Abstract
Numerous studies have reported that c-Src is highly expressed with high tyrosine kinase activity in a variety of tumors. However, it remains unclear whether c-Src contributes to the miRNA pathway. Here, we report that c-Src can interact with and phosphorylate AGO2, a core component of RISC complex, at tyr 393, tyr 529 and tyr749. Mechanistically, it is confirmed that c-Src phosphorylation of AGO2 at tyr393 reduces its binding to DICER, thereby suppressing the maturation of long-loop pre-miR-192. However, the other two phosphorylation sites don’t work on this function. Significantly, Ectopic expression of wild-type AGO2, but not the three tyrosine site mutants, has an obvious tumor-promoting effect in vitro and in vivo, which function could be blocked thoroughly by treatment with c-Src kinase inhibitor, Saracatinib. Our findings identify AGO2 as c-Src target and c-Src phosphorylation of AGO2 may therefore play a potential role during tumor progress.
Abbreviations: AGO2, Argonaute-2; EGFR, epidermal growth factor receptor; GST, glutathione S-transferase; IP, immunoprecipitation; miRNAs, microRNAs; P4H, prolyl-4-hydroxylase; PTMs, post-translational modifications; RISC, RNA-induced silencing complex; Y, tyr, tyrosine; F, phenylalanine
Keywords: c-Src, AGO2; Tyrosine phosphorylation; Tumorigenesis; Saracatinib
Introduction
c-Src, an non-receptor tyrosine kinase, is proved to be the first of several proto-oncogenes discovered in the vertebrate genome [1]. The c-Src protein promotes cell growth and migration that leads to tumor cell proliferation and metastasis [2]. After decades of basic scientific research, c-Src has been identified as an important oncogene in human cancer, which has had a profound impact on different types of cancer, including prostate cancer, lung cancer, breast cancer, colorectal cancer and neuronal tumor [3], [4], [5], [6], [7]. Although there is evidence that Src kinase can regulate certain specific microRNAs (miRNAs) thereby affecting tumorigenesis, for example, v-Src (the viral Src) suppresses miR-126 to promote tumorigenesis [8], the role of c-Src in regulating miRNA pathway has not been reported yet.
AGO2 (Argonaute-2), also termed as EIF2C2, is composed of four major domains including N domain, PAZ domain, MID domain and PIWI domain in addition with linker 1 and linker 2 [9]. As a critical regulator, AGO2 plays an important role in small RNAs guided gene silencing processes. AGO2 is a key component of RISC-loading complex containing DICER and TRBP for miRNA maturation [10]. In mammals, miRNAs or siRNAs guide the RNA-induced silencing complex (RISC) to perfectly complementary target sites in mRNAs, where endo-nucleolytically active AGO2 proteins cleave the RNA [11], [12], [13]. It is well-known that many miRNAs are critical in tumor progress. Consistently, AGO2 has also been found over-expressed in many kinds of cancers [14].
Multiple post-translational modifications (PTMs) can regulate AGO2 activities and functions. Enhanced hydroxylation at proline 700 of AGO2 by prolyl-4-hydroxylase (P4H) contributes to AGO2 stability and function [15]. Ubiquitination of AGO2 mediated by mLin41, an E3 ubiquitin ligase, regulates AGO2 turnover [16]. Recently, acetylation of AGO2 by P300/CBP was found to increase its recruitment of pre-miR-19b-1 to enhance miR-19b maturation [17]. In 2013, Shen et al. identified that epidermal growth factor receptor (EGFR) could phosphorylate AGO2 at Tyr393, and thus attenuate its affinity toward DICER and impede the maturation of long-loop miRNA in response to hypoxic stress [18]. It is reasonable that there may be other tyrosine kinases that can also contribute to phosphorylation of AGO2. In this study, we identify that c-Src kinase could interact with AGO2 directly, and phosphorylate AGO2 at Tyr393, Tyr529 and Tyr749 residues. Each phosphorylation of the three sites is essential to AGO2 mediated tumor-promotion effect, as determined by cell proliferation, migration, anchorage-independent growth and xenograft tumor growth. Moreover, the oncogenic function of AGO2 could be blocked thoroughly by treatment with c-Src kinase inhibitor, Saracatinib, which is likely to have a potential clinical implication.
Materials and methods
Antibodies and reagents
Anti-Src rabbit monoclonal (#2109), anti-phospho-Src (Tyr 416) rabbit polyclonal (#2101; used for detecting human pY419 Src), anti-AGO2 (#2897) and anti-Dicer (#3363) antibodies were obtained from Cell Signaling Technology. Anti-AGO2 (ab57113), anti-phospho-AGO2 (pY393 AGO2) (ab215746), anti-Tubulin (#66031-1-Ig), and anti-GAPDH polyclonal (#ab37168) antibodies were acquired from Abcam. Anti-Flag (M2), anti-HA and anti-Myc antibodies were purchased from Sigma-Aldrich. Protein A/G PLUS Agarose beads (#K0115) were obtained from Santa Cruz Biotechnology. Glutathione Sepharose 4B beads (#17-0756-01) were from GE Healthcare Life Sciences. Puromycin (#P8833) and EDTA-free Protease Inhibitor Cocktail were acquired from Sigma. Saracatinib (AZD0530) was purchased from Selleck.
Plasmids
The Flag-c-Src, HA-c-Src and GST-c-Src plasmids were described previously [19], [20]. The human c-Src CDS was cloned from pCMV-Tag2B-Src plasmid [19], digested with EcoR I and Not I and then subcloned into vector pCMV-Myc. The Flag-AGO2, HA-AGO2, Myc-AGO2 and GST-AGO2 plasmids were described previously [17]. Domain-deletion of HA-c-Src, Myc-AGO2 and point mutations of Myc-AGO2 were generated by using the KOD-plus-mutagenesis Kit (TOYOBO). The AGO2 cDNA was cloned from pEF-5HA-AGO2 into the vector pEGFP-C1 for the expression of GFP-AGO2 protein. The Flag-tagged full-length or tyrosine -mutated AGO2 was cloned into the lentiviral expression vector CD513B for establishing stable cell lines with the packaging plasmids pMD2G and pCMV-dR8. The AGO2 shRNA oligo sequences were gained from Sigma and sub-cloned into the lentiviral vector pLKO.1. The pre-miR-192, pre-miR-let7a-1 and pre-miR-34a were cloned into the pGREENpuro vector. The HA-EGFR plasmid was a gift from Prof. Jiong Deng in Shanghai Jiao Tong University School of Medicine.
Cell cultures and transfection
Human embryonic kidney 293T, 293FT, lung cancer A549 and prostate cancer DU145 cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (Hyclone) at 37 °C in 5% CO2 humidified incubator. Cell transfection was performed using Lipofectamine 2000 (Invitrogen).
Co-Immunoprecipitation (co-IP)
HEK293T cells transfected with indicated plasmids were lysed in the RIPA buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1%NP-40, 1 mM Na3VO4, 10 mM NaF with a protease inhibitor cocktail) on ice for 30 min. Lysates were immunoprecipitated with appropriate antibody overnight at 4 °C and then washed at least three times with RIPA buffer followed by Western blot analysis. Specially, for AGO2 domain mapping experiment, the protocol is modified by washed 5 times with modified RIPA buffer containing 500 mM NaCl.
In vitro kinase assay
Recombinant GST-Ago2 protein was incubated with or without Purified Flag-c-Src in HTScan 1× tyrosine kinase buffer (Cell Signaling) supplemented with 200 μM cold ATP and 2.5 mM DTT for 40 min at 30 °C. Kinase assay was stopped by 8.5% phosphoric acid, subjected to SDS–PAGE followed by Western blot analysis.
Northern blot analysis and qRT-PCR
The method for Northern blotting was described previously with a brief modification [17]. Briefly, total RNAs extracted by TRIZOL reagent (Invitrogen) from cells were denatured and fractionated by electrophoresis on a 20% polyacrylamide–8 M urea gel, then transferred to a nylon membrane (Roche) following cross-linkage. The membrane was pre-hybridized by North2South® Hybridization Buffer at 55 °C for 30 min, and hybridized with biotinylated probe at 55 °C overnight. After washed twice with North2South® Hybridization Stringency Wash Buffer at 55 °C for 15 min, the membrane was incubated in Streptavidin–HRP Blocking Buffer at room temperature for 1 h, and washed three times with wash buffer for 5 min following with substrate equilibration buffer for 5 min. Lastly, the signaling on membrane was detected by using Amersham Imager 600 (GE) instrument.
The method for miRNA qRT-PCR was described previously with a brief modification [21], [22]. Briefly, reverse transcription was performed by using PrimeScript™ RT-PCR Kit (TAKARA). For miRNA detection, specific miRNA reverse primers and U6 reverse primer were used to reverse transcript mature miRNAs and U6 snRNA, respectively.
| Northern miR-192-Biotin: |
| GGCTGTCAATTCATAGGTCAG |
| pre-miR-192 forward primer: GATCCGCUGACCUAUGAAUUGACAGCCAGUGCUCUCGUCUCCCCUCUGGCUGCCAAUUCCAUAGGUCACAGCTTTTTG |
| pre-miR-192 reverse primer: AATTCAAAAAGCTGTGACCTATGGAATTGGCAGCCAGAGGGGAGACGAGAGCACTGGCTGTCAATTCATAGGTCAGCG |
| qRT-PCR miR-192 forward primer: |
| GCCTGCTGACCTATGAATTG |
| qRT-PCR miR-192 reverse primer: |
| GTGCAGGGTCCGAGGT |
| Northern pre-miR-19b-Biotin: |
| TCAGTTTTGCATGGATTTGCACA |
| pre-miR-19b forward primer: |
| GATCCGAGTTTTGCAGGTTTGCATCCAGCTGTGTGATATTCTGCTGTGCAAATCCATGCAAAACTGACTTTTTG |
| pre-miR-19b reverse primer: |
| AATTCAAAAAGTCAGTTTTGCATGGATTTGCACAGCAGAATATCACACAGCTGGATGCAAACCTGCAAAACTCG |
| Northern miR-34a-Biotin: |
| ACAACCAGCTAAGACACTGCCA |
| pre-miR-34a forward primer: |
| GATCCGUGGCAGUGUCUUAGCUGGUUGUUGUGAGCAAUAGUAAGGAAGCAAUCAGCAAGUAUACUGCCCUCTTTTTG |
| pre-miR-34a reverse primer: |
| AATTCAAAAAGAGGGCAGTATACTTGCTGATTGCTTCCTTACTATTGCTCACAACAACCAGCTAAGACACTGCCACG |
| Northern let-7a-Biotin: |
| AACTATACAACCTACTACCTCA |
| pre-let-7a-1 forward primer: |
| GATCCGTGAGGTAGTAGGTTGTATAGTTTTAGGGTCACACCCACCACTGGGAGATAACTATACAATCTACTGTCTTTCCTTTTTG |
| pre-let-7a-1 reverse primer: |
| AATTCAAAAAGGAAAGACAGTAGATTGTATAGTTATCTCCCAGTGGTGGGTGTGACCCTAAAACTATACAACCTACTACCTCACG |
All the probe and primer sequences targeting pre-miR-19b and U6 were described previously [17].
Cell counting kit 8 (CCK-8) assay, wound healing assay and transwell assay
2 × 103 cells/well (for A549 or DU145 stable cell lines) were seeded in triplicate in a 96-well plate with complete growth medium. Cell proliferation was measured using CCK-8 assay. Absorbance at 450 nm was measured using a Microtiter plate reader (Promega).
Wound healing was performed as described previously [23]. 2 × 105 cells/ well (for A549 stable cell lines) were seeded in triplicate in a six-well plate and cultured overnight to ensure that they had adhered. Then, monolayers were removed with a 200 μL pipette tip and photographs were taken at the indicated times until the wound was healed.
For transwell assay, cells were pre-treated with saracatinib (5 μM) for 4 h before trypsinization. DMEM containing 10% FBS with saracatinib was added to the bottom chamber. Cell suspensions (2 × 1044 stable DU145 cells) in serum-free DMEM with saracatinib were added to the upper chamber. The average number of migrated cells per field was calculated based on five randomly selected fields per membrane in triplicate.
Colony formation assay and soft-agar colony assay
For colony formation assay, 100 cells/well (for DU145 stable cell lines) were seeded in triplicate in a six-well plate. Cells were cultured in DMEM medium containing 10% FBS for 2–3 weeks. Then they were fixed with 10% formaldehyde and stained with Giemsa stain and photographs were taken for counting colonies.
The soft-agar colony assay was performed as described previously [24]. This assay was performed in six-well plates with a base of 2 mL of medium containing 10% FBS with 1.2% Bacto agar (Amresco Solon, OH, USA). Cells were seeded in 2 mL of medium containing 10% FBS with 0.7% agar at 2 × 103 cells/ well (for A549 stable cell lines) and layered onto the base. The photographs of the cells growing in the plate and the colonies developed in soft agar were taken at 2–3 weeks. Three independent experiments were performed in triplicate.
Mouse xenograft models
Mouse xenograft models were established as described previously [20]. Briefly, 2 × 106 cells suspended in 100 μL medium (for A549 stable cell lines) were harvested and injected subcutaneously into 5-week-old male BALB/c nude mice individually. About a month later, at the experimental endpoint, mice were sacrificed and the tumors were dissected, weighed and photographed. Animal procedures were carried out according to a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Jiao Tong University School of Medicine.
Statistical analysis
Data are expressed as mean + SD. Statistical differences between groups were analyzed by the two-tailed Student’s t test. P < 0.05 was considered statistically significant. (*P < 0.05, **P < 0.01, ***P < 0.001.)
Bioinformatics analysis
Protein-protein interaction analysis between human AGO2 and c-Src (SRC) was performed by GeneMANIA (http://genemania.org).
Kaplan-Meier curves were created using GEPIA (http://gepia.cancer-pku.cn). AGO2 expression level was measured by Transcripts Per Kilobase Million (TPM). Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) were selected as datasets. A total of 481 high AGO2 TPM patients and 481 low AGO2 TPM patients were compared for median overall survival analysis.
AGO2 and c-Src (SRC) expression correlation analysis was performed by GEPIA. TCGA tumor (LUAD tumor, LUSC tumor), TCGA normal (LUAD normal, LUSC normal) and GTEx (lung) were set as used expression datasets for Spearman correlation coefficient analysis.
Immunofluorescence staining
The method for Immunofluorescence staining was described previously with a brief modification [25]. HeLa cells cultured on glass coverslips in six-well plates at 30–40% of confluence were fixed for 15 min with 4% PFA. After washed three times with washing buffer (PBS), the coverslips were blocked with blocking solution (PBS containing 5% BSA, 0.5% Triton X-100) for 1 h and then incubated with mouse anti-HA, anti-AGO2 (ab57113) and anti-c-Src antibodies in blocking solution overnight. Next, cells were rinsed three times (5 min each) with washing buffer, followed by incubation with appropriate secondary antibodies conjugated with Alexa Fluorescence 488, 568 in blocking solution for 1.5 h at room temperature in the dark. Finally, the slides were stained with DAPI for 1 h in the dark, washed three times, mounted with 90% glycerol and sealed with nail polish. Images were captured using a Zeiss Laser Scanning Microscope (LSM) 780 at pixels of 1024 × 1024.
Results
AGO2 interacts with c-Src in vitro and in vivo
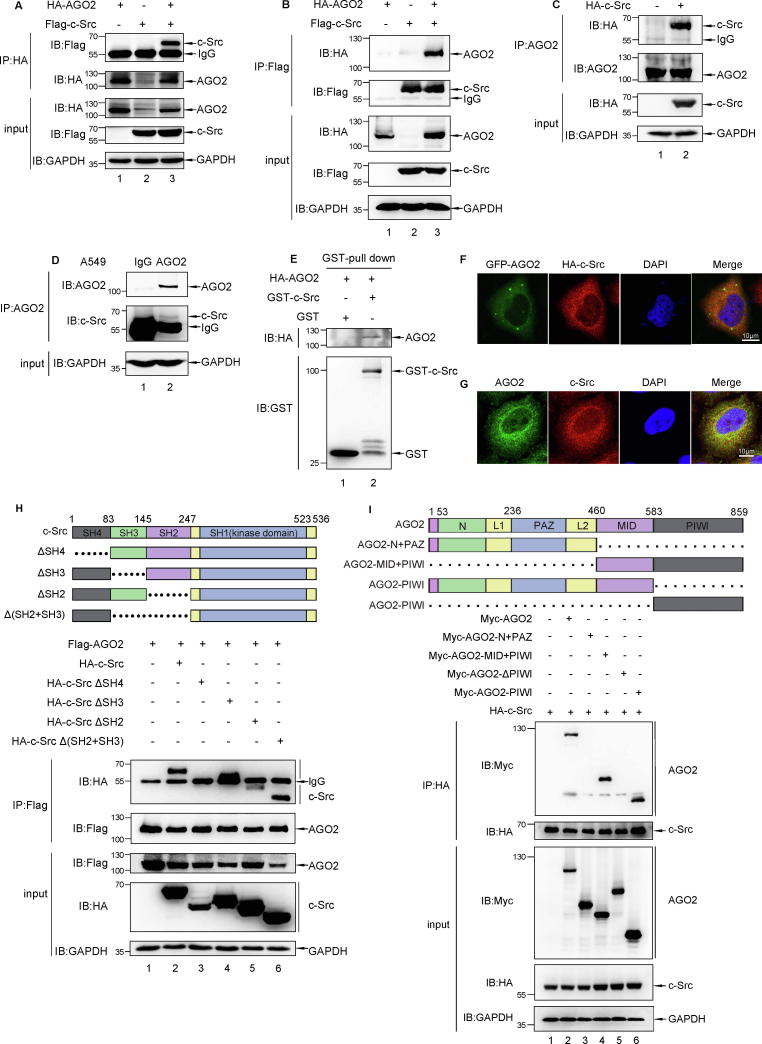
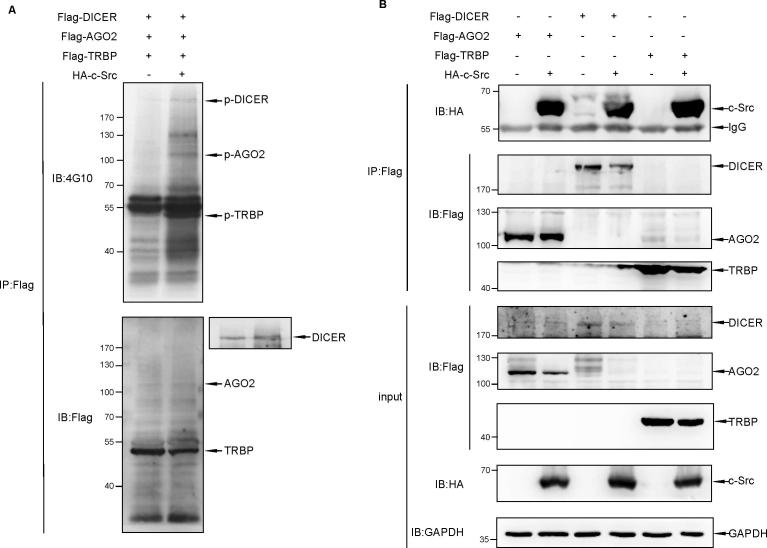
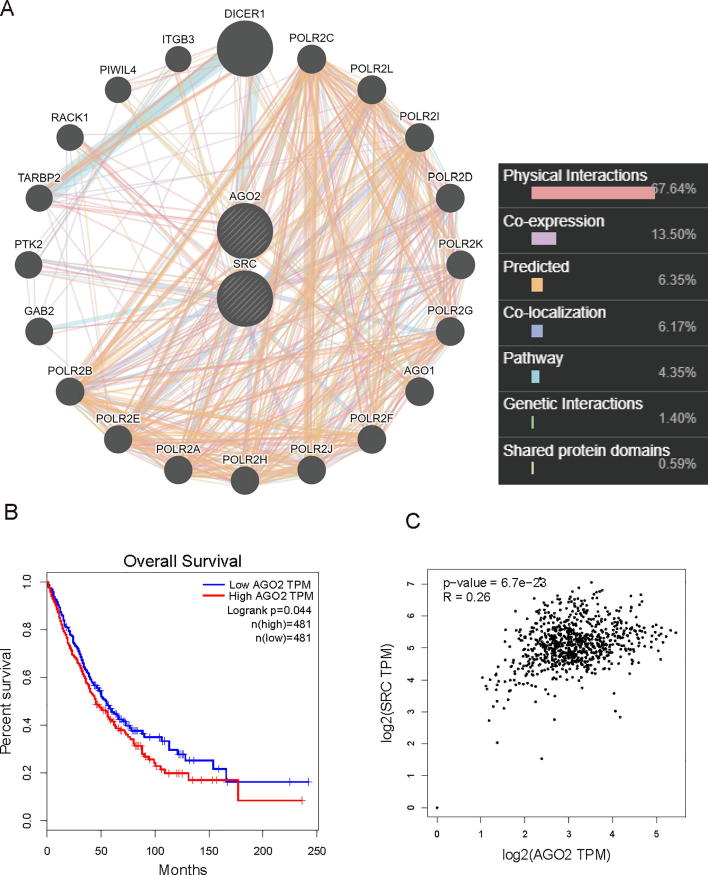
To test whether c-Src kinase contributes to microRNA pathway, we performed co-immunoprecipitation (co-IP) assay to seek for potential c-Src-interacting proteins involved in the miRNA pathway. By transient transfection in HEK293T cells, we found that c-Src binds to AGO2, DICER, TRBP (Fig. S1B), and all the three proteins could be tyrosine-phosphorylated by c-Src kinase (Fig. S1A). Because of the prominent role of AGO2 in the miRNA pathway, we focused our attention on the interaction between AGO2 and c-Src in further analyses. This interaction was confirmed with reciprocal co-IP assays by transient transfected with Flag-c-Src and HA-AGO2 in HEK293T cells (Fig. 1A and B). To further validate this interaction in vivo, immunopricipitation (IP) of endogenous AGO2 followed by immunoblotted with HA or c-Src antibody revealed the specificity of the AGO2-c-Src interaction (Fig. 1C and D). Furthermore, GST pull-down assay in vitro showed the direct interaction between HA-AGO2 and GST-c-Src, as indicated by Western blotting (Fig. 1E). We also found strong co-localization between c-Src and AGO2 in cytosol of HeLa cells using immunofluorescence staining with Ectopic or endogenous c-Src and AGO2 (Fig. 1F and G). A series of c-Src and AGO2 truncations was generated, and the domain mapping experiment demonstrated that SH4 domain of c-Src and PIWI domain of AGO2 were responsible for their mutual interaction (Fig. 1H, lane 3 and Fig. 1I, lane 4 and 6). Finally, we performed a bioinformatics analysis by GeneMANIA [26]. Human AGO2 and c-Src (SRC) were set as the input core protein. A network showed the relationships between AGO2, c-Src and their related proteins. The outcome showed a 67.64% physical interaction among these networks (Fig. S3A). Taken together, these data suggest a strong association between AGO2 and c-Src.
Fig. 1.
AGO2 interacts with c-Src in vitro and in vivo. (A, B) 293T cells were transfected with HA-AGO2 and/or Flag-c-Src as indicated. Reciprocal co-IP was carried out to immunoprecipitate with anti-HA or anti-Flag antibody, followed by Western blot analysis with anti-Flag or anti-HA antibody. (C) Semi-endogenous Co-IP assay. 293T cells were transfected with HA- c-Src. Cell lysates were used for immunoprecipitation (IP) with anti-AGO2 antibody, followed by Western blot analysis with anti-HA antibody. (D) Endogenous Co-IP assay. A549 cells lysates were precipitated with anti-AGO2 antibody, followed by Western blot analysis with anti-Src antibody. (E) GST pull-down assay was performed with HA-AGO2 immunoprecipitated with anti-HA antibody from 293T cells transfected with HA-AGO2, and GST-c-Src purified from E. coli transformed with GST-c-Src. (F) HeLa cells were co-transfected with HA-c-Src and GFP-AGO2. After 24 h of transfection, immunofluorescence staining was performed to observe the co-localization of c-Src and AGO2. Scale bars, 10 μm. (G) In HeLa cells, endogenous AGO2 and c-Src showed mainly co-localization in cytosol. Scale bars, 10 μm. (H) Schematic of c-Src protein is shown. 293T cells were transfected with Flag-AGO2 along with full-length or domain-deletion truncated HA-c-Src. cell lysates were used for immunoprecipitation (IP) with anti-Flag antibody, followed by Western blot analysis with anti-HA antibody to verify the interaction between c-Src and AGO2. (I) Schematic of AGO2 protein is shown. 293T cells were transfected with HA-c-Src along with full-length or domain-deletion truncated Myc-AGO2. Cell lysates were used for immunoprecipitation (IP) with anti-HA antibody, followed by Western blot analysis with anti-Myc antibody to verify the interaction between c-Src and AGO2.
AGO2 is phosphorylated at tyr393, tyr529 and tyr749 by c-Src kinase
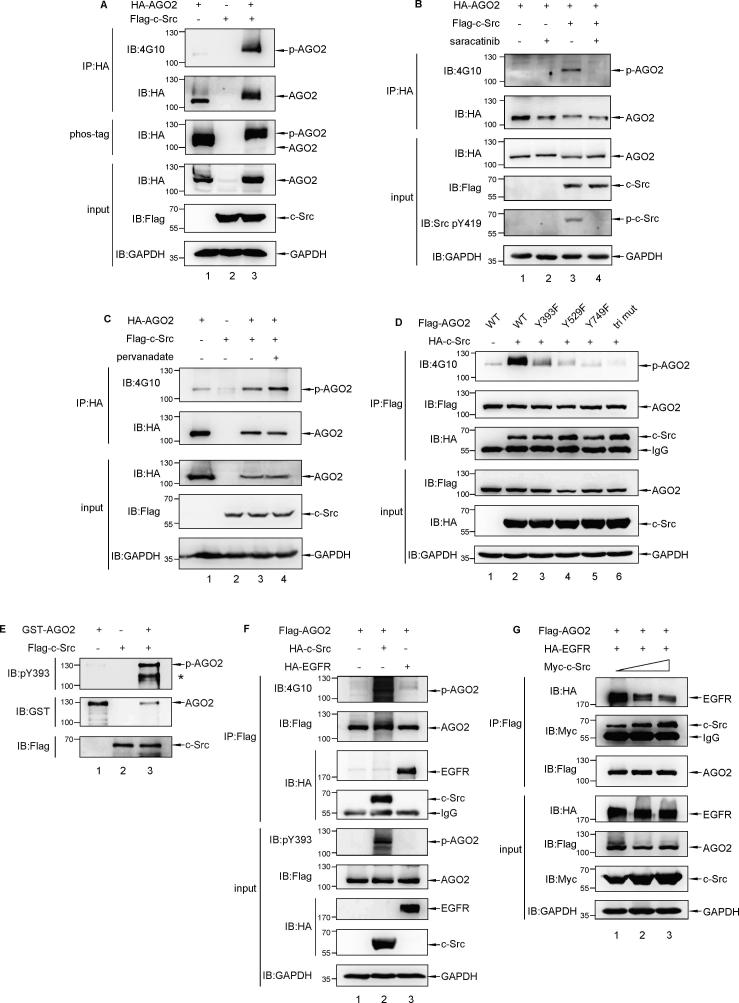
To investigate whether AGO2 is a phosphorylation substrate of c-Src kinase, transfection with Flag-c-Src and HA-AGO2 in HEK293T cells was performed, and immunoprecipitation (IP) with anti-HA antibody following by immunoblotted with a pan anti-phosphotyrosine antibody 4G10 showed that AGO2 is phosphorylated clearly (Fig. 2A, upper lane 3). This specific modification was further confirmed by the phos-tag gel analysis, in which c-Src induced a noticeable mobility shift band (Fig. 2A, middle lane 3 versus 1). Moreover, phosphorylated AGO2 (pYAGO2) was abolished by a c-Src kinase inhibitor, saracatinib (AZD0530) (Fig. 2B, lane 4) and enhanced by a tyrosine phosphatase inhibitor, pervanadate (Na3VO4) (Fig. 2C, lane 4). All the results revealed that AGO2 is a substrate for c-Src kinase.
Fig. 2.
AGO2 is phosphorylated by c-Src kinase. (A) 293T cells were co-transfected with Flag-c-Src and HA-AGO2. Cell lysates were used for immunoprecipitation with anti-HA antibody, followed by Western blot analysis with a pan anti-phospho-tyrosine antibody (4G10). The Cell lysates were also isolated by Phos-tag gel electrophoresis, followed by Western blot analysis with anti-HA antibody. (B) 293T cells were transfected with HA-AGO2 and/or Flag-c-Src as indicated. Transfected 293T cells were treated with or without 10 µM saracatinib for 4 h before harvest. Cell lysates were used for immunoprecipitation with anti-HA antibody, followed by Western blot analysis with 4G10. (C) 293T cells were transfected with HA-AGO2 and/or Flag-c-Src as indicated. Transfected 293T cells were treated with 0.1 mM pervanadate (a protein tyrosine phosphatase inhibitor) for 10 min before harvest. Cell lysates were used for immunoprecipitation with anti-HA antibody, followed by Western blot analysis with 4G10. (D) 293T cells were transfected with HA-c-Src along with full-length or Y/F mutated Flag-AGO2 as indicated. cell lysates were used for immunoprecipitation (IP) with anti-Flag antibody, followed by Western blot analysis with 4G10, or anti-HA, or anti-Flag antibody, respectively, to identify the tyrosine residues in AGO2 phosphorylated by c-Src kinase. (E) In vitro kinase assay showed GST-AGO2 was phosphorylated at Y393 by c-Src kinase directly. (F) 293T cells were transfected with Flag-AGO2 along with HA-c-Src or HA-EGFR. Cell lysates were used for immunoprecipitation with anti-Flag antibody, followed by Western blot analysis with 4G10. The Cell lysates were by Western blot analysis with anti-pY393 AGO2 antibody. (G) Competitive experiment was conducted through co-transfecting equal amount of Flag-AGO2 and HA-EGFR along with Myc-c-Src gradient (1, 2.5 and 5 μg, respectively) in 293T cells.
Based on https://www.genecards.org and published literatures, three tyrosine residues in AGO2 protein have been reported as phosphorylation sites, including tyr749 firstly identified with mass spectrometry in 2009 [27], and tyr393/tyr529 identified with mass spectrometry in 2011 [28]. Thus, we tried to determine which of the three tyrosine residues is responsible for the c-Src phosphorylation. Tyr → phe (Y → F) mutational analysis showed that each of the single point mutation (Y393F, Y529F, Y749F) has dramatically attenuated the c-Src-induced AGO2 phosphorylation (Fig. 2D, lane 3–5). Notably, tri-mutation AGO2 had hardly any phosphorylation modification by c-Src (Fig. 2D, lane 6). One result (Fig. 2E) from in vitro kinase assay further demonstrated tyr393 to be a direct phosphorylation site target for c-Src kinase using commercialized pY393 Ago2 antibody (no commercialized pY529 Ago2 or pY749 Ago2 antibody by now). Collectively, we conducted that AGO2 could be phosphorylated at tyr393, tyr529, and tyr749 by c-Src kinase.
In 2013, it was reported EGFR can phosphorylate AGO2 at tyr393 in response to hypoxic stress [18]. Because of close correlation between EGFR and c-Src, we intended to verify the different roles of the two tyrosine kinases in the phosphorylation of AGO2. After transfection with Flag-AGO2, alone or together with HA-c-Src or HA-EGFR in HEK293T cells, cells were lysed in RIPA buffer and immunoprecipitation (IP) with anti-Flag antibody following by immunoblotted as antibodies indicated. As shown in Fig. 2F, the degree of total tyrosine phosphorylation (upper lane 2) or pY393 (middle lane 2) of AGO2 by c-Src kinase was much stronger than those by EGFR (upper and middle lane 3). Furthermore, competitive experiment was conducted through co-transfecting equal amount of Flag-AGO2 and HA-EGFR along with Myc-c-Src gradient in HEK293T cells. As expected, decreased EGFR bound to AGO2 was accompanied by increased c-Src gradient transfection, which indicated that there is likely to be a direct competition between c-Src and EGFR for binding with AGO2. Together, we believe that c-Src kinase has a stronger effect on AGO2 phosphorylation than EGFR at least under normoxia.
c-Src-induced AGO2 phosphorylation at tyr393, but not tyr529 or tyr749, suppresses the maturation of the long-loop pre-miR-192
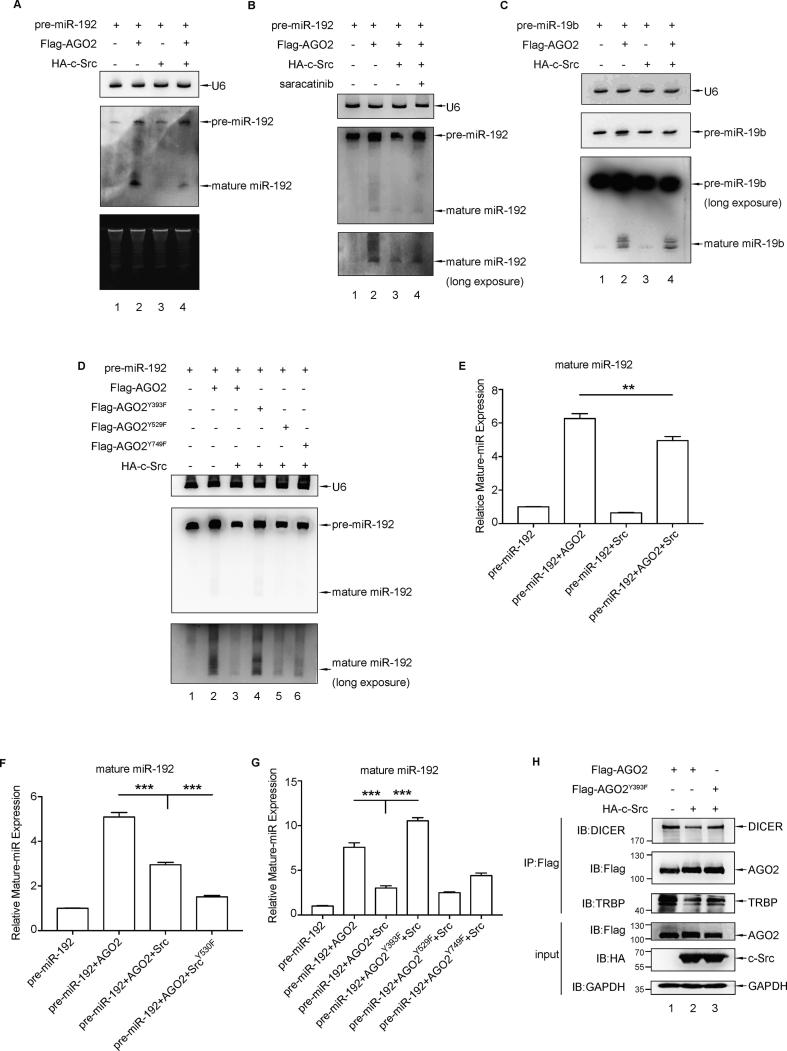
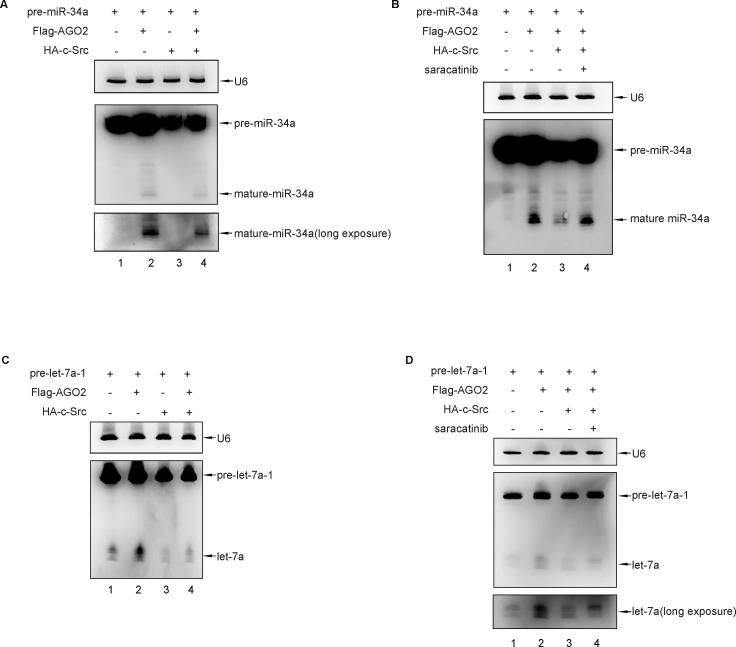
It’s reported that EGFR-induced AGO2 phosphorylation at tyr393 specifically suppresses the maturation of pre-miRNAs with long-loop structures under hypoxia [18]. Thus, we examined the possibility that c-Src-induced AGO2 phosphorylation may also have a role in miRNA maturation. The Northern Blot analysis for detecting mature miRNA from pre-miRNA has been established in our previous study [17]. Northern Blot results showed that the amount of miR-192 from pre-miR-192, a pre-miRNA with a long-loop structure [18], was significantly increased when transfection with AGO2 (Fig. 3A, lane 2), whereas this increasing effect was inhibited when co-transfection with c-Src (Fig. 3A, lane 4). And this inhibition was rescued effectively by treatment with saracatinib, a c-Src kinase inhibitor (Fig. 3B, lane 4). These results from maturation of pre-miR-192 suggest that c-Src-induced AGO2 phosphorylation may suppressed the maturation of pre-miRNAs with long-loop structures. This effect was further confirmed with another two pre-miRNAs with long-loop structures, pre-miR-34a and pre-let-7a-1. Northern Blot results also showed that the amount of miR-34a (Fig. S2A, lane 2) or let-7a (Fig. S2C, lane 2) was significantly increased when transfection with AGO2, whereas inhibited when co-transfection with c-Src (Fig. S2A or S2C, lane 4). And this inhibition was rescued effectively by treatment with saracatinib (Fig. S2B or S2D, lane 4). Moreover, the above results from pre-miR-192 were confirmed further by qRT-PCR analysis (Fig. 3E and F). As expected, c-Src Y530F, the active form, has a stronger inhibitory effect than c-Src (Fig. 3F, column 3 and 4). Conversely, there was no significant difference in maturation of miR-19b from pre-miR-19b, a pre-miRNA without a long-loop structure [17], with or without c-Src transfection (Fig. 3C, lane 2 and 4).
Fig. 3.
c-Src phosphorylates AGO2 at tyr393 to suppress the maturation of the long-loop miRNA. (A) Pre-miR-192 and Flag-AGO2 with or without HA-c-Src were transfected into 293T cells. Total RNA was extracted from 293T cells, the miR-192 expression were detected by Northern blotting. (B) Protocol as (A), except that transfected 293T cells were treated with or without 10 µM saracatinib for 4 h before harvest. (C) Pre-miR-19b and Flag-AGO2 with or without HA-c-Src were transfected into 293T cells. Total RNA was extracted from 293T cells for detecting the miR-19b expression by Northern blotting. (D) 293T cells were transfected with Pre-miR-192 and HA-c-Src and Y/F mutated Flag-AGO2 as indicated. Total RNA was extracted from 293T cells, the miR-192 expression were detected by Northern blotting. (E–G) qRT-PCR was conducted to examine the expression of miR-192 in 293T cells with different treatments as indicated. Results were normalized to U6 and shown as n-fold change relative to the corresponding control cells. Bars ± SD showed the median of three independent experiments, differences in expression levels were analysed by t-test (**P < 0.01, ***P < 0.001). (H) 293T cells were transiently transfected with Flag-AGO2 or Flag-AGO2Y393F along with HA-c-Src. Cell lysates were immunoprecipitated with anti-Flag antibody followed by Western blot analysis with anti-DICER or anti-TRBP antibody for detection of bindings between AGO2 and DICER/TRBP induced by c-Src.
Next, we try to verify which tyrosine residue in AGO2 is responsible for inhibiton of miRNA maturation. By Northern Blot analysis (Fig. 3D), we characterized that AGO2Y393F, but not AGO2Y529F or AGO2Y749F can recover the c-Src inhibition on miR-192 maturation. This conclusion was further supported by qRT-PCR result (Fig. 3G). Accordingly, since AGO2 associates with DICER and TRBP to form the RISC-loading complex, which is responsible for the miRNAs maturation from pre-miRNAs, we detected the binding pattern between AGO2 and DICER/TRBP under the action of c-Src. Western blot analysis indicated that AGO2 WT, not AGO2Y393F, reduced its binding to DICER and TRBP accompanied with c-Src phosphorylation (Fig. 3H), which suggests that pY393 is a negative regulator for AGO2-dependent miRNA maturation. Overall, it is likely that AGO2 phosphorylation at tyr393 by c-Src, similar to that by EGFR under hypoxia, selectively suppresses the processing and maturation of long-loop pre-miRNAs.
Each of AGO2 phosphorylations at tyr393, tyr529 and tyr749 has an oncogenic function
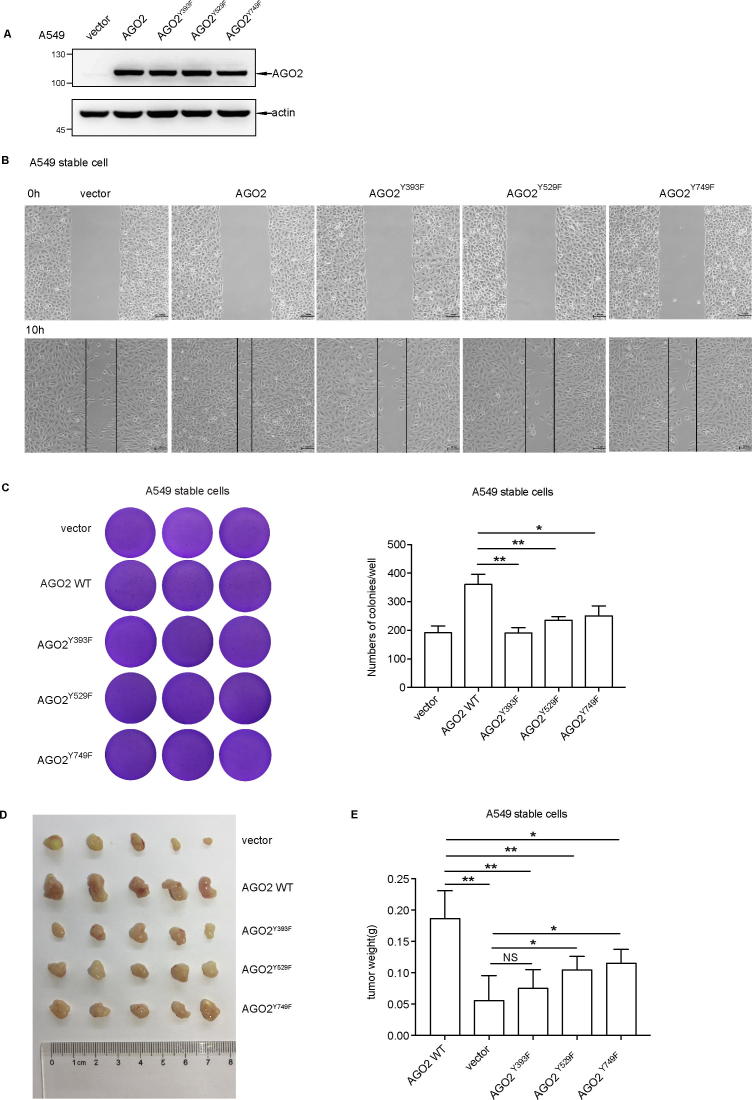
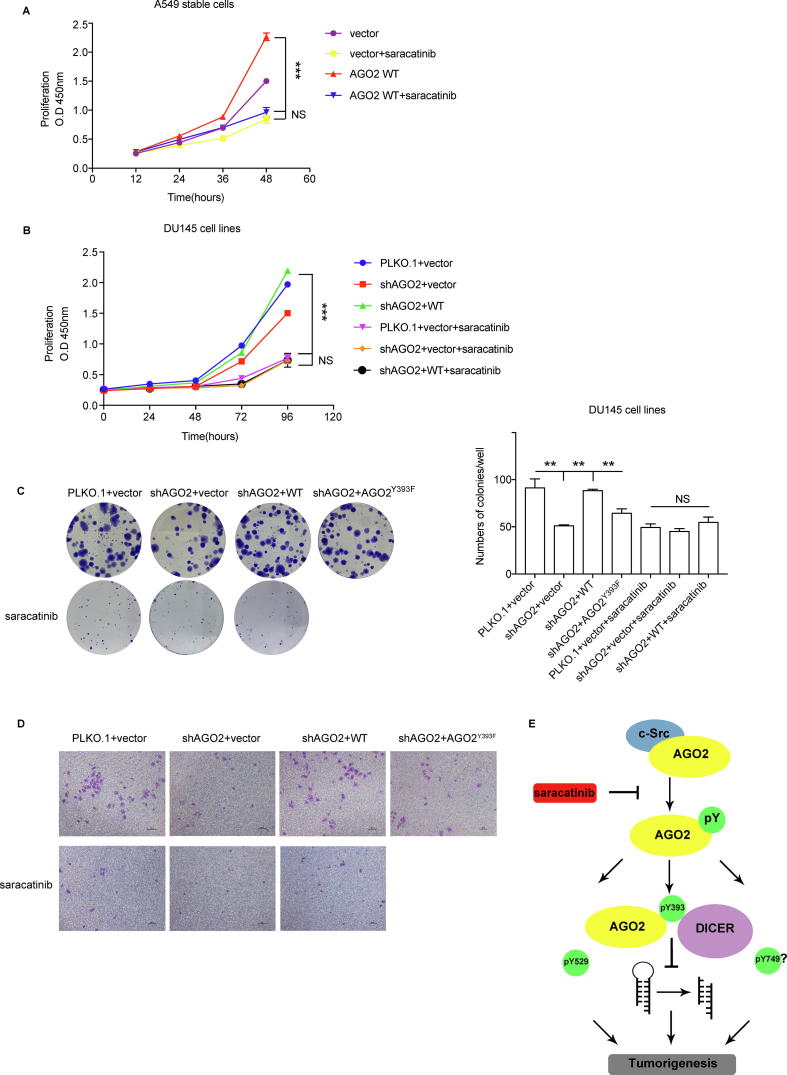
To explore whether phosphorylation of AGO2 has a biological significance in tumors, we generated stable A549 lung cancer cell lines by polyclonal lentiviral infections with vector, AGO2 WT, AGO2Y393F, AGO2Y529F, or AGO2Y749F. All the four ectopic AGO2 expression levels were equally high indicated by Western blot analysis (Fig. 4A). In a wound healing assay, ectopic AGO2 WT, but none of three Y/F mutants, could significantly increase cell migration compared with vector control (Fig. 4B). To further verify the function of AGO2 phosphorylation, anchorage-independent soft-agar colony-forming assay was conducted. As shown in Fig. 4C, cells transfected with AGO2 WT showed more numbers and sizes of colonies than those transfected with vector. However, compared with vector, the three mutants had no statistical difference in anchorage-independent growth although AGO2Y529F or AGO2Y749F had a very slight increase in colony numbers. Finally, we performed a xenograft tumor formation experiments by injecting five A549 stable cells subcutaneously into nude mice. One month after the injection, all the tumors were gathered for photo recording (Fig. 4D) and a statistical analysis on tumor weight was conducted (Fig. 4E). Interestingly, over-expressed AGO2 WT in A549 cell significantly increased the size and weight of tumor compared with both vector control and three AGO2 mutants (**P < 0.01 between AGO2 WT and vector, AGO2Y393F, AGO2Y529F, *P < 0.05 between AGO2 WT and AGO2Y749F). However, compared with vector, AGO2Y749F or AGO2Y529F also significantly increased the size and weight of tumor (*P < 0.05), and AGO2Y393F had a slight increasing effect with no statistical difference. These data suggest that phosphorylation of AGO2 promotes tumorigenesis, and each phosphorylation at the three tyrosine residues is essential to this oncogenic function. In addition, perhaps Y393 is the most important functional site among these three tyrosine residues.
Fig. 4.
Tyrosine phosphorylation of AGO2 promotes tumorigenesis. (A) Ectopic AGO2 was introduced into the lung cancer cell line A549 by lentiviral system. Western blot analysis showed the expression level of AGO2 WT and Y/F mutants. (B) The cell motility of stable A549 cell lines was determined by wound healing assay and representative images were taken at the time points 0 h and 12 h. (C) Soft-agar colony assay. Stable A549 cell lines were seeded in 2 mL of medium containing 5% FBS with 0.35% agar at 2 × 103 cells per well and layered onto the base. The photographs were taken and numbers of colonies were calculated one month later. Images are representative of three independent experiments. All data are mean ± SD. Differences in expression levels were analysed by t-test (*P < 0.05, **P < 0.01). (D) The image for the tumor is shown. The male BALB/c nude mice (n = 5) were given a suspension A549 stable cells subcutaneously (2 × 106 cells/each). Xenografted tumors were taken out after one month, and tumors were dissected and weighed. (E) Tumor weight was measured at the experimental endpoint. All data are. Mean ± SD. Differences in expression levels were analysed by t-test (*P < 0.05, **P < 0.01).
Saracatinib, c-Src inhibitor, thoroughly blocks AGO2-promoting oncogenic phenotype
To further estimate whether and how c-Src-mediated AGO2 phosphorylation promotes tumorigenesis, saracatinib (AZD0530), one small molecule inhibitor to the c-Src kinase, was used for cell experiments. To avoid contingency in cell line and experimental method, besides A549 cell line, we generated other stable DU145 prostate cancer cell lines by polyclonal lentiviral infections firstly with AGO2 shRNA, then following with vector, ectopic AGO2 WT or AGO2 Y393F. A Cell Counting Kit 8 (CCK-8) assay was used to examine the A549 and DU145 stable cells proliferation. The rate of proliferation was increased in A549 transfected with AGO2 WT stable cells compared with cells with vector, but both stable cells proliferation was inhibited to a same lower level by treatment with saracatinib (Fig. 5A). A similar pattern of results was observed in DU145 stable cell lines. As shown in Fig. 5B, compared with vector control, the rate of proliferation was increased in cells transfected with AGO2 shRNA and WT, but decreased in cells transfected only with AGO2 shRNA. However, all the three stable cells proliferation were inhibited to a same lower level by treatment with saracatinib. Furthermore, colony formation assay on DU145 stable cell lines showed cells transfected with AGO2 shRNA could reduce the numbers of colonies, but this suppression could be rescued fully by re-expression of AGO2 WT rather than AGO2Y393F. Moreover, saracatinib dramatically inhibited the sizes of colonies and reduced the numbers of colonies in three stable cell lines (Fig. 5C). In addition, transwell assay showed cells transfected with AGO2 shRNA decreased cell migration, which could be rescued by re-expression of AGO2 WT rather than AGO2Y393F. As expected, saracatinib remarkably inhibited cell migration in three stable cell lines (Fig. 5D). It is worth noting that treatment with saracatinib had a equal inhibition effect for all the stable cell lines no matter what AGO2 is. Overall, these data imply that saracatinib is responsible not only for AGO2 dephosphorylation but also for other substrates of c-Src, and AGO2-mediated oncogenic function could be blocked thoroughly by c-Src inhibitor.
Fig. 5.
Saracatinib blocks AGO2-promoting oncogenic phenotype. (A and B) Cell proliferation of stable A549 cell lines (A) and stable DU145 cell lines (B) was examined by Cell Counting Kit 8 (CCK-8). All data are mean ± SD. Differences in proliferation rate were analysed by two-way ANOVA (***P < 0.001). (C) Colony formation assay. 100 stable DU145 cells per well were seeded and cultured in DMEM medium containing 10% FBS in 6-well plates for 2–3 weeks until counting. The final concentration of saracatinib is 5 μM. All data are mean ± SD. Differences in numbers of colonies were analysed by t-test (**P < 0.01). (D) Transwell assay. Cells were pre-treated with saracatinib (5 μM) for 4 h before trypsinization. DMEM containing 10% FBS with saracatinib was added to the bottom chamber. Cell suspensions (2 × 104 stable DU145 cells) in serum-free DMEM with saracatinib added to the upper chamber. The average number of migrated cells per field was calculated based on five randomly selected fields per membrane in triplicate. (E) A schematic model. AGO2 interacts with c-Src and is phosphorylated at its tyr393, tyr529 and tyr749 sites by c-Src kinase, which phosphorylation could be blocked by treatment with saracatinib, the c-Src kinase inhibitor. Phosphorylation of AGO2 at tyr393 reduces its binding to DICER, thereby inhibiting miRNA maturation. Each of the three phosphorylations in AGO2 shows an obvious tumor-promoting effect.
Discussion
As known to all, c-Src is a classic abnormal high expressed oncogene in multiple cancers. Recent works illustrate AGO2 being a hotspot in cancer research. In this study, we identify a correlation between c-Src kinase and AGO2 by a series of molecular and cellular experiments. Moreover, a protein–protein interaction network for AGO2, c-Src and their related proteins was performed by GeneMANIA (Multiple Association Network Integration Algorithm), which is fast enough to predict gene function while achieving state-of-the-art accuracy [29]. However, it should be noticed that although GeneMANIA is a useful tool for any biologist, this approach is limited by evidence-based studies, such as IP combining with mass spectrometry, and the real interaction between protein and protein, for example, c-Src and AGO2 in our study, needs more evidences from cell experiment and animal experiment. Accordingly, AGO2 and c-Src expression correlation was analyzed by GEPIA (http://gepia.cancer-pku.cn). For example, in lung cancer, there was a significant positive correlation between AGO2 and c-Src according to their log2(TPM) levels (Fig. S3C). Furthermore, we referred to GEPIA to determine whether there is a correlation between AGO2 expression and overall survival [30]. Our results showed that AGO2 up-regulation was associated with faster disease progression and worse overall survival in 962 patients with lung cancer (Fig. S3B). Median overall survival of patients with lung cancer decreased from 100 to 25 months after the up-regulation of AGO2 expression. Thus, increased AGO2 expression in human lung cancer is associated with the reduced survival. The bioinformatics consequence revealed a tumor-promoting role of AGO2, probably with/by c-Src in cancer.
Of note, Shen et al. first reported EGFR is a tyrosine kinase for phosphorylation of AGO2. And now we have identified c-Src functions as another tyrosine kinase for AGO2. Here we tried to characterize the similarity and difference between them. First, EGFR-AGO2 association is hypoxia-dependent because EGFR is a trans-membrane protein, and transfers to late endosomes where it is co-localized with AGO2 only under hypoxia. However, c-Src is a widely-distributed cytosol protein, and c-Src-AGO2 association is easy to demonstrate under normal condition (Fig. 1A–D, F–I and Fig. 2A–D). Second, only one tyrosine residue Y393 in AGO2 is confirmed to be phosphorylated by EGFR. However, not only tyr393, but also tyr529 and tyr749, have been identified as phosphorylation sites for c-Src kinase (Fig. 2D and E). Functionally, Y393 phosphorylation by c-Src under nomoxia is identical to that by EGFR under hypoxia, which can reduce the binding of DICER to AGO2 and inhibits pre-miR-192 maturation. Third, Shen et al. did not observe any significant changes among parental, vector, AGO2 WT and AGO2Y393F stable cell lines (HeLa and MDA-MB-231) in cell proliferation (under normoxia or hypoxia) or anchorage-independent growth (Ref. [18], Fig. S36), which are completely different from our present results and previous study [17]. This contradiction should be clarified in further studies. Collectively, c-Src has a stronger binding affinity to AGO2 than EGFR, and we consider that under normoxia, c-Src is the major kinase for phosphorylation of AGO2.
Importantly, some previous studies reported the cooperative tumorigenesis by c-Src and EGFR interplay. Parsons et al. first showed c-Src overexpression in EGFR-expressing metastatic breast and other cancer cells promoted cell migration [31]. In a review, Parsons et al. described the physical and functional interactions between c-Src and EGFR. It is evidenced that EGFR can be phosphorylated on multiple sites by c-Src, most notably tyr845, which is required for EGF-induced cell proliferation and cell survival [32]. Dimri et al. showed c-Src can modify EGFR-mediated mammary epithelial oncogenesis [33]. Mader et al. showed that EGF stimulation leads to EGFR autophosphorylation, providing potential binding site for both Arg and c-Src SH2 domains, which binding relieves their autoinhibited conformation, enabling Src and Arg to autophosphorylate for downstream signaling pathway [34]. Our result that competitive roles of c-Src and EGFR in binding with AGO2 is based on overexpression of c-Src and EGFR plasmids (Fig. 2G), which might be different from physiological condition. Functionally, it is reasonable that c-Src and EGFR kinases, alone or in concert in vivo, can phosphorylate AGO2 for tumorigenesis.
Results showed that only pY393 AGO2 inhibits processing maturation of long-loop miRNAs, whereas pY529 AGO2 or pY749 AGO2 does not function through this mechanism. Although all the three phosphorylations show a similar oncogenic phenotype, we propose that each mechanism of action of these sites is different because of their different domain location. It has been reported pY529 could prevent AGO2 binding to 5′ phosphate of the small RNA [28]. Since tyr749 located in the PIWI domain, we propose that pY749 AGO2 may be responsible for its endonucleolytic activity. Further functional experiments are required to elucidate the detailed mechanism.
In domain mapping experiment, we couldn’t detect the expression of ΔSH1 c-Src mutant in 293T cells for uncertain reasons. Saracatinib (AZD0530) is a potent, orally available ATP‐competitive inhibitor of Src kinase that has high selectivity when compared with a range of protein tyrosine kinases involved in signal transduction [35]. We have noticed that saracatinib with a usual concentration (5 μM) has a very strong anti-tumor effect (Fig. 5A-D), especially, colony formation assay showed that the clones treated with the inhibitor decreased not only in number but also significantly in size compared with AGO2 stable cells (Fig. 5C), which imply that the functions of c-Src kinase versus AGO2 tyrosine phosphorylation are not equal in tumor progress. Despite c-Src being a classic oncogene and AGO2 over-expressed in many cancers, it also has been reported that AGO2 has a reduced expression in melanoma [36], and that over-expression of AGO2 could inhibit cell and tumor growth [37], [38]. More studies will be needed to identify the exact role of AGO2 and its tyrosine phosphorylation in tumorigenesis.
Conclusion
On the basis of our current findings, we present a schematic model (Fig. 5E). AGO2 interacts with c-Src and is phosphorylated at its tyr393, tyr529 and tyr749 sites by c-Src kinase, which phosphorylation could be blocked by treatment with saracatinib, the c-Src kinase inhibitor. Phosphorylation of AGO2 at Tyr393 reduces its binding to DICER, thereby inhibiting miRNA maturation. Each of the three phosphorylations in AGO2 shows an obvious tumor-promoting effect. Given that AGO2 plays an important role in tumorigenesis that could be regulated by c-Src phosphorylation, our study might offer a novel strategy for clinical treatment of cancer.
Acknowledgments
Acknowledgements
The authors thank Rui Dong, Zi Liang and Xinmin Zheng for expert technical assistance. The authors also thank Dr Imre Janszky for manuscript revision. This study was supported by grants from the National Natural Science Foundation of China (No. 81672709 and 81372190 to J.H, 81630075 to J.Y.) and the Science and Technology Commission of Shanghai (17DZ2260100 to S.G.). The authors declare that they have no conflict of interest.
Author contributions
J.H. and J.Y. supervised the project. J.H. and T.L. designed the experiments. T.L. and H.Z. performed most experiments. Z.Y. performed bioinformatics analysis. J.F., X.Z., R.C., Y.W., and S.G. helped with all the experiments. J.H., J.Y., T.L. and H.Z. discussed the results. J.H. and T.L. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.12.004.
Contributor Information
Jianxiu Yu, Email: Jianxiu.Yu@gmail.com.
Jian Huang, Email: jyhuanj@shsmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. S1.
(A) Components of RISC-loading complex could be phosphorylated by c-Src kinase. 293T cells were co-transfected with Flag-DICER, Flag-AGO2 and Flag-TRBP along with or without HA-c-Src. Cell lysates were used for immunoprecipitation with anti-Flag antibody, followed by Western blot analysis with 4G10. (B) Bindings between AGO2/DICER/TRBP with c-Src. 293T cells were transfected with Flag-AGO2/DICER/TRBP with or without HA-c-Src as indicated. Cell lysates were used for immunoprecipitation with anti-Flag antibody, followed by Western blot analysis with anti-HA antibody.
Supplementary Fig. S2.
(A) Pre-miR-34a and Flag-AGO2 with or without HA-c-Src were transfected into 293T cells. Total RNA was extracted from 293T cells, the miR-34a expression were detected by Northern blotting. (B) Protocol as (A), except that transfected 293T cells were treated with or without 10 µM saracatinib for 4 h before harvest. (C) Pre-let-7a-1 and Flag-AGO2 with or without HA-c-Src were transfected into 293T cells. Total RNA was extracted from 293T cells, the let-7a expression were detected by Northern blotting. (D) Protocol as (C), except that transfected 293T cells were treated with or without 10 µM saracatinib for 4 h before harvest.
Supplementary Fig. S3.
(A) A protein-protein interaction network for AGO2, c-Src and their related proteins. This is a composite of all of the networks chosen from the database in a way that best connects related proteins. Nodes represented proteins and links represented networks. (B) Kaplan-Meier curves, log-rank test, showing the correlation between AGO2 expression and clinical outcome in human lung cancer, as analyzed for overall survival. Red represents high AGO2 TPM, and blue represents low AGO2 TPM. n(high) = 481, n(low) = 481. (C) AGO2 and c-Src expression correlation analysis. Spearman R and P-value for log2 (c-Src TPM) versus log2 (AGO2 TPM) in human lung cancer.
References
- 1.Martin G.S. The hunting of the Src. Nat Rev Mol Cell Biol. 2001;2:467–475. doi: 10.1038/35073094. [DOI] [PubMed] [Google Scholar]
- 2.Frame M.C. Newest findings on the oldest oncogene; how activated src does it. J Cell Sci. 2004;117:989–998. doi: 10.1242/jcs.01111. [DOI] [PubMed] [Google Scholar]
- 3.Fizazi K. The role of Src in prostate cancer. Ann Oncol. 2007;18:1765–1773. doi: 10.1093/annonc/mdm086. [DOI] [PubMed] [Google Scholar]
- 4.Song L., Morris M., Bagui T., Lee F.Y., Jove R., Haura E.B. Dasatinib (BMS-354825) selectively induces apoptosis in lung cancer cells dependent on epidermal growth factor receptor signaling for survival. Cancer Res. 2006;66:5542–5548. doi: 10.1158/0008-5472.CAN-05-4620. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs C., Rubsamen H. Expression of pp60c-src protein kinase in adult and fetal human tissue: high activities in some sarcomas and mammary carcinomas. Cancer Res. 1983;43:1696–1702. [PubMed] [Google Scholar]
- 6.Talamonti M.S., Roh M.S., Curley S.A., Gallick G.E. Increase in activity and level of pp60c-src in progressive stages of human colorectal cancer. J Clin Invest. 1993;91:53–60. doi: 10.1172/JCI116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maness P.F. Nonreceptor protein tyrosine kinases associated with neuronal development. Dev Neurosci. 1992;14:257–270. doi: 10.1159/000111670. [DOI] [PubMed] [Google Scholar]
- 8.Li X., Shen Y., Ichikawa H., Antes T., Goldberg G.S. Regulation of miRNA expression by Src and contact normalization: effects on nonanchored cell growth and migration. Oncogene. 2009;28:4272–4283. doi: 10.1038/onc.2009.278. [DOI] [PubMed] [Google Scholar]
- 9.Schirle N.T., MacRae I.J. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond S.M., Bernstein E., Beach D., Hannon G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 12.Martinez J., Patkaniowska A., Urlaub H., Luhrmann R., Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 13.Hutvagner G., Zamore P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 14.Ye Z., Jin H., Qian Q. Argonaute 2: A Novel Rising Star in Cancer Research. J Cancer. 2015;6:877–882. doi: 10.7150/jca.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi H.H., Ongusaha P.P., Myllyharju J., Cheng D., Pakkanen O., Shi Y., Lee S.W., Peng J., Shi Y. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–424. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rybak A., Fuchs H., Hadian K., Smirnova L., Wulczyn E.A., Michel G., Nitsch R., Krappmann D., Wulczyn F.G. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H., Wang Y., Dou J., Guo Y., He J., Li L., Liu X., Chen R., Deng R., Huang J. Acetylation of AGO2 promotes cancer progression by increasing oncogenic miR-19b biogenesis. Oncogene. 2019;38:1410–1431. doi: 10.1038/s41388-018-0530-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen J., Xia W., Khotskaya Y.B., Huo L., Nakanishi K., Lim S.O., Du Y., Wang Y., Chang W.C., Chen C.H. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai X., Chen Q., Zhu C., Deng R., Zhao X., Chen C., Wang Y., Yu J., Huang J. Regulation of RPTPalpha-c-Src signalling pathway by miR-218. FEBS J. 2015;282:2722–2734. doi: 10.1111/febs.13314. [DOI] [PubMed] [Google Scholar]
- 20.Wang J., Deng R., Cui N., Zhang H., Liu T., Dou J., Zhao X., Chen R., Wang Y., Yu J. Src SUMOylation inhibits tumor growth via decreasing FAK Y925 phosphorylation. Neoplasia. 2017;19:961–971. doi: 10.1016/j.neo.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X., Wang Y., Sun Q., Yan J., Huang J., Zhu S., Yu J. Identification of microRNA transcriptome involved in human natural killer cell activation. Immunol Lett. 2012;143:208–217. doi: 10.1016/j.imlet.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu Y., Chen Q., Lai X., Zhu C., Chen C., Zhao X., Deng R., Xu M., Yuan H., Wang Y. SUMOylation of Grb2 enhances the ERK activity by increasing its binding with Sos1. Mol Cancer. 2014;13:95. doi: 10.1186/1476-4598-13-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui N., Liu T., Guo Y., Dou J., Yang Q., Zhang H., Chen R., Wang Y., Zhao X., Yu J. SUMOylation of Csk negatively modulates its tumor suppressor function. Neoplasia. 2019;21:676–688. doi: 10.1016/j.neo.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., Wang S., Jiang B., Huang L., Ji Z., Li X., Zhou H., Han A., Chen A., Wu Y. c-Src phosphorylation and activation of hexokinase promotes tumorigenesis and metastasis. Nat Commun. 2017;8:13732. doi: 10.1038/ncomms13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Ren Z., Kang X., Zhang L., Li X., Wang Y., Xue T., Shen Y., Liu Y. Identification of tyrosine-phosphorylated proteins associated with metastasis and functional analysis of FER in human hepatocellular carcinoma cells. BMC Cancer. 2009;9:366. doi: 10.1186/1471-2407-9-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudel S., Wang Y., Lenobel R., Korner R., Hsiao H.H., Urlaub H., Patel D., Meister G. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Research. 2011;39:2330–2343. doi: 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mostafavi S., Ray D., Warde-Farley D., Grouios C., Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biscardi J.S., Ishizawar R.C., Silva C.M., Parsons S.J. Tyrosine kinase signalling in breast cancer: epidermal growth factor receptor and c-Src interactions in breast cancer. Breast Cancer Res. 2000;2:203–210. doi: 10.1186/bcr55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishizawar R., Parsons S.J. c-Src and cooperating partners in human cancer. Cancer cell. 2004;6:209–214. doi: 10.1016/j.ccr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Dimri M., Naramura M., Duan L., Chen J., Ortega-Cava C., Chen G., Goswami R., Fernandes N., Gao Q., Dimri G.P. Modeling breast cancer-associated c-Src and EGFR overexpression in human MECs: c-Src and EGFR cooperatively promote aberrant three-dimensional acinar structure and invasive behavior. Cancer Res. 2007;67:4164–4172. doi: 10.1158/0008-5472.CAN-06-2580. [DOI] [PubMed] [Google Scholar]
- 34.Mader C.C., Oser M., Magalhaes M.A., Bravo-Cordero J.J., Condeelis J., Koleske A.J., Gil-Henn H. An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 2011;71:1730–1741. doi: 10.1158/0008-5472.CAN-10-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hannon R.A., Clack G., Rimmer M., Swaisland A., Lockton J.A., Finkelman R.D., Eastell R. Effects of the Src kinase inhibitor saracatinib (AZD0530) on bone turnover in healthy men: a randomized, double-blind, placebo-controlled, multiple-ascending-dose phase I trial. J Bone Miner Res. 2010;25:463–471. doi: 10.1359/jbmr.090830. [DOI] [PubMed] [Google Scholar]
- 36.Voller D., Reinders J., Meister G., Bosserhoff A.K. Strong reduction of AGO2 expression in melanoma and cellular consequences. Br J Cancer. 2013;109:3116–3124. doi: 10.1038/bjc.2013.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X., Graves P., Zeng Y. Overexpression of human Argonaute2 inhibits cell and tumor growth. Biochim Biophys Acta. 2013;1830:2553–2561. doi: 10.1016/j.bbagen.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 38.Tattikota S.G., Rathjen T., McAnulty S.J., Wessels H.H., Akerman I., van de Bunt M., Hausser J., Esguerra J.L., Musahl A., Pandey A.K. Argonaute2 mediates compensatory expansion of the pancreatic beta cell. Cell Metab. 2014;19:122–134. doi: 10.1016/j.cmet.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.