The phytopathogenic bacterium Xanthomonas vesicatoria is the causative agent of bacterial spot disease in various Solanaceae family members. Here, we describe the complete genome sequence of XaF13, a novel filamentous phage that infects the phytopathogenic bacterium X. vesicatoria. The 7,045-bp genome is predicted to encode 14 open reading frames, 7 of which are related to those of other filamentous Xanthomonas phages.
ABSTRACT
The phytopathogenic bacterium Xanthomonas vesicatoria is the causative agent of bacterial spot disease in various Solanaceae family members. Here, we describe the complete genome sequence of XaF13, a novel filamentous phage that infects the phytopathogenic bacterium X. vesicatoria. The 7,045-bp genome is predicted to encode 14 open reading frames, 7 of which are related to those of other filamentous Xanthomonas phages.
ANNOUNCEMENT
Mexico is the second most important producer of peppers worldwide. The production of peppers faces many phytosanitary problems such as bacterial spot disease (1–3). Bacterial spot disease control is difficult because Xanthomonas species can survive for long periods in seeds and crop debris (4). The use of bacteriophages against bacteria is considered a possible alternative to agrochemicals (5).
The bacteriophage XaF13 (family Inoviridae) was isolated from a soil sample collected in Yurécuaro, Mexico, from a pepper field affected with bacterial spot disease. A soil sample was suspended in peptone yeast glycerol (NYG) medium (100 ml) (6), inoculated with Xanthomonas vesicatoria (laboratory strain BV865), and incubated for 24 h at 28°C. The resulting slurry was centrifuged (8,000 × g; 20 min), and the supernatant was filtered (0.22 μm). Phages in the supernatant were isolated in double-layer plaque assays (0.7% agar) (7). Lysis plaques of ∼1 mm were observed in the bacterial lawn; then, a single plaque was isolated and purified two times.
DNA extraction from the bacteriophage (>1010 PFU/ml) was performed using a phage DNA extraction kit (Norgen-Biotek, Canada). The nature of the genome was characterized using DNase I (Roche, Germany), RNase A (Sigma-Aldrich, USA), and S1 nuclease (Promega, USA) according to the manufacturers’ instructions. The DNA of XaF13 was amplified using the Illustra TempliPhi amplification kit (GE Healthcare, UK); next, the DNA library was prepared with the Nextera XT DNA sample preparation kit (Illumina, USA), and the library quality was analyzed with a Bioanalyzer 2010 instrument (Agilent Technologies). High-throughput sequencing was performed by synthesis protocol (MiSeq; Illumina) with a 2 × 300-bp paired-end run at the sequencing laboratory at the Universidad Autónoma de Zacatecas (UAZ). The raw reads were filtered using Trimmomatic v. 0.38, the genome assembly was performed using SPAdes v. 3.11.0 (8), and the quality of the assembly was analyzed using QUAST v. 5.0.0 (9). To annotate the XaF13 genome, first, potential coding sequences were searched using BLAST-X; next, Pfam and HMMer were used to find homologues and conserved domains, and the best hits were grouped as a single locus. The specialized online tools PHANOTATE (10) and PHASTER (11) were used to predict specific phage genes, while tRNAs and rRNAs were predicted using tRNAscan-SE (12) and Rfam (13); transcriptional promoters were predicted using PromoterHunter (14). Default parameters were used for all software and tools except BLAST-X and PromoterHunter.
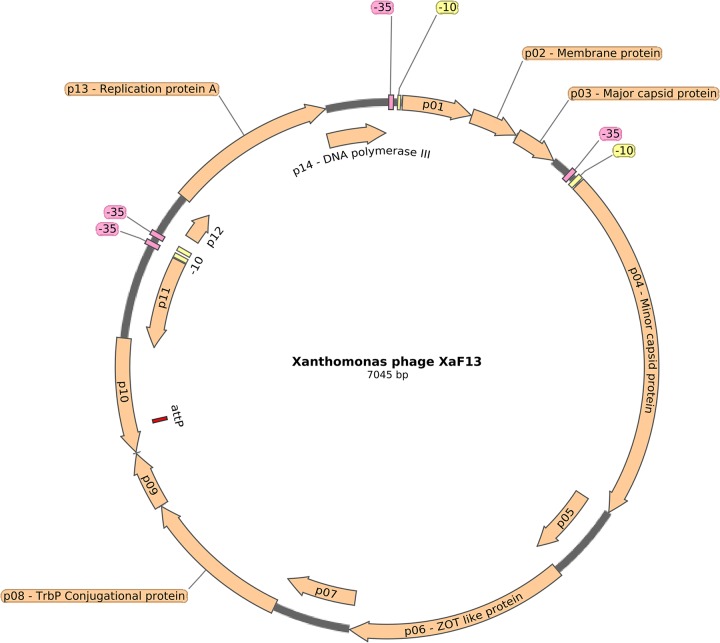
A total of 394,491 reads were obtained, and the genome of XaF13 was assembled into a single contig with a median coverage of 33,597×. The genome is composed of a single-stranded DNA molecule, as shown by the DNase I, RNase A and S1 nuclease results, with a length of 7,045 bases and a G+C content of 60.3%. Bioinformatic analysis revealed 14 open reading frames (ORFs), 12 in the positive strand and 2 in the negative strand (replicative form); no tRNAs or rRNAs were found. Seven ORFs were annotated as hypothetical proteins, and 7 were annotated as having virus-related functions, namely, ORF-2 (membrane protein), ORF-3 (major capsid protein), ORF-4 (minor capsid protein), ORF-6 (ZOT-like protein), ORF-8 (TrbP conjugational protein), ORF-13 (replication protein A), and ORF-14 (DNA polymerase III). Moreover, an attachment site (15 bp) was identified by nucleotide comparison between XaF13 and Xanthomonas phage Xf109, similar to the attP sequences of lysogenic Xanthomonas phages Cf1c, XacF1, and φLf, which use the XerC/D recombinases from their hosts to integrate into the dif locus of the bacterial genome (15). Finally, regulatory sequences, located at positions −10 and −35, suggest that the XaF13 genome is organized into 4 functional regions, 3 of which are associated with structural functions, conjugation, and replication; the fourth is formed by ORFs 10 and 11, whose functions have not been defined so far (Fig. 1). The comparison of DNA sequences between the XaF13 and 40 Inoviridae phages using BLASTN showed that XaF13 is related to the filamentous phage Xf109 of Xanthomonas oryzae (57% query coverage and 86% nucleotide identity).
FIG 1.
XaF13 phage genome organization. Arrows represent predicted genes and transcription direction. Promoter positions are indicated by yellow (−10) and pink (−35) boxes and the attP site by a red box. The illustration was prepared using SnapGene software (GSL Biotech).
Data availability.
The complete genome sequence of phage XaF13 is available in the GenBank database under accession number MN335248. The raw sequence reads are available in the SRA database under accession number SRX6866382 (BioProject number PRJNA566170).
ACKNOWLEDGMENTS
This research was supported by a CONACyT grant (APN-2015-01-338) to G.R.-E. We thank CONACyT for the postdoctoral scholarship granted to G.A.S.-S. (Currículum Vitae Único [CVU] 204332).
REFERENCES
- 1.Kenyon L, Kumar S, Tsai WS, Hughes JD. 2014. Virus diseases of peppers (Capsicum spp.) and their control. Adv Virus Res 90:297–354. doi: 10.1016/B978-0-12-801246-8.00006-8. [DOI] [PubMed] [Google Scholar]
- 2.Lee KJ, Kamala-Kannan S, Sub HS, Seong CK, Lee GW. 2008. Biological control of Phytophthora blight in red pepper (Capsicum annuum L.) using Bacillus subtilis. World J Microbiol Biotechnol 24:1139–1145. doi: 10.1007/s11274-007-9585-2. [DOI] [Google Scholar]
- 3.Potnis N, Timilsina S, Strayer A, Shantharaj D, Barak JD, Paret ML, Vallad GE, Jones JB. 2015. Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol Plant Pathol 16:907–920. doi: 10.1111/mpp.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kocks CG, Ruissen MA, Zadoks JC, Duijkers MG. 1998. Survival and extinction of Xanthomonas campestris pv. campestris in soil. Eur J Plant Pathol 104:911–923. doi: 10.1023/A:1008685832604. [DOI] [Google Scholar]
- 5.Buttimer C, McAuliffe O, Ross RP, Hill C, O’Mahony J, Coffey A. 2017. Bacteriophages and bacterial plant diseases. Front Microbiol 8:34. doi: 10.3389/fmicb.2017.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels MJ, Barber CE, Turner PC, Cleary WG, Sawczyc MK. 1984. Isolation of mutants of Xanthomonas campestris pv. campestris showing altered pathogenicity. Gen Microbiol 130:2447–2455. doi: 10.1099/00221287-130-9-2447. [DOI] [Google Scholar]
- 7.Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. 2009. Enumeration of bacteriophages by double agar overlay plaque assay, p 69–76. In Clokie MRJ, Kropinski AM (ed), Bacteriophages, methods and protocols. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 8.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNair K, Zhou C, Dinsdale EA, Souza B, Edwards RA. 2019. PHANOTATE: a novel approach to gene identification in phage genomes. Bioinformatics 35:4537–4542. doi: 10.1093/bioinformatics/btz265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arndt D, Grant J, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: search and contextual analysis of transfer RNA genes. Nucleic Acids Res 44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI. 2018. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res 46:D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klucar L, Stano M, Hajduk M. 2010. phiSITE: database of gene regulation in bacteriophages. Nucleic Acids Res 38:D366–D370. doi: 10.1093/nar/gkp911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh TY. 2017. Complete nucleotide sequence of a new filamentous phage, Xf109, which integrates its genome into the chromosomal DNA of Xanthomonas oryzae. Arch Virol 162:567–572. doi: 10.1007/s00705-016-3105-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of phage XaF13 is available in the GenBank database under accession number MN335248. The raw sequence reads are available in the SRA database under accession number SRX6866382 (BioProject number PRJNA566170).