Abstract
The Accelerating Medicines Partnership (AMP) Lupus Network was established as a partnership between the NIH, pharmaceutical companies, non-profit stakeholders and lupus investigators across multiple academic centers to apply high throughput technologies to the analysis of renal tissue, urine and blood from patients with lupus nephritis (LN). The AMP network provides publicly accessible data to the community with the goal of generating new scientific hypotheses and improving diagnostic and therapeutic tools so as to improve disease outcomes. We present here a description of the structure of the AMP Lupus Network and a summary of the preliminary results from the Phase 1 studies. The successful completion of Phase 1 sets the stage for analysis of a large cohort of LN samples in Phase 2 and provides a model for establishing similar discovery cohorts.
INTRODUCTION
Lupus nephritis (LN) is a serious complication of systemic lupus erythematosus (SLE) that affects nearly 40% of patients, with even higher rates in minority populations. Despite intense research efforts, treatment options remain inadequate and the development of novel therapies has been slow. End stage renal disease (ESRD) and death are common complications in patients with LN (1, 2). While histologic classification drives the choice of treatment for LN, this classification is only loosely correlated with patient outcome (3–5). The presence of tubular injury, tubulointerstitial inflammation and/or interstitial fibrosis are associated with a poorer prognosis of LN (6–8); however, these are late manifestations of LN that reflect the inability to detect early disease and to treat effectively. This failure likely reflects our limited knowledge of the molecular mechanisms driving kidney damage. Thus, there is a critical need for a comprehensive and high-resolution analysis of tissue and immune cells in LN to identify new drug targets and disease biomarkers.
A central challenge of LN has been identifying disease subsets among patients that can be therapeutically targeted. Pathogenic mechanisms inferred from genetic studies have not yet led to effective therapeutic interventions. Animal models are also imperfect because their relationship to human disease is not well-defined and successful interventions have not yet translated to improved patient outcomes. Some progress has been made in stratifying lupus patients based on molecular analyses of whole blood and lymphocyte subsets. Longitudinal monitoring of whole blood gene expression in 158 pediatric patients identified seven lupus sub-groups as well as a distinct neutrophil signature that is enriched in patients with LN and decreases after treatment. Abnormalities in cell activation remain even after treatment, with differences among nephritis subclasses that suggest differences in the underlying pathogenic mechanisms (9). A CD8 T cell exhaustion signature in the peripheral blood is associated with a better overall prognosis of lupus patients but not with disease activity per se (10). Nevertheless, how pathogenic mechanisms drive molecular stratification of LN remains poorly understood both because whole blood profiling yields insufficient molecular resolution for mechanistic inferences and because changes in the peripheral blood may not reflect the disease processes in the tissue.
The primary goal of the Accelerating Medicines Partnership (AMP) Lupus Network is to improve our understanding of LN pathogenesis by applying new technologies to the analysis of renal tissue, urine and blood in order to identify novel targets for drug development and improve diagnostic classification. Supported by the National Institutes of Allergy and Infectious Diseases, National Institute of Arthritis and Musculoskeletal and Skin Diseases, pharmaceutical companies, and non-profit organizations across the US, the AMP Lupus Network is comprised of academic centers and investigators focused on directly studying patient samples. The Network applies single cell molecular profiling and other high throughput approaches to generate disease-specific, publicly accessible data to the greater biomedical community for further investigations. The driving questions include:
-
(1)
Which cell-types, cell-states, and molecular programs are associated with LN disease activity and responsiveness to therapy?
-
(2)
Can surrogate molecular markers (e.g. from urine, blood leukocytes and/or skin biopsies) be leveraged for diagnostic or prognostic purposes?
STRUCTURE OF THE AMP
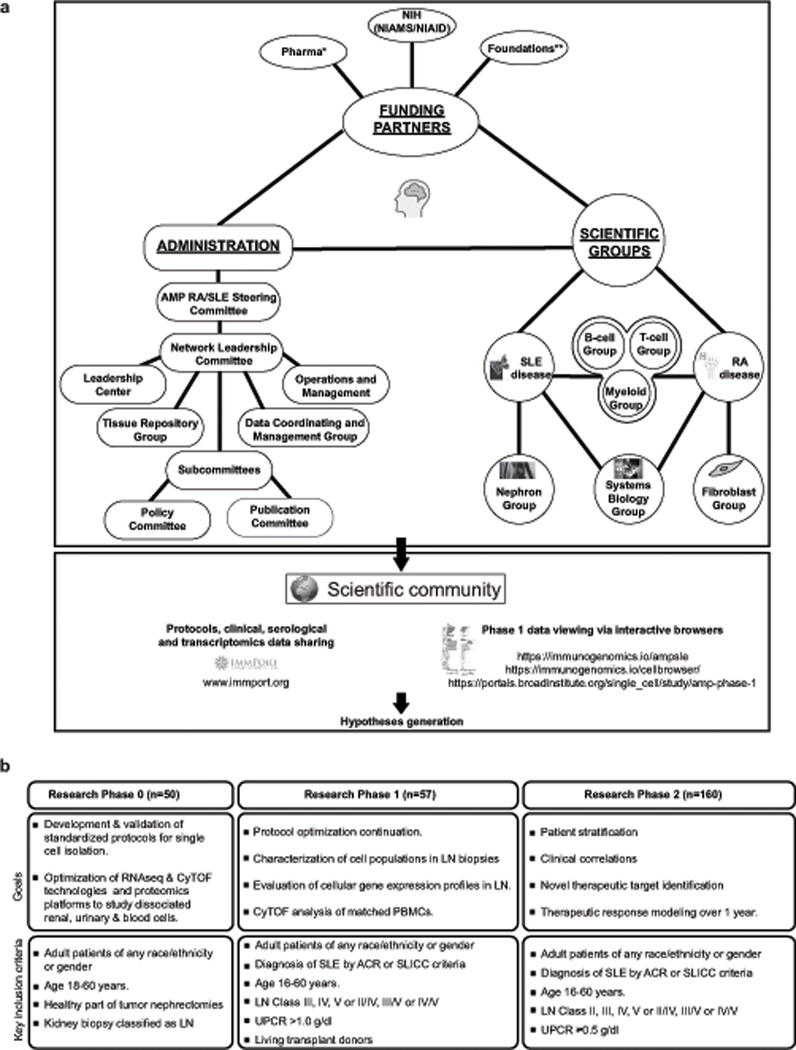
The AMP Lupus Network consists of 5 technology and clinical centers and a network of clinicians, who are collecting patient data and tissues. These centers are supported by an administrative arm, shared with the AMP Rheumatoid Arthritis Network, that oversees data collection, tissue storage and other logistics and by a network of scientific subcommittees each focused on a particular cell subtype or analytic approach (Figure 1a, Table 1). Regularly scheduled conference calls ensure the cohesiveness of the geographically diverse groups and face-to-face meetings occur as needed. Shared data has been loaded into ImmPort (www.immport.org – SDY997).
Figure 1: The structure and goals of the AMP.
a. Overall structure and integration of the AMP Lupus Network. See Table 1 for footnotes. b. Goals and enrolment criteria for each phase of the lupus nephritis studies.
Table 1:
Footnotes to Figure 1
| Funding Partners |
| *Pharma |
| AbbVie Inc. |
| Bristol Myers Squibb Company |
| Merck Sharp and Dohme Corp. |
| Pfizer Inc. |
| Sanofi |
| Takeda Pharmaceuticals International, Inc. |
| Janssen Pharmaceuticals Inc. |
| **Foundations |
| Foundation for the National Institutes of Health |
| Arthritis Foundation |
| Lupus Research Alliance |
| Rheumatology Research Foundation |
| Lupus Foundation of America |
| NIH (NIAMS/NIAID) |
| ImmPort (sponsored by NIAID) |
| Committee Chairs |
| AMP RA/SLE Steering Committee (SC) - Martin Hodge (Pfizer) and Robert Carter (NIAMS) |
| Network Leadership Committee (NLC) and Executive Committee (EC) - Michael Brenner and Jennifer Anolik |
| Policy Committee - Betty Diamond and Michael Weisman |
| Publication Committee - Betty Diamond and Michael Weisman |
| SLE Disease Focus Group - Jill Buyon and Betty Diamond |
| RA Disease Focus Group - Jennifer Anolik and Vivian Bykerk |
| Network Operations - William Apruzzese and Jennifer Goff |
| Leadership Committee – Paul J Utz and Rong Mao |
| Tissue Repository Group - Judith James and Joel Guthridge |
| Data Coordinating and Management Group - Holden Maecker and Rohit Gupta |
| T Cell Group - Deepak Rao |
| B Cell Group - Jennifer Anolik |
| Fibroblast Group - Michael Brenner |
| Nephron Group - Matthias Kretzler |
| Myeloid Group - Laura Donlin |
| Systems Biology Group - Soumya Raychaudhuri |
| Clinical and Technology Sites |
| NYU, Rockefeller Univerity, Albert Einstein College of Medicine (METRO) - Jill Buyon, Chaim Putterman and Tom Tuschl |
| The Feinstein Institutes for Medical Research, The Broad Institute, University of Michigan, University of Cincinatti (PEARL) - Betty Diamond and Nir Hacohen |
| Johns Hopkins University - Michelle Petri |
| Stanford University - Paul Utz |
| University of Oklahoma - Judith James |
| Contributing Sites |
| NYU |
| Brigham and Women’s Hospital |
| University of North Carolina |
| University of California Los Angeles |
| University of California San Diego |
| University of California San Francisco |
| Johns Hopkins University |
| University of Rochester |
| Albert Einstein College of Medicine (Bronx, NY) |
| University of Cincinnati |
| Medical University of South Carolina |
| Zucker School of Medicine - Northwell Health |
| Texas Tech University (El Paso) |
| University of Michigan |
| Temple University |
| University of Texas |
| Cedars-Sinai Hospital |
CLINICAL DESIGN OF THE AMP AND SPECIMEN COLLECTION
The goals and clinical design of each phase of the AMP Lupus Network are shown in Figure 1b. Patients recruited into Phase I of AMP for the SLE component met the following criteria: ACR or SLICC classification criteria for SLE; clinical and laboratory data consistent with LN; the need for a kidney biopsy to guide clinical care regardless of whether this was a first or repeat biopsy. For Phase 1, only patients with urine protein:creatinine ratios (UPCR) >1.0g/dl were included; however, for Phase 2, patients with UPCR ≥ 0.5g/dl are being included. Adult patients of any race/ethnicity or gender were enrolled. Only individuals with histologic RPS/ISN class III, IV, or V (or a mixed class which included one of these) were included in the pipeline analyses for Phase 1 (Figure 2). Patients received standard of care therapy at the discretion of the treating physician. Clinical correlations will be performed in Phase 2; clinical follow up was performed and blood and urine samples were obtained as per protocol at 3, 6 and 12 months. If patients underwent a 2nd biopsy, this was also collected. As expected, some technical variation was present across the various sites, including the size of the biopsy needle and biopsy lengths.
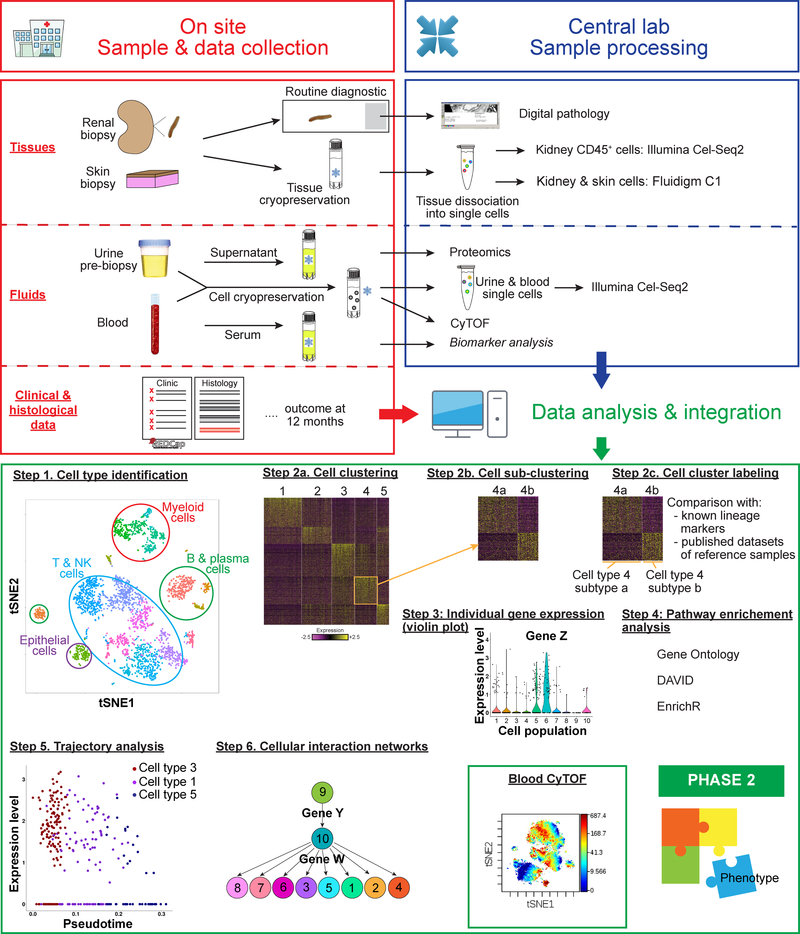
Figure 2: The AMP Lupus Network Pipeline.
Samples and clinical data are collected at point of care (red box). Patient data is loaded into RedCap and samples are processed according to optimized protocols and shipped to the Sample and Tissue Repository for distribution to the technical sites. Proteomic analyses, CYTOF and scRNAseq are each performed at different technical sites (blue box) and data analyses and integration (green box) are performed by the scientific groups. Examples of analyses and integration methods are shown in the bottom panel. Profiled cells are grouped into clusters sharing similar gene expression patterns (Step 1) (19). The dimensionality of the expression data of these genes is reduced using PCA (principal component analysis) and the resulting low-dimensional data analyzed using graph-based clustering (Step 2a). Further subclustering reveals cell subtypes (Step 2b). Cluster labeling is performed by taking into account the distribution of known lineage markers across clusters; the identity of genes specifically upregulated in each cluster; and by comparing the gene expression data of each cluster to those of published datasets of reference samples (Step 2c). Individual gene expression (violin plot) in each cell subtype can be generated (Step 3). Pathway enrichment analysis using curated databases and gene ontology analysis provides information about which genes are active in each pathway (Step 4). Developmental trajectories are constructed by linking cell types to progenitor populations (Step 5) and regulatory relationships can be inferred between genes using cellular interdependency networks (Step 6). Cluster analysis of CyTOF data-displayed as a tSNE plot (Box). Phase 2 will integrate multimodal data to address the goals shown in Figure 1b and to generate hypotheses.
For the Phase 1 program, 57 LN and 15 living transplant donor (LD) renal biopsies from unperfused freshly removed organs were collected from 10 sites over 15 months. 45 individuals with Class III, IV or V pathology and 12 LD controls were included in the Phase 1 analytic pipeline. Data collected for each enrolled participant included demographic information (Table 2), age at SLE diagnosis, ACR classification criteria, SLICC classification criteria, autoantibody titers, clinical laboratory values (such as C3, C4, creatinine, albumin), urinary parameters (urinalysis, UPCR), SELENA-SLEDAI, Physician Global Assessment (PGA), PROMIS-29, medications and adverse events. RedCap data capture forms were used for data entry; a study-specific database was created and maintained by the AMP Leadership Center.
Table 2.
AMP SLE Phase 1 Demographic information.
| Cases n=57* | Controls n=15* |
|
|---|---|---|
| Gender | ||
| Female | 52 (92%) | 11 (73%) |
| Male | 5 (9%) | 4 (27%) |
| Race | ||
| Asian | 7 (12%) | 0 |
| Black/African American | 23 (40%) | 3 (20%) |
| Unknown or not reported | 3 (5%) | 1 (7%) |
| White | 25 (43%) | 11 (73%) |
| Ethnicity | ||
| Hispanic or Latino | 17 (30%) | 2 (13%) |
| Not Hispanic | 40 (70%) | 13 (87%) |
| Age at biopsy (years) | 31.93 ± 10.50 | 35.54 ± 6.27 |
| Medication | ||
| Belimumab | 3 (5%) | |
| Prednisone | 39 (68%) | |
| Hydroxychloroquine | 51 (89%) | |
| Methotrexate | 1 (2%) | |
| Mycophenolic Acid | 1 (2%) | |
| Mycophenolate mofetil | 14 (25%) | |
| dsDNA+ (n=48) | 40 (83%) | |
| Low C3 (n=56) | 42 (75%) | |
| Low C4 (n=56) | 37 (66%) | |
| ISN Class** | ||
| [I] | 1 (2%) | |
| [II] | 2 (4%) | |
| [II][V] | 1 (2%) | |
| [III] | 10 (17%) | |
| [III][V] | 9 (16%) | |
| [IV] | 9 (16%) | |
| [IV][V] | 10 (17%) | |
| [V] | 15 (26%) | |
| Activity (n=37) | 4.69 (SEM: 0.78; Range: 0–16) † | |
| Chronicity (n=37) | 1.95 (SEM: 0.29; Range: 0–7) ‡ | |
| ACR 1997 Score (n=57) | 5.84 | |
| SLICC Score (n=51) | 7.80 | |
| SELENA-SLEDAI (n=57) | 12.93 | |
| SLICC Damage Index (n=51) | 3.99 | |
45 LN and 12 LD controls analyzed in Phase 1 and the others rolled over to Phase 2
Class VI biopsies were excluded
Maximum 24
Maximum 12
Renal biopsies were collected and stored as described (11, 12). Although some fresh samples were individually processed in the early phases of the study, protocol optimization performed in Phase 0 and Phase 1 showed that immediate freezing of tissues followed by later thawing and dissociation at a single technology site yielded high quality RNA, ample for downstream applications without a freezing associated molecular signature (11). This protocol has therefore been adopted for all AMP tissue samples. Blood was processed for serum, plasma, PBMCs, and total blood leukocytes and urine was collected and processed using optimized protocols (11, 12). For Phase 2, all samples will be shipped to a single site for storage and subsequent redistribution to the technology sites.
SINGLE CELL RNA SEQUENCING METHODS AND REPRODUCIBILITY
Single-cell RNA sequencing (scRNAseq) is transforming biomedicine by uncovering new cell types and cellular functions in complex biologic tissues (13). Thousands of single cells from individual tissue samples can now be processed in parallel for deep molecular profiling by next-generation sequencing (NGS). Unbiased bioinformatic analysis enables the identification, characterization, and molecular relationships among individual cells. Because scRNAseq is rapidly evolving, the AMP Lupus Network has adopted contemporary scRNAseq approaches to enable state-of-the-art cellular profiling.
The Phase 1 studies used both plate-based (Cel-Seq2, (11)) and Fluidigm platforms (C1 chip, (12)) to profile transcriptomes of single cells. Plate-based technology enabled deep gene profiling of sorted CD45+ cells for improved cellular characterization but was labor intensive and low-throughput; the Fluidigm platform was agnostic, easy to use and improved throughput but captured fewer genes. These methods were applied to 45 patient samples (21 by Fluidigm and 24 by Cel-Seq2) to reveal for the first time the molecular details of diseased renal parenchymal cells and activated immune cells from tissue at unprecedented resolution (11, 12).
Recently, droplet-based approaches have dramatically increased the number of cells that can be profiled in parallel as well as the number of genes detected (14). Single cells are partitioned into nano-liter scale droplets containing barcoded beads that capture gene transcripts for NGS. Droplet based scRNAseq (10X Genomics) will be applied to patient samples in Phase 2. Thousands of renal cells will be analyzed per sample, thus allowing discovery of rare cell populations and enabling new molecular insights while presenting new challenges in data analysis. Several bioinformatic tools have recently been developed that enable analysis of multiple data sets by minimizing the effects of combined analysis of different scRNAseq technologies (15, 16).
The general data analysis pipeline for the Phase 1 studies is shown in Figure 2. First, major cell types were identified by grouping the profiled cells into clusters sharing similar gene expression patterns and then further subclustering was performed to reveal cell subtypes. Cluster labeling was done using a combined approach, taking into account the distribution of known lineage markers across clusters; the identity of genes specifically upregulated in each cluster; and by comparing the gene expression data of each cluster to those of published reference datasets. Understanding which pathways are active in each cell type was elucidated through pathway enrichment and gene ontology analyses (20, 21) and enrichment programs such as DAVID (22) or enrichR (23). Developmental trajectories were revealed by linking cell types to progenitor populations (24). Importantly, Phase 1 established the feasibility for a much larger Phase 2 study of 160 LN patients that is currently underway, with initial sample collection almost complete (Figure 1b).
Single cell RNAseq analysis of dissociated tissue raises powerful new hypotheses, but has several important technical limitations (13). First, tissue disaggregation destroys spatial context among cell types and may deplete some cell populations for downstream analyses or introduce stress signatures. We have found, for example, that kidney epithelial cells are particularly sensitive to cell death and/or cell stress upon disaggregation. Second, low abundance RNAs may not be detected so that important information about cell function may be missed. Third, scRNAseq profiles RNA transcriptomes, which are only an indirect readout of protein expression and cellular function. Recent advances include multiplexed fluorescent in situ hybridization of tissue sections that gives critical information about the spatial context of multiple cell types, and multimodal analysis of single cells to add information about cell surface markers, protein abundance and epigenomic state (13). These advances are occurring in parallel with the development of new methods to integrate multimodal data and compare datasets from different experiments. While these technologies are too new to be applied in the AMP studies, it is expected that they will soon become possible in the setting of cohort studies of disease such as those described here. Finally, construction of a Human Cell Atlas (https://www.humancellatlas.org) will allow easier comparisons of disease states with normal tissue.
SUMMARY OF scRNAseq DATA FROM THE PHASE 1 STUDIES
A. Studies of whole kidney and skin by the METRO group
Both kidney and skin were analyzed by the METRO group. Biomarkers available from skin biopsies would be a desirable option, given the easier accessibility of skin as compared to kidney tissue. The concept that skin can reflect the immunologic milieu of SLE dates back to the original demonstration of immunoglobulin and complement deposition at the dermal-epidermal junction in both lesional and non-lesional skin (23). Activation of the microvasculature is found even in non-sun exposed, non-lesional skin of patients with active lupus (24–26), and endothelial changes in the kidneys of patients with LN predict poor responses to therapy (27). Thus, serial analysis of non-involved skin, though distant from the primary affected organ, may provide an opportunity to explore surrogates for renal tissue analyses so as to facilitate early identification critical to renal survival and follow treatment responses. Accordingly, 2 mm biopsies from non-lesional non-sun-exposed skin (buttocks) were performed at the METRO sites on patients donating renal tissue as part of AMP.
Using the Fluidigm C1 Autoprep system, LN and healthy control subject skin and renal biopsies were interrogated by scRNAseq without presorting or cell type selection. 21 LN kidney biopsies and 17 skin biopsies were analyzed in Phase 1 (12). Graph-based clustering and t-Distributed Stochastic Neighbor Embedding visualization (tSNE) (28) resolved major skin and kidney cell populations including tubular cells, keratinocytes, endothelial cells, fibroblasts, and leukocytes. Subtypes of skin and kidney epithelial cells were further resolved including melanocytes, sweat gland cells, proximal and distal tubular cells, and collecting duct cells. This agnostic approach allowed us to focus on the epithelial cells of both tissues which were analyzed for prognostic and diagnostic markers.
Previously published Phase 0 studies of LN keratinocytes (29) demonstrated an upregulated IFN response signature compared to healthy control subject keratinocytes. This finding was replicated in Phase 1 and further extended to tubular cells. Preliminary analysis of small numbers of LN patients with available follow-up data found that tubular cells from patients who did not respond to conventional therapy at six months post biopsy showed a higher IFN score and increased expression of genes encoding extracellular matrix (ECM) proteins and ECM interaction proteins, suggesting a fibrotic process. A trend towards upregulation of both ECM pathways was also observed in keratinocytes of non-responders (30). Exploration of the cellular interactions between various cell types in the kidney and skin suggest that the fibrotic process may be mediated through fibroblast growth factor receptors on the tubular cells whose ligand is expressed in leukocytes. Further preliminary analyses suggested that there might be gene signatures that distinguish histologic subclasses of disease. These findings need to be confirmed in the Phase 2 studies. Resident renal cells also expressed high levels of chemokines whose receptors were expressed by leukocytes, indicating a potential mechanism for immune cell infiltration into the glomeruli and tubulointerstitium (12).
B. Studies of infiltrating immune cells by the PEARL group
Viable CD45+ immune cells were sorted from renal biopsies from 24 LN and 12 unperfused living renal transplant donors (LD) and scRNAseq was performed using Cel-Seq2 (11). Batch effects were minimal, allowing comparison of data from all the samples in a single analysis. We identified 21 immune cell clusters in the LN patients including 10 subsets of NK and T cells, 4 clusters of B cells, 6 clusters of macrophages and DCs and one mixed cluster of dividing cells. Memory CD4 T cells and resident macrophages, were the most frequently identified subsets in LD. Saturation analysis indicated that this initial cohort size was sufficient to identify most cell clusters. Comparisons between LN and LD cells indicated that an IFN signature is present in most cell types from the LN patients.
Analysis of the transcriptome of each subset yielded several novel findings (11). Most of the dividing cells were CD8+ T cells and NK cells; these cells also expressed the most IFNγ. By contrast, Th1 and Th17 cytokine expressing CD4 T cells were present in lower abundance without skewing to either subset. Novel CD8 T cell subsets were identified in the tissue but exhausted CD8 T cells were not detected although these were readily identified in the peripheral blood. B cells of naive and activated phenotypes were detected, including B cells with an “age-associated” phenotype and plasma cells. TFH-like CD4 T cells were also found, confirming previous data that shows that T and B cell activation occur in situ (31). Macrophages, myeloid dendritic cells and plasmacytoid dendritic cells were all found. Analysis of the macrophage subsets showed 3 subpopulations (CM0, CM1 and CM4) that appeared related by trajectory analysis. These cells most resembled CD16+ peripheral monocytes (32). Of these, the subset most similar to peripheral blood monocytes (CM0) had an inflammatory phenotype which was lost as the cells progressed along the trajectory; instead these cells first acquired a phagocytic (CM1) and then an alternatively activated (CM4) phenotype. These alternatively activated cells were also a major immune cell source of chemokines, suggesting that they may help orchestrate immune cell infiltration and/or organization. CXCR4 and CX3CR1 were the most commonly expressed chemokine receptors among the immune cells (11).
The question of non-invasive methods for evaluation of renal status was addressed by analyzing urine samples from 8 LN patients. Of note, not all of the renal immune cells have access to the urinary space or survive in the urine; compared to kidney, urine cells had a lower frequency of T cells, and instead were dominated by a single cluster of CD16+ macrophages (cluster CM1). Despite the limited diversity of urine immune cells, their transcriptome faithfully reflected that of the kidneys, indicating that the urine can be used to estimate gene expression of the related kidney cells (11).
ANALYSIS OF URINE USING PROTEOMICS
Numerous proteins that participate in the pathophysiology of LN can be measured in the urine and several distinguish the urine of patients with active LN from that of patients with inactive disease. Nevertheless, longitudinal studies have been few and there is as yet no biomarker panel that is superior to standard clinical parameters for predicting LN outcome (33, 34). High throughput proteomic analysis of urines from patients enrolled in the AMP and followed longitudinally for a year will accelerate the pace of discovery of useful LN biomarkers, identify proteomic signatures with greater specificity and sensitivity than a single protein, and help provide additional insights into the underlying biology of the disease process. Furthermore, the ability to correlate proteomic signatures with molecular signatures will greatly enhance the power of this approach.
Two urine proteomics platforms were tested in Phase 1 to demonstrate feasibility and identify potential biomarker targets. The first is an array-based multiplex ELISA system (Quantibody- Raybiotech, Norcross, GA) for simultaneous quantitative measurement of 1000 proteins from small urine volumes including multiple cytokines, growth factors, proteases and soluble receptors. This assay is highly reproducible and combines the high specificity and sensitivity of ELISA with the high throughput of the glass chip-based array. For some molecules, the sensitivity of the arrays far exceeds that of ELISA assays (35, 36). The second approach is a Capillary Electrophoresis/Mass Spectroscopy platform that separates and identifies up to 5000 peptides in the urine with high resolution, sensitivity and reproducibility. This technology can differentiate chronic kidney disease from LN using a classification panel of peptides (37). Preliminary screens of Phase 1 urine samples have shown a large number of elevated proteins and peptides in the urine of LN patients compared to urine from healthy controls, demonstrating the feasibility of using these two proteomic methods in AMP (M. Petri, personal communication).
Questions that can now be addressed in Phase 2 are whether it is possible to differentiate histologic classes or to identify treatment responders. In addition, with the large number of proteins identified in the urine, it may be possible to perform pathway analyses similar to those performed using transcriptomic data. Integration of the two data sets would likely expand our understanding of the pathophysiology of LN.
ANALYSIS OF PERIPHERAL BLOOD SUBSETS USING CYTOF
CyTOF mass cytometry (CyTOF) is a method for comprehensive and accurate multidimensional single-cell phenotyping that employs rare earth metal isotope-tagged rather than fluorescent-tagged antibodies (36). This technology provides the opportunity to simultaneously stain cells with up to 45 different metal-tagged antibodies without major concern for signal spillover or background.
In Phase 1 studies, optimization of cell processing and cryopreservation of both total leukocytes (TL) and PBMC was followed by building of CyTOF antibody panels designed to detect major immune cell subset markers. 3 PBMC and 2 TL panels were developed for Phase 1 to inform the development of phase 2 AMP CyTOF panels. During the development period, pilot CyTOF antibody panel stains were performed to assure proper staining antibody concentrations and to validate staining accuracy. Other approaches to improve data quality included flow cytometry confirmation of cell counts, an assessment of cell viability, use of platinum isotope barcoding reagents for batched sample acquisition and normalization of signal intensity during data acquisition. To reduce batch effects, the same Helios instrument was used for the entire Phase 1 project and samples were randomized into groups to include mixtures of controls and patient samples.
The entire AMP Phase 1 blood phenotyping project analyzed 34 control, 44 SLE, and 33 RA PBMC samples and 17 control, 36 SLE, and 21 RA TL samples (uploaded as a shared dataset to ImmPort (www.immport.org – SDY997 archive). No significant batch effects were detected by the AMP Systems Biology Group. CyTOF staining data has been analyzed for significant immune cell cluster changes and to determine which markers provided the most useful information for single-cell phenotyping. In general, we observed that circulating immune cell subsets from SLE patients were significantly more different from healthy controls than were blood immune cells from RA patients. Several interesting findings included a significant increase in circulating activated CD57+ CD8 T cells, altered ratios of Vδ1 and Vδ2 TCRγδ T cells, and reduced NK cell percentages in SLE patients compared with controls (J. Lederer, personal communication).
The next generation Phase 2 CyTOF panels have removed uninformative antibodies and expanded marker detection on those immune cell subsets showing significant differences between SLE and RA patients and healthy controls. The Phase 2 AMP panels will include 45 markers per panel with new advances in metal isotope antibody labeling methods. In Phase 2, blood immune cells from as many as 400 SLE and RA patients will be profiled using newly-designed antibody panels that are T cell, B cell, innate cell, and neutrophil centric. We anticipate that the results from AMP Phase 2 will identify immune cell phenotypes that could be used to diagnose, predict, or better understand the pathobiology of the SLE disease process.
CONCLUSIONS
Phase 1 lupus AMP studies have identified novel inflammatory cell populations and their origins, have begun to identify possible molecular biomarkers for disease response and have suggested that it may eventually be possible to use non-invasive cell collections to longitudinally study the renal landscape. Together, the Phase 1 studies set the stage for Phase 2 analysis of renal tissue from 160 well-characterized LN patients from which both renal resident cells and immune cells will be analyzed using 10X technology. This will allow us to correlate peripheral blood cell phenotype by CyTOF, the renal transcriptome and the urine proteome with patient histologic subclass, response to therapy and outcome at 12 months. New hypotheses can then be examined using more focused molecular and histologic analyses in new cohorts and examination of specific molecules and pathways in relevant mouse models.
The AMP studies have been completed within the prescribed time frame and with organized input from many investigators, including multiple clinicians who are providing a rich clinical data set to accompany the OMICS studies. The Phase 1 studies have demonstrated good patient safety and the preliminary analyses have confirmed that the data is of high quality. Central storage and the ability to freeze and batch samples has been a key component in maintaining quality. The application and development of new bioinformatics tools such as trajectory analysis and Harmony (15) should enable further novel molecular insights from the larger cohort. As advanced technologies become available, such as histologic immunophenotyping with large numbers of markers, T and B cell repertoire analysis, barcoding and epigenetic profiling, the AMP organizational model can be used as a template for new discovery cohorts.
SIGNIFICANCE AND INNOVATION.
Successful completion of Phase 1 of the Accelerating Medicines Partnership Lupus Network has yielded an optimized set of protocols for state-of-the-art high throughput analysis of renal tissue, urine and blood
The Phase 1 studies have identified novel inflammatory renal cell populations and their origins and have begun to identify possible molecular biomarkers for disease response
Exploratory studies have revealed the potential of using non-invasive cell collections (urine and skin) to longitudinally study the renal landscape
Acknowledgments
Financial support: This work was supported by the Accelerating Medicines Partnership (AMP) in Rheumatoid Arthritis and Lupus Network. AMP is a public-private partnership (AbbVie Inc., Arthritis Foundation, Bristol-Myers Squibb Company, Foundation for the National Institutes of Health, Lupus Foundation of America, Lupus Research Alliance, Merck Sharp & Dohme Corp., National Institute of Allergy and Infectious Diseases, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Pfizer Inc., Rheumatology Research Foundation, Sanofi, and Takeda Pharmaceuticals International, Inc.) created to develop new ways of identifying and validating promising biological targets for diagnostics and drug development. Funding was provided through grants from the National Institutes of Health (UH2-AR067676, UH2-AR067677, UH2-AR067679, UH2-AR067681, UH2-AR067685, UH2-AR067688, UH2-AR067689, UH2-AR067690, UH2-AR067691, UH2-AR067694, and UM2-AR067678). M.P. was supported by AR069572. N.H. was supported by the David P. Ryan, MD Endowed Chair in Cancer Research. We thank participating Lupus Nephritis Trials Network clinical sites and participants, and ImmPort for maintaining the data archive.
REFERENCES
- 1.Jorge A, Wallace ZS, Zhang Y, Lu N, Costenbader KH, Choi HK. All-Cause and Cause-Specific Mortality Trends of End-Stage Renal Disease Due to Lupus Nephritis From 1995 to 2014. Arthritis Rheumatol. 2019;71(3):403–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tektonidou MG, Dasgupta A, Ward MM. Risk of End-Stage Renal Disease in Patients With Lupus Nephritis, 1971–2015: A Systematic Review and Bayesian Meta-Analysis. Arthritis Rheumatol. 2016;68(6):1432–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz MM, Lan SP, Bernstein J, Hill GS, Holley K, Lewis EJ. Irreproducibility of the activity and chronicity indices limits their utility in the management of lupus nephritis. Lupus Nephritis Collaborative Study Group. Am J Kidney Dis 1993;21(4):374–7. [DOI] [PubMed] [Google Scholar]
- 4.Rovin BH, Parikh SV, Alvarado A. The kidney biopsy in lupus nephritis: is it still relevant? Rheum Dis Clin North Am 2014;40(3):537–52, ix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandepapeliere J, Aydin S, Cosyns JP, Depresseux G, Jadoul M, Houssiau FA. Prognosis of proliferative lupus nephritis subsets in the Louvain Lupus Nephritis inception Cohort. Lupus. 2014;23(2):159–65. [DOI] [PubMed] [Google Scholar]
- 6.Hsieh C, Chang A, Brandt D, Guttikonda R, Utset TO, Clark MR. Predicting outcomes of lupus nephritis with tubulointerstitial inflammation and scarring. Arthritis Care Res (Hoboken). 2011;63(6):865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broder A, Mowrey WB, Khan HN, Jovanovic B, Londono-Jimenez A, Izmirly P, et al. Tubulointerstitial damage predicts end stage renal disease in lupus nephritis with preserved to moderately impaired renal function: A retrospective cohort study. Semin Arthritis Rheum 2018;47(4):545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill GS, Delahousse M, Nochy D, Mandet C, Bariety J. Proteinuria and tubulointerstitial lesions in lupus nephritis. Kidney Int 2001;60(5):1893–903. [DOI] [PubMed] [Google Scholar]
- 9.Banchereau R, Hong S, Cantarel B, Baldwin N, Baisch J, Edens M, et al. Personalized Immunomonitoring Uncovers Molecular Networks that Stratify Lupus Patients. Cell. 2016;165(6):1548–50. [DOI] [PubMed] [Google Scholar]
- 10.McKinney EF, Lee JC, Jayne DR, Lyons PA, Smith KG. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 2015;523(7562):612–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arazi A, Rao DA, Berthier CC, Davidson A, Liu Y, Hoover PJ, et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol 2019;20(7):902–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Der E, Suryawanshi H, Morozov P, Kustagi M, Goilav B, Ranabathou S, et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stuart T, Satija Rrno. Integrative single-cell analysis. Nature reviews Genetics. 2019;20(5):257–72. [DOI] [PubMed] [Google Scholar]
- 14.Macosko EZegmhe, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korsunsky I, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, et al. Fast, sensitive, and accurate integration of single cell data with Harmony. bioRxiv 461954v2. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck III WM, et al. Comprehensive integration of single cell data. bioRxiv 460147v1. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 2015;33(5):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mkkk-uaj Kanehisa, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic acids research. 2017;45(D1):D353–D61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie MU, Garapati P, et al. The Reactome Pathway Knowledgebase. Nucleic acids research. 2018;46(D1):D649–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic acids research. 2007;35(Web Server issue):W169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research. 2016;44(W1):W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu XU, Mao Q, Tang YC, Wang L, Chawla RU, Pliner HU, et al. Reversed graph embedding resolves complex single-cell trajectories. Nature methods. 2017;14(10):979–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burnham TK, Neblett TR, Fine G. The application of the fluorescent antibody technic to the investigation of lupus erythematosus and various dermatoses. The Journal of investigative dermatology. 1963;41:451–6. [DOI] [PubMed] [Google Scholar]
- 24.Belmont HM, Buyon J, Giorno R, Abramson S. Up-regulation of endothelial cell adhesion molecules characterizes disease activity in systemic lupus erythematosus. The Shwartzman phenomenon revisited. Arthritis and rheumatism. 1994;37(3):376–83. [DOI] [PubMed] [Google Scholar]
- 25.Belmont HM, Levartovsky D, Goel A, Amin A, Giorno R, Rediske J, et al. Increased nitric oxide production accompanied by the up-regulation of inducible nitric oxide synthase in vascular endothelium from patients with systemic lupus erythematosus. Arthritis and rheumatism. 1997;40(10):1810–6. [DOI] [PubMed] [Google Scholar]
- 26.Izmirly PM, Shvartsbeyn M, Meehan S, Franks A, Braun A, Ginzler E, et al. Dysregulation of the microvasculature in nonlesional non-sun-exposed skin of patients with lupus nephritis. The Journal of rheumatology. 2012;39(3):510–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izmirly PM, Barisoni L, Buyon JP, Kim MY, Rivera TL, Schwartzman JS, et al. Expression of endothelial protein C receptor in cortical peritubular capillaries associates with a poor clinical response in lupus nephritis. Rheumatology (Oxford). 2009;48(5):513–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jamieson ARaue, Giger ML, Drukker K, Li H, Yuan Y, Bhooshan N Exploring nonlinear feature space dimension reduction and data representation in breast Cadx with Laplacian eigenmaps and t-SNE. Medical physics. 2010;37(1):339–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, Morozov P, et al. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI insight. 2017;2(9):e93009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Der, Suryawanshi H, Morozov P, Kustagi M, Goilav B, Ranabathou S, et al. Insights from Comparison of the Renal and Skin Single Cell Transcriptomes in Lupus Nephritis. bioRxiv 382846v1. 2018. [Google Scholar]
- 31.Chang A, Henderson SG, Brandt D, Liu N, Guttikonda R, Hsieh C, et al. In situ B cell-mediated immune responses and tubulointerstitial inflammation in human lupus nephritis. J Immunol 2011;186(3):1849–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356(6335). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soliman S, Mohan CUSEaccue. Lupus nephritis biomarkers. Clin Immunol 2017;185:10–20. [DOI] [PubMed] [Google Scholar]
- 34.Brunner HI, Gulati G, Klein-Gitelman MS, Rouster-Stevens KA, Tucker L, Ardoin SP, et al. Urine biomarkers of chronic kidney damage and renal functional decline in childhood-onset systemic lupus erythematosus. Pediatr Nephrol 2019;34(1):117–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson JJ, Burgess R, Mao YQ, Luo S, Tang H, Jones VS, et al. Antibody arrays in biomarker discovery. Advances in clinical chemistry. 2015;69:255–324. [DOI] [PubMed] [Google Scholar]
- 36.Roybal CN, Velez G, Toral MA, Tsang SH, Bassuk AG, Mahajan VBavmse. Personalized Proteomics in Proliferative Vitreoretinopathy Implicate Hematopoietic Cell Recruitment and mTOR as a Therapeutic Target. American journal of ophthalmology. 2018;186:152–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pejchinovski M, Siwy J, Mullen W, Mischak H, Petri MA, Burkly LC, et al. Urine peptidomic biomarkers for diagnosis of patients with systematic lupus erythematosus. Lupus. 2018;27(1):6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]