Abstract
Staphylococcus aureus is a major human pathogen that causes an array of infections ranging from minor skin infections to more serious infections including osteomyelitis, endocarditis, necrotizing pneumonia and sepsis1. These more serious infections usually arise from an initial bloodstream infection and are frequently recalcitrant to antibiotic treatment 1. Phagocytosis by macrophages and neutrophils is the primary mechanism by which S. aureus infection is controlled by the immune system2. Macrophages have been shown to be a major reservoir of S. aureus in vivo3 but the role of macrophages in the induction of antibiotic tolerance has not been explored. Here we show that macrophages not only fail to efficiently kill phagocytosed S. aureus but also induce tolerance to multiple antibiotics. Reactive oxygen species (ROS) generated by respiratory burst attack iron-sulfur (Fe-S) cluster containing proteins, including TCA cycle enzymes, resulting in decreased respiration, lower ATP and increased antibiotic tolerance. We further show that during a murine systemic infection, respiratory burst induces antibiotic tolerance in the spleen. These results suggest that a major component of the innate immune response is antagonistic to the bactericidal activities of antibiotics.
Failure to eradicate intracellular S. aureus cannot be fully explained by poor antibiotic penetration3–5. Phagocytes can induce antibiotic tolerance of internalized Salmonella Typhimurium (through vacuolar acidification, nutritional deprivation and the activation of toxin-antitoxin modules6), and Mycobacterium tuberculosis (through nitrosative stress7). In this study, we aimed to determine the antibiotic susceptibility of phagocytosed S. aureus and examine the impact of intracellular stresses on the outcome of antibiotic therapy in vitro and during a systemic infection in mice.
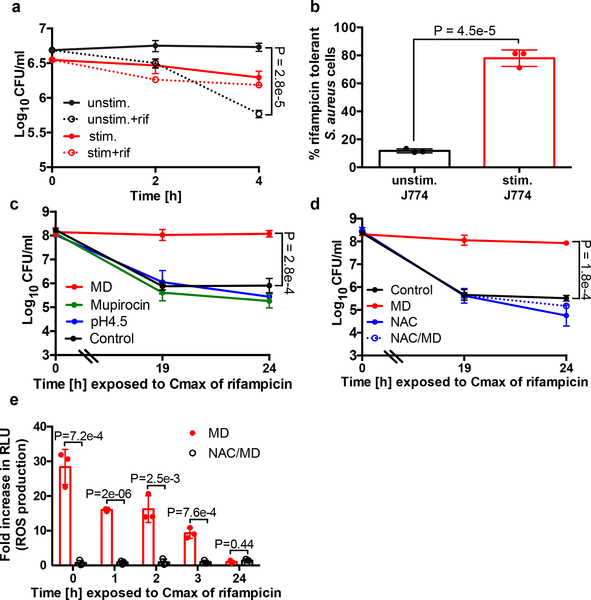
To assess the susceptibility of intracellular S. aureus to antibiotics, we incubated S. aureus with unstimulated J774 macrophages or macrophages that were stimulated overnight with lipopolysaccharide and interferon-𝛄 (LPS/IFNγ) to enhance their microbiocidal activity. Cells were challenged with rifampicin, a bactericidal antibiotic used in the treatment of S. aureus infection, which penetrates the macrophage rapidly by passive diffusion8. Interestingly, the majority of S. aureus cells that were internalized by unstimulated macrophages remained susceptible to rifampicin killing (Fig. 1a–b). In contrast, stimulated macrophages were more efficient at reducing the S. aureus burden, but the surviving bacteria were significantly more tolerant to rifampicin (Fig. 1a–b).
Fig. 1. Induction of antibiotic tolerance by stimulated macrophages.
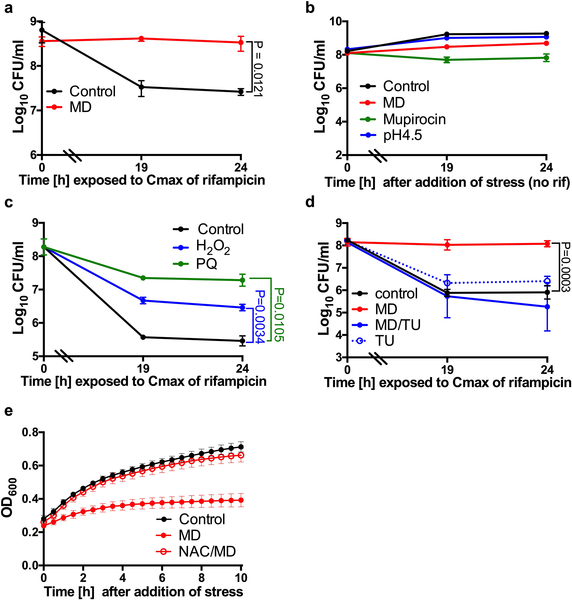
(a) Survival of S. aureus cells after internalization by unstimulated J774 macrophages (unstim.) or macrophages that were stimulated overnight with LPS/IFNγ (stim.). 10μg/ml rifampicin (rif) was added at Time 0 (dotted lines). (b) % Survival of S. aureus cells treated with rifampicin for 4h compared to the untreated control (extrapolated from a). (c-e) S. aureus strain HG003 was grown to mid-exponential phase in vitro and exposed to 80μM menadione (MD), 0.125μg/ml mupirocin, acidified media (pH4.5) or 4mM N-acetyl cysteine (NAC) for 20min prior to the addition of rifampicin at Time 0. At indicated times, an aliquot was removed, (c, d) washed in 1% NaCl and plated to enumerate survivors or (e) ROS production was measured using L-012 luminescent probe and expressed as a fold-increase compared to the untreated control. RLU denotes relative luminescence units. Averages of n=3 biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using (a, c-d) One-Way ANOVA with Sidak’s multiple comparison test or (b, e) Student’s t-test (unpaired, two-tailed). See also Extended Data Fig. 1.
We hypothesized that specific stresses encountered in the phagolysosome induce antibiotic tolerance in S. aureus. We examined the role of phagolysosome-associated stressors in the induction of antibiotic tolerant S. aureus cells in vitro. We exposed an exponential phase population of S. aureus to a variety of reagents to emulate oxidative stress, phagosome acidification and nutrient deprivation for 20min prior to treatment with rifampicin (Fig. 1c). Mupirocin mimics nutrient deprivation by inducing the stringent response; it had no effect on rifampicin susceptibility (Fig. 1c). Acidification of the media to pH4.5 to emulate acidification of the phagolysosome6 also had no effect on rifampicin susceptibility (Fig. 1c). Menadione is a redox-cycling agent which transfers electrons from NADH or NADPH to oxygen to produce superoxide and generates an oxidative stress similar to that encountered during oxidative burst in phagocytes. Remarkably, menadione treatment caused the bacteria to become completely tolerant to rifampicin (Fig. 1c). Addition of menadione also induced rifampicin tolerance in the MRSA strain COL (Extended Data Fig. 1a). Addition of these stresses in the absence of rifampicin did not cause bacterial cell death (Extended Data Fig. 1b).
Other ROS-generating compounds, paraquat, (another redox cycling agent), and hydrogen peroxide also significantly induced antibiotic tolerance. The impact of hydrogen peroxide on tolerance was less pronounced than that of the redox cycling agents, menadione and paraquat, likely due to the rapid degradation of hydrogen peroxide by catalase. (Extended Data Fig. 1c). Addition of the antioxidants N-acetyl cysteine or thiourea in the presence of menadione restored antibiotic susceptibility, ROS production and growth (Fig. 1d–e, Extended Data Fig. 1d–e). Together, these results suggest that ROS coerce S. aureus cells into an antibiotic tolerant state.
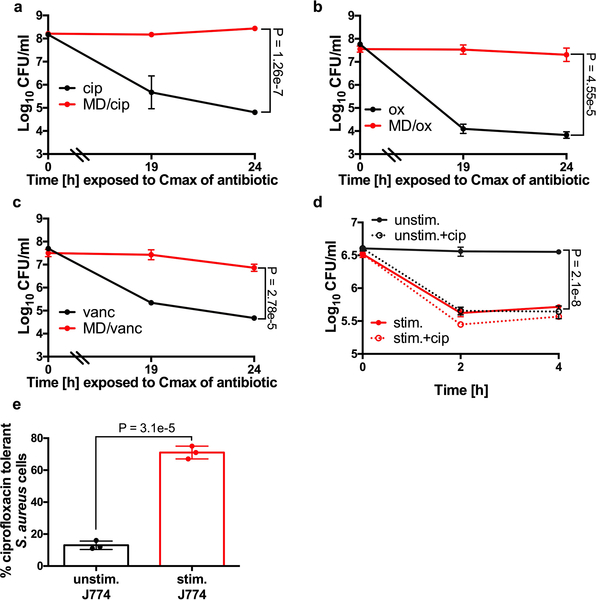
Next, we aimed to determine if ROS can impact killing by other classes of antibiotics in vitro and in our intracellular assays. Menadione caused a dramatic induction of antibiotic tolerance to ciprofloxacin, oxacillin and vancomycin in vitro (Extended Data Fig. 2a–c). In tissue culture, stimulated macrophages induced tolerance to ciprofloxacin in a manner analogous to that observed with rifampicin (Extended Data Fig. 2d–e). Both oxacillin and vancomycin penetrate poorly into macrophages35, so we did not test these antibiotics in our intracellular assays. Together, these results indicate that ROS coerce S. aureus into a multidrug tolerant state.
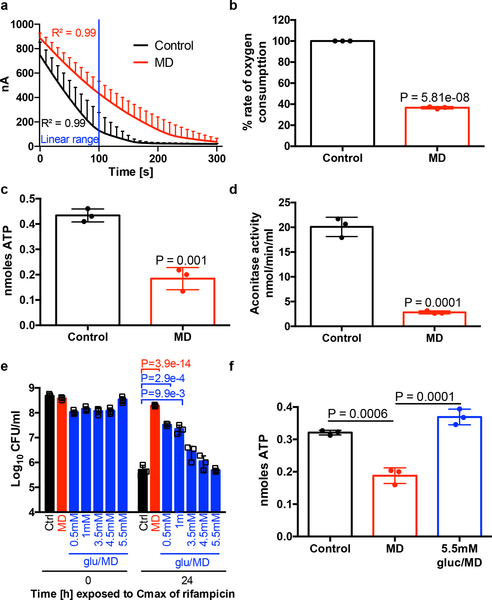
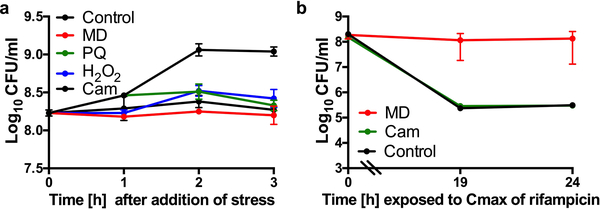
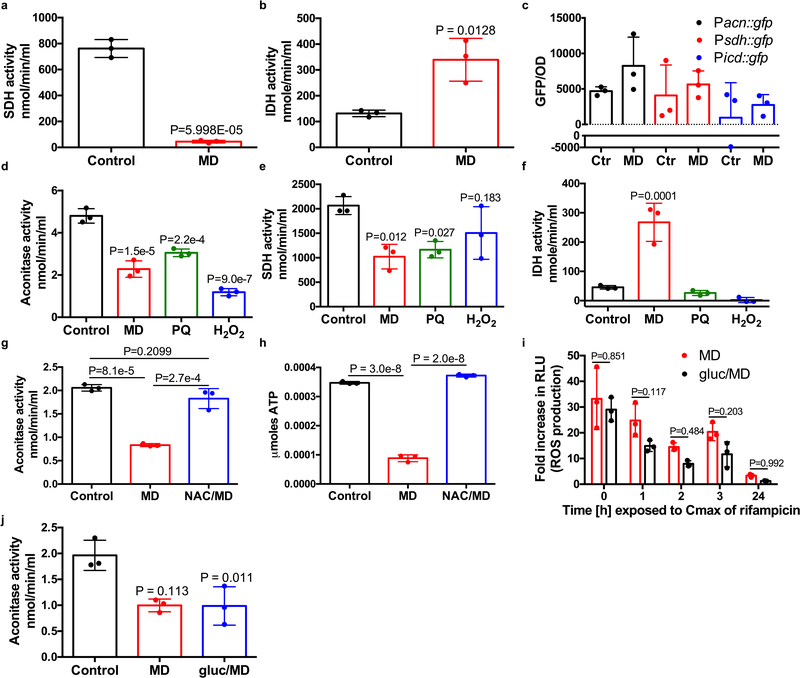
Reactive oxygen species may be growth inhibitory, which has long been associated with antibiotic tolerance9,10. However, Lopatkin et al. recently showed that metabolic activity is more predictive of antibiotic lethality than growth rate11. Additionally, several studies have shown that antibiotic tolerance of bacterial populations or sub-populations of persister cells are associated with reduced respiration, ATP levels or metabolic activity12–17. We find that although all of the ROS-generating compounds inhibited growth, growth inhibition alone is insufficient to induce rifampicin tolerance as the protein synthesis inhibitor, chloramphenicol, inhibits growth but did not induce tolerance to rifampicin (Extended Data Fig. 3a–b). We therefore focused on the potential of ROS to corrupt S. aureus central metabolism. Indeed, we found that cultures treated with menadione displayed reduced respiration and reduced levels of intracellular ATP (Fig. 2a–c). These results show that ROS reduce the metabolic activity of S. aureus.
Fig. 2. ROS decreases S. aureus metabolic activity.
(a-d) HG003 was grown to mid-exponential phase and treated with or without 80μM menadione (MD) for 2h. (a-b) Oxygen consumption was measured for 300s (nA denotes nanoamphere) and (b) the rate of oxygen consumption was calculated using the linear range (indicated by blue line in a), (c) intracellular ATP was measured using a BactiterGlo Cell Viability Assay, (d) aconitase activity was measured. (e-f) HG003 was grown to mid-exponential phase and exposed to MD and/or indicated concentrations of glucose. Cultures were either (e) challenged with 10μg/ml rifampicin and survivors were enumerated after 24h, (f) or ATP was quantified after 30min using a BactiterGlo Cell Viability Assay. Averages of n=3 biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using (b-d) Student’s t-test (unpaired, two-tailed), (e) One-Way ANOVA with Dunnett’s multiple comparison test or (f) One-Way ANOVA with Sidak’s multiple comparison test. See also Extended Data Fig. 4.
We aimed to decipher the mechanism of ROS-induced respiration inhibition. ROS (in the form of superoxide, hydrogen peroxide and peroxynitrite) target Fe-S cluster containing enzymes, two of which, aconitase and succinate dehydrogenase, participate in the S. aureus TCA cycle18–20. Inactivation of the TCA cycle leads to a decrease in available reducing equivalents (NADH and FADH2) to fuel the electron transport chain, which would lead to a reduction in respiration and intracellular ATP. We hypothesized that ROS induce antibiotic tolerance by inactivating essential TCA cycle enzymes. Using colorimetric assays, the activity of aconitase and succinate dehydrogenase were determined. Menadione decreased aconitase activity (Fig. 2d) and succinate dehydrogenase activity (Extended Data Fig. 4a), while another TCA cycle enzyme, isocitrate dehydrogenase, which does not contain Fe-S clusters was highly active in the presence of menadione (Extended Data Fig. 4b). Menadione did not significantly affect transcription of acn or sdh (Extended Data Fig. 4c), confirming that menadione affects aconitase and succinate dehydrogenase at the protein level. Paraquat and hydrogen peroxide also caused a significant reduction in aconitase activity (Extended Data Fig. 4d), while only paraquat and not hydrogen peroxide reduced succinate dehydrogenase activity (Extended Data Fig. 4e) indicating that aconitase is likely more sensitive to ROS than succinate dehydrogenase. Importantly, isocitrate dehydrogenase was unaffected by either paraquat or hydrogen peroxide (Extended Data Fig. 4f). Both aconitase activity and ATP levels in menadione treated cultures were complemented by the antioxidant N-acetyl cysteine (Extended Data Fig. 4g–h). Together these results suggest that ROS inactivates the TCA cycle, leading to reduced respiration and ATP production, and increased antibiotic tolerance.
We predicted that addition of excess glucose would circumvent the TCA cycle and restore antibiotic susceptibility to menadione treated cultures by generating ATP through substrate level phosphorylation. Fig. 2e shows that addition of excess glucose restored rifampicin susceptibility to menadione treated cultures, and this was associated with increased ATP (Fig. 2f), despite abundant ROS production and reduced aconitase activity (Extended Data Fig. 4i–j). These results further substantiate that ROS-induced antibiotic tolerance is associated with collapse of the TCA cycle and associated ATP depletion.
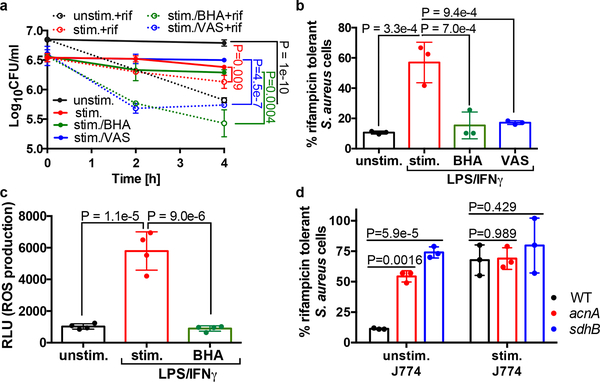
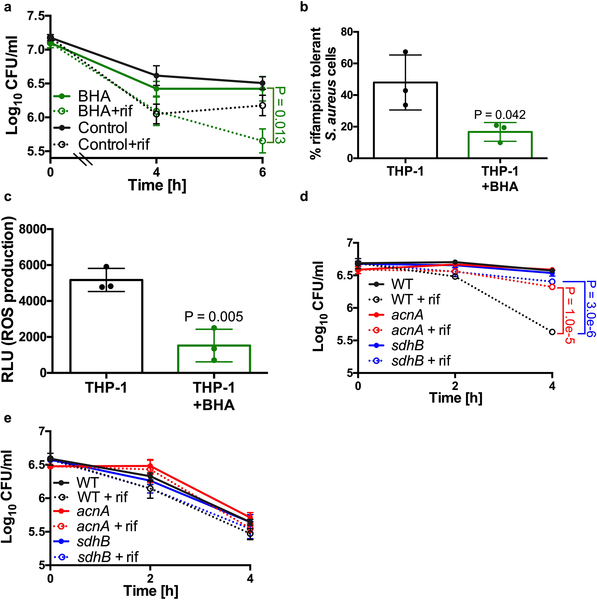
As ROS is capable of inducing antibiotic tolerance in vitro, we aimed to determine if blocking ROS production by macrophages would restore antibiotic susceptibility of phagocytosed S. aureus cells. Addition of the antioxidant butylated hydroxanisole (BHA) or the Phox inhibitor VAS2870 (VAS) to the stimulated J774 macrophages restored rifampicin susceptibility of intracellular S. aureus (Fig. 3a–b) and this corresponded to ROS production (Fig. 3c). To confirm that these findings were not isolated to J774 macrophages, we infected human THP-1 monocyte derived-macrophages with S. aureus. S. aureus cells internalized by THP-1 macrophages were more susceptible to rifampicin killing in the presence of BHA, which correlated with decreased ROS production (Extended Data Fig. 5a–c). Together these data support our findings that macrophage-produced ROS coerce S. aureus into an antibiotic tolerant state.
Fig. 3. Macrophage derived-ROS induces antibiotic tolerance in S. aureus.
(a-b, d) Survival of S. aureus strain HG003 (WT), HG003acnA::erm (acnA) and HG003sdhB::erm (sdhB) mutants following internalization by unstimulated J774 macrophages (unstim.) or macrophages that were pre-stimulated overnight with LPS/IFNγ (stim.). Where indicated, macrophages were treated with 20μM butylated hydroxanisole (BHA) or 10μM VAS2870 (VAS) for 1h prior to the addition of bacteria. (a) 10μg/ml rifampicin (rif) was added at Time 0 (dotted lines). (b, d) % survival was determined by comparing survivors after 4h (J774) of rifampicin treatment to survivors to the corresponding untreated timepoint. (c) ROS production by macrophages was quantified using the luminescent probe L-012. Where indicated stimulated macrophages were pre-treated with butylated hydroxanisole (BHA) prior to the addition of L-012. RLU denotes relative luminescence units. Averages of n=3 or (n=4 for c) biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using (a-c) One-Way ANOVA with Sidak’s multiple comparison test or (d) Two-Way ANOVA with Tukey’s multiple comparison test. See also Extended Data Fig. 5.
Our in vitro data suggests that ROS induces antibiotic tolerance by inactivating the S. aureus TCA cycle. To determine if this mechanism is also responsible for the induction of tolerance in macrophages, we examined antibiotic susceptibility of S. aureus strains harboring mutations in TCA cycle genes acnA and sdhB. Indeed, we find that both mutants display increased antibiotic tolerance compared to the wild type strain when internalized by unstimulated J774 macrophages (where ROS is low) (Fig. 3c–d, Extended Data Fig. 5d). In contrast, these mutants displayed comparable survival to the wild type strain when used to infect stimulated J774 macrophages (where ROS is high) (Fig. 3c–d, Extended Data Fig. 5e). Together these results show that ROS inactivates the TCA cycle and that this is sufficient to induce antibiotic tolerance of S. aureus in macrophages, although we cannot rule out ROS having an additional impact on other components of central metabolism, such as the electron transport chain.
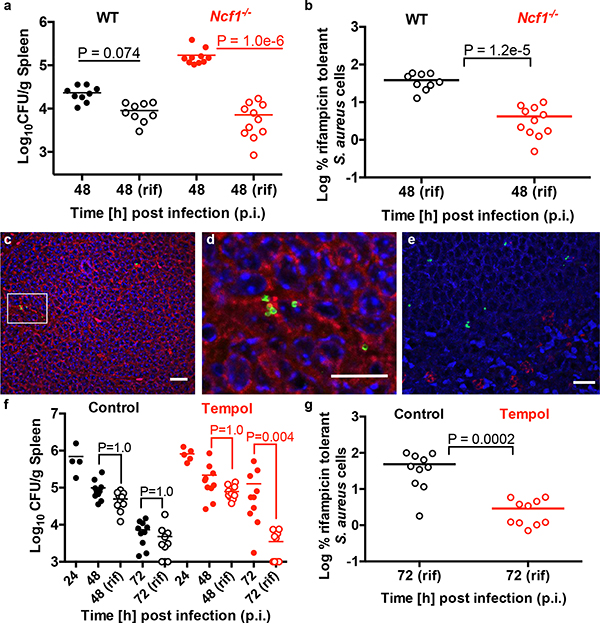
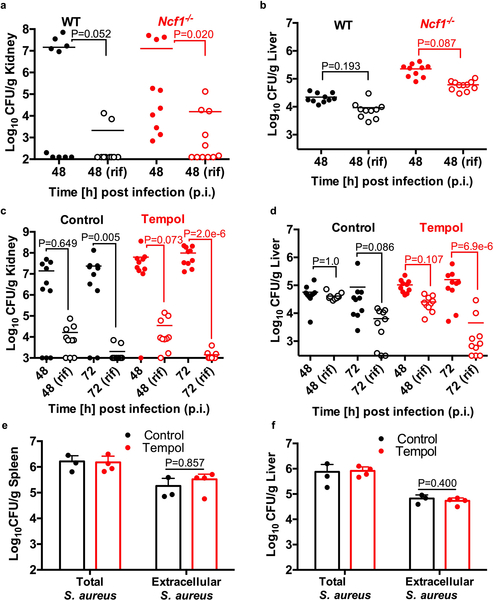
To address if ROS-induced antibiotic tolerance is relevant in vivo, we examined the outcome of antibiotic treatment of a systemic S. aureus infection in wild type or Ncf1−/− mutant mice which lack p47phox (a component of Phox) and are deficient in respiratory burst. Sepsis was induced by I.V. injection of S. aureus and mice were treated with rifampicin or the vehicle control at 24h post-infection (p.i.). At 48h p.i., S. aureus cells were enumerated from the spleen (Fig. 4a–b), kidneys and liver (Extended Data Fig. 6a–b). In the absence of respiratory burst, S. aureus achieved higher titers in the spleen, emphasizing the importance of ROS in controlling S. aureus survival and/or proliferation in this organ (Fig. 4a). Interestingly, S. aureus cells in the spleen of Ncf1−/− mice were far more susceptible to rifampicin than S. aureus in the spleen of wild-type mice (Fig. 4a–b). These results demonstrate that while respiratory burst is beneficial in controlling S. aureus burden, it also inhibits the bactericidal activity of antibiotics against infection.
Fig. 4. Host derived ROS induce antibiotic tolerance in a mouse model of sepsis.
Sepsis was induced by I.V. injection of S. aureus strain HG003 into (a-b) C57BL/6J wild type (WT) and Ncf1−/− mutant mice or (f-g) WT mice treated with or without 25mM tempol. At 24h p.i., mice were treated daily with 25mg/kg rifampicin or the vehicle control. CFU were enumerated from the spleen at 24h, 48h or 72h p.i. The mean is indicated by a horizontal line. The limit of detection is indicated by the x-axis. (a-b) Wild type mice: vehicle/rifampicin n=9 animals, Ncf1−/− mice: vehicle n=10 / rifampicin n=11 animals. (f-g) control n=4 / tempol n=5 animals (at 24h p.i,) and control/tempol mice: vehicle/rifampicin n=10 animals (at 48h and 72h p.i.). Statistical significance was determined using (a, f) Kruskal Wallis One-Way ANOVA with Dunn’s multiple comparison test or (b, g) the Mann Whitney two-tailed test. Immunofluorescence microscopy of wild type mouse spleen at 48h p.i. S. aureus cells (green in c-e) are associated with F4/80 macrophage marker (red in c-d) and not Ly6G neutrophil marker (red in e). Scale bars represent 10μm, 5μm and 10μm in c, d and e, respectively. Microscopy results are representative of three independent experiments. See also Extended Data Fig. 6.
The spleen is the largest secondary immune organ and contains large numbers of macrophages. Immunofluorescence staining suggests that S. aureus in the spleen 48h p.i. are associated with macrophages and not neutrophils (Fig. 4c–e), further suggesting that macrophages are the major reservoir of antibiotic tolerant S. aureus populations during sepsis. However, we cannot rule out the possibility that other cell types contribute to S. aureus persistence.
S. aureus in the kidney remained largely susceptible to killing by rifampicin suggesting a less prominent role of respiratory burst in this organ at 48h p.i. (Extended Data Fig. 6a). As previously reported3, antibiotic tolerance was observed in the liver, however mutation of Ncf1−/− did not increase antibiotic susceptibility of this population, suggesting an induction of antibiotic tolerance independent of Phox (Extended Data Fig. 6b). This was somewhat surprising as S. aureus associates with Kupffer macrophages in the liver during sepsis3 and would be expected to experience ROS from phagocytic burst.
It has been recently shown that mitochondrial ROS is important for controlling S. aureus in the phagosome21,22 and mutation of Ncf1−/− will not impact this ROS generation. We administered tempol, a superoxide dismutase mimetic and potent ROS scavenger23 to the drinking water of wild type mice prior to I.V. infection with S. aureus. Starting at 24h p.i., mice were treated daily with rifampicin or the vehicle control. At 48h and 72h p.i., S. aureus was enumerated from the spleen, liver and kidneys (Fig. 4f, Extended Data Fig. 6c–d). Interestingly, tempol treatment increased the antibiotic sensitivity of S. aureus in the spleen and liver (Fig. 4f–g, Extended Data Fig. 6d) suggesting that during infection of the liver, mitochondrial derived ROS may play a more prominent role than Phox-derived ROS. Importantly, the spleens and livers of both the control and tempol treated mice had similar numbers of extracellular bacteria at 24h p.i. (when rifampicin was administered) indicating that extracellular expansion did not contribute to the improved antibiotic susceptibility in the tempol treated mice (Extended Data Fig. 6e–f). Together, these data further support a role for ROS in the induction of S. aureus antibiotic tolerance during infection.
Our results show that the host immune system coerces S. aureus into a metabolic state that is incompatible with the killing mechanism of most bactericidal antibiotics. We hypothesize that S. aureus can exploit this mechanism of ROS-induced tolerance to rapidly adapt without the need for genetic selection. Subsequently, cells repair/replace damaged enzymes to re-enter a growing state and become capable of causing a relapse in infection. We would predict that patients deficient in respiratory burst, though more susceptible to bacterial infection, may actually respond better to antibiotics. Strikingly, there is already evidence to suggest this. S. aureus cells were more susceptible to rifampicin when internalized within polymorphonuclear leukocytes (PMNs) isolated from patients with chronic granulomatous disease (CGD)(which have an impaired respiratory burst) compared to wild type PMNs, although the significance of this result may have been overlooked by the authors24. In addition, a recent study showed that Nox2-deficient mice responded better to antibiotics and had increased survival compared to wild type mice following influenza/S. aureus co-infection25. The authors attribute this to reduced oxidative stress-induced tissue damage and inflammation but our results suggest that the metabolic state of S. aureus in the Nox2-deficient mice may also be a contributing factor.
Previous studies examining ROS and antibiotic activities have been extremely polarized. They include studies suggesting that antibiotics kill cells through ROS generation26, studies challenging these findings27, and studies in Gram-negative bacteria suggesting that ROS can induce antibiotic tolerance in vitro28–30. Here we show that in the case of the major Gram-positive pathogen, S. aureus, ROS are inhibitory to antibiotic mediated killing and their generation by the host increases S. aureus antibiotic tolerance during infection.
Recent work by Lehar et al. identified a rifamycin analogue capable of killing intracellular S. aureus4. It is unclear how this rifalogue targets intracellular S. aureus but our work suggests that it maintains bactericidal activity in the presence of ROS. The use of antioxidants, scavengers and oxidase inhibitors to restore antibiotic susceptibility is an exciting prospect, however, the cost to immune-mediated clearance, at least in the case of sepsis, will likely complicate this approach. It will be interesting to examine the potential of immune modulators in combination with antibiotics to maximize the clearance of S. aureus and achieve efficient eradication in patients.
Methods
Macrophage growth and infection
J774A.1 murine macrophage-like cells (ATCC TIB-67) were cultured in Dulbecco’s Modified Essential Media, high glucose (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Milipore), non-essential amino acids (NEAA) (Gibco), sodium pyruvate (Gibco) and L- glutamine (Gibco) in a humidified incubator at 37°C and 5% CO2. For infection, macrophages were seeded at a density of 2×105 per well in 24 well plates in Minimum Essential Media (MEM) (Gibco) supplemented with 10% FBS and L-glutamine. We found respiratory burst to be sensitive to the media the cells were cultured in as well as the confluency history of the cells. Cells that had become more than 90% confluent no longer reliably generated reactive species in response to S. aureus. To stimulate the J774A.1 cells, 500ng/ml lipopolysaccharide from Escherichia coli O55:B5 (Sigma) and 20ng/ml recombinant murine interferon-γ (Peprotech) (LPS/IFNγ) were added to supplemented MEM for 20h31.
THP-1 human monocytes were cultured in RPMI (Gibco) supplemented with 10% FBS, L- glutamine and 0.05mM 2-mercaptoethanol. To differentiate monocytes to macrophage-like cells, THP-1 cells were cultured at a density of 2×105 per well in 24 well plates in supplemented RPMI with 100nM phorbol 12-myristate 13-acetate (PMA) (Sigma) for 3 days. For infection, media containing PMA was removed and cells were washed with PBS. Supplemented RPMI (without PMA) was added to cells 16 hours prior to infection.
~5×106 CFU/well S. aureus strain HG003 wild type or mutant strains were used to infect macrophages at a multiplicity of infection (MOI) of 25. Where indicated, cells were pre-treated with 20μM butylated hydroxanisole (BHA) (Sigma) or 10μM VAS2870 (VAS) (Cayman Chemicals) for 1h prior to infection. 24-well plates were centrifuged at 1200xg for 2min to bring bacteria into contact with the macrophages. 30min p.i., the cells were washed once with PBS. Fresh media containing 30μg/ml gentamicin (Fisher) was added to kill extracellular bacteria32 and 10μg/ml rifampicin (Fisher) or 40μg/ml ciprofloxacin (Acros Organics) was added to the appropriate wells. At indicated times, 200μl 0.1% Triton X-100 was added to each well for 5min at 37°C to selectively lyse the macrophages and release the bacteria. 800μl PBS was added to each well, lysates were suspended by pipetting, serially diluted in 1% NaCl and plated to enumerate surviving bacteria. % survival after rifampicin treatment was determined by comparing survivors after 4h (J774) or 6h (THP-1) rifampicin treatment to survivors of the corresponding untreated 4h/6h timepoint. Averages and standard deviations of 3 biological replicates are shown (n=3). Cell lines were obtained from UNC Lineberger Comprehensive Cancer Center’s Tissue Culture Facility. We did not authenticate or test cells for mycoplasma contamination. Statistical significance was calculated using the Student’s t-test (unpaired, twotailed) or One-Way ANOVA or Two-Way ANOVA with Sidak’s multiple comparison test as described in the figure legends.
Antibiotic survival assays
S. aureus strains HG003 or COL were cultured aerobically in Mueller-Hinton broth (MHB) (Oxoid) at 37°C with shaking at 225 rpm for ~16h. For cell wall acting antibiotics, stationary cultures were diluted 1:1000 in MHB and grown to ~5×107 CFU/ml. For all other antibiotics, stationary cultures were diluted 1:100 in MHB and grown to ~2×108 CFU/ml (HG003) or ~5×108 CFU/ml (COL). An aliquot was plated to enumerate CFU (Time 0) before the addition of antibiotics. Where indicated, the culture was incubated with 80μM menadione (MD) (MP Biomedicals), 0.25μg/ml mupirocin, acid, 4mM N-acetyl cysteine (NAC), 150mM thiourea (TU), 5mM paraquat (PQ), 120mM hydrogen peroxide (H2O2) or 30μg/ml chloramphenicol (Cam) for 20min prior to antibiotic challenge. In the case of acid stress, the media was acidified to pH4.5 with HCl for 20min and then neutralized to pH7 with NaOH prior to antibiotic addition. The following antibiotics were added at concentrations similar to the Cmax in humans at recommended dosing; 10μg/ml rifampicin33 2.34μg/ml ciprofloxacin (Acros Organics)34, 50μg/ml oxacillin (Acros Organics)35, 50μg/ml vancomycin (MP Biomedicals)36. At indicated times, an aliquot was removed and washed with 1% NaCl. Cells were serially diluted and plated on tryptic soy agar (TSA) to enumerate survivors or TSA containing 10μg/ml rifampicin to enumerate rifampicin resistant mutants (rifR). rifR mutants were detected at lower levels than the antibiotic tolerant population (Extended Data Fig. 7) indicating that resistance did not contribute to the tolerance phenotype. We routinely used two time points to enumerate survivors, 19h and 24h after antibiotic challenge as we previously found that, in S. aureus, susceptible cells are killed and a stable sub-population of survivors emerges between 16 and 24h of exposure to antibiotics12,37. Averages and standard deviations of 3 biological replicates are shown (n=3). Statistical significance was calculated using the Student’s t-test (unpaired, two-tailed) or OneWay ANOVA with Sidak’s multiple comparison test or One-Way ANOVA with Dunnett’s multiple comparison test as described in the figure legends.
ROS measurement
To measure ROS, the luminescent probe L-012 (Wako Chemical Corporation) was used. For in vitro ROS measurements, HG003 was grown to ~2×108 CFU/ml in MHB. Where indicated, cells were incubated with 80μM menadione, 4mM N-acetyl cysteine (NAC) or 5.5mM glucose for 20min. 10μg/ml rifampicin was added and 100μl aliquot was removed immediately and added to the wells of a white 96 well plate containing a final concentration of 1mM L-012. Luminescence was recorded after 5min in a Biotek Synergy H1 microplate reader. Aliquots were removed at additional time points (1h, 2h, 3h and 24h) and luminescence was recorded as described above. Relative luminescence units (RLU) of menadione (or menadione + NAC/glucose) treated samples were divided by the untreated (control) samples at each time point and expressed as fold increase in ROS compared to the control. Averages and standard deviations of 3 biological replicates are shown (n=3). Statistical significance was calculated using the Student’s t-test (unpaired, two-tailed) at each time point or Two-Way ANOVA with Sidak’s multiple comparison test as described in the figure legends. For tissue culture cell assays, macrophages were seeded at 2×104 cells/well in white tissue culture treated 96 well plates. J774 cells were treated with LPS/IFNγ as described above. Where indicated 20μM BHA was added for 1h prior to the addition of L-012. After incubation with BHA, the cells were washed 3x with PBS. L-012 was diluted to 150μM in Hanks Balanced Salt Solution (Gibco). Luminescence was read immediately and recorded every 2min for 1.5h in a Biotek Synergy H1 microplate reader. Data shown are from the time point of peak luminescence of the control (untreated THP-1 or LPS/IFNγ stimulated J774). Averages and standard deviations of at least 3 replicates are shown (J774 n=4), (THP-1 n=3). Statistical significance was calculated using One-Way ANOVA with Sidak’s multiple comparison test or a Student’s t-test (unpaired, two-tailed) as described in the figure legends.
Growth curves
S. aureus strain HG003 was cultured aerobically in Mueller-Hinton broth (MHB) (Oxoid) at 37°C with shaking at 225 rpm for ~16h. Stationary cultures were diluted 1:100 in MHB and grown to ~2×108 CFU/ml (HG003). Growth curves were established by CFU or OD600 at indicated times after the addition of stresses (80μM menadione (MD), 0.25μg/ml mupirocin, acid, 4mM N-acetyl cysteine (NAC), 5mM paraquat (PQ), 120mM hydrogen peroxide (H2O2) or 30μg/ml chloramphenicol (Cam)). In the case of acid stress, the media was acidified to pH4.5 with HCl for 20min and then neutralized to pH7 with NaOH.
Oxygen consumption
HG003 was grown to ~2×108 CFU/ml in MHB and incubated with or without 80μM menadione. After 2h, cells were pelleted and concentrated 3x in MHB. The samples were vortexed vigorously and immediately added to the borosilicate glass sample vessel of an Oxygraph Plus (Hansatech Instruments). Oxygen levels were allowed to equilibrate for 30 seconds (s) at a stir speed of 5. Readings were recorded for the subsequent 300s. The linear part of the graph (the first 100s) was used to calculate the rate of oxygen consumption as follows: [(T0 - T100)/100] and normalized to CFU. % rate of oxygen was calculated by dividing by the rate of oxygen consumption in the control and multiplying by 100. Averages and standard deviations of 3 biological replicates are shown (n=3). Statistical significance was calculated using the Student’s t-test (unpaired, two-tailed) as described in the figure legends.
ATP assays
HG003 was grown to ~2×108 CFU/ml in MHB. Where indicated cells were exposed to 80μM menadione, 5.5mM glucose or 4mM N-acetyl cysteine (NAC). After 0.5h or 2h, ATP levels were measured in 100μl cells as described previously12 using a BacTiter Glo kit (Promega) according to the manufacturer’s instructions. Averages and standard deviations of 3 biological replicates are shown (n=3). Statistical significance was calculated using the Student’s t-test (unpaired, two-tailed) or One-Way ANOVA with Sidak’s multiple comparison test as described in the figure legends.
Aconitase assay
HG003 was grown to ~2×108 CFU/ml in MHB and incubated with or without 80μM menadione, 4mM N-acetyl cysteine (NAC), 5mM paraquat (PQ), 120mM hydrogen peroxide (H2O2) or 5.5mM glucose. After 0.5 or 2h, 2–10ml cells respectively, were pelleted and resuspended in 200–300μl assay buffer and lysed with 50μg/ml lysostaphin at 37°C for 5min. Samples were pelleted and the supernatant was assayed for aconitase activity using an Aconitase Assay Kit (Abcam) as per the manufacturer’s instructions. Aconitase activity was normalized to CFU. Averages and standard deviations of 3 biological replicates are shown (n=3). Statistical significance was calculated using the Student’s t-test (unpaired, two-tailed), One-Way ANOVA with Sidak’s multiple comparison test or One-Way ANOVA with Dunnett’s multiple comparison test as described in the figure legends.
Succinate dehydrogenase assay
HG003 was grown to ~2×108 CFU/ml in MHB and incubated with or without 80μM menadione, 5mM paraquat (PQ) or 120mM hydrogen peroxide (H2O2). After 2h, 2–10ml cells were pelleted and resuspended in 200μl SDH sample buffer and lysed with 50μg/ml lysostaphin at 37°C for 5min. Samples were pelleted and the supernatant was assayed for succinate dehydrogenase activity using a Succinate Dehydrogenase Activity Colorimetric Assay Kit (Sigma) as per the instructions from Biovision Incorporated. SDH activity was normalized to CFU. Averages and standard deviations of 3 biological replicates are shown (n=3). Statistical significance was calculated using the Student’s t-test (unpaired, two-tailed) or One-Way ANOVA with Dunnett’s multiple comparison test as described in the figure legends.
Isocitrate dehydrogenase assay
HG003 was grown to ~2×108 CFU/ml in MHB and incubated with or without 80μM menadione, 5mM paraquat (PQ) or 120mM hydrogen peroxide (H2O2). After 2h, 2.4ml cells were pelleted and resuspended in 400μl IDH assay buffer and lysed with 50μg/ml lysostaphin at 37°C for 5min. Samples were pelleted and the supernatant was assayed for isocitrate dehydrogenase activity using an Isocitrate Dehydrogenase Activity Assay kit (Sigma) as per the manufacturer’s instructions. Isocitrate dehydrogenase activity was normalized to CFU. Averages and standard deviations of 3 biological replicates are shown (n=3). Statistical significance was calculated using the Student’s t-test (unpaired, two-tailed) or One-Way ANOVA with Sidak’s multiple comparison test as described in the figure legends.
Strain construction
Transcriptional reporter plasmids were constructed by amplifying −755 to +76bp of the start codon of acnA 38, 222bp and 209bp upstream of the coding regions of sdhC and icdgenes, respectively, from HG003 genomic DNA using primers flanked with EcoRI and XbaI sites and inserted upstream of gfpuvr into the EcoRI and XbaI sites of plasmid pALC143439 to yield transcription gfp-reporter plasmids Pacn::gfp, Psdh::gfp and Picd::gfp. The HG003acnA::erm (acnA) and HG003sdhB::erm (sdhB) strains were constructed by transducing the transposon mutants from the NARSA library40 into HG003.
Transcriptional reporter assays
S. aureus strain HG003 harboring gfp promoter plasmids or the promoterless gfp plasmid pALC1434 were grown to ~2×108 CFU/ml in MHB containing chloramphenicol 10μg/ml. Where indicated 80μM menadione was added. After 2h, 200μl of each culture was added to the wells of a clear bottom, black side 96-well plate and absorbance (OD600) and GFP fluorescence (emission 528 nm and excitation 485 nm) was measured in a Biotek Synergy H1 microplate reader. GFP values were divided by OD600 and then values obtained for the promoter gfp plasmid were subtracted. Averages and standard deviations of 3 biological replicates are shown (n=3). Statistical significance was calculated using the Student’s t-test (two-tailed) as described in the figure legends.
Mouse infections
Mice were housed in a pathogen-specific free facility: wild type (WT) C57BL/6J (Jackson # 000664), Ncf1−/− (Jackson # 027331). All animal protocols were approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill and met guidelines of the US National Institutes of Health for the humane care of animals. For mouse infections, 8–10 week old female mice were infected with ~5×106 CFU of S. aureus strain HG003 in 100μl PBS by intravenous (I.V.) injection. Where indicated, mice were treated with tempol (4-Hydroxy-TEMPO) (Sigma) at 25mM in the drinking water starting 24h prior to infection. 4% sucrose was added to tempol water to increase the volume consumed by the mice. Control groups for the tempol experiments also received drinking water with 4% sucrose. Rifampicin was dissolved in vehicle (6.25% DMSO + 12.5% PEG300) at a final concentration of 6.25mg/ml. At 24h p.i., mice were treated daily with an intraperitoneal (I.P.) injection of 100μl/25g rifampicin or vehicle. At 24h, 48h or 72h p.i., organs were homogenized, serially diluted and plated on BHI agar plates for enumeration of bacterial CFU (and BHI agar plates containing rifampicin 10μg/ml for the enumeration of rifR mutants). % rifampicin tolerant cells was determined by comparing survivors after rifampicin treatment to survivors of the vehicle treated group. To quantify extracellular bacteria, at 24h p.i., samples of homogenized livers and spleens were treated with a RPMI-based digest buffer containing 2mg/ml Collagenase V (Invitrogen), 3mM CaCl2, 5U/ml DNase I (Fisher), 5U/ml dispase (Sigma), 5% FBS (Millipore) and 10mM HEPES for 30min. The homogenates were then centrifuged at a low speed (300xg) to pellet host cells. Aliquots of the supernatant from the low speed spin was serially diluted and plated on BHI agar to quantify extracellular bacteria. Ncf1−/− infections: Wild type mice: vehicle n=9 and rifampicin n=9. Ncf1−/− mice: vehicle n=10 and rifampicin n=11. For tempol inhibitor experiments, at 24h p.i.: control n=4, tempol n=5, at 48h p.i. and 72h p.i., control and tempol mice: vehicle n=10 and rifampicin n=10. The mean is indicated by a horizontal line. Statistical significance was calculated using the Kruskal Wallis One-Way ANOVA or the Mann-Whitney test as described in the figure legends. Sample size was chosen based on the number of mice required to reach statistical significance using the Mann Whitney test from previous experiments using the systemic model of infection. Blinding or randomization was not necessary as all outputs (CFU/g tissue) are objective.
Histopathology
Mice were infected as above. At 48h p.i., organs were harvested and fixed in 10% neutral buffered formalin for 72h and transferred to 70% ethanol. Fixed organs were paraffin embedded and sectioned at 5μm.
Immunofluorescence Microscopy
Sections were deparaffinized and rehydrated with the following washes: 3x xylene, 3× 100% ethanol, 2× 95% ethanol, 1× 80% ethanol, 1x water. Slides were boiled for 20min and cooled to RT in citrate buffer (pH 6) to unmask antigens. Samples were blocked with 1.5% normal goat serum in PBS for 40min. Samples were incubated with block solution (control), rat monoclonal anti-mouse F4/80 (clone BM8) (eBiosciences Cat# 14–4801-82) (1:100) or rat monoclonal antimouse Ly6G (clone 1A8) (eBiosciences Cat# 16–9668-82) (1:100) overnight at 4°C. Samples were washed (3x in PBS) and permeabilized with 0.5% Tween 20 in PBS for 30min to allow the S. aureus antibody to enter cells. Samples were washed 3x with PBS and incubated for 30min with 1.5% normal goat serum. Samples were incubated with block solution (control) or anti-S. aureus antibody (Abcam ab20920) (1:400) overnight at 4°C. Samples were washed 3x in PBS and incubated for 1h at RT in secondary antibodies goat anti-rat conjugated to Alexa 594 (Invitrogen Cat# A11007) and goat anti-rabbit conjugated to Alexa 488 (Invitrogen Cat# A32731) (both at 1:1000). Samples were washed 3x in PBS, and mounted with 1.5 coverslips over tissue sections with ProLong Diamond with DAPI (Invitrogen). All antibodies were diluted in block solution. Images were obtained on a Zeiss LSM 700 Confocal Laser Scanning Microscope using a 63X/1.4 Plan Apo Oil objective lens and Zeiss ZEN 2011 software. DeconvolutionLab2 and ImageJ were used to deconvolve the images. Representative images shown are the maximum intensity projections of the Z stack.
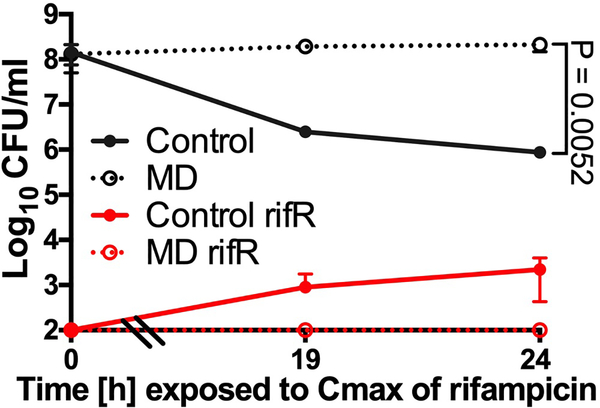
Rifampicin stability test
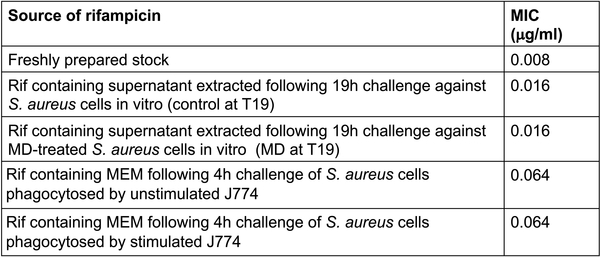
We aimed to determine if rifampicin is inactivated by ROS in our in vitro (Fig. 1c) and tissue culture assays (Fig. 1a). Minimum inhibitory concentration (MIC) assays for rifampicin were determined using a microdilution method. ~5×105 S. aureus strain HG003 in MHB was incubated with various concentrations of rifampicin, either from a freshly prepared stock solution or following extraction from in vitro or tissue culture antibiotic survival assays as described below. An aliquot of rifampicin-containing cultures (from Fig. 1c) were collected, pelleted to remove cells and filter sterilized following 19h of exposure to S. aureus with or without 80μM menadione. This supernatant was used as the source of rifampicin and underwent MIC testing. Similarly, sterilized rifampicin-containing MEM from either unstimulated or LPS/IFNγ stimulated J774 cells incubated with S. aureus for 4h (Fig. 1a) were used in MIC assays. MICs were determined following 24h incubation at 37oC. Extended Data Fig. 8 shows that in vitro after 19h, when all of the killing had occurred, rifampicin activity was reduced by 2-fold in both the control-treated and menadione-treated samples. Similarly, supernatants extracted from tissue culture experiments revealed the rifampicin MIC decreased by 6-fold in both stimulated and unstimulated J774 macrophages. This data indicates that while rifampicin activity is reduced over the course of our assays, it is not affected by ROS.
Statistical information
Statistical method and sample size (n) are indicated in the methods for each experiment. Statistical analysis was performed using Excel (Microsoft) or Prism 8 (GraphPad) software. For in vivo studies, n represents the number of mice per group and statistical significance was determined using the Kruskal Wallis One-Way ANOVA with Dunn’s multiple comparison test or the Mann-Whitney test. For in vitro studies, n represents the number of biological replicates and statistical significance was determined using a Student’s t-test (unpaired, two-tailed), One-Way ANOVA or Two-Way ANOVA with Tukey’s multiple comparison test, Sidak’s multiple comparison test, Dunnett’s multiple comparison test (indicated in the figure legends). Statistical significance was defined as p<0.05. Error bars on figures show standard deviations of the mean.
Extended Data
Extended Data Fig. 1: Induction of antibiotic tolerance by ROS.
(a) S. aureus strain COL or (b-e) HG003 was grown to mid-exponential phase in vitro and exposed to 80μM menadione (MD), 0.125μg/ml mupirocin, acidified media (pH4.5), 5mM paraquat (PQ), 120mM hydrogen peroxide (H2O2), 4mM N-acetyl cysteine (NAC) or 150mM thiourea (TU) for 20min and either (a, c-d) challenged with 10μg/ml rifampicin or (b) plated for cfu or (e) OD600 was recorded every 30min for 10h. (a-d) At indicated times, an aliquot was removed and plated on tryptic soy agar (TSA) to enumerate cfu. Averages of n=3 biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using (a, c) Student’s t-test (unpaired, two-tailed) or (d) One-Way ANOVA with Dunnett’s multiple comparison test.
Extended Data Fig. 2: ROS induces multidrug tolerance in S. aureus.
HG003 was grown to exponential phase in vitro and exposed to 80μM menadione (MD) for 20min prior to challenge with (a) 2.34μg/ml ciprofloxacin (cip), (b) 50μg/ml oxacillin (ox) or (c) 50μg/ml vancomycin (vanc). (d) Survival of S. aureus cells after internalization by unstimulated J774 macrophages (unstim.) or macrophages that were stimulated overnight with LPS/IFNγ (stim.). 40μg/ml cip was added at Time 0 (dotted lines). At the indicated times, an aliquot was removed, washed and plated to enumerate survivors. (e) % Survival of S. aureus cells treated with ciprofloxacin for 4h compared to the untreated control (data extrapolated from d). Averages of n=3 biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using (a-c, e) the Student’s t-test (unpaired, two-tailed) or (d) One-Way ANOVA with Sidak’s multiple comparison test.
Extended Data Fig. 3: Growth cessation is insufficient to induce tolerance to rifampicin.
S. aureus strain HG003 was grown to mid-exponential phase in vitro and exposed to 80μM menadione (MD), 5mM paraquat (PQ), 120mM hydrogen peroxide (H2O2) or 30μg/ml chloramphenicol (Cam) for 20min and either (a) plated for cfu or (b) challenged with 10μg/ml rifampicin. At indicated times, an aliquot was removed and plated on tryptic soy agar (TSA) to enumerate cfu. Averages of n=3 biologically independent samples. Error bars represent standard deviation.
Extended Data Fig. 4: ROS decreases S. aureus metabolic activity.
HG003 was grown to mid-exponential phase and treated with 80μM menadione (MD), 5mM paraquat (PQ), 120mM hydrogen peroxide (H2O2), 4mM N-acetyl cysteine (NAC) or 5.5mM glucose (gluc) for (a-f, j) 2h or (g-h) 0.5h and then assayed for (a, e) succinate dehydrogenase (SDH) activity, (b, f) isocitrate dehydrogenase (IDH) activity and (d, g, j) aconitase activity. (c) Transcriptional activity of acn, sdh and icd genes was determined using promoter-gfp reporters. (h) ATP was quantified using a BactiterGlo Cell Viability Assay. (i) At indicated times, ROS production was measured using L-012 luminescent probe and expressed as a fold-increase compared to the untreated control. RLU denotes relative luminescence units. Averages of n=3 biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using (a-c) Student’s t-test (unpaired, two-tailed) or (d-e, j) One-Way ANOVA with Dunnett’s multiple comparison test, (f-h) One-Way ANOVA with Sidak’s multiple comparison test or (i) Two-Way ANOVA with Sidak’s multiple comparison test.
Extended Data Fig. 5: Macrophage-derived ROS induces antibiotic tolerance in S. aureus.
(a-b) Survival of S. aureus strain HG003 after internalization by THP-1 macrophages. Where indicated, macrophages were pre-treated with 20μM butylated hydroxanisole (BHA) for 1h prior to the addition of bacteria. (a) 10μg/ml rifampicin (rif) was added at Time 0 (dotted lines). (b) % survival was determined by comparing survivors after 6h of rifampicin treatment to survivors of the corresponding untreated timepoint (extrapolated from a). (c) ROS production by THP-1 macrophages with or without BHA treatment was quantified using the luminescent probe L-012. RLU denotes relative luminescence units. (d-e) Survival of S. aureus strain HG003 (WT), HG003acnA::erm (acnA) and HG003sdhB::erm (sdhB) mutants after internalization by (d) unstimulated J774 macrophages or macrophages that were (e) pre-stimulated overnight with LPS/IFNγ. 10μg/ml rifampicin (rif) was added at Time 0 (dotted lines). Averages of n=3 biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using (a, d-e) One-Way ANOVA with Sidak’s multiple comparison test or (b-c) Student’s t-test (unpaired, two-tailed).
Extended Data Fig. 6: Host derived-ROS induces antibiotic tolerance in a mouse model of sepsis.
Sepsis was induced by I.V. injection of S. aureus strain HG003 into (a-b) C57BL/6J wild type (WT) and Ncf1−/− mutant mice or (c-d) wild type mice treated with or without 25mM tempol. At 24h p.i., mice were treated daily with 25mg/kg rifampicin or the vehicle control. S. aureus cfu were enumerated from the kidney and liver at 48h p.i. or 72h p.i. The mean is indicated by a horizontal line and the limit of detection is indicated by the x-axis (a-d). (a-b) Wild type mice: vehicle/rifampicin n=9 animals, Ncf1−/− mice: vehicle n=10 / rifampicin n=11 animals. (c-d) control n=4 / tempol n=5 animals (at 24h p.i,) and control/tempol mice: vehicle/rifampicin n=10 animals (at 48h and 72h p.i.). (e-f) Extracellular bacterial numbers were enumerated in the spleen and liver of control (n=3 animals) and tempol treated (n=4 animals) mice at 24h p.i. prior to antibiotic challenge and error bars represent standard deviation. Statistical significance was determined using (a-d) Kruskal Wallis One-Way ANOVA with Dunn’s multiple comparison test or the (e-f) Mann-Whitney test.
Extended Data Fig. 7: Resistance does not contribute to menadione-induced antibiotic tolerance.
HG003 was grown to mid-exponential phase in vitro and exposed to 80μM menadione (MD) for 20min prior to addition of 10μg/ml rifampicin. At indicated times, an aliquot was removed and plated on tryptic soy agar (TSA) to enumerate survivors or (f) TSA containing 10μg/ml rifampicin to enumerate rifampicin resistant mutants (rifR). Averages of n=3 biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using Student’s t-test (unpaired, two-tailed).
Extended Data Fig. 8: ROS does not inactivate rifampicin.
At indicated times the minimum inhibitory concentration (MIC) of rifampicin (rif) was determined following exposure to ROS in the in vitro antibiotic survival assays (Fig. 1c) and in tissue culture intracellular survival assays (Fig. 1a).
Acknowledgments
This work was supported primarily by the NIH grant R01AI137273 and K22AI125501 to B.P.C. The Microscopy Services Laboratory is supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center. The oxygraph experiments were supported by grant R01CA211732 to Q.Z. Animal experiments were supported by R01AI137273 and K22AI125501 to B.P.C and AI133236, AI139304, AI119073, and AI136920 and the Yang Biomedical Scholars Award to E.A.M. Animal experiments were assisted by the UNC Lineberger Animal Studies Core Facility, which is supported in part by an NCI Center Core Support Grant (CA16086) to the UNC Lineberger Comprehensive Cancer Center. We are grateful to the Gastrointestinal Biology and Disease (CGIBD) Imaging and Histology Core for processing histology samples, supported by NIH grant P30-DK 034987 awarded to the CGIBD Imaging and Histology Core. We are grateful to Grant Jones and Janelle Arthur for technical advice and equipment.
Footnotes
Competing interests
The authors declare no competing interests.
Data availability
Additional data that support the findings of this study are available from the corresponding author, Brian P. Conlon, upon request (brian_conlon@med.unc.edu).
References
- 1.Lowy FD Staphylococcus aureus infections. N Engl J Med 339, 520–532, doi: 10.1056/NEJM199808203390806 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Rogers DE Studies on bacteriemia. I. Mechanisms relating to the persistence of bacteriemia in rabbits following the intravenous injection of staphylococci. J Exp Med 103, 713–742 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Surewaard BG et al. Identification and treatment of the Staphylococcus aureus reservoir in vivo. J Exp Med 213, 1141–1151, doi: 10.1084/jem.20160334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehar SM et al. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature 527, 323–328, doi: 10.1038/nature16057 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Barcia-Macay M, Seral C, Mingeot-Leclercq MP, Tulkens PM & Van Bambeke F Pharmacodynamic evaluation of the intracellular activities of antibiotics against Staphylococcus aureus in a model of THP-1 macrophages. Antimicrob AgentsChemother 50, 841–851, doi: 10.1128/AAC.50.3.841-851.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helaine S et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343, 204–208, doi: 10.1126/science.1244705 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y et al. Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo. J Exp Med 213, 809–825, doi: 10.1084/jem.20151248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acocella G, Carlone NA, Cuffini AM & Cavallo G The penetration of rifampicin, pyrazinamide, and pyrazinoic acid into mouse macrophages. Am Rev Respir Dis 132, 1268–1273, doi: 10.1164/arrd.1985.132.6.1268 (1985). [DOI] [PubMed] [Google Scholar]
- 9.Pontes MH & Groisman EA Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci Signal 12, doi: 10.1126/scisignal.aax3938 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hobby GL The Antibacterial Action of Penicillin against Gram Negative Organisms. Science 100, 500–501, doi: 10.1126/science.100.2605.500 (1944). [DOI] [PubMed] [Google Scholar]
- 11.Lopatkin AJS, J.M.; Zheng EJ; Yang JH; Takahashi MK; You L; Collins JJ Bacterial metabolic state more accurately predicts antibiotic lethality than growth rate. Nature Microbiology (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conlon BP et al. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1, 16051, doi: 10.1038/nmicrobiol.2016.51 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Sonderholm M et al. The Consequences of Being in an Infectious Biofilm: Microenvironmental Conditions Governing Antibiotic Tolerance. Int J Mol Sci 18, doi: 10.3390/ijms18122688 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart PS Antimicrobial Tolerance in Biofilms. Microbiol Spectr 3, doi: 10.1128/microbiolspec.MB-0010-2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y et al. Inactivation of TCA cycle enhances Staphylococcus aureus persister cell formation in stationary phase. Sci Rep 8, 10849, doi: 10.1038/s41598-018-29123-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobritz MA et al. Antibiotic efficacy is linked to bacterial cellular respiration. ProcNatl AcadSci U S A 112, 8173–8180, doi: 10.1073/pnas.1509743112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood TK, Knabel SJ & Kwan BW Bacterial persister cell formation and dormancy. Appl Environ Microbiol 79, 7116–7121, doi: 10.1128/AEM.02636-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner PR & Fridovich I Inactivation-reactivation of aconitase in Escherichia coli. A sensitive measure of superoxide radical. J Biol Chem 267, 8757–8763 (1992). [PubMed] [Google Scholar]
- 19.Jang S & Imlay JA Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem 282, 929–937, doi: 10.1074/jbc.M607646200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro L, Rodriguez M & Radi R Aconitase is readily inactivated by peroxynitrite, but not by its precursor, nitric oxide. J Biol Chem 269, 29409–29415 (1994). [PubMed] [Google Scholar]
- 21.Cohen TS et al. S. aureus Evades Macrophage Killing through NLRP3-Dependent Effects on Mitochondrial Trafficking. Cell Rep 22, 2431–2441, doi: 10.1016/j.celrep.2018.02.027 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abuaita BH, Schultz TL & O’Riordan MX Mitochondria-Derived Vesicles Deliver Antimicrobial Reactive Oxygen Species to Control Phagosome-Localized Staphylococcus aureus. Cell Host Microbe 24, 625–636 e625, doi: 10.1016/j.chom.2018.10.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell JB et al. A low molecular weight antioxidant decreases weight and lowers tumor incidence. Free Radic Biol Med 34, 93–102 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Jacobs RF & Wilson CB Activity of antibiotics in chronic granulomatous disease leukocytes. Pediatr Res 17, 916–919, doi: 10.1203/00006450-198311000-00016 (1983). [DOI] [PubMed] [Google Scholar]
- 25.Sun K et al. Nox2-derived oxidative stress results in inefficacy of antibiotics against post-influenza S. aureus pneumonia. J Exp Med 213, 1851–1864, doi: 10.1084/jem.20150514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA & Collins JJ A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810, doi: 10.1016/j.cell.2007.06.049 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Keren I, Wu Y, Inocencio J, Mulcahy LR & Lewis K Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339, 1213–1216, doi: 10.1126/science.1232688 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Mosel M, Li L, Drlica K & Zhao X Superoxide-mediated protection of Escherichia coli from antimicrobials. Antimicrob Agents Chemother 57, 5755–5759, doi: 10.1128/AAC.00754-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y, Vulic M, Keren I & Lewis K Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother 56, 4922–4926, doi: 10.1128/AAC.00921-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T, El Meouche I & Dunlop MJ Bacterial persistence induced by salicylate via reactive oxygen species.Sci Rep 7, 43839, doi: 10.1038/srep43839 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang X, Li Y, Fu M & Xin HB Polarizing Macrophages In Vitro. Methods Mol Biol 1784, 119–126, doi: 10.1007/978-1-4939-7837-3_12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall JD, Craven RR, Fuller JR, Pickles RJ & Kawula TH Francisella tularensis replicates within alveolar type II epithelial cells in vitro and in vivo following inhalation. Infect Immun 75, 1034–1039, doi: 10.1128/IAI.01254-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peloquin CA et al. Population pharmacokinetic modeling of isoniazid, rifampin, and pyrazinamide. Antimicrob Agents Chemother 41, 2670–2679 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szalek E, Kaminska A, Gozdzik-Spychalska J, Grzeskowiak E & Batura-Gabryel H The PK/PD index (CMAX/MIC) for ciprofloxacin in patients with cystic fibrosis. Acta Pol Pharm 68, 777–783 (2011). [PubMed] [Google Scholar]
- 35.Nesseler N et al. High-dose continuous oxacillin infusion results in achievement of pharmacokinetics targets in critically ill patients with deep sternal wound infections following cardiac surgery. Antimicrob Agents Chemother 58, 5448–5455, doi: 10.1128/AAC.02624-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barcia-Macay M, Lemaire S, Mingeot-Leclercq MP, Tulkens PM & Van Bambeke F Evaluation of the extracellular and intracellular activities (human THP-1 macrophages) of telavancin versus vancomycin against methicillin-susceptible, methicillin-resistant, vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus. J Antimicrob Chemother 58, 1177–1184, doi: 10.1093/jac/dkl424 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Radlinski L et al. Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol 15, e2003981, doi: 10.1371/journal.pbio.2003981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding Y et al. Metabolic sensor governing bacterial virulence in Staphylococcus aureus. Proc Natl Acad Sci U S A 111, E4981–4990, doi: 10.1073/pnas.1411077111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheung AL, Nast CC & Bayer AS Selective activation of sar promoters with the use of green fluorescent protein transcriptional fusions as the detection system in the rabbit endocarditis model. Infect Immun 66, 5988–5993 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fey PD et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 4, e00537–00512, doi: 10.1128/mBio.00537-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]