Summary
The increasing rising of multiple drug-resistant Staphylococcus aureus has become a major public health concern, underscoring a pressing need for developing therapies essentially based on the understanding of host defensive mechanism. In the present study, we showed that microRNA (miR)-127 played a key role in controlling bacterial infection and conferred a profound protection against staphylococcal pneumonia. The protective effect of miR-127 was largely dependent on its regulation of macrophage bactericidal activity and the generation of IL-22, IL-17, and anti-microbial peptides (AMPs), the pathway primarily driven by STAT3. Importantly, we revealed that the ubiquitin-editing enzyme A20, a genuine target of miR-127, specifically interacted with and repressed K63-ubiquitination of STAT3, thereby compromising its phosphorylation upon bacterial infection. Thus, our data not only identify miR-127 as a non-coding molecule with anti-bacterial activity but also delineate an unappreciated mechanism whereby A20 regulates STAT3-driven anti-microbial signaling via modulating its ubiquitination.
Subject Areas: Molecular Mechanism of Behavior, Immunology, Microbiology, Bacteriology
Graphical Abstract
Highlights
-
•
miR-127 confers the protection against staphylococcal pneumonia
-
•
miR-127 augments macrophage anti-microbial responses by regulating STAT3 activity
-
•
A20 directly interacts and represses STAT3 K63-ubiquitination
-
•
The A20/STAT3 axis mediates the anti-microbial role of miR-127
Molecular Mechanism of Behavior; Immunology; Microbiology; Bacteriology
Introduction
Staphylococcus aureus (S. aureus) remains to be one of the leading causes of iatrogenic and community-associated infections, causing high mortality and limited therapeutics (Tong et al., 2015, Zheng et al., 2017). The increasing emergence and spreading of antibiotic-resistant bacteria such as methicillin-resistant Staphylococcus aureus (MRSA) make its treatment more challenging, thereby underscoring a pressing need for developing antimicrobial therapies. A critical tenant in the battle against staphylococcal infections is to further understand host defensive machinery (Parker and Prince, 2012, Miller and Cho, 2011).
Macrophages are the frontline of host defense with the ability to recognize, uptake, and finally eliminate the invading pathogens. Also, they are required for tissue repair, regeneration, and maintaining barrier integrity (West et al., 2011, Preston et al., 2019). The importance of macrophages in combating against staphylococcal infection was highlighted by the study using Rag1−/− mice, which exhibited the intact anti-bacterial immune responses despite lack of mature T and B cells (Schmaler et al., 2011). However, the depletion of macrophages substantially affected host defensive response and impeded bacterial clearance, indicating that the innate rather than the adaptive immune cells were required for controlling infection (Chan et al., 2018). Furthermore, the adoptive experiments revealed that protection against staphylococcus re-infection was primarily mediated by macrophages, which would release copious pro-inflammatory cytokines, chemokines, and anti-microbial peptides (AMPs) and recruit immune cells to coordinately combat pathogens (Vaishnava et al., 2011).
Among these proinflammatory cytokines, interleukin (IL)-17 and IL-22 have gained much attention due to their leading roles in mucosal immunity and tissue repair (Li et al., 2018, Dudakov et al., 2015). Evidences have shown that IL-17 and IL-22 can induce the expression of antimicrobial proteins such as regenerating islet-derived protein 3 (Reg3) and defensins, which would potentially kill or inactivate microorganisms and simultaneously promote tissue repairing and regeneration (Loonen et al., 2014, Sechet et al., 2018). Accordingly, loss of IL-17 or IL-22 led to higher lung bacterial burdens and exaggerated staphylococcus pneumonia (Cho et al., 2010, Treerat et al., 2017). IL-17 and IL-22 have been thought to release from lymphoid cells, but recent data indicated that macrophages also produced IL-22, IL-23, and subsequently IL-17, which played an indispensible role in regulating macrophages function. As a result, the inactivation of the IL-23-IL-22 signaling caused the defection in macrophages activation and differentiation, leading to exacerbated bacterial pneumonia and higher mortality (Longman et al., 2014). Given the preponderance of the IL-17/IL-22/IL-23 pathway in host defensive response, it is vital to clarify the mechanism involved and the factors with the regulatory potential.
Signal transducer and activator of transcription 3 (STAT3) is a master factor that has a key role in both immune and inflammatory responses. Activation of STAT3 is the key signaling event required for the generation of IL-17 and IL-22 (Villarino et al., 2017). In response to infectious or inflammatory signals, STAT3 undergoes phosphorylation, homo-dimerization, and nuclear translocation to initiate the transcription of IL-22 and IL-17 (Ciofani et al., 2012). Loss-of-functional mutation of STAT3 led to impaired IL-17 and IL-22 expression and rendered hosts more susceptible to candidiasis and staphylococcal infection (Abusleme et al., 2018, Choi et al., 2013). In mice, myeloid STAT3 deficiency was causatively associated with chronic enterocolitis and endotoxic shock (Gao et al., 2018). Due to its central importance in anti-bacterial responses and other vital biological processes, STAT3 activity is reasonably fine-tuned by multiple layers of mechanisms. The activation of STAT3 is primarily initiated by phosphorylation of its C-terminal at Tyr 705 under the effect of Janus-activated kinases (JAK) or phosphorylation at Ser 727 by protein kinase C (PKC), mitogen-activated protein kinases (MAPKs), and CDK5 (Kosack et al., 2019). In addition to the phosphorylation, other post-translational modifications such as methylation, acetylation, and ubiquitination were recently discovered and played essential roles in controlling STAT3 activity. For example, the histone-modifying enzymes, SET domain containing lysine methyltransferase 9 (SET9) and enhancer of zeste homolog 2 (EZH2), exhibited to methylate and regulate STAT3 activity (Yuan et al., 2005, Dasgupta et al., 2015). The deacetylase Sirtuin1 can also deacetylate and hence inhibit STAT3 activity, resulting in the restricted Th17 cell differentiation (Limagne et al., 2017). Ubiquitination is a highly conserved posttranslational modification that emerges as a regulatory mode for STAT3 activity. Ubiquitin is a 76-amino acid protein conjugated to a wide variety of substrates and thereby influences vital signaling pathways (Malynn and Ma, 2010). Among the most studied polyubiquitinations, Lys48 (K48)-linked ubiquitination enables proteasome-mediated degradation of targeted proteins, whereas K63-linked ubiquitination contributes to protein stabilization and signaling transduction (Heaton et al., 2016). STAT3 has been conjugated with K63-ubiquitin by the E3 ligase tumor-necrosis-factor-receptor-associated factor 6 (TRAF6) during the response to Salmonella infection (Ruan et al., 2017). Also, Hectd3-mediated non-degradative ubiquitination constituted the prerequisite for STAT3 activation and contributed to Th17 responses and the resultant neuroinflammatory diseases (Cho et al., 2019). These data highlight the importance of the ubiquitination in modulating STAT3 activity, but the exact role and action mode of STAT3-Ub remain to be addressed.
In the present study, we identified a small non-coding RNA, miR-127 (Ying et al., 2015, Shi et al., 2017), as a regulator of host defensive response, which conferred a profound protection against staphylococcal pneumonia. Specifically, miR-127 remarkably enhanced macrophage bactericidal activity and the generation of IL-22, IL-17, and anti-microbial peptides, the responses essentially driven by STAT3 pathway. Importantly, we demonstrated that the ubiquitin-editing enzyme A20, a genuine target of miR-127, physically interacted with STAT3 and downregulated K63-ubiquitination and hence phosphorylation of STAT3, leading to the compromised anti-bacterial responses and the exacerbated pneumonia. Thus, our study not only identifies miR-127 as a modulator of host innate immunity but also elucidates a regulatory circuitry that integrates STAT3 phosphorylation, ubiquitination, and non-coding miRNA to shape the optimized anti-microbial response.
Results
MiR-127 Promotes Bacterial Clearance and Protects Mice from Staphylococcal Pneumonia
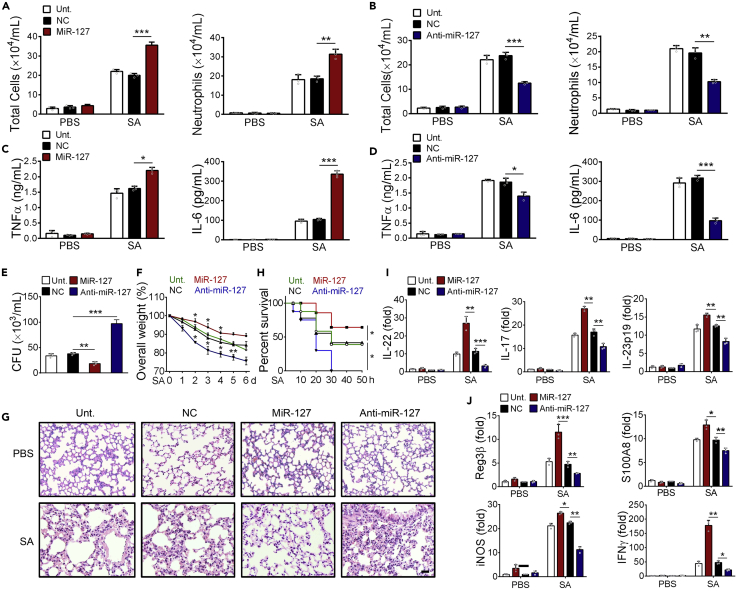
To explore the potential role for miR-127 in host defense against bacterial infection, we exploited murine staphylococcal pneumonia model. The result showed that intratracheal delivery of miR-127 mimic caused the enhanced proinflammatory reaction upon infection, as manifested by a remarkable increase in cellular infiltration and bronchoalveolar lavage fluid (BALF) levels of inflammatory cytokines such as IL-6 and tumor necrosis factor (TNF) α. By contrast, miR-127 inhibition led to the decreased cellular infiltration and lower levels of inflammatory cytokines (Figures 1A–1D). Bacterial loads, however, were reduced upon miR-127 administration and increased with anti-miR-127 treatment (Figure 1E). Consistently, mice with miR-127 administration displayed less body weight loss and the improved tissue pathology, whereas those treated with anti-miR-127 exhibited more weight loss and exacerbated tissue damage (Figures 1F and 1G), as evidenced by increased inflammatory cell infiltration, more prominent alveolar edema, and lung tissue disintegrity. Accordingly, the survival rate of mice was elevated upon miR-127 administration while decreased upon anti-miR-127 treatment when challenged with lethal dose of S. aureus (Figure 1H). The results thus supported a protective role of miR-127 against staphylococcal pneumonia.
Figure 1.
MiR-127 Confers the Protection against Staphylococcal Pneumonia
Mice (n = 5/group) were intratracheally administrated with miR-127, anti-miR-127, and non-specific control (NC) respectively for 24 h, and the untreated animals were used as controls. All the animals were then challenged with 5 × 106 colony-forming units (CFUs) of S. aureus and sacrificed 12 h later for the subsequent functional analysis.
(A and B) Mice BALF were collected and shown are counts of total cells and neutrophils in BALF.
(C and D) The levels of BALF cytokines (TNFα and IL-6) were detected by ELISA assay.
(E) BALF bacterial burden.
(F) Body weight loss.
(G) H&E staining of lung sections. Scale bar, 100 μm.
(H) Survival rate of mice challenged with lethal dose of S. aureus (1×108 CFU/mice).
(I and J) Quantitative PCR (qPCR) analysis of lung levels of the indicated cytokines (IL-22, IL-17 and IL-23p19), AMPs (Reg3b, S100A8 and iNOS), and other effector molecules associated with bactericidal activity.
Results are expressed as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 by Student's t test.
Considering that IL-22 and the associated effector molecules are essential for host dense against bacterial infection (Cho et al., 2010, Treerat et al., 2017), we then wondered whether miR-127 impacted the generation of these cytokines. Remarkably, miR-127 administration boosted while anti-miR-127 treatment suppressed lung levels of IL-22 and the related cytokine IL-17 and IL-23 in S. aureus-infected mice (Figure 1I). Congruent with this, the expression of anti-microbial molecules such as regenerating islet-derived protein 3β (Reg3β) and S100 calcium binding protein A8 (S100A8) as well as inducible nitric oxide synthase (iNOS) and interferon-γ (IFNγ) was enhanced by miR-127 overexpression and reduced by miR-127 inhibition (Figure 1J). Taken together, we herein provided the evidences to demonstrate a protective role of miR-127 during staphylococcal pneumonia, which was likely associated with its promotion of pathogen clearance and maintenance of tissue integrity.
miR-127 Increases Macrophages Inflammatory Responses in Response to Staphylococcal Infection
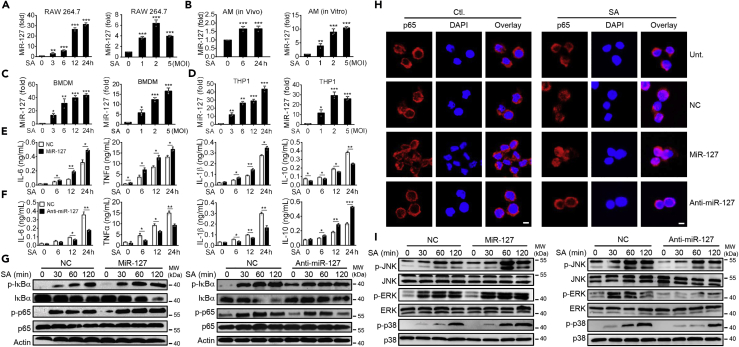
Given the importance of macrophages in host defense against bacterial infection (Preston et al., 2019, Schmaler et al., 2011, Chan et al., 2018), we next assessed the influence of miR-127 on macrophage activity during staphylococcus infection. Firstly, we noted that miR-127 was induced, in a time-dependent and does-dependent manner, in murine macrophage cell lines—RAW264.7 cells—upon infection (Figure 2A). This expressive profile was also observed in alveolar macrophages (AMs), either in vivo or in vitro (Figure 2B). Also, miR-127 was induced in murine bone-marrow-derived macrophages (BMDMs) (Figure 2C) and human monocytic cell line THP1 (Figure 2D), implying the involvement of miR-127 in macrophage anti-bacterial responses. Indeed, administration of miR-127 significantly increased the expression of pro-inflammatory cytokines IL-6, TNFα, and IL-1β but decreased the expression of anti-inflammatory cytokine IL-10 (Figure 2E). By contrast, the interference of miR-127 expression enabled macrophages to produce lower level of pro-inflammatory cytokines, whereas higher level of IL-10 (Figure 2F). In agreement with this, the activation of nuclear factor-kappa B (NF-κB), as manifested with both p-p65 level (Figure 2G) and p65 nuclear translocation (Figure 2H). The results further demonstrate that the level of phosphorylated p65 was enhanced in miR-127-expressing macrophages but decreased in miR-127-silenced cells upon infection. The similar regulatory effect of miR-127 was also observed in the activation of mitogen-activated protein kinases (MAPKs), the other major pro-inflammatory pathway involved in the anti-bacterial response (Figure 2I). Together, our data indicated that miR-127 boosted macrophages inflammatory response at the early stage of staphylococcus infection.
Figure 2.
Regulation of Pro-inflammatory Signaling by miR-127 in Response to Staphylococcal Infection
(A) QPCR analysis of miR-127 in RAW264·7 cells infected with S. aureus for the indicated time periods or for the indicated multiplicity of infection (MOI).
(B) QPCR analysis of miR-127 in alveolar macrophages (AMs) isolated from S. aureus-infected mice (left) or in vitro cultured AMs infected with S. aureus at the indicated MOI (right).
(C and D) QPCR analysis of miR-127 in bone-marrow-derived macrophages (BMDMs) (C) or in human monocytic cell line THP1 (D) upon staphylococcal infection at the indicated periods or at the indicated MOI.
(E–I) RAW264·7 cells were transfected with miR-127, anti-miR-127, or their non-specific controls (NC) for 24 h and then stimulated with S. aureus (MOI = 1) for the indicated time periods. The production of the indicated cytokines in cell culture supernatant was examined by ELISA (E and F). The levels of the indicated proteins involved in the NF-κB signaling were detected by immunoblotting (G). The nuclear translocation of p65 was detected by the confocal methods. Scale bar, 10 μm (H). Levels of indicated proteins involved in the MAPK signaling were detected by immunoblotting (I).
Results are from at least three independent experiments and expressed as mean ± SD. *p < 0.05, **p < 0.01 by Student's t test. Representative images are shown in G–I.
miR-127 Enhances Bactericidal Activity and the Generation of Anti-microbial Molecules in Macrophages
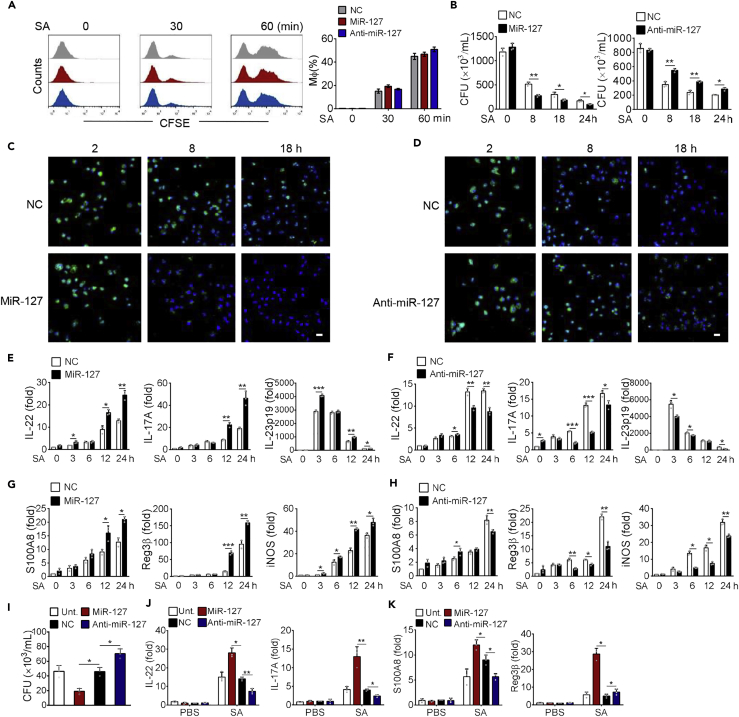
We next examined the impact of miR-127 on bacterial clearance ability in macrophages. For this purpose, we initially evaluated phagocytic activity of macrophages using fluorescein isothiocyanate (FITC)-labeled S. aureus. The result showed that neither miR-127 overexpression nor miR-127 inhibition substantially affected the amounts of the internalized bacterium (Figure 3A), indicating that miR-127 had no apparent impact on macrophage phagocytosis. However, the enumeration of bacterial colony-forming unit (CFU) revealed that miR-127 administration significantly decreased bacterial loads, whereas anti-miR-127 treatment elevated bacterial burden during the infectious course of 18 h (Figure 3B). To further assess bactericidal capability of macrophages, we carried out the lysostaphin protection assay wherein the survival of internalized bacteria was visualized with fluorescent microscopy (West et al., 2011). Remarkably, the amounts of alive bacteria were less in miR-127-treated macrophages but increased in anti-miR-127-treated macrophages compared with that in non-specific (NC)-treated cells (Figures 3C and 3D), indicating that miR-127 enhanced macrophage ability to eliminate invading bacteria.
Figure 3.
MiR-127 Enhances Macrophage Bacterial Clearance Ability
RAW264·7 cells were transfected with miR-127, anti-miR-127, or their non-specific controls for 24 h, followed by staphylococcus infection (MOI = 1) for the indicated time periods.
(A) Phagocytosis capability was examined by the percent of macrophages uptaking carboxyfluorescein succinimidyl ester (CFSE)-labeled S. aureus. Shown are the representative histograms and bar charts from five independent experiments of flow cytometry.
(B) Bacterial burden was enumerated in RAW 264.7 cells at the indicated time periods postinfection.
(C and D) Shown are intracellular bacteria measured by immunofluorescent microscopy. Uptake of S. aureus remained in RAW264·7 cells at 2, 8, and 18 h post-infection was labeled by CFSE (green). Nuclei were stained with DAPI (blue). Representative images of five per group are depicted. Scale bar, 30 μm.
(E–H) QPCR analysis of levels of the indicated cytokines (E and F) and anti-microbial molecules (G and H) in RAW264·7 cells upon infection.
(I–K) THP1 cells were transfected with miR-127, anti-miR-127, or their non-specific controls for 24 h and then infected with S. aureus (MOI = 1) for 12 h. Bacterial loads (I) and the levels of the indicated cytokines or AMPs (J and K).
All results are from three independent experiments and presented as mean ± SD. *p < 0.05, **p < 0.01 by Student's t test.
Accumulating evidences have demonstrated that anti-microbial peptides (AMPs) played a central role in limiting bacterial replication and preventing tissue damage (Madouri et al., 2018, Berger et al., 2018). We therefore assessed the effect of miR-127 on the production of AMPs and initially analyzed the level of IL-22, the presumed inducer of AMPs such as Reg3β and S100A8. Remarkably, the level of IL-22, as well as the related cytokine IL-17 and IL-23 was enhanced upon miR-127 treatment while reduced by miR-127 inhibition in S. aureus-infected macrophages (Figures 3E and 3F). Consistently, the expression of the effector molecules with anti-bacterial property, such as S100A8, Reg3β, and iNOS, was increased in miR-127-expressing macrophages but decreased in miR-127-silenced cells (Figures 3G and 3H). To further test the regulatory role of miR-127 in human anti-bacterial innate immunity, we then applied human monocytic cell line THP1. The results consistently demonstrated that miR-127 treatment reduced while anti-miR-127 application increased bacterial loads (Figure 3I), conversely correlating with the expression of anti-bacterial cytokine IL-22 and IL-17 (Figure 3J) and anti-microbial peptide S100A8 and Reg3β (Figure 3K). Collectively, our data supported that miR-127 potentially enhances bacterial clearing ability in macrophage and strengthens their production of anti-microbial effector molecules.
MiR-127 Boosts Anti-bacterial Signaling through Regulating STAT3 Activity
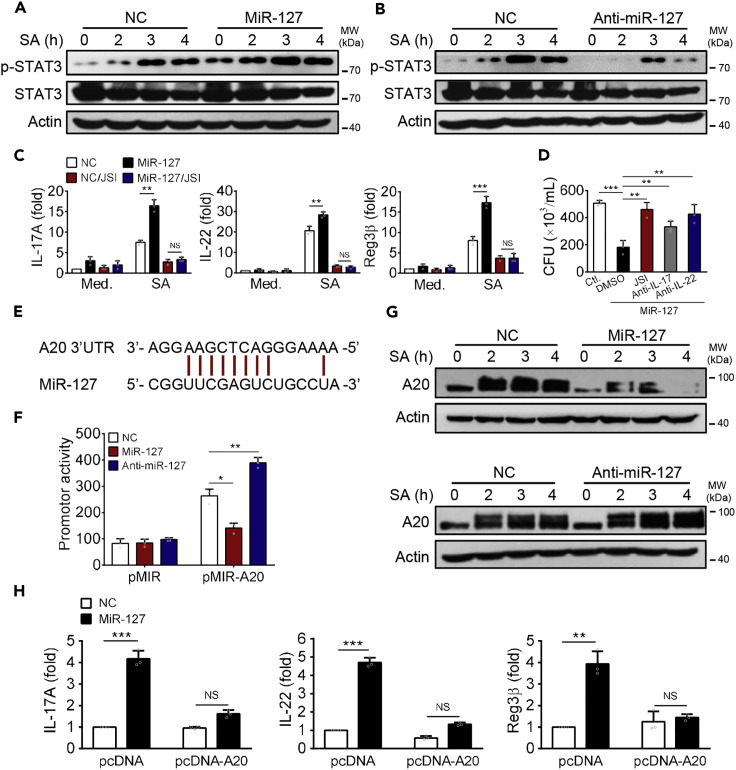
Next, we sought to understand the mechanism exploited by miR-127 to regulate the production of anti-microbial effector molecules by macrophages. Because STAT3 signaling played an essential role in triggering IL-22 and subsequently AMPs production (Abusleme et al., 2018, Choi et al., 2013, Frey-Jakobs et al., 2018, Zhao et al., 2018), we wondered whether miR-127 would enhance anti-microbial response via modulating STAT3 activity. Indeed, our data demonstrated that miR-127 treatment significantly enhanced phosphorylation of STAT3, whereas knockdown of miR-127 suppressed its activation (Figures 4A and 4B). To confirm the functional importance of the miR-127/STAT3 axis, we then utilized STAT3-specific inhibitor JSI-124 (JSI) to evaluate its role in anti-bacteria response. As shown in Figure 4C, miR-127-induced elevation in IL-17, IL-22, and Reg3β expression was largely abrogated by JCI treatment. Associated with this, the reduced bacterial burden upon miR-127 treatment was abolished either by STAT3 inhibition or by the neutralization of IL-22 or IL-17 (Figure 4D). Thus, the augmented anti-bacterial activity of miR-127 was largely mediated by its regulation of STAT3 signaling.
Figure 4.
A20 Serves as the Genuine Target of miR-127 Involved in the Regulation of Anti-bacterial Signaling
(A and B) RAW264·7 cells were transfected with miR-127, anti-miR-127, or their respective controls for 24 h, followed by S. aureus infection for the indicated time periods. Cells were then lysed and examined levels of the p-STAT3 and STAT3 by immunobloting.
(C) MiR-127- or non-specific control (NC)-transfected RAW264·7 cells were pretreated with STAT3 inhibitor JSI-124 (JCI) or DMSO and then infected with S. aureus for 12 h. The expression of the indicated molecules was examined by qPCR.
(D) Bacterial burdens were enumerated in miR-127-expressing macrophages pre-treated with JSI, anti-IL-17, or anti-IL-22 neutralizing antibody, respectively.
(E) The miR-127 targeting site at the 3′ UTR of the A20 gene.
(F) The promoter activity containing A20 3′UTR was analyzed upon miR-127 or anti-miR-127 treatment.
(G) The abundance of A20 protein was detected in RAW264·7 cells that were transfected with miR-127, anti-miR-127, or their respective non-specific controls respectively, followed by S. aureus challenge for the indicated time periods.
(H) A20-expressing or control plasmids were introduced into RAW264·7 cells that were treated with miR-127 or non-specific control (NC). The levels of representative anti-microbial molecules were analyzed 12 h after staphylococcal infection.
All results are from three independent experiments and presented as mean ± SD. *p < 0.05, **p < 0.01 by Student's t test.
Then the question arose on how miR-127 modulated STAT3 activity during macrophage response to bacterial infection. It appeared that STAT3 was not directly targeted by miR-127 because its expressive level was unaltered upon the treatment of miR-127 or anti-miR-127. To find out the causative link between miR-127 and STAT3, we turned to the Targetscan database and focused on the ubiquitin-editing enzyme A20, which harbored a putative site complementary to the seed sequences of miR-127 gene within its 3′ un-translated region (UTR) (Figure 4E). To confirm the direct regulation of A20 by miR-127, we then constructed the A20 3′UTR-containing reporter plasmid and found that A20-driven luciferase activity was markedly reduced by miR-127 treatment but increased upon miR-127 inhibition, substantiating the specification regulation of A20 by miR-127 (Figure 4F). To be supportive, A20 protein level was decreased by miR-127 treatment and elevated by anti-miR-127 administration (Figure 4G). MiR-127-mediated increased expression of anti-bacterial molecules IL-17, IL-22, and Reg3β was largely abrogated upon A20 overexpression, further supporting the functional relevance of the miR-127/A20 in anti-bacterial signaling (Figure 4H). Together, the ubiquitin-editing enzyme A20 was identified as a bona fide target of miR-127 and mediates the regulation of anti-bacterial reaction.
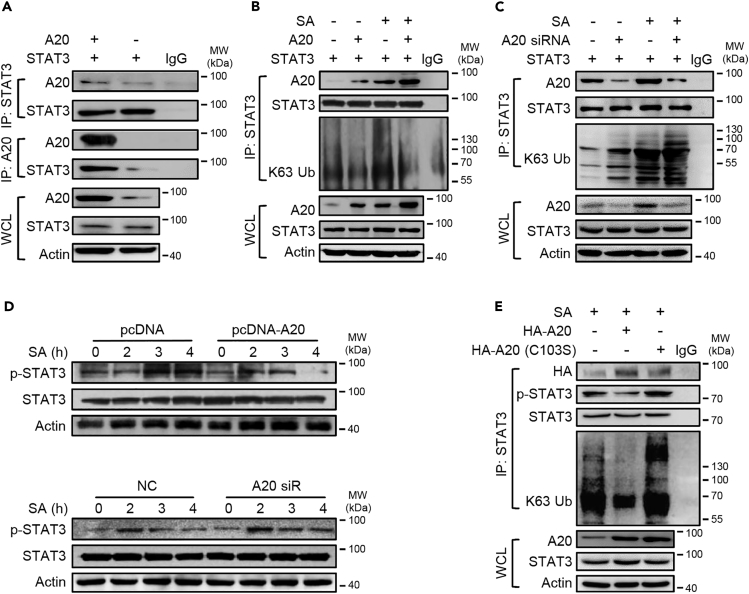
A20 Physically Interacts with STAT3 and Represses Its Ubiquitination and Hence Activation
A20 is an ubiquitin-editing enzyme with the ability to potentially regulate the immune and inflammatory responses mostly through the modulation of ubiquitination of key signaling molecules such as TRAF6 upstream of NF-κB (Ito et al., 2017, Vande Walle et al., 2014, Zhou et al., 2016). Because STAT3 has been subjected to the ubiquitinative modification (Cho et al., 2019, Ruan et al., 2017), we postulated that miR-127 might regulate STAT3 signaling through A20-mediated ubiquitination. To test this, we detected the direct interaction between STAT3 and A20 by Co-IP test. Prominently, A20 was recruited and bond to STAT3 and as a result, repressed the K63-ubiquitination of STAT3 (Figures 5A and 5B). By contrast, the knockdown of A20 led to the enhanced ligation of K63-linked polyubiquitin chains to STAT3 (Figure 5C). Congruently, A20 overexpression suppressed, whereas A20 knockdown increased phosphorylation of STAT3 in response to staphylococcal infection (Figure 5D), suggesting that A20 regulated STAT3 activation via its de-ubiquitinase activity. To further confirm this, we generated the mutation A20-expression plasmid with the C103S substitution within the ovarian tumor (OUT) domain, the structure responsible for A20 de-ubiquitinase activity (Voet et al., 2018). The result showed that A20-mediated de-ubiquitination of STAT3 was substantially impaired when transfected with the OUT-mutant plasmids, and the repressed STAT3 phosphorylation was accordingly resumed (Figure 5E). Together, we demonstrated that the ubiquitin-editing enzyme A20 specifically interacted with STAT3 and suppressed STAT3 ubiquitination and hence activation in response to bacterial infection.
Figure 5.
A20 Physically Interacts with STAT3 and Regulates Its Ubiquitination and Hence Activation in Response to Staphylococcal Infection
(A) 293T cells were transfected with STAT3-and/or A20-expressing plasmids for 48 h. Cells were then lysed for co-immunoprecipitation (co-IP) of STAT3 with anti-A20 antibody or for co-IP of A20 with anti-STAT3 antibody.
(B and C) 293T cells were transfected with STAT3-and/or A20-expressing plasmids (B) or transfected with A20-specific siRNA (C) for 48 h. The level of K63-ubiquitinated STAT3 was detected by co-IP.
(D) The level of phosphorylated STAT3 was detected in RAW264·7 cells transfected with A20-expressing plasmids or A20-specific siRNA or their controls.
(E) RAW264·7 cells were transfected with the intact or mutant (C103S) A20-expressing plasmids. The levels of total and phosphorylated STAT3 and K63-ubiquitinated STAT3 were examined. Shown are the representatives of two or three similar experiments. IgG was used as a negative control.
The Involvement of the miR-127/A20/STAT3 Axis in the Development of Staphylococcal Pneumonia
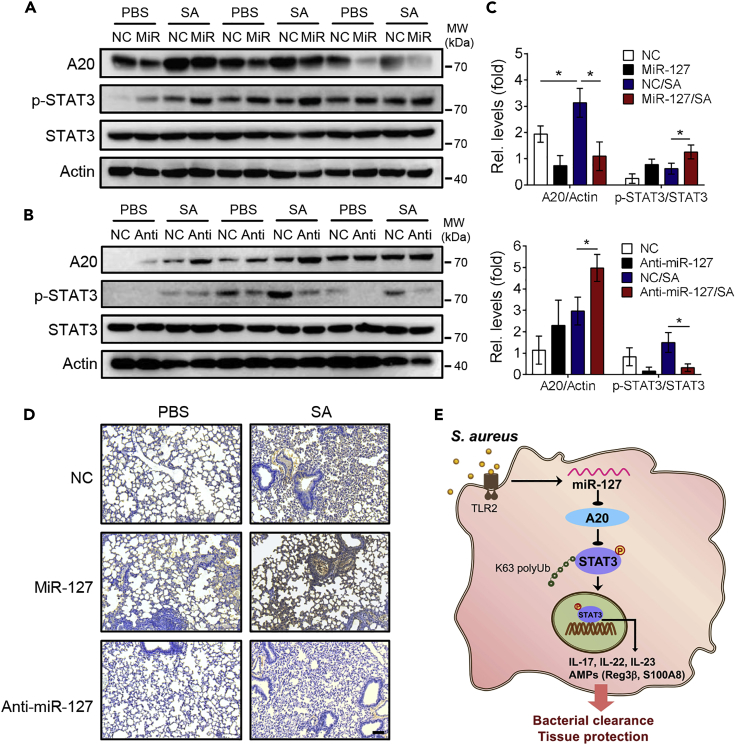
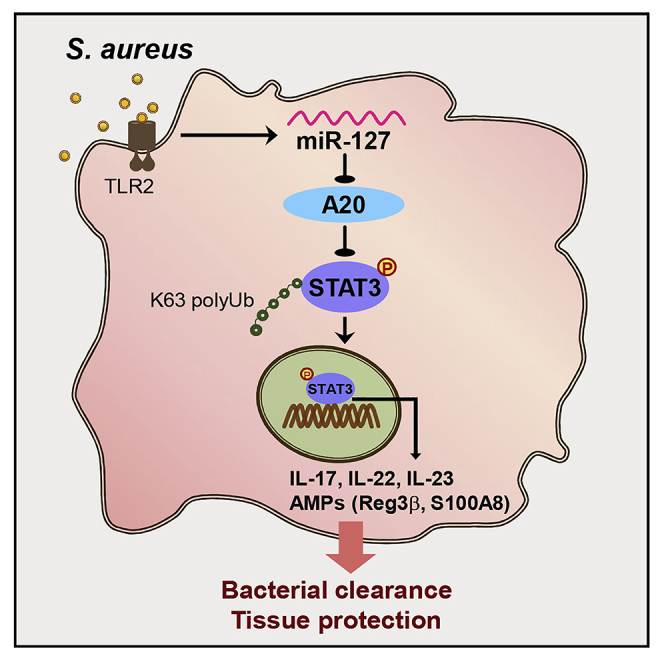
We next valuated the in vivo functional relevance of the miR-127/A20/STAT3 pathway by exploiting murine model of staphylococcal pneumonia. To this end, miR-127, anti-miR-127, or their non-specific control nucleotides (NC) were intratracheally instilled into mice 24 h prior to S. aureus infection, and the levels of A20 and total and phosphorylated STAT3 were analyzed in murine lung tissues. The result showed that the intratracheal administration of miR-127 reduced, whereas anti-miR-127 treatment elevated the level of A20 upon staphylococcus infection (Figures 6A and 6B). Meanwhile, the intensity of phosphorylated STAT3 was augmented upon miR-127 treatment but weakened on miR-127 inhibition, correlating inversely with A20 expression (Figure 6C). The positive correlation between the levels of miR-127 and phosphorylated STAT3 was also observed in murine lung tissues (Figure 6D). Combined with the protective role of miR-127 during staphylococcal infection observed above (Figures 1A–1J), we thus proposed a regulatory paradigm wherein A20, a genuine target of miR-127, restrained STAT3 activity through the repression of its K63-conjugated ubiquitination, and the induction of miR-127 upon infection led to the de-repression of this restraint and the facilitation of the anti-bacterial signaling (Figure 6E).
Figure 6.
The In Vivo Functional Relevance of the miR-127/A20/STAT3 Regulatory Circuitry
Age- and sex-matched mice (3 or 4 mice/group) were intratracheally administrated with miR-127, anti-miR-127, or their non-specific control (NC) respectively for 24 h, followed by staphylococcal infection for 12 h.
(A–C) Immunoblotting of A20, total and phosphorylated STAT3 at murine lung tissues (A and B); band intensities of A20 and phosphorylated STAT3 were normalized and quantitated (C). Data are presented as mean ± SD. *p < 0.05, **p < 0.01 by Student's t test.
(D) Immunohistological staining of phosphorylated STAT3 in the recovered lung tissues. Representative images are shown. Scale bar, 40 μm.
(E) The proposed working model showing the regulatory role of the miR-127/A20/STAT3 pathway in anti-bacterial response.
Discussion
The growing problem of antibiotic resistance in obstinate pathogens such as MRSA and the poor pipeline of antimicrobial molecules ignite the enthusiasm to develop therapeutic strategies, which demands more knowledge about host defensive machinery in addition to pathogenic factors. In this study, we identify miR-127 as a regulator of host anti-bacterial innate immunity and illustrate its functional importance and action mode in combating drug-resistant staphylococcus. Specifically, our data demonstrated that miR-127, induced upon infection, expedited clearance of bacteria and protected mice against pneumonia, whereas inhibition of miR-127 rendered mice more prone to staphylococcal infection with higher bacterial burden and exacerbated pneumonia. Importantly, we showed that A20, a genuine target of miR-127, repressed STAT3-mediated anti-bacterial signaling through constraining K63-ligated ubiquitination and hence activation of STAT3. We thus discover an unappreciated regulatory paradigm linking non-coding miRNA and STAT3 ubiquitination and phosphorylation that has a key role in coordinating anti-microbial immunity. The findings extend our understanding about host-pathogen interaction and provide potential targets for combating highly drug-resistant bacteria such as MRSA (Baral et al., 2018, Liu et al., 2018).
MicroRNAs (miRNAs) are short, conserved, non-coding RNA molecules with the potential to regulate mRNA stability and/or translation (Seeley et al., 2018). A number of miRNAs such as miR-302, miR-155, miR-223, miR-146, and miR-122 have demonstrated to be essential in the regulation of macrophage activity and innate immune responses (Rothchild et al., 2016, Liu et al., 2012, Wang et al., 2019a, Wang et al., 2019b). Nevertheless, previous studies mainly center on the effect of miRNA on inflammatory signaling. Evidences are emerging to delineate the direct effect of miRNAs, such as miRNA-328, on anti-microbial responses (Tay et al., 2015), but the related clues are relatively scarce. Our discovery that a small non-coding RNA, miR-127, plays a key role in regulating anti-bacterial pathway through the A20/STAT3 axis confers insights into microRNA biology in the field of innate immunity. The finding however seems odd since miRNA is generally considered as the regulator of gene expression rather than as the “on-off switcher.” In this regard, we show the de-ubiquitinase A20 exerts a central role that translates the miRNA-mediated gene regulation into the signaling modulation dependent on its de-ubiquitinase activity. In particular, our data indicate that A20 specifically interacted with STAT3 and inhibited the conjugation of K63-ubiquitin to STAT3, which in turn promoted STAT3 activation in response to staphylococcal infection. Although phosphorylation of STAT3 at Tyr705 or Ser727 has been identified as essential step for STAT3 activity, other post-translation modification and their relevance to STAT3 activation remain to be explored (Cho et al., 2019, Dallavalle et al., 2016, Ruan et al., 2017). We herein prove that de-repression of A20-mediated restraint of STAT3 ubiquitination is required for STAT3 activation and hence anti-bacterial signaling. The finding supports that integration of multiple layers of regulation such as ubiquitination and phosphorylation are essential for fine-tuning of STAT3, given its pleotropic function and its implication in pathophysiological conditions when dysregulated. In addition, we note that miR-127 is induced by staphylococcal infection in a dose- and time-dependent manner. Our previous study indicated that the induction of miR-127 was essentially dependent on NF-κB (Ying et al., 2015), the pathway known to be suppressed by A20. Thus, A20 likely serves as a molecular rheostat to be overcome to induce miR-127 production, which is essentially dependent on the infectious or inflammatory signals sensed and transduced into cells (Huynh et al., 2019).
As is known, the inflammatory response is indispensable for immune cells recruitment and anti-microbial response, but uncontrolled inflammatory signaling results in tissue damage, inflammapathology, and lethality (Branchett and Lloyd, 2019). In the present study, we found that in addition to anti-microbial response, miR-127 also promoted inflammatory cytokines production and inflammatory cells infiltration. Intriguingly, the amplified inflammatory response did not jeopardize but improve disease manifestation, as evidenced by alleviated lung immunopathology, lessened weight body loss, and decreased mortality upon intratracheal miR-127 administration. This is likely due to the tissue-protective effect of IL-17, IL-22, and AMPs such as Reg3β and S100A8, the molecules with particular importance in tissue repairing, regeneration, and mucosal barrier integrity (Li et al., 2018, Dudakov et al., 2015, Loonen et al., 2014, Sechet et al., 2018, Cho et al., 2010). Indeed, pro-inflammatory cytokines such as IL-6, TNFα, and IL-1β have demonstrated to play an instructive role in the generation of anti-microbial cytokines or other effector molecules and contribute to infection resolution. For instance, TNFα, induced by the recruited monocytes, was required for the production of IL-17, macrophage phagocytosis, and bacterial clearance, and hence recovery from pneumonia (Xiong et al., 2016). Likewise, IL-6 exhibited the ability to promote IL-17/IL-22-mediated anti-microbial responses through stimulating STAT3 signaling (Saint-Criq et al., 2018). The defective production of IL-6, induced by pathogenic component LasB of Pseudomonas aeruginosa, resulted in defective STAT3 activation and compromised anti-bacterial reactivity. These data, combined with our current findings, demonstrate a coherent link between NF-κB-driven proinflammatory signaling and STAT3-mediated anti-microbial pathway, which ensures defensive response at appropriate magnitude and kinetics to avoid unresolved infection (Ji et al., 2019). As an additional support, a recent study demonstrated that TRAF6, the E3 ligase required for NF-κB activation, exerted a boosting role in STAT3 activation through enhancing K63 ubiquitin (Ruan et al., 2017). Interestingly, it has been demonstrated that the de-ubiquitinase A20 can counteract the activity of TRAF6 through removing K63-ubiquitin from the subsets and feedback regulate vital signal pathways driven by NF-κB and TGF-β1/Smad6 (Shembade et al., 2010, Jung et al., 2013). Whether the A20/TRAF6 axis is involved in the regulation of STAT3 activity or not merits further investigation.
Taken together, our study identifies miR-127 as a key regulator of host defense against staphylococcus infection and delineates a regulatory circuitry that integrates non-coding miRNA and STAT3 ubiquitination/phosphorylation to shape the optimized anti-bacterial response. The findings are believed to have both biological and clinical implications and may provide therapeutic options for highly anti-antibiotic pathogens such as MRSA.
Limitations of the Study
Our current study elucidates a regulatory mechanism mediated by the miR-127/A20/STAT3 axis, which plays a key role in modulating macrophage anti-bacterial responses. However, as STAT3-driven anti-microbial signaling is also triggered by other immune cell subsets such as innate lymphoid cells and γδ T cells (Baral et al., 2018, Xiong et al., 2016, Paudel et al., 2019), we currently cannot exclude the effect of miR-127 on these cells’ response to bacterial infection, the topic of which is currently under investigation in our lab.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81470210, 81770014 and 81270066), the National Key Research and Development Program Project (2018YFC1705900), the Priority Academic Program Development of Jiangsu Higher Education Institutions, and by the final support for Jiangsu Key Laboratory for Pharmacology and Safety Evaluation of Chinese Materia Medica.
Author Contributions
X.Y. L., Y. M., Y. H. K., and L. H. carried out experiments, collected, and analyzed data. X.Y. L., B. Z., and W. Z. assisted with manuscript preparation. Y. L., Q. N. W., and D. K. X. contributed to experimental material and provided the insightful advices. L. Y. S. designed experiments and prepared the manuscript.
Declarations of Interests
The authors declare no competing interests.
Published: January 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.100763.
Supplemental Information
References
- Abusleme L., Diaz P.I., Freeman A.F., Greenwell-Wild T., Brenchley L., Desai J.V., Ng W.I., Holland S.M., Lionakis M.S., Segre J.A. Human defects in STAT3 promote oral mucosal fungal and bacterial dysbiosis. JCI Insight. 2018;3:122061. doi: 10.1172/jci.insight.122061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral P., Umans B.D., Li L., Wallrapp A., Bist M., Kirschbaum T., Wei Y., Zhou Y., Kuchroo V.K., Burkett P.R. Nociceptor sensory neurons suppress neutrophil and gammadelta T cell responses in bacterial lung infections and lethal pneumonia. Nat. Med. 2018;24:417–426. doi: 10.1038/nm.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C.N., Crepin V.F., Roumeliotis T.I., Wright J.C., Serafini N., Pevsner-Fischer M., Yu L., Elinav E., Di Santo J.P., Choudhary J.S. The Citrobacter rodentium type III secretion system effector EspO affects mucosal damage repair and antimicrobial responses. PLoS Pathog. 2018;14:e1007406. doi: 10.1371/journal.ppat.1007406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchett W.J., Lloyd C.M. Regulatory cytokine function in the respiratory tract. Mucosal Immunol. 2019;12:589–600. doi: 10.1038/s41385-019-0158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L.C., Rossetti M., Miller L.S., Filler S.G., Johnson C.W., Lee H.K., Wang H., Gjertson D., Fowler V.G., Jr., Reed E.F. Protective immunity in recurrent Staphylococcus aureus infection reflects localized immune signatures and macrophage-conferred memory. Proc. Natl. Acad. Sci. U S A. 2018;115:E11111–E11119. doi: 10.1073/pnas.1808353115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.S., Pietras E.M., Garcia N.C., Ramos R.I., Farzam D.M., Monroe H.R., Magorien J.E., Blauvelt A., Kolls J.K., Cheung A.L. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J. Clin. Invest. 2010;120:1762–1773. doi: 10.1172/JCI40891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.J., Xu Z., Parthasarathy U., Drashansky T.T., Helm E.Y., Zuniga A.N., Lorentsen K.J., Mansouri S., Cho J.Y., Edelmann M.J. Hectd3 promotes pathogenic Th17 lineage through Stat3 activation and Malt1 signaling in neuroinflammation. Nat. Commun. 2019;10:701. doi: 10.1038/s41467-019-08605-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.M., Mcaleer J.P., Zheng M., Pociask D.A., Kaplan M.H., Qin S., Reinhart T.A., Kolls J.K. Innate Stat3-mediated induction of the antimicrobial protein Reg3gamma is required for host defense against MRSA pneumonia. J. Exp. Med. 2013;210:551–561. doi: 10.1084/jem.20120260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkhurst C.N., Muratet M. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallavalle C., Albino D., Civenni G., Merulla J., Ostano P., Mello-Grand M., Rossi S., Losa M., D'ambrosio G., Sessa F. MicroRNA-424 impairs ubiquitination to activate STAT3 and promote prostate tumor progression. J. Clin. Invest. 2016;126:4585–4602. doi: 10.1172/JCI86505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta M., Dermawan J.K., Willard B., Stark G.R. STAT3-driven transcription depends upon the dimethylation of K49 by EZH2. Proc. Natl. Acad. Sci. U S A. 2015;112:3985–3990. doi: 10.1073/pnas.1503152112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakov J.A., Hanash A.M., van den Brink M.R. Interleukin-22: immunobiology and pathology. Annu. Rev. Immunol. 2015;33:747–785. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey-Jakobs S., Hartberger J.M., Fliegauf M., Bossen C., Wehmeyer M.L., Neubauer J.C., Bulashevska A., Proietti M., Frobel P., Noltner C. ZNF341 controls STAT3 expression and thereby immunocompetence. Sci. Immunol. 2018;3:eaat4941. doi: 10.1126/sciimmunol.aat4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Basile J.I., Classon C., Gavier-Widen D., Yoshimura A., Carow B., Rottenberg M.E. STAT3 expression by myeloid cells is detrimental for the T- cell-mediated control of infection with Mycobacterium tuberculosis. PLoS Pathog. 2018;14:e1006809. doi: 10.1371/journal.ppat.1006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton S.M., Borg N.A., Dixit V.M. Ubiquitin in the activation and attenuation of innate antiviral immunity. J. Exp. Med. 2016;213:1–13. doi: 10.1084/jem.20151531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J., Chand A., Gough D., Ernst M. Therapeutically exploiting STAT3 activity in cancer - using tissue repair as a road map. Nat. Rev. Cancer. 2019;19:82–96. doi: 10.1038/s41568-018-0090-8. [DOI] [PubMed] [Google Scholar]
- Ito T., Hirose K., Saku A., Kono K., Takatori H., Tamachi T., Goto Y., Renauld J.C., Kiyono H., Nakajima H. IL-22 induces Reg3gamma and inhibits allergic inflammation in house dust mite-induced asthma models. J. Exp. Med. 2017;214:3037–3050. doi: 10.1084/jem.20162108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., He L., Regev A., Struhl K. Inflammatory regulatory network mediated by the joint action of NF-kB, STAT3, and AP-1 factors is involved in many human cancers. Proc. Natl. Acad. Sci. U S A. 2019;116:9453–9462. doi: 10.1073/pnas.1821068116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S.M., Lee J.H., Park J., Oh Y.S., Lee S.K., Park J.S., Lee Y.S., Kim J.H., Lee J.Y., Bae Y.S. Smad6 inhibits non-canonical TGF-beta1 signalling by recruiting the deubiquitinase A20 to TRAF6. Nat. Commun. 2013;4:2562. doi: 10.1038/ncomms3562. [DOI] [PubMed] [Google Scholar]
- Kosack L., Wingelhofer B., Popa A., Orlova A., Agerer B., Vilagos B., Majek P., Parapatics K., Lercher A., Ringler A. The ERBB-STAT3 axis drives tasmanian devil facial tumor disease. Cancer Cell. 2019;35:125–139.e9. doi: 10.1016/j.ccell.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Casanova J.L., Puel A. Mucocutaneous IL-17 immunity in mice and humans: host defense vs. excessive inflammation. Mucosal Immunol. 2018;11:581–589. doi: 10.1038/mi.2017.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limagne E., Thibaudin M., Euvrard R., Berger H., Chalons P., Vegan F., Humblin E., Boidot R., Rebe C., Derangere V. Sirtuin-1 activation controls tumor growth by impeding Th17 differentiation via STAT3 deacetylation. Cell Rep. 2017;19:746–759. doi: 10.1016/j.celrep.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Liu P.T., Wheelwright M., Teles R., Komisopoulou E., Edfeldt K., Ferguson B., Mehta M.D., Vazirnia A., Rea T.H., Sarno E.N. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat. Med. 2012;18:267–273. doi: 10.1038/nm.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bai P., Woischnig A.K., Charpin-El Hamri G., Ye H., Folcher M., Xie M., Khanna N., Fussenegger M. Immunomimetic designer cells protect mice from MRSA infection. Cell. 2018;174:259–270.e11. doi: 10.1016/j.cell.2018.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman R.S., Diehl G.E., Victorio D.A., Huh J.R., Galan C., Miraldi E.R., Swaminath A., Bonneau R., Scherl E.J., Littman D.R. CX(3)CR1(+) mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J. Exp. Med. 2014;211:1571–1583. doi: 10.1084/jem.20140678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonen L.M., Stolte E.H., Jaklofsky M.T., Meijerink M., Dekker J., van Baarlen P., Wells J.M. REG3gamma-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 2014;7:939–947. doi: 10.1038/mi.2013.109. [DOI] [PubMed] [Google Scholar]
- Madouri F., Barada O., Kervoaze G., Trottein F., Pichavant M., Gosset P. Production of Interleukin-20 cytokines limits bacterial clearance and lung inflammation during infection by Streptococcus pneumoniae. EBioMedicine. 2018;37:417–427. doi: 10.1016/j.ebiom.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn B.A., Ma A. Ubiquitin makes its mark on immune regulation. Immunity. 2010;33:843–852. doi: 10.1016/j.immuni.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L.S., Cho J.S. Immunity against Staphylococcus aureus cutaneous infections. Nat. Rev. Immunol. 2011;11:505–518. doi: 10.1038/nri3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D., Prince A. Immunopathogenesis of Staphylococcus aureus pulmonary infection. Semin. Immunopathol. 2012;34:281–297. doi: 10.1007/s00281-011-0291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel S., Baral P., Ghimire L., Bergeron S., Jin L., Decorte J.A., Le J.T., Cai S., Jeyaseelan S. CXCL1 regulates neutrophil homeostasis in pneumonia-derived sepsis caused by Streptococcus pneumoniae serotype 3. Blood. 2019;133:1335–1345. doi: 10.1182/blood-2018-10-878082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J.A., Bewley M.A., Marriott H.M., Houghton A.M., Mohasin M., Jubrail J., Morris L., Stephenson Y.L., Cross S., Greaves D.R. Alveolar macrophage apoptosis-associated bacterial killing helps prevent murine pneumonia. Am. J. Respir. Crit. Care Med. 2019;200:84–97. doi: 10.1164/rccm.201804-0646OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothchild A.C., Sissons J.R., Shafiani S., Plaisier C., Min D., Mai D., Gilchrist M., Peschon J., Larson R.P., Bergthaler A. MiR-155-regulated molecular network orchestrates cell fate in the innate and adaptive immune response to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U S A. 2016;113:E6172–E6181. doi: 10.1073/pnas.1608255113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H.H., Zhang Z., Wang S.Y., Nickels L.M., Tian L., Qiao J.J., Zhu J. Tumor necrosis factor receptor-associated factor 6 (TRAF6) mediates ubiquitination-dependent STAT3 activation upon Salmonella enterica serovar typhimurium infection. Infect. Immun. 2017;85 doi: 10.1128/IAI.00081-17. e00081–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Criq V., Villeret B., Bastaert F., Kheir S., Hatton A., Cazes A., Xing Z., Sermet-Gaudelus I., Garcia-Verdugo I., Edelman A., Sallenave J.M. Pseudomonas aeruginosa LasB protease impairs innate immunity in mice and humans by targeting a lung epithelial cystic fibrosis transmembrane regulator-IL-6-antimicrobial-repair pathway. Thorax. 2018;73:49–61. doi: 10.1136/thoraxjnl-2017-210298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaler M., Jann N.J., Ferracin F., Landmann R. T and B cells are not required for clearing Staphylococcus aureus in systemic infection despite a strong TLR2-MyD88-dependent T cell activation. J. Immunol. 2011;186:443–452. doi: 10.4049/jimmunol.1001407. [DOI] [PubMed] [Google Scholar]
- Sechet E., Telford E., Bonamy C., Sansonetti P.J., Sperandio B. Natural molecules induce and synergize to boost expression of the human antimicrobial peptide beta-defensin-3. Proc. Natl. Acad. Sci. U S A. 2018;115:E9869–E9878. doi: 10.1073/pnas.1805298115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley J.J., Baker R.G., Mohamed G., Bruns T., Hayden M.S., Deshmukh S.D., Freedberg D.E., Ghosh S. Induction of innate immune memory via microRNA targeting of chromatin remodelling factors. Nature. 2018;559:114–119. doi: 10.1038/s41586-018-0253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N., Ma A., Harhaj E.W. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Wang Y., Lu Z., Zhang H., Zhuang N., Wang B., Song Z., Chen G., Huang C., Xu D. miR-127 promotes EMT and stem-like traits in lung cancer through a feed-forward regulatory loop. Oncogene. 2017;36:1631–1643. doi: 10.1038/onc.2016.332. [DOI] [PubMed] [Google Scholar]
- Tay H.L., Kaiko G.E., Plank M., Li J., Maltby S., Essilfie A.T., Jarnicki A., Yang M., Mattes J., Hansbro P.M. Antagonism of miR-328 increases the antimicrobial function of macrophages and neutrophils and rapid clearance of non-typeable Haemophilus influenzae (NTHi) from infected lung. PLoS Pathog. 2015;11:e1004549. doi: 10.1371/journal.ppat.1004549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S.Y., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G., Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treerat P., Prince O., Cruz-Lagunas A., Munoz-Torrico M., Salazar-Lezama M.A., Selman M., Fallert-Junecko B., Reinhardt T.A., Alcorn J.F., Kaushal D. Novel role for IL-22 in protection during chronic Mycobacterium tuberculosis HN878 infection. Mucosal Immunol. 2017;10:1069–1081. doi: 10.1038/mi.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S., Yamamoto M., Severson K.M., Ruhn K.A., Yu X., Koren O., Ley R., Wakeland E.K., Hooper L.V. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Walle L., van Opdenbosch N., Jacques P., Fossoul A., Verheugen E., Vogel P., Beyaert R., Elewaut D., Kanneganti T.D., van Loo G., Lamkanfi M. Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature. 2014;512:69–73. doi: 10.1038/nature13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino A.V., Kanno Y., O'shea J.J. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat. Immunol. 2017;18:374–384. doi: 10.1038/ni.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet S., Mc Guire C., Hagemeyer N., Martens A., Schroeder A., Wieghofer P., Daems C., Staszewski O., Vande Walle L., Jordao M.J.C. A20 critically controls microglia activation and inhibits inflammasome-dependent neuroinflammation. Nat. Commun. 2018;9:2036. doi: 10.1038/s41467-018-04376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Y., Zhang P., Baker S.T., Wolfson M.R., Weiser J.N., Tian Y., Shen H. Regenerative therapy based on miRNA-302 mimics for enhancing host recovery from pneumonia caused by Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U S A. 2019;116:8493–8498. doi: 10.1073/pnas.1818522116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liang H., Jin F., Yan X., Xu G., Hu H., Liang G., Zhan S., Hu X., Zhao Q. Injured liver-released miRNA-122 elicits acute pulmonary inflammation via activating alveolar macrophage TLR7 signaling pathway. Proc. Natl. Acad. Sci. U S A. 2019;116:6162–6171. doi: 10.1073/pnas.1814139116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P., Brodsky I.E., Rahner C., Woo D.K., Erdjument-Bromage H., Tempst P., Walsh M.C., Choi Y., Shadel G.S., Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H., Keith J.W., Samilo D.W., Carter R.A., Leiner I.M., Pamer E.G. Innate lymphocyte/Ly6C(hi) monocyte crosstalk promotes Klebsiella pneumoniae clearance. Cell. 2016;165:679–689. doi: 10.1016/j.cell.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H., Kang Y., Zhang H., Zhao D., Xia J., Lu Z., Wang H., Xu F., Shi L. MiR-127 modulates macrophage polarization and promotes lung inflammation and injury by activating the JNK pathway. J. Immunol. 2015;194:1239–1251. doi: 10.4049/jimmunol.1402088. [DOI] [PubMed] [Google Scholar]
- Yuan Z.L., Guan Y.J., Chatterjee D., Chin Y.E. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Chen F., Wu W., Sun M., Bilotta A.J., Yao S., Xiao Y., Huang X., Eaves-Pyles T.D., Golovko G. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018;11:752–762. doi: 10.1038/mi.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Xu H., Li A., Ji J., Wang Y., Xiao Y., Li L. Community-associated meticillin-resistant Staphylococcus aureus pneumonia in China. Lancet Infect. Dis. 2017;17:26. doi: 10.1016/S1473-3099(16)30554-0. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Wang H., Schwartz D.M., Stoffels M., Park Y.H., Zhang Y., Yang D., Demirkaya E., Takeuchi M., Tsai W.L. Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early-onset autoinflammatory disease. Nat. Genet. 2016;48:67–73. doi: 10.1038/ng.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.