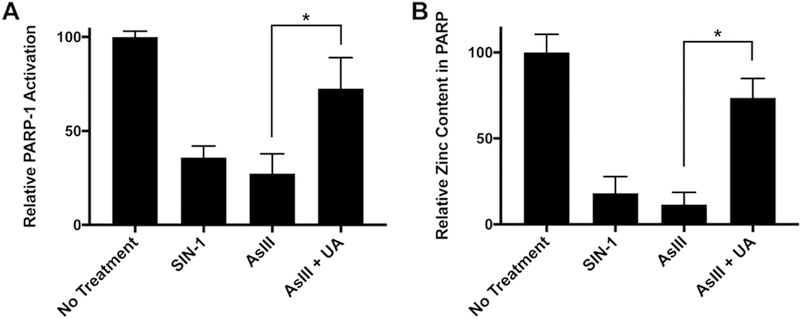
A) PARP-1 activity restoration by scavenging peroxynitrite. HEKn cells were treated by 10 μM SIN-1 (peroxynitrite donor), 1 μM AsIII, or co-treated with 100 μM Uric Acid for 24 h. PARP-1 activity was then measured as described in the methods section. Scavenging peroxynitrite significantly rescued AsIII-inhibited PARP-1 activity. Bar charts show mean ± SD. *, p < 0.05, n = 3. B) Scavenging peroxynitrite restored AsIII-induced zinc loss on PARP-1 protein. HEKn cells were treated as described in panel A. Zinc content in PARP-1 protein was then measured as described in the methods section. Scavenging peroxynitrite restored zinc loss induced by AsIII. *, p < 0.05, n = 3.
Peroxynitrite caused S-nitrosation and conformational change of the PARP-1 zinc finger
ROS/RNS is reported to cause PARP-1 zinc deficiency through a redox-dependent post-translational modification of Cys residues (12,13). However, direct S-nitrosation of Cys residues by nitric oxide is slow and inefficient (14) and zinc protects zinc finger motifs from redox-modifications (12). Therefore, we asked whether peroxynitrite, rather than nitric oxide, is responsible for S-nitrosation modification to the PARP-1 zinc finger domain. To this end, we utilized the biotin-switch method to test S-nitrosation of PARP-1 (13,17,18,20). HEKn cells were treated with 10 μM SIN-1, 1 μM AsIII, or co-treated with 1 μM AsIII and 100 μM UA for 24 h. Densitometric analysis of PARP-1 S-nitrosation using the modified biotin-switch assay paired with immunoblotting (Fig. 4A) revealed a 10-fold increase in S-nitrosation of PARP-1 by SIN-1 or AsIII, while scavenging of peroxynitrite by UA decreased PARP-1 S-nitrosation by approximately 20% (Fig. 4B). This data indicates AsIII-induced S-nitrosation of PARP-1 is indeed a result of peroxynitrite production. Our data indicates peroxynitrite is a product of superoxide and nitric oxide generation, and as such is responsible for nitrosative modification of PARP-1 and subsequent zinc loss and protein dysfunction. These findings suggest that AsIII-induced ROS/RNS generation work in concert through peroxynitrite to prevent DNA repair by inhibiting PARP activity.
Figure 4. AsIII-induced PARP-1 S-nitrosation through peroxynitrite production.
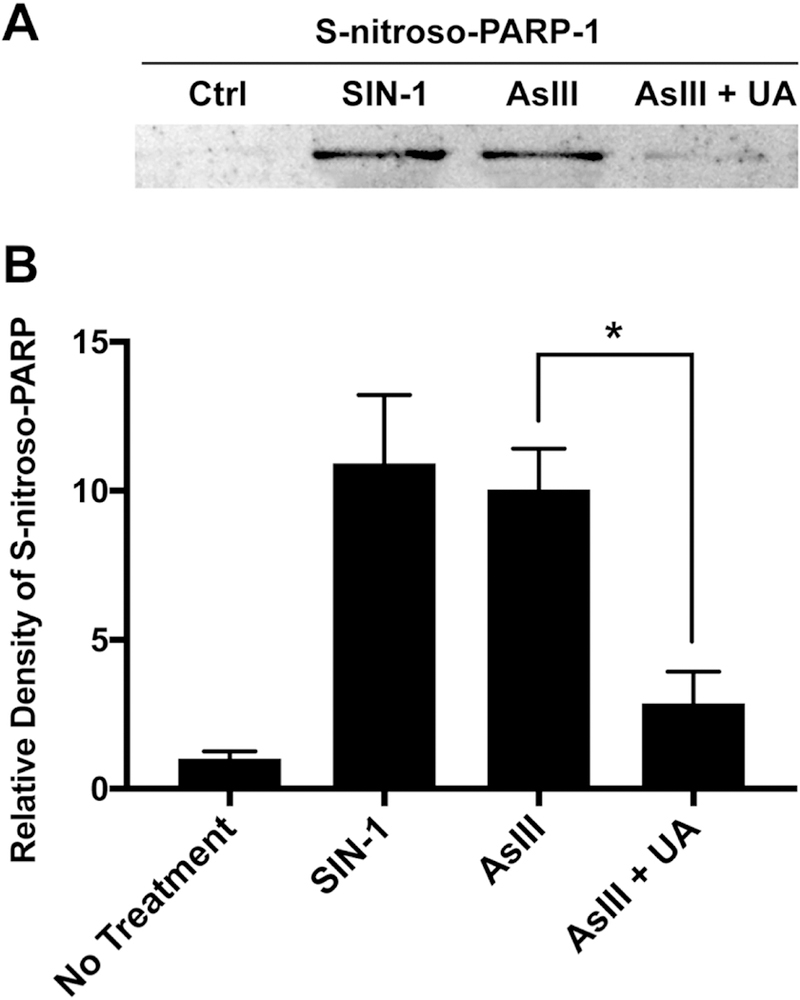
A) AsIII-induced PARP-1 S-nitrosation in a peroxynitrite-dependent manner. HEKn cells were treated for 24 h with 10 μM SIN-1 (peroxynitrite donor), 1 μM AsIII, or co-treated with 100 μM Uric Acid. PARP-1 S-nitrosation was measured by modified biotin-switch assay and immunoblotting. B) Densitometric analysis of immunoblotting for PARP-1 S-nitrosation. SIN-1 and AsIII induced PARP-1 S-nitrosation. Scavenging peroxynitrite significantly reduced PARP-1 S-nitrosation. Bar chart shows mean ± SD. *, p < 0.05, n = 3.
The zinc finger motifs on PARP-1 are responsible for DNA recognition and binding (8,11,21). Therefore, maintaining correct zinc finger conformation is critical for PARP-1 activity. To test whether peroxynitrite causes conformational changes to the PARP-1 zinc finger domain as a mechanism of peroxynitrite inhibition of PARP-1 activity, we used SIN-1 as a source of peroxynitrite and performed fluorescence analysis investigating changes of PARP-1 zinc finger tertiary structure. A synthesized peptide representing the PARP-1 zinc finger was treated with zinc chloride or SIN-1. Upon addition of zinc, PARP-1 zinc finger fluorescence was increased, while SIN-1 treatment resulted in decreased fluorescence (Fig. 5A). These results indicate zinc binding helps the zinc finger motif form a tighter tertiary structure, while SIN-1 treatment results in a less compact PARP-1 zinc finger conformation. Furthermore, the representative fluorescent intensity at 340 nm was decreased in a SIN-1 concentration dependent manner (Fig. 5B). These results suggest peroxynitrite induces conformational changes to the tertiary structure of the PARP-1 zinc finger in a concentration dependent manner.
Figure 5. Peroxynitrite induced conformational change on the PARP-1 zinc finger.
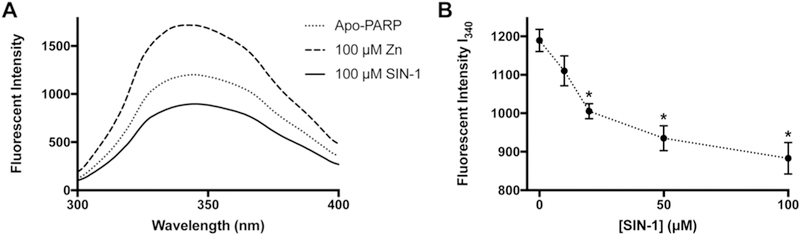
A) Fluorescent measurement of PARP-1 zinc finger peptide with zinc or SIN-1. 100 μM PARP-1 zinc finger peptide was incubated with equal concentration of zinc or SIN-1 for 30 min at room temperature in 10 mM Tris buffer, pH=7.4. Emission scan was made from 300 to 400 nm with excitation at 280 nm. B) SIN-1 concentration dependence of fluorescent intensity at 340 nm. 100 μM PARP-1 zinc finger peptide was incubated with 10, 20, 50, 100 μM SIN-1, Samples were prepared and measured as described for panel A. *, p < 0.05 in student’s t-test vs. control group ([SIN-1] = 0), n = 3.
Peroxynitrite contributes to AsIII-enhanced DNA damage by UV
Finally, in order to assess the functional impact of AsIII-generated peroxynitrite on DNA repair, we tested whether AsIII-generated peroxynitrite contributes to AsIII-enhanced oxidative DNA damage by UV in HEKn cells. We measured the formation of 8-OHdG, a biomarker of oxidative DNA damage (22) through immunocytochemistry. HEKn cells were treated with 1 μM AsIII, or co-treated with 1 μM AsIII and the peroxynitrite scavenger UA for 24 h. Thereafter, cells were exposed to 1 kJ/m2 dose of solar-simulated ultraviolet radiation (ssUVR). Analysis of immunocytochemistry images (Fig. 6A) revealed 1 μM AsIII treatment had no significant effect on 8-OHdG production. Conversely, UVR increased 8-OHdG, and a combined exposure of AsIII and UVR markedly increased 8-OHdG production as compared to UVR alone. Importantly, the presence of UA significantly decreased 8-OHdG signal intensity, almost to the level of UVR alone. Statistical analysis of 8-OHdG production via immunocytochemistry confirmed AsIII enhances UV-induced oxidative DNA damage (Fig. 6B) and demonstrates peroxynitrite contributes to AsIII enhancement of UVR-induced oxidative DNA damage resulting from DNA repair inhibition.
Figure 6. Effects of AsIII-generated peroxynitrite on the level of 8-OHdG production by UVR.
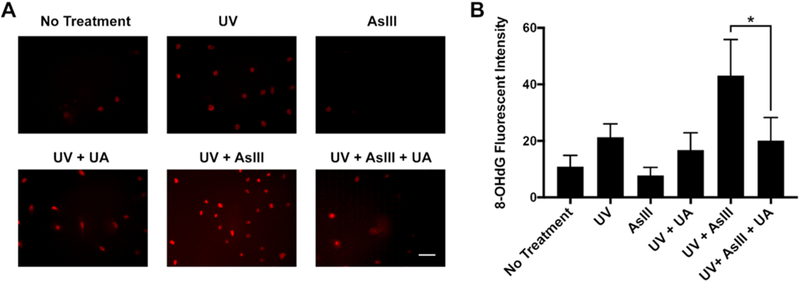
A) HEKn cells were treated by 1 μM AsIII, or co-treated with 100 μM Uric Acid for 24 h, then exposed to 1 kJ/m2 solar-simulated ultraviolet radiation (ssUVR). 8-OHdG production was measured by immunocytochemistry. Scale bar = 100 μM. B) Statistical summary of 8-OHdG production in immunocytochemistry. Bar chart shows mean ± SD. *, p < 0.05, n = 6.
Discussion
The data from the present study demonstrate AsIII treatment at environmentally relevant concentrations induces generation of peroxynitrite (Fig. 1), which is consistent with previous observations in other systems (9,23). Although it is well-established that superoxide and nitric oxide produce peroxynitrite rapidly (23–25), previously studies have not addressed peroxynitrite generation by AsIII in HEKn cells. In this study, we not only demonstrate AsIII is capable of generating peroxynitrite in HEKn cells, but also peroxynitrite generation is dependent on superoxide and nitric oxide production. Moreover, we substantiated these findings through peroxynitrite fluorescent labeling following treatment with a peroxynitrite scavenger. As expected, UA treatment of HEKn cells decreased AsIII-induced peroxynitrite production (Fig. 2).
Several studies report AsIII interferes DNA repair by inhibiting certain DNA repair proteins including PARP-1. In the present study, we assessed whether PARP-1 is a molecular target of AsIII generated peroxynitrite. Along this line, we analyzed PARP-1 activity in the presence of the peroxynitrite scavenger UA. We observed significant inhibition of PARP-1 activity in HEKn cells treated with 1 μM AsIII, consistent with previous reports (6,7). In addition, 10 μM SIN-1 (used as a peroxynitrite donor) decreased PARP-1 activity, to the equivalent of AsIII treatment. Co-treatment with AsIII and UA rescued PARP-1 activity (Fig 3A), indicating peroxynitrite was responsible for AsIII inhibition of PARP-1. The zinc ion in PARP-1 stabilizes zinc finger conformation and is thus responsible for maintaining the structure and function of PARP-1 (8). We found AsIII-induced PARP-1 zinc deficiency was restored by the reduction of peroxynitrite through the use of a peroxynitrite scavenger (Fig. 3B). These findings suggest AsIII-generated peroxynitrite targets the zinc finger domain of PARP-1, resulting in zinc loss and dysfunction.
Utilizing intrinsic fluorescent analysis, we showed peroxynitrite directly caused conformational change on PARP-1 zinc finger (Fig. 5). Not only did peroxynitrite loosen the tertiary structure of the PARP-1 zinc finger peptide, but also the effect was enhanced with increasing the concentration of SIN-1. These results explained the molecular mechanism of peroxynitrite inhibiting PARP-1 through S-nitrosation and conformational change. Furthermore, these results suggested the effect of nitric oxide induced by AsIII is through peroxynitrite.
Our results demonstrated peroxynitrite functionally contributes to DNA damage amplification by AsIII. The presence of UA significantly decreased UVR-generated 8-OHdG level in cells treated with AsIII (Fig. 6B), confirming peroxynitrite was generated by AsIII and indicating peroxynitrite was contributed to AsIII-enhanced DNA damage. Here we observed scavenging peroxynitrite reduced 8-OHdG production to the level similar to UVR alone, suggesting ROS and RNS may amplify DNA damage through peroxynitrite. It has been reported that peroxynitrite could directly cause oxidative DNA damage (26,27). Therefore, in combination with DNA repair inhibition, peroxynitrite should play an important role in ROS/RNS effect of enhancing DNA damage by AsIII.
There is evidence that both ROS and RNS are generated by AsIII and contribute to PARP-1 inhibition; however, there are some distinctions between ROS and RNS mechanisms. AsIII rapidly stimulates superoxide production in keratinocytes, and cysteine oxidation of PARP-1 is evident within 10–20 minutes and increases over time (28). The loss of zinc from the PARP-1 DNA binding domain corresponds closely to PARP-1 oxidation status (10). In contrast, AsIII-dependent iNOS expression and S-nitrosation of PARP-1 protein is detected by 18h after AsIII exposure (13). Co-treatment of AsIII with an iNOS inhibitor, nitric oxide scavenger, inhibitor of NADPH oxidase or a superoxide scavenger each significantly rescued the effect of AsIII on PARP-1 inhibition and zinc loss (9–11). These findings highlight the contributions of ROS and NOS to disruption of PARP-1 activity. In this study, co-treatment of AsIII with a superoxide scavenger or nitric oxide scavenger for 24 h each inhibited peroxynitrite production (Fig. 2). Peroxynitrite induced S-nitrosation on PARP-1 protein (Fig. 4), and scavenging peroxynitrite almost eliminated S-nitrosation on PARP-1, suggesting peroxynitrite is a key molecule to S-nitrosate PARP-1, and nitric oxide may impact PARP-1 zinc finger through peroxynitrite. These results are consistent with previous reports that peroxynitrite is an active intermediate for protein S-nitrosation (29,30).
PARP-1 sensitivity to AsIII may be due to AsIII generation of ROS and RNS leading to production of peroxynitrite with each reactive species able to modify PARP-1 zinc finger motifs. It is important to note that oxidative and nitrosative modifications have different chemical characteristics. Oxidative modifications, such as Cysteine sulfenic acid (-SOH), are rapidly formed and reversible (31,32). In contrast, Cysteine nitrosation is slowly and inefficiently formed by nitric oxide directly (15,29,33). In this study, we demonstrate ROS work together with RNS through peroxynitrite at an extended AsIII treatment time point (24 h). Rapid and sustained modifications to the PARP-1 zinc finger domain are likely to collectively be important to the observed inhibition of PARP-1 function by AsIII. Several important questions remain: 1) Is peroxynitrite responsible for an irreversible impact on PARP-1? 2) What are the time courses of ROS/RNS generation and various redox modifications? 3) What are the causal relationships among various species and modifications? Answering these questions could further clarify the precise mechanism of DNA repair inhibition by AsIII-generated ROS/RNS.
In summary, AsIII exposure generates both ROS and RNS, produces peroxynitrite as an active molecule, modifies PARP-1 zinc finger, leading to zinc loss, PARP-1 dysfunction and DNA repair inhibition (Fig. 7). These results demonstrate ROS and RNS mechanisms work synergistically in PARP-1 inhibition by AsIII. The findings not only deepen our understanding of the ROS/RNS mechanism of AsIII inhibition of DNA repair, but also suggest the potential of more effective preventions and interventions to AsIII co-carcinogenesis.
Figure 7.
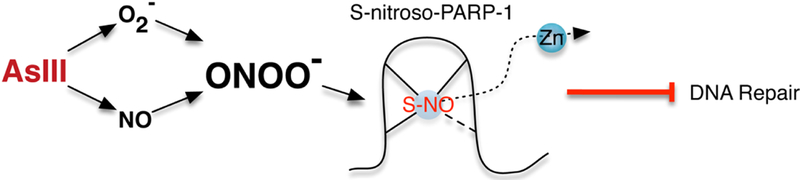
Schematic illustration of peroxynitrite mechanism on AsIII inhibition of DNA repair through PARP-1 inhibition.
Highlights.
AsIII generates peroxynitrite through superoxide and nitric oxide production.
Peroxynitrite contributes to PARP-1 inhibition by AsIII.
Peroxynitrite contributes to AsIII enhancement of UVR-induced DNA damage.
Acknowledgements
This work was supported by the grants from NIH/National Cancer Institute (R01CA182969) and National Institute for Environmental Health Sciences (R01ES021100).
Abbreviations
- ROS
reactive oxygen species
- RNS
reactive nitrogen species
- UVR
ultraviolet radiation
- PTM
post-translational modifications
- iNOS
inducible nitric oxide synthase
- PARP-1
poly (ADP-ribose) polymerase 1
- UA
uric acid
- c-PTIO
carboxy-PTIO
- PAR
4-(2-pyridylazo) resorcinol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Ding W, Hudson LG, Sun X, Feng C, and Liu KJ (2008) As(III) inhibits ultraviolet radiation-induced cyclobutane pyrimidine dimer repair via generation of nitric oxide in human keratinocytes. Free Radic Biol Med 45, 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin XJ, Hudson LG, Liu W, Timmins GS, and Liu KJ (2008) Low concentration of arsenite exacerbates UVR-induced DNA strand breaks by inhibiting PARP-1 activity. Toxicol Appl Pharmacol 232, 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossman TG, and Klein CB (2011) Genetic and epigenetic effects of environmental arsenicals. Metallomics 3, 1135–1141 [DOI] [PubMed] [Google Scholar]
- 4.Kitchin KT, and Wallace K (2008) The role of protein binding of trivalent arsenicals in arsenic carcinogenesis and toxicity. J Inorg Biochem 102, 532–539 [DOI] [PubMed] [Google Scholar]
- 5.Muenyi CS, Ljungman M, and States JC (2015) Arsenic Disruption of DNA Damage Responses-Potential Role in Carcinogenesis and Chemotherapy. Biomolecules 5, 2184–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding W, Liu W, Cooper KL, Qin XJ, de Souza Bergo PL, Hudson LG, and Liu KJ (2009) Inhibition of poly(ADP-ribose) polymerase-1 by arsenite interferes with repair of oxidative DNA damage. J Biol Chem 284, 6809–6817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X, Sun X, Cooper KL, Wang F, Liu KJ, and Hudson LG (2011) Arsenite interacts selectively with zinc finger proteins containing C3H1 or C4 motifs. J Biol Chem 286, 22855–22863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ali AAE, Timinszky G, Arribas-Bosacoma R, Kozlowski M, Hassa PO, Hassler M, Ladurner AG, Pearl LH, and Oliver AW (2012) The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol 19, 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen SC, and Chen WC (2008) Vascular leakage induced by exposure to arsenic via increased production of NO, hydroxyl radical and peroxynitrite. Microvasc Res 75, 373–380 [DOI] [PubMed] [Google Scholar]
- 10.Kumagai Y, and Pi J (2004) Molecular basis for arsenic-induced alteration in nitric oxide production and oxidative stress: implication of endothelial dysfunction. Toxicol Appl Pharmacol 198, 450–457 [DOI] [PubMed] [Google Scholar]
- 11.Wang F, Zhou X, Liu W, Sun X, Chen C, Hudson LG, and Jian Liu K (2013) Arsenite-induced ROS/RNS generation causes zinc loss and inhibits the activity of poly(ADP-ribose) polymerase-1. Free Radic Biol Med 61, 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou X, Cooper KL, Sun X, Liu KJ, and Hudson LG (2015) Selective Sensitization of Zinc Finger Protein Oxidation by Reactive Oxygen Species through Arsenic Binding. J Biol Chem 290, 18361–18369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Cooper KL, Huestis J, Xu H, Burchiel SW, Hudson LG, and Liu KJ (2016) S-nitrosation on zinc finger motif of PARP-1 as a mechanism of DNA repair inhibition by arsenite. Oncotarget 7, 80482–80492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hess DT, Matsumoto A, Kim SO, Marshall HE, and Stamler JS (2005) Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 15.Foster MW, Hess DT, and Stamler JS (2009) Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med 15, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng T, Wong NK, Chen X, Chan YK, Sun Z, Hu JJ, Shen J, El-Nezami H, and Yang D (2014) Molecular imaging of peroxynitrite with HKGreen-4 in live cells and tissues. J Am Chem Soc 136, 11728–11734 [DOI] [PubMed] [Google Scholar]
- 17.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, and Snyder SH (2001) Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol 3, 193–197 [DOI] [PubMed] [Google Scholar]
- 18.Huang B, and Chen C (2010) Detection of protein S-nitrosation using irreversible biotinylation procedures (IBP). Free Radic Biol Med 49, 447–456 [DOI] [PubMed] [Google Scholar]
- 19.Qin XJ, Hudson LG, Liu W, Ding W, Cooper KL, and Liu KJ (2008) Dual actions involved in arsenite-induced oxidative DNA damage. Chem Res Toxicol 21, 1806–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li R, and Kast J (2017) Biotin Switch Assays for Quantitation of Reversible Cysteine Oxidation. Methods Enzymol 585, 269–284 [DOI] [PubMed] [Google Scholar]
- 21.Buelow B, Uzunparmak B, Paddock M, and Scharenberg AM (2009) Structure/function analysis of PARP-1 in oxidative and nitrosative stress-induced monomeric ADPR formation. PLoS One 4, e6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dizdaroglu M (1992) Oxidative damage to DNA in mammalian chromatin. Mutat Res 275, 331–342 [DOI] [PubMed] [Google Scholar]
- 23.Bunderson M, Coffin JD, and Beall HD (2002) Arsenic induces peroxynitrite generation and cyclooxygenase-2 protein expression in aortic endothelial cells: possible role in atherosclerosis. Toxicol Appl Pharmacol 184, 11–18 [PubMed] [Google Scholar]
- 24.Radi R, Beckman JS, Bush KM, and Freeman BA (1991) Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J Biol Chem 266, 4244–4250 [PubMed] [Google Scholar]
- 25.Ding W, Hudson LG, and Liu KJ (2005) Inorganic arsenic compounds cause oxidative damage to DNA and protein by inducing ROS and RNS generation in human keratinocytes. Mol Cell Biochem 279, 105–112 [DOI] [PubMed] [Google Scholar]
- 26.Islam BU, Habib S, Ahmad P, Allarakha S, Moinsuddin, and Ali A (2015) Pathophysiological Role of Peroxynitrite Induced DNA Damage in Human Diseases: A Special Focus on Poly(ADP-ribose) Polymerase (PARP). Indian J Clin Biochem 30, 368–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin Y, Meng F, Sui C, Jiang Y, and Zhang L (2018) Arsenic enhances cell death and DNA damage induced by ultraviolet B exposure in mouse epidermal cells through the production of reactive oxygen species. Clin Exp Dermatol [DOI] [PubMed]
- 28.Cooper KL, Yager JW, and Hudson LG (2014) Melanocytes and keratinocytes have distinct and shared responses to ultraviolet radiation and arsenic. Toxicol Lett 224, 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landino LM (2008) Protein thiol modification by peroxynitrite anion and nitric oxide donors. Methods Enzymol 440, 95–109 [DOI] [PubMed] [Google Scholar]
- 30.Hlaing KH, and Clement MV (2014) Formation of protein S-nitrosylation by reactive oxygen species. Free Radic Res 48, 996–1010 [DOI] [PubMed] [Google Scholar]
- 31.Devarie-Baez NO, Silva Lopez EI, and Furdui CM (2016) Biological chemistry and functionality of protein sulfenic acids and related thiol modifications. Free Radic Res 50, 172–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poole LB (2015) The basics of thiols and cysteines in redox biology and chemistry. Free Radic Biol Med 80, 148–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toledo JC Jr., and Augusto O (2012) Connecting the chemical and biological properties of nitric oxide. Chem Res Toxicol 25, 975–989 [DOI] [PubMed] [Google Scholar]


