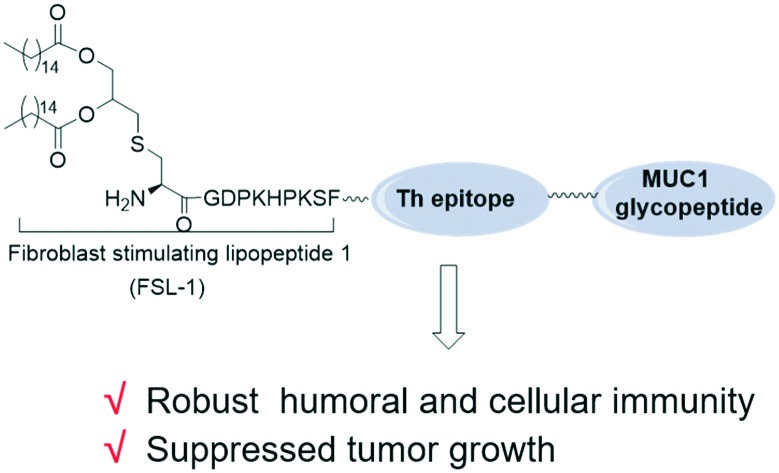
 The designed tricomponent MUC1-based vaccine induced robust immune responses and exhibited a significant reduction in tumor burden.
The designed tricomponent MUC1-based vaccine induced robust immune responses and exhibited a significant reduction in tumor burden.
Abstract
MUC1 is an attractive target for cancer vaccines as a result of its over-expression and aberrant glycosylation pattern on many tumor cells. However, the low immunogenicity of MUC1 and immune tolerance have limited its application. Herein, we designed MUC1-based tricomponent antitumor vaccines adjuvanted with fibroblast stimulating lipopeptide 1 (FSL-1). Immunological results indicate that the glycosylated tricomponent vaccine candidate has elicited both humoral and cellular immune responses. The induced antibodies could effectively bind to MCF-7. Furthermore, the vaccine exhibited an obvious reduction in tumour burden.
Introduction
Recent years have witnessed the rapid development of cancer immunotherapy, which overcomes the shortcomings of traditional methods to treat cancer.1 Therapeutic cancer vaccines have emerged as a highly attractive class of cancer immunotherapeutics. The transmembrane glycoprotein MUC1 is an important target of cancer vaccines.2 The extracellular domain of MUC1 consists of variable number of tandem repeats (VNTRs). A VNTR is composed of 20 amino acids, and the sequence is HGVTSAPDTRPAPGSTAPPA.3 Five Ser/Thr residues of this sequence may be O-glycosylated.4 Compared with normal cells, MUC1 is overexpressed on tumor cells and characterized by a truncated and over-sialylated glycosylation pattern. Tumor-associated carbohydrate antigens (TACAs), such as the T antigen, Tn antigen, and their sialylated forms, are glycan side chains expressed on MUC1.5 Both the peptide epitopes and saccharide epitopes can be recognized by the immune system, therefore, MUC1 becomes a target of cancer vaccines.
However, MUC1 glycopeptide-based antigens are T-cell independent and they are tolerated by immune systems, so they are weakly immunogenic.6 To solve these problems, many studies have been conducted to develop effective MUC1-based cancer vaccines.7–14 The Payne group constructed tricomponent vaccines utilizing a per-glycosylated MUC1 peptide as a B epitope, Pam3CysSer as an immune stimulator, and a tetanus toxin peptide as a T-helper epitope. The three components were connected through covalent bonds. Immunological evaluation proved that these vaccines elicited high IgG antibody levels in mice.15 The Boons group has also constructed a tripartite vaccine composed of Pam3CysSK4, a peptide T helper epitope KLFAVWKITYKDT derived from polio virus and an aberrantly glycosylated MUC1 peptide SAPDT(αGalNAc)RPAP. This tripartite vaccine elicited robust IgG antibody levels in wild-type mice.16 Using the humanized mouse model with mammary cancer, they found that this vaccine produced CTLs and ADCC-mediating antibodies, and immunization with this tripartite vaccine exhibited superior antitumor effects.17 The Fukase group has developed a fully synthetic N-propionyl STn trimer vaccine possessing a helper T-cell epitope and Pam3CSK4, and it can induce high triSTn specific IgGs with minimal autoimmunity and immunosuppression.18 Although many reported vaccines have shown decent immunogenicity, the development of novel MUC1-based vaccines which can be used for cancer immunotherapy is still desperately needed.
Fibroblast stimulating lipopeptide 1 (FSL-1, Pam2CGDPKHPKSF) derived from Mycoplasma salivarium can activate macrophages.19 FSL-1 promotes the generation of cytokines including interleukin (IL)-6, IL-8, IL-10, and IL-12 and tumor necrosis factor (TNF)-α through recognition by TLR2 and TLR6.20 In our previous work, we have constructed MUC1–FSL-1 conjugates as potential anti-tumor vaccines. Both the relatively higher IgG1 level and IL4 level induced by the designed vaccines have shown robust T cell dependent immune responses. Strong specific binding affinities between antisera and MCF-7 tumor cells further proved that these vaccines were effective.21
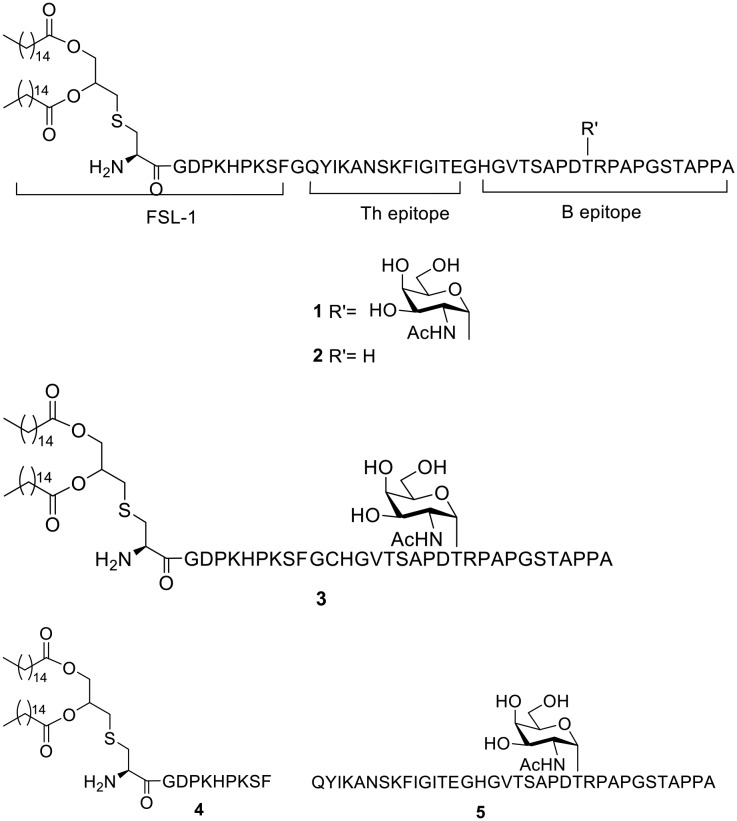
As is known, the intensity of immune responses is quite dependent on the efficacy of the T-helper epitope in most cases.10 To improve the effectiveness of a MUC1-based vaccine adjuvanted with FSL-1 and deepen the research on its therapeutic effects, herein, we develop a three-component cancer vaccine candidate, composed of a glycosylated MUC1-derived glycopeptide connected with the immunoadjuvant FSL-1 and T helper epitope through covalent bonds. We designed the sequence Pam2CysGDPKHPKSFGQYIKANSKFIGITEGHGVTSAPDT(α-GalNAc)RPAPGSTAPPA (compound 1). QYIKANSKFIGITE, a peptide from the tetanus toxoid, was introduced as a helper T-cell epitope.22 Glycine was used as a linker to combine the separate components (Fig. 1). To explore the importance of the carbohydrate group in the constructed vaccine, compound 2 was designed, and it is similar to compound 1 in structure apart from the O-glycosylated threonine residue in the MUC1 B epitope. The vaccine candidate designed in our previous report, i.e. a peptide sequence excluding the T helper epitope (compound 3), was introduced as a control to study whether the tricomponent design is more effective than the two-component design. To research whether covalent bonds between epitopes and adjuvants are necessary for immune responses, a mixture of lipopeptide FSL-1 (compound 4) and QYIKANSKFIGITEGHGVTSAPDT(α-GalNAc)RPAPGSTAPPA (compound 5) in PBS was also prepared. A tricomponent design of a vaccine composed of an adjuvant, the sequence QYIKANSKFIGITE and the MUC1 peptide HGVTSAPDT(α-GalNAc)RPAPGSTAPPA has been reported before.23 However, this is the first time that FSL-1 was conjugated with a T epitope and a tumor associated antigen through covalent linkages in a tripartite manner.
Fig. 1. Structures of the synthetic compounds.
Results and discussion
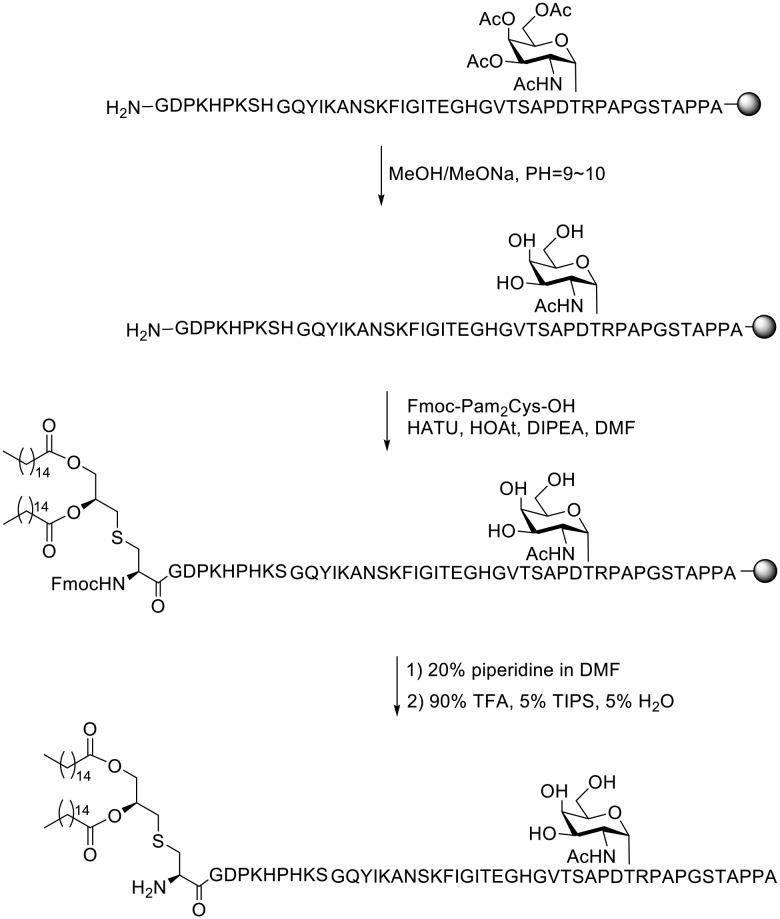
As is known, the synthesis and purification of the designed compounds are very challenging due to their amphipathic character and the difficulty in deprotection of the acetyl groups. In order to overcome these problems, a variety of strategies were reported.24–26 Herein, the preparation of the above peptides was carried out as we previously reported.21 The solid-phase peptide synthesis (SPPS) for preparing compound 1 was performed with 2-chlorotrityl resin preloaded with Fmoc-alanine. Amino acids protected with Fmoc groups were introduced with HBTU/HOBt. The glycosylated amino acid was introduced with HATU/HOAt and its acetyl groups were removed with sodium methoxide dissolved in methanol at a pH between 10 and 11. Fmoc-Pam2Cys-OH was coupled to the sequence with HATU/HOAt and DIPEA. The acid-sensitive side chain protecting groups were removed and the glycopeptide was detached from the resin with a mixture of 90% TFA, 5% TIPS, and 5% H2O (Scheme 1). The glycopeptide was purified with a C18 column by semi-preparative HPLC. Compounds 2, 3, 4 and 5 were prepared in a similar way. The structures of the synthesized peptides were confirmed with the HPLC spectrum and MS data (see the ESI†).
Scheme 1. Solid-phase synthesis of vaccine candidate 1.
The immunological evaluation of the designed vaccines was then conducted. Female C57BL/6 mice were divided into groups and there were 6 mice per group. The mice were immunized every two weeks four times via intraperitoneal injection of the vaccine candidates. Each mouse was immunized with 3.2 nmol vaccine candidates dissolved in 100 μL sterile PBS solution, while mice injected with PBS were taken as control groups. Sera were collected one week after the last immunization.
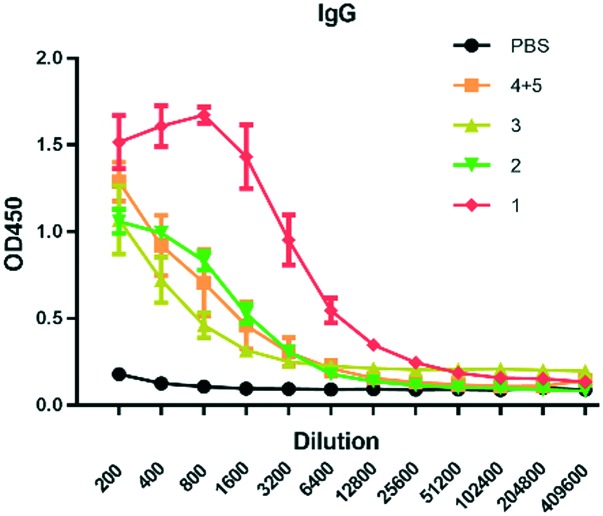
The antibody level was evaluated with MUC1-antigen-specific-ELISA. Compared with other groups, the glycosylated tricomponent vaccine candidate 1 elicited the highest levels of IgG (Fig. 2). It elicited a higher titer of IgG than vaccine 2, showing the importance of glycosylation in MUC1-based vaccines. The comparison of 1 and 3 demonstrated that the tricomponent vaccine was more effective than the two-component vaccine. The mixture of compounds 4 and 5 elicited low antibody levels, illustrating that the covalent linkage of FSL-1 with the T helper epitope and MUC1 epitope was critical for robust antigenic responses.
Fig. 2. Analysis of the antisera induced by the vaccine candidates through ELISA. Microtiter plates were coated with MUC1 glycopeptides. Data are shown as mean ± SD (n = 6).
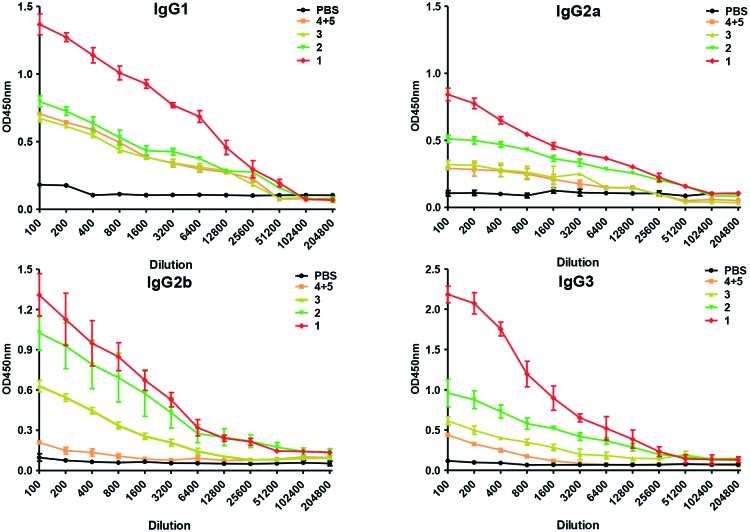
We further examined the vaccine candidates by titration against dilution of different IgG isotypes (Fig. 3). It's proven that generation of IgG1 reflects the effects of type 2 T-helper (Th2) cells, and IgG2a signifies Th1 responses.27,28 Vaccine 1 elicited the generation of both IgG1 and IgG2a, indicating mixed Th1/Th2 responses.29 High IgG3 antibody levels were induced by vaccine 1. This isotype is specific against carbohydrates.30 The strong IgG3 titer elicited by vaccine 1 may indicate its capability to generate anti-glycopeptide humoral immunity.31 The robust production of IgG isotypes induced by vaccine 1 confirmed its advantageous immunogenicity.
Fig. 3. Analysis of antibody isotypes of the antisera induced by the vaccine candidates through ELISA. Data are shown as mean ± SD (n = 6).
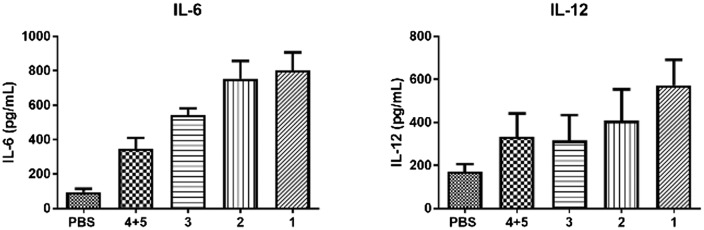
We also tested the release of cytokines in mice sera induced by the vaccine candidates by ELISA. Production levels of IL-6 and IL-12 are shown in Fig. 4. As is known to us, the generation of IL-6 and IL-12 indicates humoral immunity and cellular immunity, respectively.32,33 Both compounds 1 and 2 elicited elevated levels of IL-6 and IL-12. The cytokine release demonstrated effective humoral immunity and cellular immunity induced by compounds 1 and 2.
Fig. 4. IL-6 and IL-12 released from mice serum were tested by ELISA kit. Data are presented as mean ± SD (n = 6).
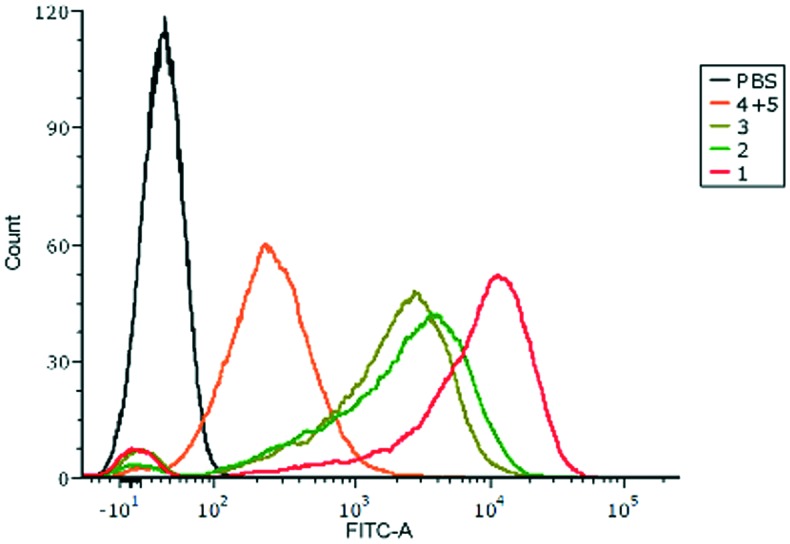
The binding affinity of the antibodies induced by the designed vaccines to MCF-7 tumor cells that express MUC1 was evaluated with flow cytometry (FACS). Antisera from immunized mice were incubated with MCF-7 cells, and then they were cultured with fluorescein isothiocyanate (FITC)-labeled rabbit anti-mouse antibodies. As shown in Fig. 5, the fluorescence peak of anti-1 sera drifted remarkably compared with the other groups. It's supposed that the anti-1 sera from the mice intensively bind to MUC1 exposed on the surface of MCF-7 cells. The FACS results between the antibodies and MUC1-expressing B16-MUC1 further supported the close binding of anti-1 sera with tumor cells (Fig. S1†).
Fig. 5. FACS analysis of the antisera binding with MCF-7 cells. The sera from mice injected with PBS were used as the control (black).
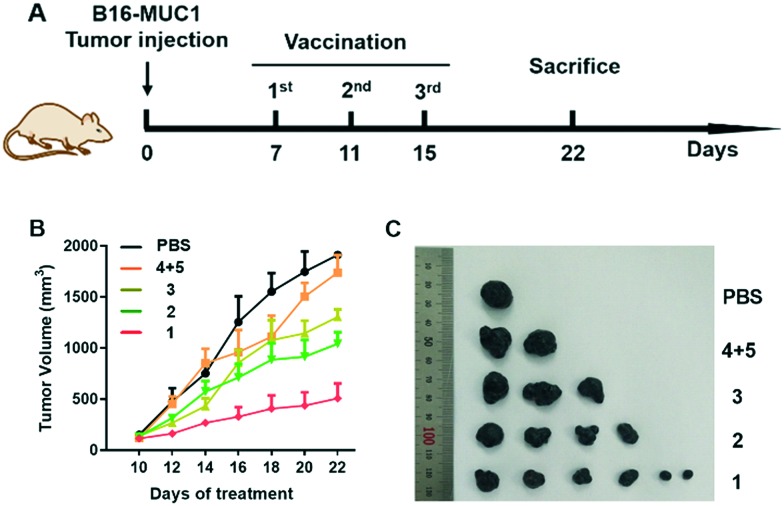
It has been reported that a MUC1-based vaccine could cause a slowdown of tumor progression and an increase in survival of breast tumor-bearing mice.34 Besides, it was supposed that active immunization of patients with their own tumor tissues might be feasible.35 Herein, we tested the in vivo anti-tumor efficacy of the designed groups. Female C57BL/6 mice were subcutaneously challenged with the B16-MUC1 tumor cells (4 × 105) into the right flank.36 One week after transplant, the diameter of the tumor reached 5 mm. Then, the designed vaccines were subcutaneously injected into the peritumoral area three times every four days and the tumor sizes were monitored every two days. As depicted in Fig. 6, injection of compound 1 exhibited significant inhibition effects compared with the other groups. Vaccination with compound 1 exhibited a significant reduction in tumor burden, reflecting its efficacy in killing tumor cells. This is consistent with the robust humoral and cellular immunity elicited by vaccine candidate 1.
Fig. 6. (A) Schematic illustration of immunization against B16-MUC1 tumors. (B) Tumor volumes were monitored. (C) Image of tumors of mice euthanized at day 22.
Conclusions
In conclusion, we have constructed a MUC1-based tricomponent antitumor vaccine adjuvanted with FSL-1. It elicited robust humoral and cellular immune responses. In particular, the constructed vaccine led to an obvious reduction in tumor burden, proving its potential as a vaccine for cancer immunotherapy. It is noteworthy that, compared with our previous design, the constructed vaccine showed much higher immunological activities and anti-tumor effects. It is demonstrated that covalent attachment of the TLR agonist FSL-1, the tricomponent design, and glycosylation of the MUC1 peptide are critical factors for the optimal immune responses. This rational design of the MUC1 glycopeptide vaccine also indicated its possible application to other carbohydrate antigens for further research on cancer immunotherapy.
Experimental section
Materials and instruments
Mouse IgG antibodies and a monoclonal antibody isotyping kit were purchased from Sigma-Aldrich (USA). ELISA kits for detecting IL-6 and IL-12 were purchased from BioLegend (San Diego, CA, USA). The MCF-7 cell line and B16-MUC1 cell line were purchased from China Infrastructure of Cell Line Resources.
Immunological studies
Vaccine immunization
C57BL/6 female mice (6–8 weeks) were fed at the Institute of Radiation Medicine, Chinese Academy of Medical Sciences. The mice were immunized with injections of 3.2 nmol vaccine in 100 μL PBS intraperitoneally. The immunization was conducted every two weeks four times (six mice per group). Blood samples were prepared by eyeball extirpation one week after the last vaccination. The anti-sera were collected for evaluation. The experimental protocols were evaluated and approved by the Animal Ethics Committee of the Institute of Radiation Medicine, Chinese Academy of Medical Sciences. All animal procedures were performed in accordance with the Institutional Animal Care and Use Committee-approved protocols (approval number IRM-DWLL-2018018).
Analysis of antibody titers
96-Well ELISA plates were coated with 20 μg mL–1 MUC1 glycopeptide (100 μL per well) dissolved in a 0.1 M NaHCO3 solution (pH = 9.6) for 12 h at 4 °C. 1% BSA solution was used to block the plates. The sera were respectively diluted and added into the plates (100 μL per well). Then, the plates were incubated for 2 h at 37 °C. After washing, IgG antibodies (goat anti-mouse, diluted to 1 : 3000) were added into the plates and incubated for 1 h at 37 °C, then HRP-labeled IgG antibodies (rabbit anti-goat, diluted to 1 : 3000) were added. TMB solution was added, followed by the addition of a 0.5 M H2SO4 solution 0.5 h later. The absorption was measured at 450 nm wavelength. All the tests were repeated three times.
Analysis of antibody isotypes
Analysis of antibody isotypes was carried out in the same way as that of antibody titers except that the IgG 1/IgG 2a/IgG 2b/IgG 3 antibodies were diluted to 1 : 1000.
Analysis of cytokine levels in anti-sera
The levels of IL-6 and IL-12 in the anti-sera were determined by ELISA using mouse IL-6 and mouse IL-12 ELISA Max™ Set Deluxe kits (BioLegend) in accordance with the manufacturer's instructions.
The binding of sera to MCF-7 cells
MCF-7 cells (2 × 104) were incubated with 100 μL sera (diluted with 1 : 50) for 1 h at 4 °C. After washing, FITC-conjugated IgG antibodies (rabbit anti-mouse, diluted with 1 : 1000) were added and incubated for 1 h at 4 °C. FACS analysis was performed on a BD LSRFortessa.
Analysis of anti-tumor immune response
B16-MUC1 tumor cells (4 × 105) were administered subcutaneously into the right flank of C57BL/6 female mice (4–6 weeks). Seven days after transplant, the mice with a 5 mm tumor diameter were grouped (six mice per group). The mice were given peritumoral injections of 3.2 nmol designed vaccine candidates every four days and a total of three injections were conducted. PBS was used as the blank control. Tumor sizes were measured every two days. According to animal ethics guidelines, the mice were euthanized in case that the length of the tumor exceeded 15 mm. The tumor volume was calculated by 0.5 × length × width.
Conflicts of interest
The authors declare no competing financial interests.
Supplementary Material
Acknowledgments
This work was supported by the National Key R&D Program of China (2018YFA0507204), the National Natural Science Foundation of China (21572105), and the Natural Science Foundation of Tianjin (17JCZDJC32800).
Footnotes
†Electronic supplementary information (ESI) available: Experimental details for chemical synthesis, HPLC and MS data and spectra, and immunological FACS results. See DOI: 10.1039/c9md00254e
References
- Chen D. S., Mellman I. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- Wilson R. M., Danishefsky S. J. J. Am. Chem. Soc. 2013;135:14462–14472. doi: 10.1021/ja405932r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acres J. L. B. Expert Rev. Vaccines. 2005;4:493–502. doi: 10.1586/14760584.4.4.493. [DOI] [PubMed] [Google Scholar]
- Gaidzik N., Westerlind U., Kunz H. Chem. Soc. Rev. 2013;42:4421–4442. doi: 10.1039/c3cs35470a. [DOI] [PubMed] [Google Scholar]
- Toyokuni T., Singhal A. K. J. C. S. R. Chem. Soc. Rev. 1995;24:231–242. [Google Scholar]
- Danishefsky S. J., Allen J. R. Angew. Chem., Int. Ed. 2000;39:836–863. doi: 10.1002/(sici)1521-3773(20000303)39:5<836::aid-anie836>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Cai H., Huang Z. H., Shi L., Sun Z. Y., Zhao Y. F., Kunz H., Li Y. M. Angew. Chem., Int. Ed. 2012;51:1719–1723. doi: 10.1002/anie.201106396. [DOI] [PubMed] [Google Scholar]
- Dziadek S., Kowalczyk D., Kunz H. Angew. Chem., Int. Ed. 2005;44:7624–7630. doi: 10.1002/anie.200501593. [DOI] [PubMed] [Google Scholar]
- Broecker F., Gotze S., Hudon J., Rathwell D. C. K., Pereira C. L., Stallforth P., Anish C., Seeberger P. H. J. Med. Chem. 2018;61:4918–4927. doi: 10.1021/acs.jmedchem.8b00312. [DOI] [PubMed] [Google Scholar]
- Palitzsch B., Hartmann S., Stergiou N., Glaffig M., Schmitt E., Kunz H. Angew. Chem., Int. Ed. 2014;53:14245–14249. doi: 10.1002/anie.201406843. [DOI] [PubMed] [Google Scholar]
- Yin Z., Wu X., Kaczanowska K., Sungsuwan S., Comellas Aragones M., Pett C., Yu J., Baniel C., Westerlind U., Finn M. G., Huang X. ACS Chem. Biol. 2018;13:1668–1676. doi: 10.1021/acschembio.8b00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Yin Z., McKay C., Pett C., Yu J., Schorlemer M., Gohl T., Sungsuwan S., Ramadan S., Baniel C., Allmon A., Das R., Westerlind U., Finn M. G., Huang X. J. Am. Chem. Soc. 2018;140:16596–16609. doi: 10.1021/jacs.8b08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar N. T., Lakshminarayanan V., Capicciotti C. J., Sirohiwal A., Madsen C. S., Wolfert M. A., Cohen P. A., Gendler S. J., Boons G.-J. ChemBioChem. 2018;19:121–125. doi: 10.1002/cbic.201700424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Aal A.-B. M., Lakshminarayanan V., Thompson P., Supekar N., Bradley J. M., Wolfert M. A., Cohen P. A., Gendler S. J., Boons G.-J. ChemBioChem. 2014;15:1508–1513. doi: 10.1002/cbic.201402077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson B. L., Day S., Malins L. R., Apostolopoulos V., Payne R. J. Angew. Chem., Int. Ed. 2011;50:1635–1639. doi: 10.1002/anie.201006115. [DOI] [PubMed] [Google Scholar]
- Ingale S., Wolfert M. A., Gaekwad J., Buskas T., Boons G. J. Nat. Chem. Biol. 2007;3:663–667. doi: 10.1038/nchembio.2007.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarayanan V., Thompson P., Wolfert M. A., Buskas T., Bradley J. M., Pathangey L. B., Madsen C. S., Cohen P. A., Gendler S. J., Boons G. J. Proc. Natl. Acad. Sci. U. S. A. 2012;109:261–266. doi: 10.1073/pnas.1115166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. C., Manabe Y., Fujimoto Y., Ohshima S., Kametani Y., Kabayama K., Nimura Y., Lin C. C., Fukase K. Angew. Chem., Int. Ed. 2018;57:8219–8224. doi: 10.1002/anie.201804437. [DOI] [PubMed] [Google Scholar]
- Shibata K. i., Hasebe A., Into T., Yamada M., Watanabe T. J. Immunol. 2000;165:6538–6544. doi: 10.4049/jimmunol.165.11.6538. [DOI] [PubMed] [Google Scholar]
- Okusawa T., Fujita M., Nakamura J. i., Into T., Yasuda M., Yoshimura A., Hara Y., Hasebe A., Golenbock D. T., Morita M., Kuroki Y., Ogawa T., Shibata K. i. Infect. Immun. 2004;72:1657–1665. doi: 10.1128/IAI.72.3.1657-1665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang W., He Q., Yu F., Song T., Liu T., Zhang Z., Zhou J., Wang P. G., Zhao W. Chem. Commun. 2016;52:10886–10889. doi: 10.1039/c6cc04623a. [DOI] [PubMed] [Google Scholar]
- Nuhn L., Hartmann S., Palitzsch B., Gerlitzki B., Schmitt E., Zentel R., Kunz H. Angew. Chem., Int. Ed. 2013;52:10652–10656. doi: 10.1002/anie.201304212. [DOI] [PubMed] [Google Scholar]
- Cai H., Sun Z. Y., Huang Z. H., Shi L., Zhao Y. F., Kunz H., Li Y. M. Chem. – Eur. J. 2013;19:1962–1970. doi: 10.1002/chem.201203709. [DOI] [PubMed] [Google Scholar]
- Brocke C., Kunz H. Synthesis. 2004;4:525–542. [Google Scholar]
- Liebe B., Kunz H. Angew. Chem., Int. Ed. Engl. 1997;36:618–621. [Google Scholar]
- McDonald D. M., Hanna C. C., Ashhurst A. S., Corcilius L., Byrne S. N., Payne R. J. ACS Chem. Biol. 2018;13:3279–3285. doi: 10.1021/acschembio.8b00675. [DOI] [PubMed] [Google Scholar]
- Skwarczynski M., Zaman M., Urbani C. N., Lin I. C., Jia Z., Batzloff M. R., Good M. F., Monteiro M. J., Toth I. Angew. Chem., Int. Ed. 2010;49:5742–5745. doi: 10.1002/anie.201002221. [DOI] [PubMed] [Google Scholar]
- Seregin S. S., Appledorn D. M., McBride A. J., Schuldt N. J., Aldhamen Y. A., Voss T., Wei J., Bujold M., Nance W., Godbehere S., Amalfitano A. Mol. Ther. 2009;17:685–696. doi: 10.1038/mt.2008.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Sun Z. Y., Chen M. S., Zhao Y. F., Kunz H., Li Y. M. Angew. Chem., Int. Ed. 2014;53:1699–1703. doi: 10.1002/anie.201308875. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Liao G., Mandal S. S., Suryawanshi S., Guo Z. Chem. Sci. 2015;6:7112–7121. doi: 10.1039/c5sc01402f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson B. L., Day S., Chapman R., Perrier S., Apostolopoulos V., Payne R. J. Chem. – Eur. J. 2012;18:16540–16548. doi: 10.1002/chem.201202629. [DOI] [PubMed] [Google Scholar]
- Diehl S. A., Schmidlin H., Nagasawa M., Blom B., Spits H. Immunol. Cell Biol. 2012;90:802–811. doi: 10.1038/icb.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz E. G., Mariani L., Radbruch A., Hofer T. Immunity. 2009;30:673–683. doi: 10.1016/j.immuni.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Stergiou N., Gaidzik N., Heimes A.-S., Dietzen S., Besenius P., Jäkel J., Brenner W., Schmidt M., Kunz H., Schmitt E. Cancer Immunol. Res. 2019;7:113–122. doi: 10.1158/2326-6066.CIR-18-0256. [DOI] [PubMed] [Google Scholar]
- Gaidzik N., Kaiser A., Kowalczyk D., Westerlind U., Gerlitzki B., Sinn H. P., Schmitt E., Kunz H. Angew. Chem., Int. Ed. 2011;50:9977–9981. doi: 10.1002/anie.201104529. [DOI] [PubMed] [Google Scholar]
- Shao Y., Sun Z.-Y., Wang Y., Zhang B.-D., Liu D., Li Y.-M. ACS Appl. Mater. Interfaces. 2018;10:9310–9314. doi: 10.1021/acsami.8b00312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.