Abstract
HOXA Transcript Antisense RNA, Myeloid-Specific 1 (HOTAIRM1) is a conserved long non-coding RNA (lncRNA) involved in myeloid and neural differentiation that is deregulated in acute myeloid leukemia and other cancers. Previous studies focused on the nuclear unspliced HOTAIRM1 transcript, however cytoplasmic splice variants exist whose roles have remained unknown. Here, we report novel functions of HOTAIRM1 in the kidney. HOTAIRM1 transcripts are induced during renal lineage differentiation of embryonic stem cells and required for expression of specific renal differentiation genes. We show that the major HOTAIRM1 transcript in differentiated cells is the spliced cytoplasmic HM1–3 isoform and that HM1–3 is downregulated in >90% of clear cell renal cell carcinomas (ccRCCs). Knockdown of HM1–3 in renal cells deregulates hypoxia-responsive and angiogenic genes, including ANGPTL4. Furthermore, HOTAIRM1 transcripts are downregulated by hypoxia-mimetic stress and knockdown of the cytoplasmic HM1–3 isoform in normoxic cells post-transcriptionally induces Hypoxia-Inducible Factor 1α (HIF1α) protein, a key activator of ANGPTL4. Our results demonstrate the pervasive downregulation of the specific HOTAIRM1 cytoplasmic isoform HM1–3 in ccRCC and suggest possible roles of HOTAIRM1 in kidney differentiation and suppression of HIF1-dependent angiogenic pathways.
Keywords: Long non-coding RNA, ccRCC, kidney lineage, cell differentiation, Hypoxia-Inducible Factor 1α (HIF1α)
1. Introduction
Kidney and renal pelvis cancers are among the most pervasive cancers found within the United States [1]. Renal cell carcinomas (RCCs) comprise >90% of kidney cancers, which have been shown to be particularly difficult to treat with conventional therapies [2–4]. Among these, the clear cell renal cell carcinoma (ccRCC) type accounts for 75% of all kidney cancers [5]. The Von Hippel-Lindau (VHL) tumor suppressor gene is the most frequently mutated gene in sporadic ccRCC and codes for the substrate recognition subunit of an E3 ubiquitin ligase complex that targets the Hypoxia-Inducible Factors 1α and 2 α (HIF1 α and HIF2 α) for proteasomal degradation [6]. Hence, the VHL-HIF axis is often altered in ccRCC tumors, which are highly vascularized due to hyperactivation of various HIF target genes involved in angiogenesis. Recently, additional genetic alterations have been associated with ccRCC development [7], and transcriptomic analyses have identified several long non-coding RNAs (lncRNAs) that are deregulated in ccRCC [8, 9]. One notable example is the lncRNA PVT1, which increases the stability of the MYC oncoprotein [10], while the PVT1 promoter locus was shown to inhibit MYC transcription [11]. PVT1 is part of a network of lncRNAs that modulates MYC activity and consequently the VHL-HIF axis by affecting the binding partners of HIF1α and HIF2α in ccRCC [12–15]. However, only few lncRNAs have been extensively explored in RCC/ccRCC [16]. In our recent bioinformatics studies examining isoform-specific transcript alterations in ccRCC tumors, we identified several candidate deregulated lncRNAs, potentially including HOTAIRM1 [17]. The HOTAIRM1 locus is located within the HOXA cluster between (and antisense to) the HOXA1 and HOXA2 genes. HOTAIRM1 is best known for its role in activation of the HOXA genes during neural differentiation of pluripotent cells and differentiation of promyelocytic leukemia cells via binding MLL and PRC2 and altering chromatin structure in cis at the HOXA locus [18–20]. However, contrary to its original designated name, HOTAIRM1 expression has now been reported in numerous developing and fully differentiated tissues and cell types, and HOTAIRM1 expression is altered in several human cancers [21–24]. While the mechanistic roles of HOTAIRM1 in cancer are largely unknown, recent evidence in NB4 promyelocytic leukemia cells suggests a function in the autophagy pathway via acting as a miRNA sponge [25].
In the current study, we report for the first time a role of HOTAIRM1 during renal lineage differentiation and the pervasive downregulation of specifically its spliced isoform HM1–3 in >90% of ccRCC tumors cataloged in the The Cancer Genome Atlas (TCGA) database. Using a model kidney proximal tubule ccRCC cell line (CAKI-1) that is VHL-positive and expresses all HOTAIRM1 isoforms, we demonstrate that the cytoplasmic isoform HM1–3 suppresses HIF1α protein levels and attenuates hypoxia-responsive target genes under normoxic conditions. Our results suggest possible pro-differentiation and tumor-suppressive roles of HM1–3 in the kidney via inhibition of the oncogenic HIF1 pathway, which is activated in the vast majority of ccRCC tumors.
2. Materials and Methods
2.1. Cell culture
The HK-2, ACHN and CAKI-1 cell lines were acquired from ATCC and were cultured as recommended. The HK-2 cell line was cultured in keratinocyte serum-free medium supplied with 0.05 mg/ml bovine pituitary extract and 5 ng/ml human recombinant epidermal growth factor (Invitrogen, Carlsbad, CA), unless otherwise indicated. The ACHN cell line was cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS (Gibco, Grand Island, USA). The CAKI-1 cell line was cultured in McCoy’s 5a Modified Medium supplemented with 10% FBS (Gibco, Grand Island, USA), unless otherwise indicated. All cultures were maintained in a humidified incubator with 5% CO2 at 37°C. Mouse embryonic stem (mES) cell line D3 (CRL-11632) and mES KH2 cell line (MESKH2 B912) were purchased from ATCC and Mirimus, respectively, and cultured in pluripotency media according to manufacturers’ specifications. Generation of KH2 mouse ES cell lines with doxycycline-inducible shRNAs was as previously described [26] (see Supplemental Information). The mES cells were grown and expanded on a monolayer of Mitomycin C treated primary mouse embryo fibroblasts (MEFs). Before each experiment, MEFs were removed and mES cells were expanded on 0.1% gelatin-coated cell culture dishes. Pluripotency media contained DMEM with high glucose, 15% FBS, 2 mM L-glutamine, 0.1mM non-essential amino acids, 0.1 U/ml penicillin, 0.1 μg/ml streptomycin, 0.55 mM 2-mercaptoethanol and 1000 U/ml of Leukemia Inhibitory Factor (LIF). For KH2 cells the medium was further complemented with 2i (1 μM PDO325901 + 2μM CHIR99621) per manufacturer’s instructions. For retinoic acid (RA)-induced differentiation towards early neural lineage, mES cells were cultured in medium without LIF or 2i and supplemented with 10−6 M all-trans retinoic acid (R625, Sigma), and medium was changed every 24 hours for the indicated times. For early renal lineage differentiation KH2 mES cells were differentiated as previously described (Nishikawa et al., 2012). Briefly, cells were seeded at a density of 6.4 × 102 cells/cm2 in medium (as above without LIF or 2i) and supplemented sequentially with differentiating factors: 10 ng/ml Activin A, 50 ng/ml BMP-4, 10 mM LiCl, and 100 nM RA. Each factor was added in succession in the above order and cumulatively, starting at the time of plating and every 48 h thereafter. Cells were collected by trypsinization after 8 days for RNA extraction and quantitative PCR analysis.
2.2. RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Cells were collected using 0.25% trypsin and total RNA was extracted using the GeneJet RNA purification kit (Thermoscientific, Carlsbad, CA) per manufacturer’s recommendations. DNA was digested using the Rnase-Free DNase set (Qiagen, Valencia, CA) for 1 hour on the column according to the manufacturer’s instructions. Total RNA from mouse ES cells was purified using the Direct-Zol RNA kit (Zymo Research). Extracted RNA was verified for quality and quantity using gel electrophoresis and the Thermoscientific Nanodrop2000 spectrophotometer. cDNA was synthesized using 1ug of total RNA using the iScript reverse transcription supermix (Biorad, Irvine, CA) according to the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using the Biorad iQ SYBR green supermix and a Biorad CFX Connect thermocylcer (Biorad, Irvine, CA) and analyzed using the CFX manager software. Using a single threshold Cq determination, the Livak method was employed for all gene expression analyses. Expression analyses in renal cells and tissues were normalized to PPIA, as PPIA was found to be the most suitable reference gene when comparing ccRCC tumors to normal adjacent tissue [27, 28], and no significant change was observed with HOTAIRM1 knockdown. The 12 ccRCC tumor/normal matched pair RNA samples were obtained from Origene (see Supplemental Information). The multiple human tissue cDNA arrays were obtained from Origene (CSRT301, HKRT102) and were normalized to ACTB. Expression analyses in mouse ES cells were normalized to three housekeeping genes (Gapdh, Rpol2, and Actb). Three technical replicates of each biological replicate were performed for every qPCR reaction using the aforementioned protocols, reagents and instrumentation.
2.3. Primer design
Primers sequences were obtained either from qPrimerDepot (https://primerdepot.nci.nih.gov/) or designed using Primer3Plus (www.primer3plus.com/) using the qPCR settings and adhered to the specifications set forth by the manufacturer of the qPCR equipment used (Bio-Rad), including primer efficiencies. (see Supplemental Information). All primers were synthesized by Integrated DNA Technologies.
2.4. Nuclear and cytoplasmic RNA expression analysis
Approximately 1 million cells were lysed in 175 ul of cytoplasmic lysis buffer (50mM TrisCl pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% P-40, 1mM DTT) on ice for 5 minutes for the HK-2 and CAKI-1 cells and 35 minutes for the ACHN cells. Following incubation, the lysate was spun at 300g for 2 minutes at 4C. Cytoplasmic supernatant and nuclear pellet were separated, and RNA was purified as above. Corresponding cell equivalents of cytoplasmic and nuclear RNA were used for reverse transcription and qPCR.
2.5. siRNA transfection and RNA-seq analyses
All custom siRNAs (Supplemental Information) were designed using the MIT Whitehead software (http://sirna.wi.mit.edu/) and synthesized as “Silencer Select siRNAs” by Ambion (Carlsbad, CA, USA). The validated Silencer Select negative siRNA #2 (Ambion) was used as control in transient knockdown assays. siRNAs were transfected using Lipofectamine3000 per manufacturer’s recommendations at a final concentration of 100 nM. Total cellular RNA was extracted 60 hours after transfection. For knockdown of mouse Hotairm1 in differentiated renal cell progenitors, a Silencer Select siRNA “e3–3.7” (Supplemental Information) which targets exon 3 of mouse Hotairm1 was transfected with Lipofectamine 3000 per manufacturer’s recommendations at a concentrations of 10 nM in differentiation medium.
For RNA-seq analysis, three biological replicates of siRNA-transfected CAKI-1 cells were trypsinized and total RNA was extracted, as above. RNA quality and quantity were evaluated with a bioanalyzer and Thermoscientific Nanodrop2000 spectrophotometer. Single-end read RNA-seq libraries were constructed using the NEBNext Ultra Directional RNA library prep kit (Illumina). Samples were multiplexed and sequenced with the NEX-seq Illumina sequencing platform at the UCR Core Genomics Facility. The RNA-seq data can be accessed at NCBI’s Gene Expression Omnibus (GEO) under number GSE136604.
2.6. Bioinformatic analyses
A total of 542 fastq RNA-seq files were downloaded from The Cancer Genome Atlas (TCGA) legacy archive website (https://portal.gdc.cancer.gov/legacy-archive/search/f). Human cDNA and ncRNA FASTA formatted transcript files (Ensembl v89 annotation) were acquired form the Ensembl ftp site (https://www.ensembl.org/info/data/ftp/index.html), and merged to create a master file of all putative coding and non-coding transcripts.
Transcript quantifications and differential expression analyses were performed using the cufflink suite (TCGA data analysis) or the kallisto-sleuth pipeline (RNA-seq analysis) [29–31]. Cufflinks was used to obtain transcript quantifications [30]. Calculated transcript quantifications were then used to generate tumor/normal ratios. A two-tailed Wilcoxon signed rank test was performed to determine statistical significance. Cuffdiff was used to confirm differential expression. Using the default settings, kallisto was used to create an index for quantification using the aforementioned FASTA master file. Subsequently, kallisto was used to quantify all putative transcripts using 50 bootstrap samples. Differential expression analysis was performed with sleuth using the Wald test with a cutoff of q-value <0.05 and beta >0.5.
For the gene-level analyses, alignment of the fastq files was performed first with HISAT2 using the hg38 human assembly [32]. Read counting was performed using the summarizeOverlaps package, with union mode [33]. Using the read counts, an edgeR analysis was performed using the default settings [34, 35]. The entire pipeline was performed within the systemPipeR package [32, 36]. Normalization of the gene counts was performed using DESeq2 and then subsequently used in consensus clustering to determine the number molecular subtypes in ccRCC [37]. Consensus clustering was performed using the ConsensusClusterPlus R package [38]. A total 1,000 of the most variable genes, based on mean absolute deviation were used in the clustering generating consensus matrices for k=2–7. Number of molecular subtypes was determined based on the consensus matrices and the cumulative distribution functions for each k.
2.7. Western blot
Cells were scraped and lysed using RIPA buffer (150mM NaCL, 5mM EDTA, 50mM Tris pH 8.0, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS). Lysate was spun in a microfuge at maximum speed and the soluble protein concentration was determined using Bradford reagent (Biorad). A total of 10–20 ug protein was subjected to SDS-PAGE. Proteins were transferred to a nitrocellulose membrane using a Trans-Blot Turbo (Biorad) for 45 minutes at 25V. Membranes were blocked with 1% non-fat dry milk for 1 hour and probed overnight with primary antibodies, β-actin at 1:7500 (sc-47778, Santa Cruz Biotech.), DDAH1 at 1:1000 (sc-514841, Santa Cruz Biotech.), VHL at 1:3000 (sc-17780, Santa Cruz Biotech.), and HIF1α at 1:3000 (ab179483, Abcam). Secondary HRP-conjugated antibodies, anti-mouse and anti-rabbit, were incubated at 1:1000 dilution for 1 hour at room temperature and chemiluminescence reactions were performed as per manufacturer’s instructions (GE Healthcare).
3. Results
3.1. The HOTAIRM1 spliced isoform HM1–3 is downregulated in ccRCC
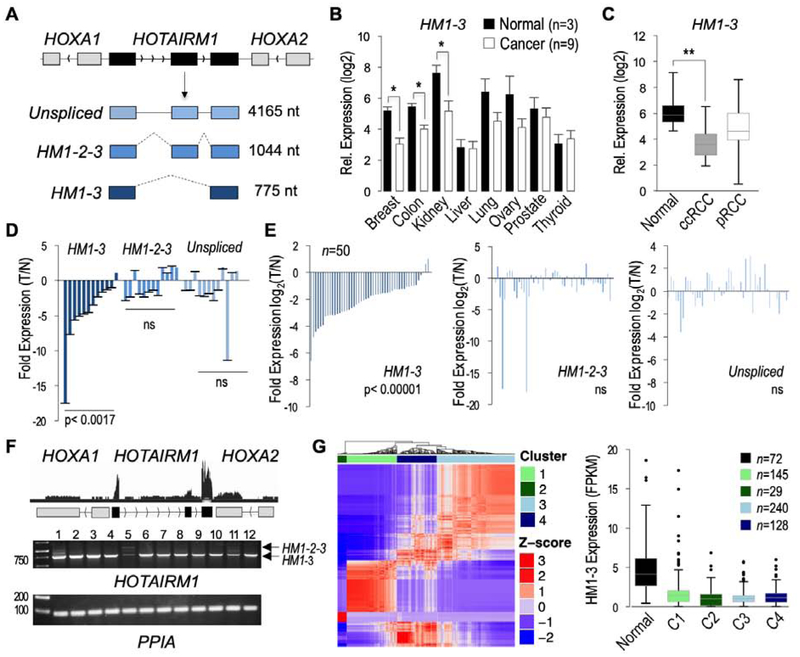
We initially performed a survey of HOTAIRM1 transcripts in 8 different cancers and cognate normal tissues by qPCR using a multiple tissue cDNA array (Figure 1A–B, Supplemental Figure 1A–B). This revealed a significant downregulation of the spliced HM1–3 isoform in kidney cancer, renal cell carcinomas (RCCs). HM1–3 downregulation was also observed in breast and colorectal cancers, consistent with previous reports [21, 39]. Examination of HM1–3 expression specifically in clear cell RCC (ccRCC) and in papillary RCC (pRCC), using an independent cDNA array, demonstrated that HM1–3 downregulation was significant in ccRCC (Figure 1C). An average ~5.5 fold downregulation in HM1–3 expression was seen when comparing 9 normal renal tissue samples to 21 ccRCC samples. No statistically significant HM1–3 downregulation was observed in the 10 pRCC tumors tested. These results were supported further using 12 ccRCC matched pair samples, which showed 11 ccRCC tumors with a HM1–3 downregulation relative to their normal adjacent tissue (Figure 1D). In contrast, there were no statistically significant differences in expression of the HM1-2-3 isoform or the unspliced HOTAIRM1 transcript (Figure 1D).
Figure 1. Reduced HM1–3 expression in ccRCC.
A. The HOTAIRM1 gene is located between HOXA1 and HOXA2 and produces an unspliced transcript and two major spliced RefSeq transcripts (HM1-3, HM1-2-3). B. Relative qPCR expression analysis of HM1–3 in a panel of eight human cancers (n=9 tumors each) and their respective normal tissues (n=3 each). HM1–3 levels were normalized to β-actin. C. Analysis of HM1–3 expression by qPCR in normal tissues (n=9) versus ccRCC (n=21) and pRCC (n=10, papillary renal cell carcinoma) tumors was performed as in panel B. D. Analysis of HM1–3 expression by qPCR in 12 ccRCC matched pair samples. Fold changes in expression in tumor vs normal (T/N) (ΔΔCt) are indicated. E. FPKM tumor/normal ratios of 50 ccRCC matched pair TCGA samples for all HOTAIRM1 transcripts. F. Quantification of HOTAIRM1 transcripts within normal adjacent tissues. Shown is the merged trace of 72 normal renal RNA-seq datasets of TCGA (top) and a validation using 12 independent normal renal RNA samples using endpoint RT-PCR with primers in exon 1 and 3 (bottom); PCR products for HM1–3 and HM1-2-3 are indicated with arrows. G. Unsupervised consensus clustering identified 4 distinct ccRCC subtypes (left). HM1-3 expression (FPKM) is decreased in each ccRCC tumor subtype (right). Statistical significance was determined by using two-tailed Student’s t-test for panels B, C, and G and the Wilcoxon signed-rank test for the match paired samples in panels D and E (* p<0.05, **p<0.005, ns p>0.01).
To further substantiate HM1–3 downregulation in ccRCC, 614 RNA-seq datasets (72 normal and 542 ccRCC samples) from TCGA were bioinformatically examined. Evaluation of HOTAIRM1 FPKM tumor/normal ratios, using 50 matched pair samples contained within these datasets, confirmed the above qPCR results showing that HM1–3 is the only significantly downregulated HOTAIRM1 transcript, as determined by Wilcoxon signed ranked test (Figure 1E). Notably, 46 of the 50 tumors (92%) had greater than 2-fold reduction in HM1–3 levels compared to their normal adjacent tissue. Data from the GTEx database (https://gtexportal.org/home/) confirmed that HOTAIRM1 is significantly expressed in normal kidney cortex relative to other tissues (Supplemental Figure 2A). We further examined the expression levels of the different HOTAIRM1 transcripts by merging the RNA-seq alignments files from the 72 normal adjacent renal tissues of TCGA, which indicated that inclusion of exon 2 is rare (Figure 1F, top), and by quantifying the different transcripts using cufflinks (Supplemental Figure 2B). This showed that the HM1–3 isoform is the most abundant HOTAIRM1 transcript in normal renal tissue. The predominance of HM1–3 over the HM1-2-3 isoform was further supported by endpoint PCR using 12 normal renal tissue samples (Figure 1F, bottom).
As ccRCC is a herterogeneous cancer, HM1–3 expression was explored within the different molecular subtypes of ccRCC. Consensus clustering was performed using gene-level read counts from the 542 ccRCC samples, which confirmed the existence of four molecularly distinct subtypes of ccRCC (Figure 1G, left, clusters 1–4; Supplemental Figure 3) corresponding to the previously reported ccA, ccB, mixed ccA/ccB and distal tubule subtypes [7, 9]. Using a two-tailed Student’s t-test, a significant HM1–3 downregulation was found within all four subtypes of ccRCC (Figure 1G, right; p<0.05). Our attempts to stratify HM1–3 expression levels according to ccRCC tumor stage or grade in the TCGA datasets did not retrieve a significant correlation nor did we find a correlation with patient survival, suggesting that HM1–3 downregulation might be an early event in ccRCC development (data not shown). Altogether, these results identify the spliced HM1–3 isoform as the major HOTAIRM1 transcript in non-tumor renal tissue and indicate its widespread downregulation in ccRCC tumors of all subtypes, including in over 90% of ccRCC tumors for which matched normal adjacent tissue data were available in TCGA.
3.2. Expression of HOTAIRM1 spliced isoforms is associated with the non-transformed and differentiated phenotypes of renal and non-renal cell types
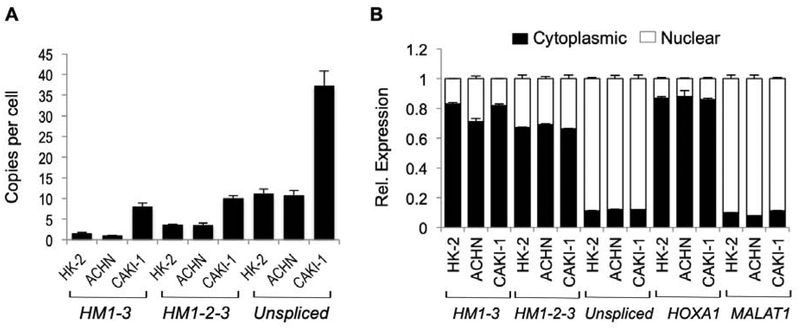
Several human renal proximal tubule epithelial cell lines were investigated for their HOTAIRM1 expression by RT-qPCR, including normal immortalized cells (HK-2) and two ccRCC cell lines (ACHN and CAKI-1). Absolute levels of the spliced transcripts were very low in all cell lines tested. The highest amounts of the spliced HM1–3 and HM1-2-3 transcripts were observed in CAKI-1 cells, which contained approximately 10 copies each per cell (Figure 2A). All the immortalized and cancer cell lines tested expressed lower levels of the spliced isoforms relative to the unspliced HOTAIRM1 transcript (Figure 2A), reminiscent of the preferential downregulation of the HM1–3 isoform in ccRCC tumors (see above and Supplemental Figure 4). Other non-renal immortalized or cancer cell lines of the breast (MCF10A and MCF7), and brain (DAOY) showed comparable or lower levels of the spliced isoforms and a predominance of the unspliced HOTAIRM1 transcript (data not shown). Subcellular fractionation showed that the spliced isoforms HM1–3 and HM1-2-3 are predominantly cytoplasmic, while the unspliced HOTAIRM1 transcript is mostly nuclear (Figure 2B).
Figure 2. Characterization of HOTAIRM1 transcripts in renal proximal tubule cell lines.
A. Copies per cell using qPCR. Error bars represent SEM across three biological replicates. B. Subcellular localization of transcripts. As controls the nuclear MALAT1 lncRNA and cytoplasmic HOXA1 mRNA were analyzed. Error bars represent SEM for three replicates.
Given the reduced expression of the spliced HM1–3 isoform in immortalized/cancer cell lines and in ccRCC tumors compared to normal renal tissue, we further analyzed HM1–3 expression during normal cell differentiation. Since HOTAIRM1 was shown to be induced during neural differentiation of pluripotent embryonal carcinoma NT2/D1 cells [20], we first characterized the different Hotairm1 isoforms and their expression during retinoic acid (RA)-induced early neural lineage differentiation of normal mouse ES cells in vitro. All Hotairm1 transcripts were rapidly induced and the mouse HM1–3 paralog (E1–3) was by far the most abundant isoform (average ~50 copies/cell after 3 days) and accumulated in the cytoplasm with kinetics similar to the induction of the Hoxa1 mRNA (~30 copies/cell after 3 days) (Supplemental Figure 5 and data not shown). Similar results were obtained during differentiation of mouse ES cells into early renal lineage progenitor cells (see further below). Altogether these results indicate that the spliced HM1–3 isoform is highly induced during normal cell differentiation (both neural and renal differentiation) but its expression is suppressed in immortalized/cancer cell lines in vitro and in ccRCC tumors in vivo.
3.3. HOTAIRM1 regulates genes involved in the hypoxia pathway and in early renal lineage differentiation.
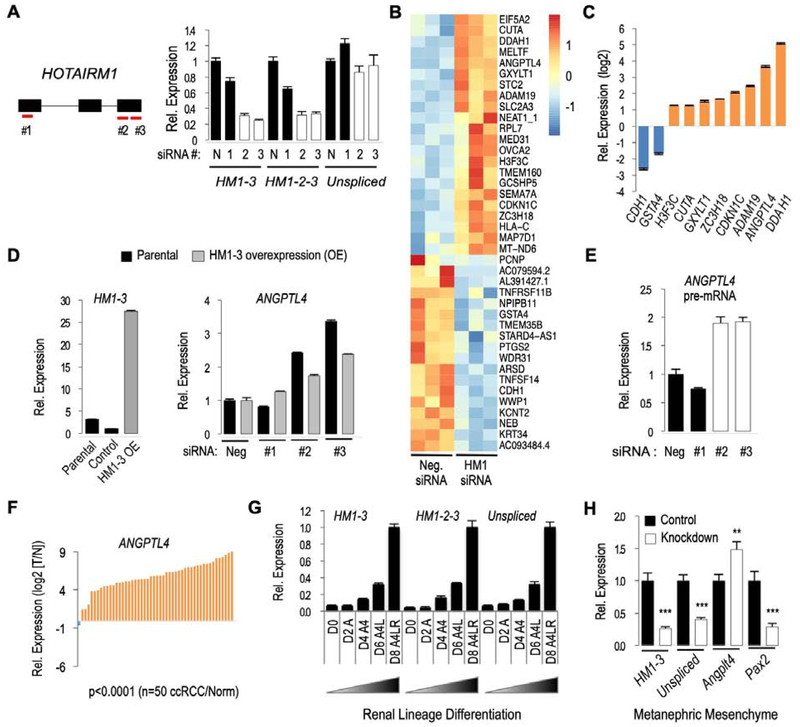
To investigate the functions of HOTAIRM1 we performed knockdown experiments in CAKI-1 cells, as this cell line had the highest levels of HOTAIRM1 transcripts among all the renal proximal tubule epithelial cell lines analyzed (see above). Of the three siRNAs tested, siRNA #1 did not efficiently reduce expression of HOTAIRM1, while siRNAs #2 and #3 were effective in knocking down both spliced isoforms but did not affect the levels of the nuclear unspliced HOTAIRM1 transcript under the conditions used (Figure 3A). This provided us with conditions to selectively test the functions of the spliced isoforms. We hypothesized that albeit cytoplasmic the spliced isoforms could nevertheless influence gene expression and the levels of specific mRNAs. To test this, we performed a stranded RNA-seq analysis of cells transfected with negative control siRNA or specific siRNA #2 (Figure 3B). Combining edgeR and kallisto-sleuth analyses - which provides increased sensitivity [17] - a total of 40 genes were found differentially expressed (Figure 3B). The edgeR analysis identified 28 differentially expressed genes (16 upregulated and 12 downregulated), with at least a 1.25 fold change (FDR ≤ 0.05). Using kallisto gene counts and sleuth for differential expression analysis, 14 differentially expressed genes (8 upregulated and 6 downregulated) were identified with a 0.5 bias estimator value (FDR ≤ 0.05). Two genes DDAH1 and MELTF were found upregulated in both the edgeR and kallisto-sleuth analyses. Among a set of 10 upregulated genes that were partially randomly selected for RT-qPCR validation, 8 genes (80%) were confirmed to be upregulated: DDAH1, ANGPTL4, ADAM19, CDKN1C, ZC3H18, GXYLT1, CUTA, and H3F3C. Conversely, of a selection of 7 downregulated genes only two genes (29%) were validated by RT-qPCR: CDH1 and GSTA4 (Figure 3C). This could suggest a predominantly inhibitory role of the cytoplasmic HOTAIRM1 spliced isoforms (HM1–3 and HM1-2-3) on expression of a limited number of specific genes in CAKI-1 cells.
Figure 3. Analysis of HOTAIRM1-dependent genes in human renal CAKI-1 cells and in differentiating mouse kidney progenitor cells identifies ANGPTL4 as a transcriptional target.
A. HOTAIRM1 siRNAs (#2 and #3) selectively reduce the levels of the spliced isoforms (HM1–3 and HM1-2-3) in CAKI-1 cells. Expression relative to control (N) was analyzed by RT-qPCR. B. Knockdown of HOTAIRM1 spliced isoforms with siRNA#2 identifies 40 deregulated genes (DEGs) by combining edgeR (fold change >1.25 and FDR <0.05) and sleuth analyses (β >0.5 and FDR <0.05). C. Validation by qPCR of 10 DEGs in CAKI-1 cells. Shown is log2 fold change expression between control siRNA (N) and specific siRNA (#2) treated cells. D. Overexpression of ectopic HM1–3 inhibits ANGPTL4 induction upon knockdown of HOTAIRM1 in CAKI-1 cells. E. RT-qPCR analysis of ANGPTL4 pre-mRNA upon HOTAIRM1 knockdown. F. ANGPTL4 expression (log2 TPM tumor/normal ratios) in 50 ccRCC matched pair TCGA samples. Statistical significance was determined using the Wilcoxon signed-rank test for the paired samples. G. Mouse Hotairm1 isoform expression by RT-qPCR during mES cell differentiation into kidney progenitor cells. D=day, A=Activin-A, 4=BMP-4, L=LiCl, R=Retionic acid. H. RT-qPCR analysis of Angpl4 and Pax2 expression upon Hotairm1 knockdown in kidney progenitor cells. Two-tailed student’s t-test used to determine statistical significance. Error bars represent SEM of technical replicates for A,C,D,E and biological replicates for G and H (**p<0.005, ***p<0.0005).
A Metascape enrichment analysis of the differentially expressed genes (DEGs) was performed to identify molecular pathways altered in the knockdown cells (http://www.metascape.org). This suggested a possible enrichment in genes related to the response to hypoxia (p=0.0014 for all 40 DEGs, or p=0.0032 for selectively the 23 upregulated genes). Further analysis of the specific set of upregulated genes using the Enrichr comprehensive database analysis tool (http://amp.pharm.mssm.edu/Enrichr) confirmed a significant enrichment in hypoxia upregulated genes (p=2.3E-5, adjusted p-value= 0.011; disease perturbations from GEO-up GSE4483 dataset; ANGPTL4, SLC2A3, HLA-C, RPL7, NEAT1). Consistent with a possible connection to hypoxia, Enrichr also retrieved ChIP-seq data from the ChEA 2016 database identifying HIF1α target genes (p=0.005; ANGPTL4, SLC2A3, STC2).
To investigate the functions of the spliced HM1–3 isoform we focused on DDAH1 and ANGPTL4, two genes that were most deregulated in knockdown cells and are linked to hypoxia and angiogenesis pathways. We confirmed that the upregulation of both ANGPTL4 and DDAH1 mRNAs in HOTAIRM1 knockdown CAKI-1 cells could be partially rescued by overexpression of the HM1–3 isoform (Figure 3D; Supplemental Figure 6). These results supported the notion that the spliced HM1–3 isoform contributes (at least in part) to the reduced steady state levels of the DDAH1 and ANGPTL4 mRNAs in these cells. Interestingly, this regulation is probably at the transcriptional level for ANGPTL4, since knockdown of HM1–3 and HM1-2-3 (with either siRNA #2 or #3) also increased the levels of the ANGPTL4 pre-mRNA (Figure 3E, see also below). In contrast, DDAH1 pre-mRNA expression was unaffected suggesting a different post-transcriptional regulation of DDAH1 mRNA levels (Supplemental Figure 6). Consistent with these results and the above observation that HM1–3 is the predominant isoform downregulated in most ccRCC tumors, ANGPTL4 was found significantly upregulated in the vast majority of TCGA ccRCC samples relative to their respective normal adjacent tissues (Figure 3F, Supplemental Figure 4B). In contrast, DDAH1 mRNA was consistently downregulated in these ccRCC tumor samples (data not shown).
Since HOTAIRM1 expression was associated with a non-transformed or differentiated cellular state, we further analyzed the gene regulatory functions of HOTAIRM1 during neural and kidney lineage differentiation. Mouse Hotairm1 transcripts, which are highly expressed during RA-induced neural differentiation of ES cells (Supplemental Figure 5), were knocked down during RA treatment in an shRNA-inducible mouse ES cell line (Supplemental Figure 7A). As expected, this selectively inhibited expression of the most anterior Hoxa genes (Hoxa1–5), although Hoxa1 was only marginally affected and only after 96h of induction with RA. Notably, Hotairm1 knockdown increased the expression of Angptl4 and reduced Ddah1 mRNA levels, concomitant with a modest induction of the pluripotency gene Sox2 (Supplemental Figure 7B). Alternatively, mouse ES cells were differentiated into early renal lineages using a stepwise protocol that mimics in vivo development up to the nephrogenic intermediate mesoderm and early metanephric mesenchyme and ureteric bud stages, as previously described [40]. The progressive differentiation was verified by the expression of lineage stage-specific gene markers (Supplemental Figure 8), Evaluation of the three main Hotairm1 isoforms showed a progressive increase in expression and a peak expression at the end of the induction process at day 8 (Figure 3G). Knockdown of Hotairm1 in these renal progenitor cells showed slight reductions in OSR1 and GDNF expression (data not shown) and a large reduction in PAX2 expression (Figure 3H). We did not observe significant changes in the expression of the other kidney lineage differentiation markers analyzed here. However, as seen above with CAKI-1 cells and ES-derived neural lineage cells, Hotairm1 knockdown also induced Angptl4 expression in these renal progenitor cells (Figure 3H).
Altogether, these results suggested a role of HOTAIRM1 during neural and renal lineage differentiation and in regulation of hypoxia signaling, including a possible indirect transcriptional regulation of ANGPTL4 by the cytoplasmic HM1–3 spliced isoform.
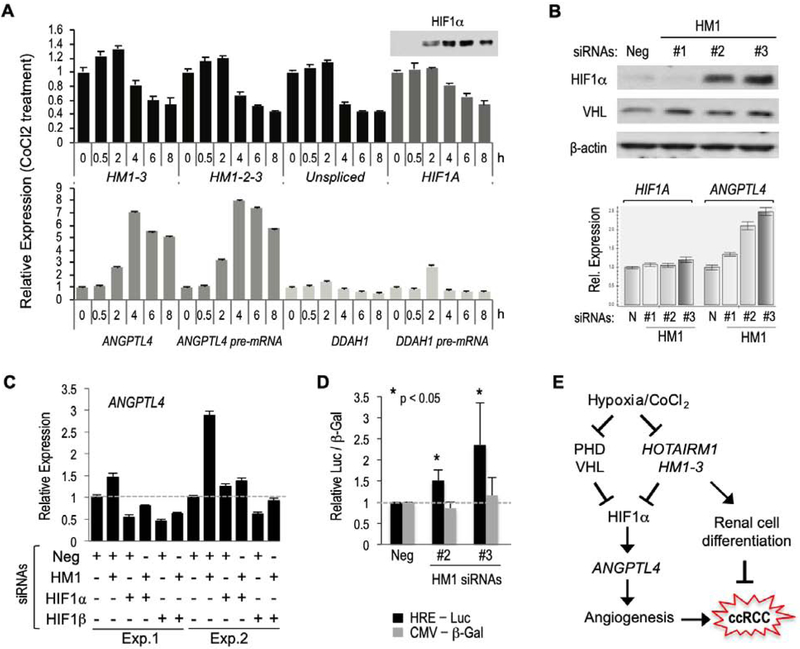
3.4. HOTAIRM1 is downregulated by hypoxia-mimetic stress and inhibits HIF1α protein expression in normoxic cells.
To explore the possible connection of HOTAIRM1 with the hypoxia pathway, we exposed CAKI-1 cells to 100 uM cobalt chloride, a hypoxia-mimetic agent that inhibits prolyl hydroxylases and thereby prevents VHL-mediated ubiqutination and proteasomal degradation of HIF1α. As anticipated, a time course analysis of cells treated with cobalt chloride showed a gradual induction of the HIF1α protein (despite slight downregulation of HIF1A mRNA) and concomitant induction of the HIF1α direct target gene ANGPTL4, peaking at approximately 4 hours of treatment (Figure 4A). In contrast, the DDAH1 mRNA was not induced but seemed to decline during the 4–8h treatment period. Interestingly, the levels of all HOTAIRM1 transcripts remained relatively constant during the first 2h of cobalt chloride treatment, and then rapidly decreased at 4h and later time points (Figure 4A). Thus, HOTAIRM1 expression inversely correlates with both HIF1α protein levels and expression of the ANGPTL4 gene during cobalt chloride treatment. This suggested a possible antagonist regulation between HOTAIRM1 and HIF1α and the possibility that HOTAIRM1 negatively regulates HIF1α expression under normoxic conditions. To test this, HOTAIRM1 spliced transcripts were knocked down in normoxic CAKI-1 cells (with siRNA #2 or #3). This resulted in stimulation of ANGPTL4, as expected, but interestingly also induced HIF1α protein levels (Figure 4B, top panel). Since HIF1A mRNA levels were not affected (Figure 4B, bottom panel) this implies a negative posttranscriptional regulation of HIF1α by the HOTAIRM1 spliced isoforms. Note also that the knockdown of HOTAIRM1 transcripts did not affect VHL protein expression (Figure 4B).
Figure 4. Hypoxia-mimetic stress agent CoCl2 downregulates HOTAIRM1 and HOTAIRM1 downregulation increases HIF1α protein levels and HIF1 activity in normoxic renal cells.
A. Gene expression analysis by RT-qPCR in CAKI-1 cells exposed to 100uM cobalt chloride for the indicated times (0–8h). HIF1A protein (HIF1α) levels were also analyzed by Western blot (inset above HIF1A). B. Western blot analysis of HIF1α and VHL proteins in cells treated with the indicated siRNAs (top) and corresponding RT-qPCR analysis of HIF1A and ANGPTL4 expression (bottom). C. RT-qPCR analysis of ANGPTL4 expression in CAKI-1 cells treated with the indicated siRNAs against HOTAIRM1 (HM1) or HIF1 complex components (HIF1α or HIF1β). D. Knockdown of HM1 in normoxic CAKI-1 cells increases hypoxia response element-dependent transcription of a luciferase reporter gene (HRE-Luc) but does not affect expression of the co-transfected CMV-β-Galactosidase reporter (CMV-b-Gal). E. Schematic model of possible role of HOTAIRM1 lncRNA HM1–3 in kidney cell differentiation and ccRCC. The model does not differentiate a role of HM1–3 within the prolyl-hydroxylase (PHD)/VHL pathway or in a parallel pathway.
To determine whether the negative regulation of ANGPTL4 by the HOTAIRM1 spliced isoforms involved suppression of the HIF1 pathway under normoxia, cells were depleted of either HIF1α or its obligatory dimerization/DNA-binding partner HIF1b with specific siRNAs. Under these conditions knockdown of HOTAIRM1 failed to induce ANGPTL4 (Figure 4C), indicating a requirement for HIF1 signaling. To further verify that HOTAIRM1 regulates the HIF1 pathway, the activity of a luciferase reporter gene under the control of three hypoxia-responsive DNA elements (HRE-Luc) was analyzed in cells transfected with either the negative control siRNA or the two HOTAIRM1-specific siRNAs (#2 and #3). Consistent with the above results, HOTAIRM1 knockdown stimulated the HRE-Luc reporter gene but not a co-transfected CMV-b-Gal reporter plasmid (Figure 4D). Altogether these analyses show that HOTAIRM1 expression is downregulated by hypoxia-mimetic stress and that its cytoplasmic spliced isoforms inhibit transcription of the ANGPTL4 gene via a posttranscriptional mechanism that prevents HIF1α protein accumulation and HIF1 signaling in normoxic cells.
4. Discussion
In the current study, we provide new insights into the functional role(s) of the HOTAIRM1 lncRNAs in kidney biology. Our analyses identify a novel and pervasive downregulation of the spliced HM1–3 isoform in ccRCCs, not previously reported. We demonstrate that HM1–3 is the only HOTAIRM1 isoform downregulated in ccRCC and is the most abundant HOTAIRM1 transcript found in normal kidney tissue. Furthermore, we show that HM1–3 is largely localized to the cytoplasm in all renal and non-renal cell lines analyzed. HOTAIRM1 is downregulated by hypoxia-mimetic stress and knockdown of its cytoplasmic isoforms in normoxic CAKI-1 cells leads to increased HIF1α protein levels and upregulation of hypoxia-responsive genes, including the angiogenic ANGPTL4 gene. Our results reveal for the first time that HOTAIRM1 transcripts are induced during early renal lineage differentiation in vitro and are required for expression of several differentiation markers suggesting a possible role of HOTAIRM1 in kidney tissue differentiation and maintenance. Altogether, our results suggest the possibility of pro-differentiation and tumor-suppressive roles of HM1–3 in the kidney via attenuation of HIF1 signaling, an oncogenic pathway that is recurrently engaged in the vast majority of ccRCC tumors (Figure 4E).
Our findings support a new, and possibly conserved, role of HOTAIRM1 during differentiation of normal ES cells into early renal and neural cell lineages that extends beyond its previously reported involvement in RA-induced myeloid and neural differentiation of cancer cell lines. The expression profiles of HOTAIRM1 during differentiation of mouse ES cells into early renal lineages mirrored the expression profiles of WNT11, GDNF and CDH11. Additionally, HOTAIRM1 appeared to be necessary to maintain the kidney progenitor state, as knockdown of HOTAIRM1 reduced expression of the kidney lineage differentiation markers, OSR1, GDNF and PAX2. Since knockdown of mouse Hotairm1 did not exclusively or preferentially affect HM1–3, it remains unclear which of the different lncRNA isoforms is responsible for this regulation. However, we suspect that HM1–3 is likely to be a major contributor as it is the most abundant Hotairm1 isoform in these early renal lineage cells and in adult kidney tissue. Notably, Angptl4 was downregulated by Hotairm1 during both neural and renal differentiation of mouse ES cells similar to human CAKI-1 cells, indicating a conserved regulatory pathway.
Mechanistically, we provide evidence that expression of both HIF1α and HIF1β, is required for the increased transcription of ANGPTL4 pre-mRNA observed upon depletion of cytoplasmic HOTAIRM1 splice variants in CAKI-1 cells. However, it remains unclear at this stage how HOTAIRM1 cytoplasmic isoforms inhibit posttranscriptionally the accumulation of the HIF1α protein in normoxic cells (i.e., without altering HIF1α mRNA levels). Future analyses will address this important question. The fact that the HIF1 pathway is recurrently activated in ccRCC via the frequent inactivation of the VHL tumor suppressor [6, 7] raises the important questions of whether HM1–3 is a bona fide tumor-suppressive lncRNA and whether it helps maintain low levels of HIF1α protein levels in normoxic cells by functioning either as an intrinsic component of the VHL pathway or in a separate/parallel pathway. We show that during hypoxia-mimetic stress all HOTARM1 transcripts are downregulated raising the possibility that HIF1α accumulation during hypoxia might inhibit transcription of the HOTAIRM1 locus. It will be important to further characterize this potential negative feedback loop between HOTAIRM1 and HIF1α and determine whether its deregulation impacts ccRCC development. Furthermore, the fact that only the HM1–3 spliced isoform but not the primary unspliced HOTAIRM1 transcript is downregulated in ccRCC may suggest a defective HOTAIRM1 splicing mechanism in this major kidney cancer that is worth further investigating. Note that while it remains unclear whether the reduced HM1–3 levels drive or are merely a consequence of ccRCC development, the reduced ratio of spliced (HM1–3) versus unspliced HOTAIRM1 observed in the vast majority of ccRCC tumors may be a new molecular indicator of this specific kidney malignancy, which is similar to - and consistent with - the recent identification of ANGPTL4 expression as a diagnostic marker of specifically ccRCC [41].
In conclusion, this study reveals a new function of HOTAIRM1 in the kidney that is associated with renal cell differentiation and the hypoxia pathway, and uncovers the pervasive downregulation of its major HM1–3 cytoplasmic spliced isoform in ccRCC tumors.
Supplementary Material
Highlights:
New role of HOTAIRM1 in the kidney
Spliced cytoplasmic isoform of HOTAIRM1 is downregulated in ccRCC
Expression of HOTAIRM1 is associated with renal lineage differentiation
HOTAIRM1 expression is inhibited by hypoxic stress signaling
HOTAIRM1 downregulates Hypoxia-Inducible Factor 1α
Acknowledgements
The results reported here are in part based upon data generated by The Cancer Genome Atlas (TCGA) managed by the NCI and NHGRI of the National Institutes of Health (NIH). Information about TCGA can be found at http://cancergenome.nih.gov. This work was supported by a grant from NIH (R01CA158540). A.T.K. was supported by a MARC U-STAR training grant from the NIH (T34GM062756).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest Statement
The authors have no conflicts of interest.
6. References
- 1.United States Cancer Statistics: 1999–2014 Incidence and Mortality Web-based Report.
- 2.Seitz W, Karcher KH, Binder W: Radiotherapy of metastatic renal cell carcinoma. Seminars in surgical oncology 1988, 4(2):100–102. [PubMed] [Google Scholar]
- 3.Ferguson RE, Jackson SM, Stanley AJ, Joyce AD, Harnden P, Morrison EE, Patel PM, Phillips RM, Selby PJ, Banks RE: Intrinsic chemotherapy resistance to the tubulin-binding antimitotic agents in renal cell carcinoma. International journal of cancer 2005, 115(1):155–163. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Facts & Figures. 2018.
- 5.Linehan WM, Walther MM, Zbar B: The genetic basis of cancer of the kidney. J Urol 2003, 170(6 Pt 1):2163–2172. [DOI] [PubMed] [Google Scholar]
- 6.Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM et al. : Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet 1994, 7(1):85–90. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research N: Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499(7456):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blondeau JJ, Deng M, Syring I, Schrodter S, Schmidt D, Perner S, Muller SC, Ellinger J: Identification of novel long non-coding RNAs in clear cell renal cell carcinoma. Clinical epigenetics 2015, 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malouf GG, Zhang J, Yuan Y, Comperat E, Roupret M, Cussenot O, Chen Y, Thompson EJ, Tannir NM, Weinstein JN et al. : Characterization of long non-coding RNA transcriptome in clear-cell renal cell carcinoma by next-generation deep sequencing. Molecular oncology 2015, 9(1):32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC et al. : PVT1 dependence in cancer with MYC copy-number increase. Nature 2014, 512(7512):82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho SW, Xu J, Sun R, Mumbach MR, Carter AC, Chen YG, Yost KE, Kim J, He J, Nevins SA et al. : Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell 2018, 173(6):1398–1412 e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton MJ, Young MD, Sauer S, Martinez E: The interplay of long non-coding RNAs and MYC in cancer. AIMS biophysics 2015, 2(4):794–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC: HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer cell 2007, 11(4):335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordan JD, Lal P, Dondeti VR, Letrero R, Parekh KN, Oquendo CE, Greenberg RA, Flaherty KT, Rathmell WK, Keith B et al. : HIF-alpha effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer cell 2008, 14(6):435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grampp S, Platt JL, Lauer V, Salama R, Kranz F, Neumann VK, Wach S, Stohr C, Hartmann A, Eckardt KU et al. : Genetic variation at the 8q24.21 renal cancer susceptibility locus affects HIF binding to a MYC enhancer. Nature communications 2016, 7:13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Hao Y, Yu W, Yang X, Luo X, Zhao J, Li J, Hu X, Li L: Long Non-Coding RNA Emergence During Renal Cell Carcinoma Tumorigenesis. Cell Physiol Biochem 2018, 47(2):735–746. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton MJ, Girke T, Martinez E: Global isoform-specific transcript alterations and deregulated networks in clear cell renal cell carcinoma. Oncotarget 2018, 9(34):23670–23680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE: A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009, 113(11):2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Weissman SM, Newburger PE: Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol 2014, 11(6):777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang XQ, Dostie J: Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan L, Kong J, Tang J, Wu Y, Xu E, Lai M, Zhang H: HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. Journal of cellular and molecular medicine 2016, 20(11):2036–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, Lui WM, Wong ST, Leung GK: Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiology of disease 2012, 48(1):1–8. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y, Gong B, Jiang ZL, Zhong S, Liu XC, Dong K, Wu HS, Yang HJ, Zhu SK: Microarray expression profile analysis of long non-coding RNAs in pancreatic ductal adenocarcinoma. International journal of oncology 2016, 48(2):670–680. [DOI] [PubMed] [Google Scholar]
- 24.Diaz-Beya M, Brunet S, Nomdedeu J, Pratcorona M, Cordeiro A, Gallardo D, Escoda L, Tormo M, Heras I, Ribera JM et al. : The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget 2015, 6(31):31613–31627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen ZH, Wang WT, Huang W, Fang K, Sun YM, Liu SR, Luo XQ, Chen YQ: The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ 2017, 24(2):212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, Park Y, Dickins RA, Hannon GJ, Lowe SW: A pipeline for the generation of shRNA transgenic mice. Nat Protoc 2012, 7(2):374–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupasquier S, Delmarcelle AS, Marbaix E, Cosyns JP, Courtoy PJ, Pierreux CE: Validation of housekeeping gene and impact on normalized gene expression in clear cell renal cell carcinoma: critical reassessment of YBX3/ZONAB/CSDA expression. BMC molecular biology 2014, 15:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung M, Ramankulov A, Roigas J, Johannsen M, Ringsdorf M, Kristiansen G, Jung K: In search of suitable reference genes for gene expression studies of human renal cell carcinoma by real-time PCR. BMC molecular biology 2007, 8:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bray NL, Pimentel H, Melsted P, Pachter L: Near-optimal probabilistic RNA-seq quantification. Nature biotechnology 2016, 34(5):525–527. [DOI] [PubMed] [Google Scholar]
- 30.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L: Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols 2012, 7(3):562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L: Differential analysis of RNA-seq incorporating quantification uncertainty. Nature methods 2017, 14(7):687–690. [DOI] [PubMed] [Google Scholar]
- 32.Kim D, Langmead B, Salzberg SL: HISAT: a fast spliced aligner with low memory requirements. Nature methods 2015, 12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ: Software for computing and annotating genomic ranges. PLoS computational biology 2013, 9(8):e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson MD, McCarthy DJ, Smyth GK: edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy DJ, Chen Y, Smyth GK: Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic acids research 2012, 40(10):4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.TW HB, Girke T: systemPipeR: NGS workflow and report generation environment. BMC bioinformatics 2016, 17:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love MI, Huber W, Anders S: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 2014, 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkerson MD, Hayes DN: ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26(12):1572–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novak P, Jensen T, Oshiro MM, Wozniak RJ, Nouzova M, Watts GS, Klimecki WT, Kim C, Futscher BW: Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer research 2006, 66(22):10664–10670. [DOI] [PubMed] [Google Scholar]
- 40.Nishikawa M, Yanagawa N, Kojima N, Yuri S, Hauser PV, Jo OD, Yanagawa N: Stepwise renal lineage differentiation of mouse embryonic stem cells tracing in vivo development. Biochemical and biophysical research communications 2012, 417(2):897–902. [DOI] [PubMed] [Google Scholar]
- 41.Verine J, Lehmann-Che J, Soliman H, Feugeas JP, Vidal JS, Mongiat-Artus P, Belhadj S, Philippe J, Lesage M, Wittmer E et al. : Determination of angptl4 mRNA as a diagnostic marker of primary and metastatic clear cell renal-cell carcinoma. PLoS One 2010, 5(4):e10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.